Abstract
It is particularly meaningful to investigate natural selection in African Americans (AfA) due to the high mortality their African ancestry has experienced in history. In this study, we examined 491,526 autosomal single nucleotide polymorphisms (SNPs) genotyped in 5210 individuals and conducted a genome-wide search for selection signals in 1890 AfA. Several genomic regions showing an excess of African or European ancestry, which were considered the footprints of selection since population admixture, were detected based on a commonly used approach. However, we also developed a new strategy to detect natural selection both pre- and post-admixture by reconstructing an ancestral African population (AAF) from inferred African components of ancestry in AfA and comparing it with indigenous African populations (IAF). Interestingly, many selection-candidate genes identified by the new approach were associated with AfA-specific high-risk diseases such as prostate cancer and hypertension, suggesting an important role these disease-related genes might have played in adapting to a new environment. CD36 and HBB, whose mutations confer a degree of protection against malaria, were also located in the highly differentiated regions between AAF and IAF. Further analysis showed that the frequencies of alleles protecting against malaria in AAF were lower than those in IAF, which is consistent with the relaxed selection pressure of malaria in the New World. There is no overlap between the top candidate genes detected by the two approaches, indicating the different environmental pressures AfA experienced pre- and post-population admixture. We suggest that the new approach is reasonably powerful and can also be applied to other admixed populations such as Latinos and Uyghurs.
Although the vast majority of human genetic variations evolve neutrally, some parts, however, have been shaped by natural selection (Kimura 2003; Balaresque et al. 2007). Studies on recent natural selection have led to the discovery of genes showing population differences in adapting to pathogens, diet, climate, and other environmental challenges. These discoveries have greatly enriched our understanding about the origins and also the evolutionary history of the human species, identified many genes with important biological functions, and will lead to further elucidation of the genetic basis of some human diseases (Balaresque et al. 2007; Nielsen et al. 2007; Sabeti et al. 2007; Hancock et al. 2008; Akey 2009). The recent availability of high-density SNPs has provided essential resources for genome-wide detection of natural selection, especially in ethnically well-defined populations with little admixture (Sabeti et al. 2007; Barreiro et al. 2008; Akey 2009). Although there have been several studies on recently admixed populations (Tang et al. 2007; Basu et al. 2008; Bryc et al. 2010), no study so far has particularly investigated the locus-specific population differentiation between the ancestral components of an admixed population and its ancestral parental population, which might reflect the natural selection since the two split. In this case, we used African Americans, a well-studied admixed population, as the object of our study.
African Americans (AfA) are residents of the United States with partial recent Sub-Saharan African ancestry. The majority of them are descendents of the Africans, probably 500,000 to 650,000 in number, who were forcibly brought to North America during the Middle Passage (Thomas 1999; Zakharia et al. 2009). However, many of those captive Africans died either during the Atlantic shipment to America due to the severe conditions, or soon upon their arrival in the New World as a result of exposure to foreign pathogens and/or the poor living conditions. Although the exact amount of life lost in this process remains a mystery, it may equal or exceed the actual amount enslaved (Stannard 1993). Therefore, the high mortality of AfA in the whole slavery era could be attributed to the overwhelming environmental challenges. These persistent selection pressures might make the frequencies of “beneficial” alleles increase continually, which should lead to higher population differentiation between African components of ancestry in AfA and indigenous Africans at these loci, in contrast to those evolved neutrally.
Meanwhile, before Africans and Europeans migrated to the New World, their ancestral parental populations had evolved independently in distinct environments for tens of thousands of years (Basu et al. 2008). The completely new environment in the New World constituted a challenge for both populations and the populations of their admixture. Therefore, it is very likely that some genomic regions in AfA show an excess of a particular ancestry as a result of selection pressures after the population admixture (Tang et al. 2007; Basu et al. 2008). For example, there are various studies in detecting signatures of selection in AfA by examining admixture proportion using a small number of available loci (Workman et al. 1963; Reed 1969; Blumberg and Hesser 1971; Adams and Ward 1973; Long 1991). Recently, Bryc et al. (2010) identified three autosomal regions showing excessive or reduced African ancestry (by at least three standard deviations [SD] of the mean) as natural selection candidates based on ∼500K SNPs genotyped in 365 AfA.
Here, we analyzed the genotype data from ∼500,000 autosomal SNPs shared by 1890 AfA and 3320 non-AfA for detecting the signatures of selection in AfA that were classified into pre-admixture and post-admixture according to the different environments they experienced (Supplemental Fig. S1). Firstly, we detected the natural selections that were more likely to occur after admixture by examining the genome-wide distribution of ancestry in AfA. Then we developed a new strategy by reconstructing an ancestral African population (AAF) from inferred African components of ancestry in AfA and comparing it with indigenous African populations (IAF) (Supplemental Fig. S2), which reflect natural selection since the African ancestors of AfA left Africa (including both pre- and post-admixture). Many candidate genes identified by our new approach could explain the challenges that AfA and their ancestry had experienced. Thus we suggest that our new approach is reasonably powerful and can also be applied to the studies of other admixed populations.
Results
African and European ancestries in AfA
The populations from West Africa and Europe, who had undoubtedly contributed to the current gene pool of AfA, were considered as ancestral parental populations of AfA in this study. However, it was practically difficult to select proper populations as genetic donors to the gene pool of AfA since the extant populations in West Africa and Europe might not necessarily represent those 300 yr ago, given the possible influence of genetic drift, selection, and demographic history. In this study, we chose the samples of Yoruba in Ibadan, Nigeria (YRI), and Utah residents with ancestry from northern and western Europe (CEU), to respectively represent those who contributed to the formation of AfA, largely due to the availability of phased genetic data for these two populations. Besides, an exploratory analysis including Human Genome Diversity Project (HGDP) populations (Li et al. 2008) also suggested that YRI and CEU are better choices for ancestral parental populations of AfA than the other populations with available genome-wide data (see Supplemental Text and Supplemental Fig. S3).
