Abstract
Diabetes is the most common metabolic disorder and is recognized as one of the most important health threats of our time. MicroRNAs (miRNAs) are a novel group of non-coding small RNAs that have been implicated in a variety of physiological processes, including glucose homeostasis. Recent research has suggested that miRNAs play a critical role in the pathogenesis of diabetes and its related cardiovascular complications. This review focuses on the aberrant expression of miRNAs in diabetes and examines their role in the pathogenesis of endothelial dysfunction, cardiovascular disease, and diabetic retinopathy. Furthermore, we discuss the potential role of miRNAs as blood biomarkers and examine the potential of therapeutic interventions targeting miRNAs in diabetes.
Keywords: MicroRNAs, Diabetes, Cardiovascular complications
1. Introduction
Diabetes mellitus (DM) is a complex, multisystem disease that represents the most common metabolic disorder,1,2 affecting around 8% of the US population.3 Type 1 diabetes mellitus (T1DM) results from insulin deficiency,4 usually secondary to autoimmune β-cell destruction; and type 2 diabetes mellitus (T2DM) is characterized by insulin resistance, with or without abnormal insulin secretion.5 Although T2DM is far more prevalent, both types can result in complications. Many of the complications of diabetes are vascular in origin, be they macrovascular and/or microvascular (nephropathy, retinopathy, and microangiopathy in several organs), and this puts diabetics at an increased risk of ischaemic heart disease, renal failure, stroke, lower limb amputations, and blindness.6
MicroRNAs (miRNAs) are a family of small (∼22 nucleotide), noncoding single-strand RNA molecules that were first discovered in the nematode Caenorhabditis elegans in 1993.7,8 Transcription of miRNAs occurs through RNA polymerase II9 and subsequent processing is mediated by the nuclear ribonuclease III (RNase III) enzyme Drosha to form precursor miRNAs (70–100 nucleotides). Following transportation to the cytoplasm by exportin 5, a further cleavage occurs via another RNase III enzyme, Dicer, to form the mature miRNA.10 miRNAs modulate both physiological and pathological pathways by post-transcriptionally inhibiting the expression of a plethora of target genes.11 Much work has been done on the role of miRNAs in human disease, especially in cancers and infections.12,13
The aim of this review is to describe the role of miRNAs in diabetes and its cardiovascular complications, with reference to the recent research. Specifically, we look at changes in miRNA expression in diabetes, as well as their role in endothelial dysfunction, angiogenesis, cardiac disease, and retinopathy. We do not discuss diabetic nephropathy, as the role of miRNAs in this complication is well-described in another review.14 We also discuss the potential role of miRNAs as biomarkers in diabetes and how aberrant pathways could be corrected therapeutically. A literature search was performed using PubMed and Embase to look for relevant papers. The last search was performed in September 2011. The key search words used were diabetes, glycaemia, microRNA, and miRNA. References of eligible papers were screened for further relevant studies.