FRAPPE (Tang et al. 2005) was applied to the genome-wide high-density SNP data (by taking K = 2), and the estimated European contribution to AfA was 21.65% at population level (Supplemental Figs. S4, S5). structure (Pritchard et al. 2000; Falush et al. 2003) analyses yielded virtually identical results. Based on the thinned data with 341,672 SNPs, the European contribution to the 1890 AfA was estimated to be 21.61% using PCA results (Supplemental Fig. S3D). These estimations were essentially consistent with those in previous studies (Smith et al. 2004; Xu et al. 2007; Bryc et al. 2010), although data sets analyzed were different.
Identification of genomic regions with biased ancestry in AfA
To estimate the distribution of genetic contributions of European and African ancestry to AfA across the genome, we used haplotypes of 88 CEU and 88 YRI (the phased parents of trios, presumably unrelated, from HapMap3 data) to represent their ancestral parental populations. The identical monomorphic SNPs in CEU and YRI samples were removed for they could not provide valuable information in the local ancestry inference. HAPMIX (Price et al. 2009) was employed to estimate the locus-specific genetic contributions of the ancestral parental populations to AfA (see Methods) by taking 21.65% as the European contribution, as estimated by FRAPPE. Based on the likelihood given by HAPMIX, generation since admixture (λ) was estimated to be λ = 7 (an essentially hybrid isolation model), which was similar to those based on other AfA data sets (Smith et al. 2004; Price et al. 2009). Detailed analysis showed that λ values for most individuals ranged from 1 to 12, and could be explained by a continuous-gene-flow (CGF) model (see Supplemental Text and Supplemental Fig. S6). The locus-specific European ancestry proportion across the genome of AfA was estimated to be 21.68% ± 0.75% (mean ± SD). The SD of locus-specific ancestral genetic contributions in this study is lower than those in any previous studies (Workman et al. 1963; Reed 1969; Blumberg and Hesser 1971; Tang et al. 2007; Basu et al. 2008; Bryc et al. 2010), which was not beyond our expectation since we used a much larger sample size.
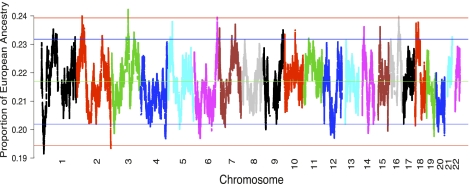
The genomic regions showing excessive or reduced ancestry in the admixed population are likely to be signatures of natural selection (Tang et al. 2007; Basu et al. 2008; Bryc et al. 2010; Oleksyk et al. 2010). The loci showing strong deviation of European ancestry (3 SDs above or below the genome-wide average) were therefore identified as candidates of natural selection in this study. Four regions (2p22, 3q13, 6q26, 16q21) with excessive European influence and two regions (1p36, 2q37) with excessive African influence were observed in AfA genomes (Fig. 1). Each of the six regions was significantly different from the genome-wide average of ancestral contributions (P < 2.2 × 10−16, t-test). The detailed annotations of the six candidate regions are presented in Table 1. Most genomic regions showing ancestry deviation can be replicated by an analysis with LAMP-ANC (see Supplemental Text; Supplemental Fig. S7), although it has lower accuracy than HAPMIX based on our simulated data.
Figure 1.
Genome-wide distribution of European ancestral contributions. Mean European ancestral contribution across 1890 African-American individuals at each SNP. (Green line) Estimated genome-wide mean European ancestral contribution (21.68%). Blue bands indicate +2 and −2 SDs from the mean ancestral contribution and red bands indicate +3 and −3 SDs from the mean ancestral contribution.
Table 1.
Regions showing excess of European or African ancestry
A close examination of SNPs in the six regions showing biased ancestry revealed neither significant deviation from the Hardy-Weinberg expectation nor an unusual fraction of missing data, suggesting that the genotyping quality is unlikely the cause of this bias. In addition, the six regions did not overlap with previously reported long-ranged linkage disequilibrium (LD) blocks including inversions which may confound genome-wide scans for selection signals in admixed populations (Price et al. 2008), even though three short inversions have been found in one European individual in 1p36. Although the estimated locus-specific ancestral contributions across the genome are generally consistent with the study by Bryc et al. (2010), the three regions showing biased ancestry identified by them were only in moderate excess of African or European ancestry in this study, possibly due to the different samples and/or sample sizes between the two studies. Our estimations were also generally consistent with the genome-wide distribution of ancestry calculated in previous admixture mappings (Reich et al. 2005; Kao et al. 2008).
The region showing the strongest bias is located on 1p36, where African ancestry is over-represented. In this region, IGSF21 and AKR7A2 are located next to the SNPs that show the strongest signals, respectively. IGSF21 belongs to the immunoglobin superfamily, while AKR7A2 is involved in the detoxification of aldehydes and ketones, and is implicated in various cancers such as pancreatic cancer (Praml et al. 2008; Cui et al. 2009). Another region with over-represented African ancestry is 2q37, in which PDCD1 is involved in signaling of the immune system and various diseases. The region showing the highest excessive European ancestry is located on 3q13, which only harbors LSAMP, a candidate of the tumor suppressor gene in human osteosarcomas, and is associated with coronary artery disease (Kresse et al. 2009; Yen et al. 2009). Interestingly, EIF2AK2, involved in influenza infection pathways (McAllister et al. 2010; Pereira et al. 2010), is located ∼200 Kb away from the peak (2p22) showing the second highest excessive European ancestry, which suggested the possible difference between African and European resistance to influenza. PACRG, located in 6q26, is associated with Parkinson's disease. However, the region in 16q21 showing excessive European ancestry did not contain any genes or known function elements.
African/European components of ancestry in AfA
The segments of African ancestry and those of the European ancestry in AfA, inferred by HAPMIX, were collectively referred to as African components of ancestry and European components of ancestry, respectively, each of which could be considered as an AAF or an ancestral European population (AEU) residing in America before admixture. The inference of AAF and AEU are credible given the accuracy of HAPMIX that is >98% based on simulated data (see Supplemental Text).