2. MicroRNAs, insulin secretion, and β-cell function
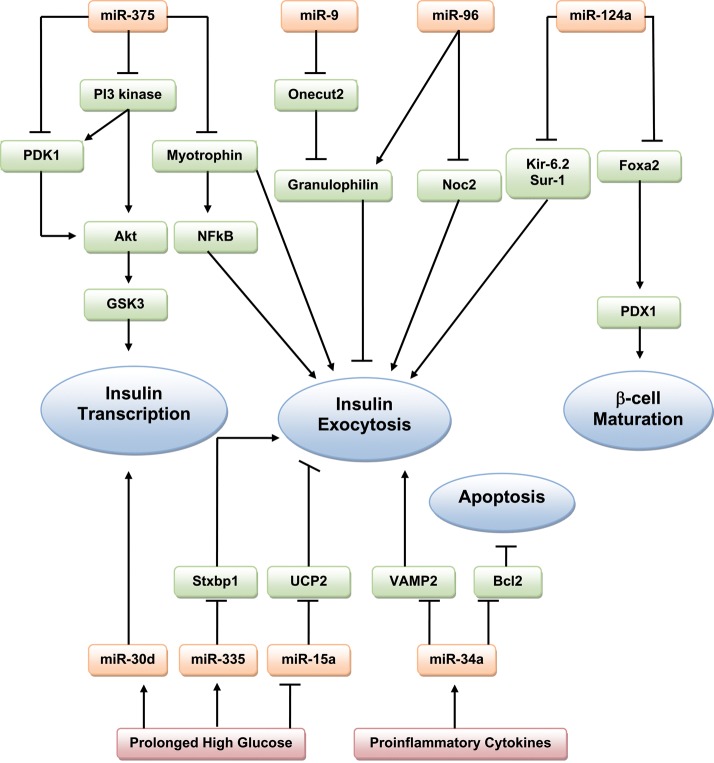
The pancreatic β-cell and its endocrine product insulin play a central role in glucose homeostasis and the pathogenesis of diabetes. A large number of miRNAs have been implicated in normal pancreatic development and function. Moreover, given the complex interplay between many miRNAs in normal pancreatic cells, it is expected that aberrant miRNA expression or mutations could result in β-cell pathology. miR-375, one of the most abundant miRNAs present in islet cells, is extensively studied in this context. miR-375 negatively regulates glucose-stimulated insulin secretion (GSIS).15 In fact, inhibition of miR-375 enhances insulin secretion, while miR-375 overexpression impairs the insulin secretory pathway by reducing expression of myotrophin (Mtpn), a protein involved in insulin–granule fusion.15,16 The effects of miR-375 on Mtpn expression may involve the transcription factor nuclear factor-kappaB (NF-kB), the activation of which is associated with improved GSIS.17 Indeed, myotrophin functions as a transcription activator of NF-kB in cardiomyocytes, suggesting that the regulation of myotrophin by miR-375 may lead to changes in NF-kB activity.18 miR-375 also targets insulin gene expression, downregulating expression of phosphoinositide-dependent protein kinase-1, a key component in the phosphatidylinositol 3-kinase (PI 3-kinase) signalling cascade, thus resulting in decreased insulin-induced phosphorylation of AKT and GSK3 (glycogen synthase kinase 3).19 High levels of miR-375 are found in the pancreatic islet of ob/ob mice (which represent a model of obesity, insulin resistance, and T2DM),20 and in patients with T2DM.21 Surprisingly, miR-375 knockout mice are hyperglycaemic and glucose intolerant; changes that occurred secondary to a decrease in β-cell mass. They also show increased numbers of α-cells and an elevated plasma glucagons.20
Several other miRNAs have been shown to have inhibitory roles in insulin secretion. miR-9 targets the transcription factor Onecut 2, which inhibits the expression of Granuphilin—a negative regulator of insulin exocytosis.22 Overexpression of miR-9 therefore decreases GSIS. miR-96 also downregulates insulin secretion by decreasing the expression of nucleolar complex protein 2 (Noc2), a Rab GTPase effector required for insulin exocytosis, as well as by upregulation of granuphilin, although this appears to occur via a distinct mechanism not involving Onecut-2.23 miR-124a, which is thought to be vital for pancreatic β-cell development,24 also modulates several components of the exocytotic system by directly targeting forkhead box protein A2 (Foxa2)—a transcription factor involved in glucose metabolism and insulin secretion.25 Modulation of miR-124a in MIN6 (mouse insulinoma) cells causes changes in Foxa2 and its downstream target gene PDX-1 (which regulates insulin transcription). miR-124a overexpression also correlates with downregulation of Kir-6.2 and Sur-1 (sulfonylurea receptor 1), both significant regulators of pancreatic development and function. Overexpression of mir-124a in MIN6 cells leads to increased insulin secretion in response to basal glucose concentrations and reduced secretion in response to stimulatory glucose concentrations.23
The destruction of pancreatic β-cells is the primary cause of T1DM. In the early stages, pancreatic islets are infiltrated by immune cells, hence β-cells are exposed to proinflammatory cytokines, resulting in altered insulin content, insulin secretion, and sensitisation to apoptosis.26 High levels of miR-34a were seen in islets from T2DM db/db mice.27 Moreover, MIN6 cells treated with proinflammatory cytokines show significant induction of miR-21, miR-34a, and miR-146.28 Subsequent blockade of these miRs prevented cytokine-induced reduction in GSIS and protected β-cells from cytokine-induced cell death. Experimental chronic exposure to the free fatty-acid palmitate mimics the adverse environmental conditions that promote failure of β-cells, arising in defective GSIS. A further study found that exposure of insulin-secreting cell lines or pancreatic islets to palmitate led to an increase in miR-34a and miR-146 expression.27 Overexpression of miR-34a in MIN6 cells resulted in a decreased GSIS, along with a reduction in the expression of the antiapoptotic protein Bcl2 and of VAMP2 (vesicle-associated membrane protein 2), which is involved in β-cell exocytosis.27 While antagonism of either miR-34a or miR-146 activity partially prevented palmitate-induced β-cell apoptosis, normal secretory activity was not restored,27 suggesting that palmitate may affect other components of the exocytotic machinery that are not targeted by the two studied miRNAs.
Diabetes results in prolonged periods of hyperglycaemia. Prolonged exposure of MIN6 cells to high glucose (HG) results in differential expression of large numbers of miRNAs, including upregulation of miR-30d.29 Overexpression of miR-30d increases insulin gene expression, whereas its inhibition attenuates glucose-stimulated insulin gene transcription.29 In contrast, miR-30d does not modulate insulin secretion.29 miR-15a promotes insulin biosynthesis by inhibiting endogenous UCP-2 (uncoupling protein-2) expression in mouse β-cells.30 UCP-2 is a member of the mitochondrial inner membrane family, and has previously been shown to inhibit GSIS.31 miR-15a levels have been shown to increase in parallel with insulin in mouse islets after short-term exposure to HG concentrations.30 However, prolonged exposure to hyperglycaemia resulted in a downregulation of both miR-15a and insulin. Transfection of MIN6 cells with miR-15a increases insulin secretion, and inhibition of miR-15a decreases insulin levels.30 Esguerra et al.32 studied the differential expression of miRNAs in the pancreatic islets of Wistar and Goto-Kakizaki (GK) rats—a non-obese model of T2DM that displays hyperglycaemia, impaired glucose tolerance (IGT), insulin resistance, and defects in insulin secretion. miR-335 was upregulated in the pancreatic islets of GK rats, and was shown to target the messenger RNA (mRNA) for the exocytotic protein Stxbp1.32 However, many of the differentially expressed miRNAs had predicted target genes that are known to be involved in insulin exocytosis.
Figure 1 summarizes the known targets of miRNAs in β-cell function.
Figure 1.
Schematic overview of the role of microRNAs in β-cell function.
3. MicroRNAs in insulin target tissues: energy metabolism and insulin resistance
miRNAs also control insulin signalling in target tissues, including the liver, skeletal muscle, and adipose tissues. Insulin resistance describes the failure of target tissues to respond adequately to circulating insulin. Insulin resistance in adipose tissue and skeletal muscle reduces glucose uptake and the local storage of triglycerides and glycogen. Insulin resistance in liver cells leads to reduced glycogen synthesis and storage and concomitant failure to suppress glucose production. The end results are elevated blood glucose and free fatty acid levels, along with raised insulin levels—hallmarks of the metabolic syndrome. If there is no sufficient compensatory increase in pancreatic β-cell function to counteract the insulin resistance, then T2DM results.33
3.1. Adipose tissue
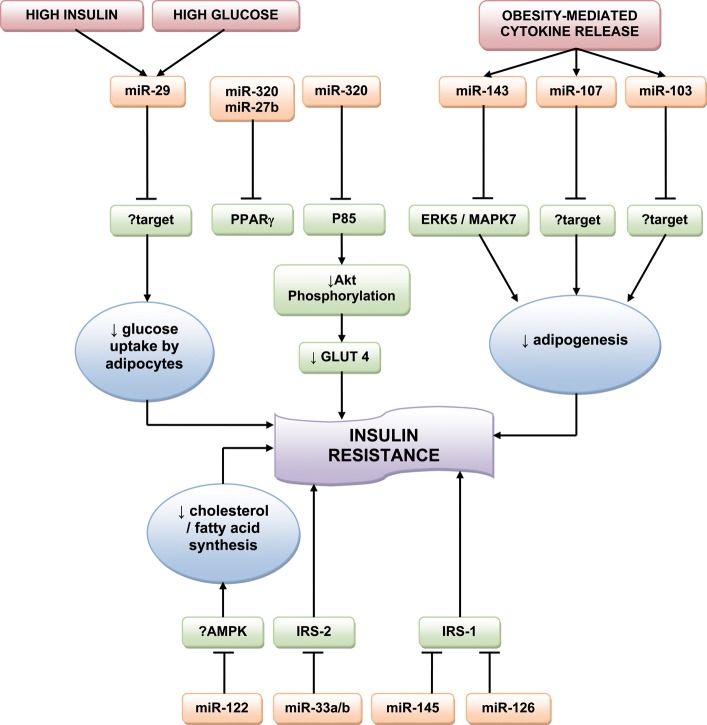
Early studies in Drosophila melanogaster implicated miRNAs in energy metabolism. Xu et al.34 found that miR-14 regulates adipocyte droplet size and triglyceride levels, with miR-14-null flies showing increased lipid droplet accumulation in adipose tissue that reverted on re-introduction of the miRNA. Another miRNA of Drosophila energy homeostasis, miR-278, may control insulin sensitivity in adipose tissue: miR-278-knockout flies are lean and display hyperglycaemia despite having an elevated insulin production, indicating a loss of insulin sensitivity.35 This idea is supported by the inappropriately high levels of insulin target genes found in the mutants. miR-278 acts by regulating the target gene expanded, and overexpression of this gene resulted in the same features. Although miR-14 and miR-278 have only been found in insects, a loss of insulin signalling in mammalian adipose tissue may result in a similar insulin-resistant phenotype. For example, mice that have had the insulin receptor gene deleted from adipose tissue are lean.36