Then population differentiation between AEU and each putative European parental population was calculated, with CEU showing the lowest FST with AEU (FST [AEU-CEU] = 0.0005) among all putative parental populations. When 2648 Caucasian from GWAS data (referred to as CAU-GWAS) were considered, FST [AEU-CAU-GWAS] was 0.0006, which was the second lowest among all values. Among all IAF, YRI showed the lowest FST with AAF (FST [AAF-YRI] = 0.0007). When the observed FST's were compared with those simulated under neutrality (see Methods), a different pattern emerged between European ancestry and African ancestry. For European ancestry, the observed FST between AEU and CEU (0.0005) was lower than that simulated (simulated FST [AEU-CEU] = 0.0006), and the genome-wide distribution of observed locus-specific FST did not deviate much from those simulated (P = 0.042, Kolmogorov–Smirnov test).
However, for African ancestry, the observed FST between AAF and YRI (0.0007) was higher than that simulated (simulated FST [AAF-YRI] = 0.0006), and genome-wide distribution of locus-specific FST [AAF-YRI] was significantly different from those simulated (P < 2.2 × 10−16, Kolmogorov–Smirnov test). In particular, a Q-Q plot between the observed and simulated locus-specific FST [AAF-YRI] showed an enrichment of SNPs with high FST at the tail of the observed locus-specific FST (Supplemental Fig. S8), suggesting a possible role of natural selection. The results remain essentially unchanged when bottlenecks for African ancestry were assumed in our simulations.
Identification of regions highly differentiated between AAF and African
Since the African immigrants left for America, they might have experienced a completely different population history compared with indigenous Africans. The high mortality of AfA during the slavery era (Meltzer 1993; Stannard 1993; Thomas 1999) suggested a possible presence of strong selective pressures. According to the theory of neutrality, FST is largely influenced by demographic history which affects all loci similarly (Weir and Cockerham 1984; Kimura 2003), if not equally. In contrast, the force of positive selection acts in a locus-specific manner and tends to increase FST (Nielsen 2005), which has been widely used to detect the positive selection in various studies (Akey et al. 2002; Oleksyk et al. 2010). In this study, we calculated the locus-specific FST between AAF and YRI (Supplemental Fig. S9). However, loci with a very low minor allele frequency (MAF) may be subjected to sampling error and statistical error; therefore, the SNPs with MAF < 0.05 in AAF or YRI were removed from further analyses.
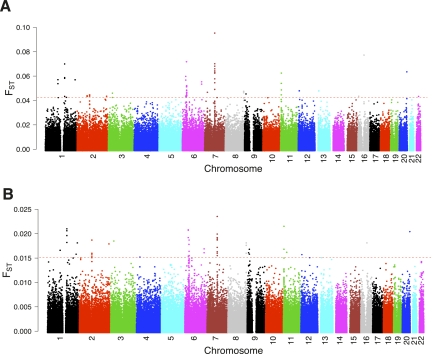
The genome-wide distribution of FST between AAF and YRI for 401,599 autosomal SNPs is presented in Figure 2A. In spite of the low differentiation between AAF and YRI, a substantial proportion of SNPs was located at the right tail of the distribution. The functional annotations of the most differentiated SNP clusters (99.99th percentile; FST > 0.0452) are listed in Table 2. Although it is reported that FST of an individual marker was too variable (Weir et al. 2005), the four most significant regions (7q21, 6p21–22, 1q22, and 11p15) carrying multiple highly differentiated SNPs should be indicative. The 7q21 region harbors two genes: CD36 and SEMA3C. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes and it binds long chain fatty acids and may function in the transport and/or as a regulator of fatty acid transport (Oquendo et al. 1989; Baruch et al. 1996; Erdman et al. 2009). It was reported that CD36 has been subjected to positive selection for malaria or some unknown selection pressures (Aitman et al. 2000; Omi et al. 2002, 2003; Fry et al. 2009). SEMA3C, induced by ADAMTS1, promotes the migration of cancer cells (Esselens et al. 2010). The 6p21–22 region, harboring human major histocompatibility complex (MHC), also showed an over-representation of African ancestry in this study (although <3 SD) and has been reported under selection in various studies (Garrigan and Hedrick 2003; Tang et al. 2007). MUC1, located in 1q22, is involved in signaling by PDGF and serves a protective function by binding to pathogens (Davila et al. 2010; Li et al. 2010). HBB and HBD, located in 11p15, have been subjected to balancing selections because their mutations protected against malaria according to numerous studies (Ashley-Koch et al. 2000; Wood et al. 2005).
Figure 2.
(A) Genomic distribution of FST between AAF and YRI. (B) Genomic distribution of FST between AfA and rAfA. Dashed red horizontal line indicates the cutoff threshold (99.99th percentile). Locus-specific FST between YRI and CEU were calculated when MAF > 0.05 in both populations. The rAfA was constituted according to the ancestry proportion of CEU and YRI under neutrality.
Table 2.
Regions with highly differentiated allele frequency between AAF and YRI (FST > 0.0452)
Ingenuity pathway analysis (IPA) was particularly helpful in exploring the function and pathways of the selection-candidate genes in the context of higher-order cellular and molecular mechanisms. The 402 SNPs with the highest FST (99.90th percentile; FST > 0.0287) were subjected to IPA analysis, whose results showed that genes involved in metabolic diseases were the most significantly enriched (P = 1.51 × 10−16) among all function classes, followed by the genes involved in endocrine system disorders (P = 2.23 × 10−16), immunological diseases (P = 9.30 × 10−12), and genetic disorders (P = 5.67 × 10−11). Antigen presentation pathway was the most significantly enriched (P = 1.95 × 10−4), followed by allograft rejection signaling (P = 4.69 × 10−3), Graft-versus-host disease signaling (P = 4.69 × 10−3), and autoimmune thyroid disease signaling (P = 5.35 × 10−3). All of the four aforementioned pathways are related to the immune system, which might reflect a great environmental differentiation between Sub-Saharan Africa and North America. We also conducted IPA on 4011 SNPs showing the highest FST (99.00th percentile; FST > 0.0162), which yielded results similar to those with FST > 0.0287 (99.90th percentile). And two additional pathways emerged: IL-9 signaling pathway (P = 8.01 × 10−3) and EGF signaling pathway (P = 6.38 × 10−3).
Reconstituted African American (rAfA) and its difference with AfA
We then compared the AfA genome with that of reconstituted African Americans (rAfA) using genotypes of YRI and CEU. The rationale is that the former have been subjected to possible natural selection, while the latter have not. In particular, the allele frequencies of rAfA were estimated at each locus using YRI and CEU for the given level of admixture (21.68%), assuming no natural selection after admixture. The candidates identified under selection in this way could avoid potential errors introduced in the inference of ancestry. The genome-wide distribution of FST between AfA and rAfA is shown in Figure 2B. Overall, 81% of the SNPs showing the highest difference between AAF and IAF could be validated in the comparison between AfA and rAfA. In particular, these main FST peaks between AAF and YRI are essentially the same as those between AfA and rAfA, which indicated that almost all the selection signals identified by comparing AfA and rAfA originated from AAF or IAF. We also used CAU-GWAS instead of CEU to construct rAfA, with the genomic distribution of FST similar to that using CEU (Supplemental Fig. S10).
Next, we reconstructed a rAfA population using a set of putative parental populations. First, we constructed an African parental population of AfA (APP) using 64% Yoruba, 19% Mandenka, and 14% Bantu according to previously reported ancestral contributions to AAF (Zakharia et al. 2009). Then we used genotypes of the APP and CAU-GWAS to reconstruct rAfA. We obtained the genome-wide distribution of FST between AfA and rAfA (Supplemental Fig. S11). Although the population differentiation between this rAfA and AfA is higher than that based on the two aforementioned rAfAs, the genome-wide distribution of FST is similar to that using only two pure parental populations.
Further evidence for positive selection in AAF and African
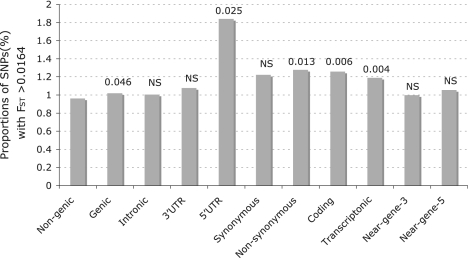
Since the force of positive selection acts in a locus-specific manner and tends to increase FST (Nielsen 2005), we hypothesized that positive selection preferentially acted upon functionally important loci over the others in the genome, thus leading to an enrichment of functional SNPs in the high FST bin (Barreiro et al. 2008). Here, we investigated the enrichment of different SNP classes among the high FST bin (top first percentile among all SNPs, FST > 0.0164) between AAF and YRI.
The SNPs, based on their location and function relative to the genes, were classified into nongenic, genic, intronic, 3′UTR, 5′UTR, synonymous, nonsynonymous, coding, transcriptonic, near-gene-3, and near-gene-5 according to UCSC annotation. We found that FST distributions of each SNP category were not significantly different from those of nongenic SNPs. However, the proportion of genic SNPs among the high FST bin (top first percentile among all SNPs, FST > 0.0164) is significantly higher than that of nongenic SNPs (χ2 test, P = 0.046) (Fig. 3). Notably, this excess is particularly marked for transcriptonic SNPs (χ2 test, P = 0.004) (Fig. 3). The proportion of synonymous SNPs in the high FST bin was 1.22-fold higher than the expectation under neutrality, which could attribute to the LD of those SNPs with loci under selection (a phenomenon known as hitchhiking). Similar observations could be made when the thresholds for the high FST bin were set at the top 5% (FST > 0.0083) or the top 0.1% (FST > 0.0304) (Supplemental Figs. S12, S13), and the conclusions still held when the SNPs with MAF > 0.05 in both YRI and AAF were examined (Supplemental Fig. S14). The significant enrichment of high FST loci in the SNP categories with genetic functions supports the presence of positive selection either in AAF or in YRI, or both.
Figure 3.
Enrichment of high FST loci for different SNP categories. Observed excess of high FST loci in different SNP classes, with respect to nongenic class, in the high FST bin (99th percentile; FST > 0.0164). The values on the bar are P-values of χ2 tests. NS, not significant.
Discussion
Admixed populations such as AfA provide a unique opportunity to study very recent natural selection, as their genomes are donated by long-diverged continental ancestries and may have been subjected to novel environmental challenges. The first strategy, detecting an excessive or decreased ancestry contribution from its ancestral parental populations, has been used in several recent studies (Tang et al. 2007; Basu et al. 2008; Bryc et al. 2010). However, natural selections before admixture cannot be detected by this approach because the distribution of ancestry across the admixed genomes would not be affected by such selections. Therefore, we developed a new strategy, which examined selections since the African ancestry left for America (including selections both pre- and post-admixture). The candidate genes under selection identified by our new strategy were also confirmed in an analysis of allele frequency differentiation between rAfA and AfA (although the two analyses are not completely independent).
Above all, we have identified six regions showing an excess of African or European ancestry using the first strategy. The highest ancestry deviation among all regions showing an excess of African and European ancestry is <0.026, which is lower than the values of any previous studies on the admixed populations in the New World (Workman et al. 1963; Reed 1969; Blumberg and Hesser 1971; Tang et al. 2007; Basu et al. 2008; Bryc et al. 2010). For example, the African ancestry deviation even exceeds 0.14 in Puerto Ricans according to a study by Tang et al. (2007), which is more than fivefold higher than that in this study. We attribute the low ancestry deviation to the much larger sample size, denser markers, more accurate phased data using trios, and more powerful statistical methods used in this study. Based on the maximum deviation regions, we estimated that the highest selection coefficient is approximately 0.002 (assuming 12 generations since admixture under a CGF model). In fact, the real selection coefficient could be much lower than 0.002 considering the statistics and evolutionary noises. These results are reasonable considering the fact that there has been a much lower mortality rate of AfA in the last 200 yr compared with that during the Atlantic slave trade.
Secondly, we identified a large number of genes with highly differentiated allele frequencies between AAF and YRI using our new approach. These genes do not overlap with those identified by the former approach, suggesting the different environmental pressures AfA experienced before and after population admixture. IPA analysis of SNPs with high population differentiation between AAF and YRI showed that genes involved in metabolic diseases (P = 1.51 × 10−16), endocrine system disorder (P = 2.23 × 10−16), immunological diseases (P = 9.30 × 10−12), and genetic disorder (P = 5.67 × 10−11) were significantly enriched. Especially, we found that genes such as PSCA, ZP4, and AKAP12 were associated with AfA-specific high-risk diseases such as hypertension and prostate cancer (Smith and O'Brien 2005; Goran 2008). Five genes (CD36, HBB, HBD, HLA-B, HLA-DR), whose mutations protect against malaria, are also located in the highly differentiated regions between AAF and IAF.