He et al.37 examined the miRNA expression profiles of skeletal muscle from healthy Wistar and GK rats.37 The miR-29 family was significantly upregulated in the context of diabetes. Further in vitro study on 3T3-L1 adipocytes suggested that miR-29a and miR-29b were upregulated by HG and insulin levels. Moreover, miR-29a/b/c overexpression reduced insulin-induced glucose import by 3T3-L1 adipocytes, signifying a role in insulin resistance, and this was paralleled by a decrease in Akt activation, suggesting that the miR-29 family acts by silencing components of the insulin signalling pathway.37 However, while in vitro inhibition of the miR-29 family improved Akt activation, it had little effect on glucose uptake. This may be due in part to a relatively low endogenous expression of miR-29, but could also be explained by miR-29 acting on other targets within the insulin signalling pathway, as yet undetermined, rather than having a direct effect on Akt.37 Two genes were also validated as direct targets of the miR-29 family: Insig1 (insulin-induced gene 1)—an endoplasmic reticulum membrane protein involved in the control of cholesterol biosynthesis; and Cav2 (caveolin 2), which is involved with lipid metabolism, cellular growth, and apoptosis.37
Insulin stimulates lipogenesis in adipose tissue, transforming blood glucose into fatty acids for storage of energy. Obesity triggers macrophage infiltration and cytokine release in adipose tissue, and many of these cytokines, such as TNFα, interfere with insulin signalling and inhibit adipogenesis.38 Some miRNAs that are induced during adipogenesis are downregulated in obesity.39 For example, miR-103 and miR-143 are upregulated during in vitro and in vivo adipogenesis, and inhibition of miR-143 inhibits adipocyte differentiation.39,40 Both miR-103 and miR-143 are downregulated in the adipocytes from ob/ob mice.39 miR-143 antagonism in adipocytes results in upregulation of the miR-143 target mitogen-activated protein kinase ERK5/MAPK7,40 which is known to promote cell proliferation and differentiation, although the role of ERK5/MAPK7 in adipocytes has not been investigated. miR-107 has also been shown to accelerate adipogenesis and is predicted to target pathways that regulate lipid levels.41 Levels of miR-143, miR-103, and miR-107 are reduced in adipocytes after treatment with TNFα, suggesting that cytokines contribute to reduced adipogenesis in obesity.39 These data indicate that obesity leads to a loss of miRNA function that is required for adipogenesis, and suggest a mechanism for obesity-induced insulin resistance. Other miRNAs that may be involved in insulin resistance include miR-320 and miR-27b. miR-320 expression in insulin-resistant adipocytes is 50-fold that of normal 3T3-L1 adipocytes. Ling et al.42 found that treatment of insulin-resistant adipocytes with anti-miR-320 increases the insulin sensitivity by targeting p85, which contributes to cell growth by increasing Akt phosphorylation and GLUT4 levels. miR-27b is downregulated during