Compared with Caucasians, AfA have a higher mortality rate for all cancers combined and for most major cancers (Jemal et al. 2006), as well as a higher risk of obesity-related diseases such as diabetes, hypertension, and prostate cancer (Goran 2008). Interestingly, many genes located in selection-candidate regions identified by the novel approach are associated with AfA ethnic high-risk diseases such as hypertension, prostate cancer, and systemic sclerosis. Especially, one of the most significantly differentiated SNPs (rs2294008; FST = 0.04561) between AAF and YRI, located in 8q24, is a missense mutation c.57T>C (p.Met1Thr) in PSCA and was reported to be associated with gastric and bladder cancer (Sakamoto et al. 2008; Matsuo et al. 2009; Wu et al. 2009). Many studies also reported that multiple loci in 8q24 were associated with prostate cancer in AfA (Freedman et al. 2006; Al Olama et al. 2009; Yeager et al. 2009). We proposed a hypothesis that most of the genes associated with AfA ethnic diseases may have played an important role in AfA's adaptation to the local environment and thus show higher population differentiation between AAF and IAF. Further analysis of the 8q24 region would provide new insights into the etiology and evolutionary history of these cancers.
Among the selection-candidate genes detected by genome-wide locus specific FST between AAF and YRI, five genes (CD36, HBB, HBD, HLA-B, HLA-DR) have been reported to be subjected to natural selection probably due to malaria (Kwiatkowski 1999, 2005). Because of the strong selection pressure of malaria, loss-of-function or abnormality of these genes was supposed to increase the survival rate of an individual living in Africa. Some mutations in these genes have reached much higher frequencies in Africans compared with those in areas of low incidence of malaria. However, these mutations that defend against malaria could become a disadvantage in AfA because malaria is no longer a strong selection pressure in North America, and these mutations could even lead to morbidity or mortality (Platt et al. 1994). We hypothesize that frequencies of these mutations would have decreased in AAF compared with those in IAF due to their disadvantage in AfA.
Next, we examined this hypothesis in the empirical data. Since the functional mutations in these genes were not genotyped in this study, we examined the SNPs strongly linked (linkage disequilibrium) with these mutations instead of these mutations themselves. It is well known that rs3211938 is a nonsense mutation c.1389T>G (p.Tyr325X) in CD36 and has been subjected to natural selection because of malaria or some other environmental factors in African (Aitman et al. 2000; Omi et al. 2003; Erdman et al. 2009; Fry et al. 2009). We found three SNPs that are highly differentiated between AAF and YRI (Supplemental Table S1) in 7q21 and are strongly linked with rs3211938 (each with r2 > 0.4 in YRI). Interestingly, we did observe that the frequencies of the alleles linked with rs3211938(G), the derived allele, were much lower in AAF compared with those in YRI (Supplemental Table S1). Another example, rs334 is a missense mutation c.70A>T (p.Glu7Val) in HBB, which leads to sickle cell anemia (MIM 603903) (Ashley-Koch et al. 2000; Winichagoon et al. 2000; Wood et al. 2005), one of the most well-studied genetic disorders. rs7952293, one of the SNPs showing high FST between AAF and YRI, was strongly linked with rs334 (r2 = 0.237 in YRI). In particular, the haplotype constructed by rs7952293(A) and rs334(T) accounted for 86.67% of the haplotypes containing rs334(T) in YRI. We observed that the frequency of rs7952293(A) in AAF (0.2261) was lower compared with that in YRI (0.3172), which also supports the hypothesis that frequencies of alleles protecting against malaria in AAF are lower than those in IAF. The other three genes were not examined using the same procedure because the frequencies of mutations on these genes are too low to find strong-linked representative SNPs.
Our study takes advantage of both large sample size and high-density genome-wide data. However, our analysis demonstrated that the maximum deviation showing an excess of African or European ancestry was small (<2.6%). Detecting such weak selection signals in an admixed population such as AfA is a big challenge, which needs a large sample size as in this study, or even a larger sample size, to distinguish the real signals from the ancestry deviation caused by genetic drift and sampling error. Therefore, we propose that any study in the future trying to detect such weak selection signals in AfA or other recently admixed populations should collect at least thousands of samples. With the new strategy, we detected a lot of genes associated with AfA-specific high-risk diseases such as hypertension and prostate cancer, and we also detected five genes whose mutations are against malaria. This new approach is powerful in detecting natural selection both before and after the establishment of AfA and can be applied to other admixed populations such as Latinos in the New World and the Uyghurs in Asia (Xu and Jin 2008; Xu et al. 2008; Xu et al. 2009).
Methods
Data assembly and quality control
The genotypic data were obtained from the International HapMap Project (HapMap; http://www.hapmap.org), the Human Genome Diversity Project (HGDP;http://www.cephb.fr/en/hgdp), and Illumina iControlDB (http://www.illumina.com), respectively. The combined data set processed with PEAS v1.0 (Xu et al. 2010) includes the genotypic data of 588 HapMap samples (87 ASW, 167 YRI, 165 CEU, 85 CHD, 84 CHB) (The International HapMap Consortium 2007; Altshuler et al. 2010), 300 HGDP samples (156 indigenous European, 102 indigenous African, and 42 Amerindian) (Li et al. 2008), 2161 AfA, and 3294 Caucasians (referred to as CAU-GWAS) from iControlDB genotyped by the Illumina 550K Beadarray. The samples from iControlDB have passed Illumina's rigorous quality control and have been used as controls in five genome-wide association studies.
ASW from HapMap and AfA from iControlDB were merged into one AfA population, and Yoruba from HGDP was collected from Nigeria and therefore was merged with YRI from HapMap in subsequent analyses. The following samples were removed from the data set: (1) the relatives based on sample information or PLINK (Purcell et al. 2007) result (IBD > 0.2), (2) individuals identified as outliers of each population (except AfA) based on the top 10 principal components of PCA analysis (SD > 6), (3) AfA individuals with >2% Native-American/East-Asian, (4) AfA individuals with >99% European contribution, which are likely to be descendents of individuals of European ancestry, (5) AfA individuals with >99% African contribution, in which recent African immigrants could not be practically identified (Bryc et al. 2010).