adipogenesis from human multipotent adipose-derived stem cells.43 Moreover, overexpression of miR-27b impairs human adipocyte differentiation and inhibits the peroxisome proliferator-activated receptor (PPARγ), the receptor target for thiazolidinediones—insulin-sensitising agents used for treating T2DM.43,44 More recently, miR-130 overexpression was also found to impair adipogenesis and to repress PPARγ.45 These connections between miRNA expression and adipogenesis may be exploited as therapeutic targets in the management of insulin resistance.
3.2. Liver
Once secreted from the islet, insulin travels via the portal circulation to the liver, to control hepatic glucose and lipid metabolism, and liver insulin resistance contributes to the development of the metabolic syndrome. miR-122 is the most abundant miRNA in the liver.46 Inhibition of miR-122 in mice results in decreased hepatic fatty acid and cholesterol synthesis, along with a reduction in plasma cholesterol.47 In addition, hepatic inhibition of miR-122 in diet-induced obese mice led to decreased plasma cholesterol, as well as a significant improvement in hepatic steatosis. While the authors noted an increase in phosphorylated AMP-activated protein kinase (AMPK) in the livers of these mice, they were unable to clarify whether miR-122 directly regulated AMPK signalling. In any case, targeting miR-122 therapeutically may correct the imbalances seen in liver insulin resistance.
miR-33a and miR-33b have been shown to regulate cholesterol homeostasis through interaction with sterol regulatory element-binding proteins.48 Davalos et al.49 have recently reported the role of these two miRNAs in regulating fatty acid metabolism and insulin signalling. miR-33a/b inhibit the expression of insulin receptor substrate-2 (IRS-2) in hepatic cells, subsequently reducing the activation of downstream insulin signalling pathways, including AKT and ERK. Antagonism of endogenous miR-33-a/b upregulates fatty acid oxidation and the response to insulin in hepatocytes, suggesting its therapeutic potential in the metabolic syndrome.49
The insulin receptor substrate-1 (IRS-1), like IRS-2, is a significant mediator of insulin signalling. Indeed, IRS-1 knockout mice are insulin-resistant.50 Mitochondrial dysfunction is associated with the development of insulin resistance as well as with downregulation of IRS-1 in myocytes.51 Ryu et al.52 found that miR-126 was upregulated in the context of mitochondrial dysfunction in SK-Hep-1 (hepatic cancer) cells, and reduced expression of IRS-1. miR-145 has been shown to downregulate IRS-1 protein expression in human colon cancer cells, resulting in adverse growth and proliferation,53 although this miRNA needs further study in the context of diabetes.
A summary of the miRNAs involved in insulin resistance is given in Figure 2.
Figure 2.
MicroRNAs involved in insulin resistance.
3.3. Skeletal muscle
Huang et al.54 found that miR-24 was significantly downregulated in the skeletal muscle of GK rats. p38 mitogen activated protein kinase (MAPK) is activated by hyperglycaemia,55 is overexpressed in GK rats,56 and is involved in early diabetic nephropathy in T2DM.57 Huang et al.54 confirmed that p38 MAPK is a direct target of miR-24 in rat tissue.
A list of miRNAs differentially expressed in diabetic models can be found in Table 1.
Table 1.