These filtered samples described above were merged and the SNPs sharing reference SNP ID (rs) and vendor-specified strands were kept in combined data. Then, the data set was further filtered for individuals with >10% missing genotypes and SNPs with >10% missing data, as well as Hardy-Weinberg disequilibrium (P < 2 × 10−6) within each population except AfA. The final data set is composed of 503,694 SNPs (491,526 autosomal SNPs) shared by 5210 individuals from 21 population groups, with a total genotyping call rate of 99.74%.
Populations and samples
Overall, 1890 unrelated AfA samples with no ancestry outside of Africa and Europe were studied, among which 1838 individuals were downloaded from iControlDB, and another 52 individuals were from the International HapMap Project (Altshuler et al. 2010). In addition, 113 YRI from HapMap (Altshuler et al. 2010) and 102 IAF in seven different groups collected from HGDP (Li et al. 2008) were merged, representing the extant Africans, while 113 CEU from HapMap (Altshuler et al. 2010) were merged with 156 Europeans in eight different groups collected from HGDP, representing the extant Europeans. In total, 2648 CAU-GWAS samples from iControlDB were taken as another representation of the extant Europeans. In addition, 84 CHB and 85 CHD represented populations from East Asia, and 24 pure Amerindians from HGDP represented Native American.
Population genetic analysis
In order to reduce the LD between markers, those with r2 > 0.5 were removed, calculated in a sliding window of 50 SNPs, and shifted every five SNPs (see Supplemental Text). This process reduced the original data set to 341,672 autosomal SNPs. Based on these thinned markers, principal component analysis (PCA) was performed at the individual level using smartpca, from the package EIGENSOFT (Patterson et al. 2006). Individual ancestry proportion was estimated using FRAPPE, which implements an expectation-maximization (EM) algorithm (Tang et al. 2005). FRAPPE was run on all 491,526 SNPs with 10,000 iterations by setting K from 2 to 4. We also ran structure (Falush et al. 2003) on SNPs with inter-marker distance >1 M (see Supplemental Text). Genetic difference between populations was measured using FST following Weir and Cockerham (Weir and Cockerham 1984), which accounts for differences in the sample size in each population. The locus-specific FST between any two populations was also calculated using the same formula.
Locus-specific ancestry inference
Various methods and software have been developed for inferring locus-specific ancestry based on high-density SNP data, such as ANCESTRYMAP (Patterson et al. 2004), SABER (Tang et al. 2006), LAMP and LAMP-ANC (Sankararaman et al. 2008b), uSWITCH and uSWITCH-ANC (Sankararaman et al. 2008a), HAPAA (Sundquist et al. 2008), and HAPMIX (Price et al. 2009). A simple analysis showed that HAPMIX outperformed other methods with implemented software based on our simulated data (see Supplemental Text). Therefore, HAPMIX was used to infer locus-specific ancestry in AfA in this study. We also used LAMP-ANC to do a similar analysis since it also performs very well.
The phased data of HapMap 3 were downloaded from the HapMap website (Altshuler et al. 2010). We used haplotypes of 88 CEU and 88 YRI (all from trio samples) representing the European and African ancestral populations, respectively, in the subsequent analysis. The mean European ancestry proportion in AfA (θ), which is a required input parameter for HAPMIX, was based on the estimation of FRAPPE. Generation since admixture (λ) with the largest likelihood was taken as its estimation. By running HAPMIX in diploid mode, we obtained the haplotypes and ancestry segments for each AfA individual. Then we reconstructed an AAF and an AEU using inferred chromosomal segments of African ancestry and those of European ancestry in AfA, respectively. In brief, we constructed AAF using only those chromosomal segments with pure African ancestry. For each given SNP, the allele frequency of AAF was calculated based on the available genotypes with African ancestry across all AfA. This procedure was also applied to the construction of AEU using chromosomal segments with pure European ancestry.
Simulation of AfA and its parental populations
Under selective neutrality, the genetic drift of the admixed population and its parental populations contribute to the variation of ancestry proportion and population differentiation among loci (Weir and Cockerham 1984; Long 1991). Therefore, we performed an extensive forward-time simulation to explore the potential impact of genetic drift on the locus-specific population differentiations between AAF and YRI, as well as that between AEU and CEU. In this simulation, recombination was introduced according to the genetic map adapted from HapMap (release #22) (The International HapMap Consortium 2007), and mutation was ignored given the short history of AfA. The effective population sizes (Ne) of each population was obtained from the HapMap website (Altshuler et al. 2010). In particular, Ne for African, European, and AfA were set to 17,094, 11,418, and 17,094, respectively. A CGF model (Supplemental Fig. S15) was used based on previous studies (Pfaff et al. 2001; Price et al. 2009).
The aforementioned phased data of 88 YRI and 88 CEU were taken respectively as the genotypes of common African ancestry and common European ancestry before population admixture. We simulated individuals of AfA by constructing their genomes from a mosaic of haploid YRI and haploid CEU genomes. The generations since admixture (λ) were set to 12 according to a CGF model based on previous reports (Pfaff et al. 2001; Price et al. 2009), which was also supported by our observations (Supplemental Fig. S6). The gene flow (α) that African ancestry received from European each generation was calculated by α = 1 − (m1)1/t, in which m1 represents the mean proportion of European ancestry in AfA. Both African and European parental populations evolved 12 generations simultaneously. Finally, genotypes of 113 European, 113 African, and 1890 AfA, as well as their SNP-specific ancestral status, were output to match the sample sizes of the data. Based on the primary simulation, we performed extended simulations by setting a bottleneck event in the first generation during which Ne for AfA were reduced to 8000, 5000, 3000, 2000, and 1000, respectively.