List of microRNAs differentially expressed in diabetic models, by tissue, and their potential targets
| Tissue | Reference | Model | miRNA changes | Up/down-regulated in diabetes? | Potential targets |
|---|---|---|---|---|---|
| Skeletal muscle | He et al.37 | GK rat | miR29 family (a/b/c) | Up | Akt |
| Huang et al.54 | GK rat | miR24 | Down | p38 MAPK | |
| Herrera et al.119 | GK rat | miR10b | Down | ||
| Gallagher et al.73 | Humans | miR133 | Down | ||
| Caporali et al.74 | Humans and diabetic mice | miR503 | Up | CDC25a,CCNE1 | |
| Liver | Herrera et al.119 | GK rat | miR195, miR103 | Up | |
| Herrera et al.119 | GK rat | miR125a | Up | MAPK pathway | |
| Adipose Tissue | Herrera et al.120 | GK rat | miR222, miR 27a | Up | |
| Herrera et al.120 | GK rat | miR125a | Up | MAPK pathway | |
| Omentum | Kloting et al.121 | Humans | miR181a | Up | |
| miR17-5p, miR132miR 134 | Down | ||||
| Subcutaneous fat | Kloting et al.121 | Humans | miR147, miR197 | Up | |
| miR27a, miR30e, miR140, miR155, miR210 | Down | ||||
| Endothelial Cells | Wang et al.64 | MMVECs | miR320 | Up | VEGF, FGF, IGF-1, IGF-1-R |
| Li et al.68 | HUVECS | miR221 | Up | ckit, p27kip1, p57kip2 | |
| Villeneuve et al.71 | VSMC | miR125b | Up | suv39h1 | |
| Caporali et al.74 | HUVECS/HMVECS | miR503 | Up | CDC, CCNE1 | |
| Cardiomyocytes | Feng et al.81 | STZ mouse | miR133a | Down | IGF-1-R and SGK1 |
| Zhang et al.84 | Diabetic rabbit | miR133, miR1 | Up | HERG (133 only) | |
| Yu et al.88 | Rat | miR1 | Up | IGF-1 | |
| Katare et al.91 | STZ mouse | miR1 | Up | Pim-1 | |
| Shan et al.89 | Rat | miR1, miR206 | Up | Hsp60 | |
| Retina | McArthur et al.95 | STZ rat | miR200b | Down | VEGF |
| Kovacs et al.96 | STZ rat | miR146, miR155, miR132, miR21 | Up | NF-kB |
4. Endothelial function and angiogenesis
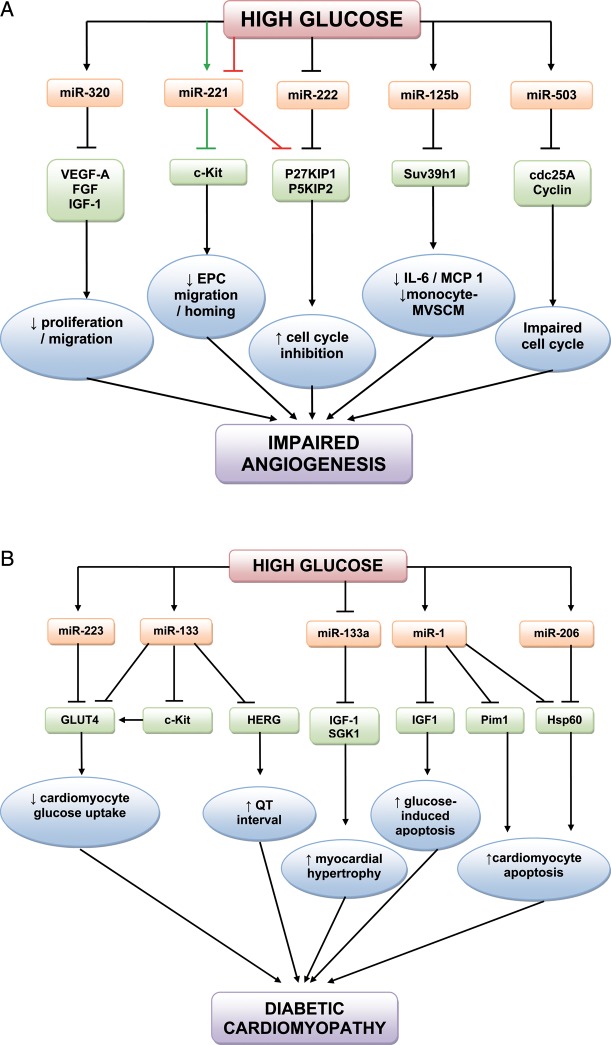
Vascular complications associated with hyperglycaemia in diabetes often begin with endothelial dysfunction.58 Altered expression of multiple factors results in capillary and arteriole rarefaction in limbs and heart modifications59 and a reduced post-ischaemic angiogenic and collateral vessel formation.60–62 Conversely, a pro-angiogenic pathogenic phenotype is found in the retina.63 Wang et al.64 were the first to describe that miRNAs are differently expressed in endothelial cells (ECs) in the presence of HG. They studied myocardial microvascular ECs (MMVEC) and compared miRNA expression in GK and Wistar rats. Of those differentially expressed, miR-320 may target several angiogenic factors and their receptors, including vascular endothelial growth factor (VEGF)-A, fibroblast growth factors (FGFs), insulin-like growth factor 1 (IGF-1), and the IGF-1 receptor.65,66 Moreover, elevated miR-320 level in diabetic MMVECs was accompanied by decreased cell proliferation and migration. Subsequent transfection of an miRNA-320 inhibitor in MMVECs of GK rats improved both proliferation and migration of these cells and increased the expression of IGF-1, which is known to promote angiogenesis.67 The correlation between elevated miR-320 and suppressed IGF-1 may play a role in the impaired angiogenesis in diabetes.