Function annotations and ingenuity pathway analysis (IPA)
Genomic regions that deviated from genome-wide ancestral contributions or with extremely high FST were annotated based on the HapMap website (Thorisson et al. 2005). The SNPs showing substantial population differentiation were interrogated for network and functional interrelatedness using the IPA version 8.5 software tools. This software searches for information on genes in the Ingenuity Pathways Knowledge Base, a repository of molecular interactions, regulatory events, gene-to-phenotype associations, and chemical knowledge, all collected from the full text of the peer-reviewed life science literature. With IPA, we can analyze data in the context of molecular mechanisms, identify key mechanistic differences between subpopulations, and further relate molecular events to higher-order cellular and disease processes.
Statistical analysis
All statistical computation and graphics were performed using R version 2.9 (Ihaka and Gentleman 1996). Kolmogorov–Smirnov tests were performed to compare the empirical distributions of locus-specific FST with those simulated, and χ2 tests were performed to test the over-representation of each SNP category compared with nongenic SNPs among the high FST bin.
Acknowledgments
S.X. was supported by the National Science Foundation of China (30971577, 31171218), Shanghai Rising-Star Program (11QA1407600), and Science Foundation of The Chinese Academy of Sciences (KSCX2-EW-Q-1-11, KSCX2-EW-R-01-05, KSCX2-EW-J-15-05). L.J. was supported by the National Science Foundation of China (30890034, 30625016) and the Science and Technology Commission of Shanghai Municipality (09540704300). S.X. is a Max-Planck Independent Junior Research Group Leader and a member of the CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the K.C. Wong Education Foundation, Hong Kong. B.W. is a senior author of the Harvard group. This work was also supported by the MoST International Cooperation Base of China.
Authors' contributions: S.X. and L.J. conceived and designed the study. H.W. collected genotype data from the Illumina iControl Database (iControlDB). W.J. performed data analysis, with contributions from S.X. Y.Y., Y.S., and B.W. performed IPA analysis. S.X. and W.J. interpreted the data. W.J., S.X., and L.J. wrote the paper. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124784.111.
References
- Adams J, Ward RH 1973. Admixture studies and the detection of selection. Science 180: 1137–1143 [DOI] [PubMed] [Google Scholar]
- Aitman TJ, Cooper LD, Norsworthy PJ, Wahid FN, Gray JK, Curtis BR, McKeigue PM, Kwiatkowski D, Greenwood BM, Snow RW, et al. 2000. Malaria susceptibility and CD36 mutation. Nature 405: 1015–1016 [DOI] [PubMed] [Google Scholar]
- Akey JM 2009. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res 19: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, et al. 2009. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 41: 1058–1060 [DOI] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley-Koch A, Yang Q, Olney RS 2000. Sickle hemoglobin (HbS) allele and sickle cell disease: A HuGE review. Am J Epidemiol 151: 839–845 [DOI] [PubMed] [Google Scholar]
- Balaresque PL, Ballereau SJ, Jobling MA 2007. Challenges in human genetic diversity: Demographic history and adaptation. Hum Mol Genet 16: R134–R139 [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L 2008. Natural selection has driven population differentiation in modern humans. Nat Genet 40: 340–345 [DOI] [PubMed] [Google Scholar]
- Baruch DI, Gormely JA, Ma C, Howard RJ, Pasloske BL 1996. Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc Natl Acad Sci 93: 3497–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Tang H, Zhu X, Gu CC, Hanis C, Boerwinkle E, Risch N 2008. Genome-wide distribution of ancestry in Mexican Americans. Hum Genet 124: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg BS, Hesser JE 1971. Loci differentially affected by selection in two American black populations. Proc Natl Acad Sci 68: 2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo J-M, Wambebe C, Tishkoff SA, et al. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci 107: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Tian M, Zong M, Teng M, Chen Y, Lu J, Jiang J, Liu X, Han J 2009. Proteomic analysis of pancreatic ductal adenocarcinoma compared with normal adjacent pancreatic tissue and pancreatic benign cystadenoma. Pancreatology 9: 89–98 [DOI] [PubMed] [Google Scholar]
- Davila S, Froeling FE, Tan A, Bonnard C, Boland GJ, Snippe H, Hibberd ML, Seielstad M 2010. New genetic associations detected in a host response study to hepatitis B vaccine. Genes Immun 11: 232–238 [DOI] [PubMed] [Google Scholar]
- Erdman LK, Cosio G, Helmers AJ, Gowda D, Grinstein S, Kain KC 2009. CD36 and TLR interactions in inflammation and phagocytosis: Implications for malaria. J Immunol 183: 6452–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselens C, Malapeira J, Colome N, Casal C, Rodriguez-Manzaneque JC, Canals F, Arribas J 2010. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J Biol Chem 285: 2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK 2003. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, et al. 2006. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci 103: 14068–14073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AE, Ghansa A, Small KS, Palma A, Auburn S, Diakite M, Green A, Campino S, Teo YY, Clark TG, et al. 2009. Positive selection of a CD36 nonsense variant in sub-Saharan Africa, but no association with severe malaria phenotypes. Hum Mol Genet 18: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW 2003. Perspective: Detecting adaptive molecular polymorphism: Lessons from the MHC. Evolution 57: 1707–1722 [DOI] [PubMed] [Google Scholar]
- Goran MI 2008. Ethnic-specific pathways to obesity-related disease: The Hispanic vs. African-American paradox. Obesity (Silver Spring) 16: 2561–2565 [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A 2008. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet 4: e32 doi: 10.1371/journal.pgen.0040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R 1996. R: A language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- The International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ 2006. Cancer statistics, 2006. CA Cancer J Clin 56: 106–130 [DOI] [PubMed] [Google Scholar]
- Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, et al. 2008. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M 2003. The neutral theory of molecular evolution. Cambridge University Press, Cambridge [Google Scholar]
- Kresse SH, Ohnstad HO, Paulsen EB, Bjerkehagen B, Szuhai K, Serra M, Schaefer KL, Myklebost O, Meza-Zepeda LA 2009. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer 48: 679–693 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski D 1999. The molecular genetic approach to malarial pathogenesis and immunity. Parassitologia 41: 233–240 [PubMed] [Google Scholar]
- Kwiatkowski DP 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet 77: 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Li Y, Dinwiddie DL, Harrod KS, Jiang Y, Kim KC 2010. Anti-inflammatory effect of MUC1 during respiratory syncytial virus infection of lung epithelial cells in vitro. Am J Physiol Lung Cell Mol Physiol 298: L558–L563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC 1991. The genetic structure of admixed populations. Genetics 127: 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Tajima K, Suzuki T, Kawase T, Watanabe M, Shitara K, Misawa K, Ito S, Sawaki A, Muro K, et al. 2009. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int J Cancer 125: 1961–1964 [DOI] [PubMed] [Google Scholar]
- McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol 84: 380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer M 1993. Slavery: A world history. Da Capo Press, New York [Google Scholar]
- Nielsen R 2005. Molecular signatures of natural selection. Annu Rev Genet 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG 2007. Recent and ongoing selection in the human genome. Nat Rev Genet 8: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksyk TK, Smith MW, O'Brien SJ 2010. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci 365: 185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, Tokunaga K 2002. Fcγ receptor IIA and IIIB polymorphisms are associated with susceptibility to cerebral malaria. Parasitol Int 51: 361–366 [DOI] [PubMed] [Google Scholar]
- Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, Tokunaga K 2003. CD36 polymorphism is associated with protection from cerebral malaria. Am J Hum Genet 72: 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquendo P, Hundt E, Lawler J, Seed B 1989. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell 58: 95–101 [DOI] [PubMed] [Google Scholar]
- Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, et al. 2004. Methods for high-density admixture mapping of disease genes. Am J Hum Genet 74: 979–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D 2006. Population structure and eigenanalysis. PLoS Genet 2: e190 doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RM, Teixeira KL, Barreto-de-Souza V, Calegari-Silva TC, De-Melo LD, Soares DC, Bou-Habib DC, Silva AM, Saraiva EM, Lopes UG 2010. Novel role for the double-stranded RNA-activated protein kinase PKR: Modulation of macrophage infection by the protozoan parasite Leishmania. FASEB J 24: 617–626 [DOI] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD 2001. Population structure in admixed populations: Effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet 68: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP 1994. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med 330: 1639–1644 [DOI] [PubMed] [Google Scholar]
- Praml C, Schulz W, Claas A, Mollenhauer J, Poustka A, Ackermann R, Schwab M, Henrich KO 2008. Genetic variation of Aflatoxin B1 aldehyde reductase genes (AFAR) in human tumour cells. Cancer Lett 272: 160–166 [DOI] [PubMed] [Google Scholar]
- Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD, et al. 2008. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet 83: 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 5: e1000519 doi: 10.1371/journal.pgen.1000519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed TE 1969. Caucasian genes in American Negroes. Science 165: 762–768 [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P 2005. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet 37: 1113–1118 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, Kato S, et al. 2008. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet 40: 730–740 [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Kimmel G, Halperin E, Jordan MI 2008a. On the inference of ancestries in admixed populations. Genome Res 18: 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, Halperin E 2008b. Estimating local ancestry in admixed populations. Am J Hum Genet 82: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, O'Brien SJ 2005. Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nat Rev Genet 6: 623–632 [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E 2004. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 74: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stannard D 1993. American Holocaust. Oxford University Press, Oxford [Google Scholar]
- Sundquist A, Fratkin E, Do CB, Batzoglou S 2008. Effect of genetic divergence in identifying ancestral origin using HAPAA. Genome Res 18: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ 2005. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol 28: 289–301 [DOI] [PubMed] [Google Scholar]
- Tang H, Coram M, Wang P, Zhu X, Risch N 2006. Reconstructing genetic ancestry blocks in admixed individuals. Am J Hum Genet 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ 2007. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet 81: 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H 1999. The slave trade. The story of the Atlantic slave trade: 1440–1870. Simon and Schuster, New York [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD 2005. The international HapMap project web site. Genome Res 15: 1592–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG 2005. Measures of human population structure show heterogeneity among genomic regions. Genome Res 15: 1468–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winichagoon P, Fucharoen S, Chen P, Wasi P 2000. Genetic factors affecting clinical severity in β-thalassemia syndromes. J Pediatr Hematol Oncol 22: 573–580 [DOI] [PubMed] [Google Scholar]
- Wood ET, Stover DA, Slatkin M, Nachman MW, Hammer MF 2005. The β-globin recombinational hotspot reduces the effects of strong selection around HbC, a recently arisen mutation providing resistance to malaria. Am J Hum Genet 77: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman PL, Blumberg BS, Cooper AJ 1963. Selection, gene migration and polymorphic stability in a U. S. white and negro population. Am J Hum Genet 15: 429–437 [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, Seminara D, Yoshida T, Saeki N, Andrew AS, et al. 2009. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet 41: 991–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jin L 2008. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet 83: 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Huang W, Wang H, He Y, Wang Y, Wang Y, Qian J, Xiong M, Jin L 2007. Dissecting linkage disequilibrium in African-American genomes: roles of markers and individuals. Mol Biol Evol 24: 2049–2058 [DOI] [PubMed] [Google Scholar]
- Xu S, Huang W, Qian J, Jin L 2008. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet 82: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jin W, Jin L 2009. Haplotype-sharing analysis showing Uyghurs are unlikely genetic donors. Mol Biol Evol 26: 2197–2206 [DOI] [PubMed] [Google Scholar]
- Xu S, Gupta S, Jin L 2010. PEAS V1.0: a package for elementary analysis of SNP data. Mol Ecol Resour 10: 1085–1088 [DOI] [PubMed] [Google Scholar]
- Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, et al. 2009. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet 41: 1055–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CC, Chen WM, Chen TH, Chen WY, Chen PC, Chiou HJ, Hung GY, Wu HT, Wei CJ, Shiau CY 2009. Identification of chromosomal aberrations associated with disease progression and a novel 3q13.31 deletion involving LSAMP gene in osteosarcoma. Int J Oncol 35: 775–788 [DOI] [PubMed] [Google Scholar]
- Zakharia F, Basu A, Absher D, Assimes TL, Go AS, Hlatky MA, Iribarren C, Knowles JW, Li J, Narasimhan B, et al. 2009. Characterizing the admixed African ancestry of African Americans. Genome Biol 10: R141 doi: 10.1186/gb-2009-10-12-r141 [DOI] [PMC free article] [PubMed] [Google Scholar]