Li et al.68 studied the role of miR-221 in diabetes-induced endothelial dysfunction. miR-221, which is present in human umbilical vein ECs (HUVECs), participates in angiogenesis regulation by altering the expression of c-kit (CD117), the receptor for stem cell factor which additionally promotes endothelial progenitor cell (EPC) migration and homing.69 Incubating HUVECs in HG increased miR-221 expression and reduced c-kit expression, while a miR-221 inhibitor reversed this inhibitory effect of HG on c-kit expression.68 Another study by Togliatto et al.70 examined the role of miR-221 and miR-222 in HG and advanced glycation end-product (AGE)-mediated vascular damage, both in HUVECs and in a model of angiogenesis (Matrigel plugs) in diabetic mice. They found that HG and high AGEs inhibited cell cycle progression and resulted in impaired EC and EPC proliferation as well as reduced angiogenesis. These conditions were also associated with downregulation of both miR-221 and miR-222 expression. Additionally, miR-221 and miR-222 were found to directly inhibit P27KIP1 and P57KIP2 (cyclin-dependent kinase inhibitor proteins that inhibit the cell cycle). Hence, these miRNAs are likely to be directly involved in AGE/HG-related cell cycle changes.70 The results regarding the regulation of miR-221 by glucose levels in HUVECs are contradictory. While there were some differences in the cell culture methods, the results in Togliatto et al.70 were confirmed in vivo. Clearly, further research is needed regarding the role of miR-221 in the context of HG. Villeneuve et al.71 examined the role of miR-125b in vascular smooth muscle cells (VSMCs) cultured from T2DM db/db mice. They found that HG upregulated miR-125b, with parallel downregulation of its predicted target Suv39h1, a histone-lysine N-methyltransferase. The reduced recruitment of Suv39h1 at inflammatory gene promoters is a key mechanism underlying the enhanced inflammatory gene expression in db/db microvascular VSMCs (MVSMCs).72 miR-125b mimics inhibited Suv39h1 protein levels and miR-125b inhibitors had the opposite effect. Furthermore, miR-125b-mediated Suv39h1 knockdown resulted in the increased expression of inflammatory proteins (interleukin 6 and monocyte chemotactic protein 1) and increased monocyte-MVSMC binding in hyperglycaemia, thus exhibiting a role for miR-125b in accelerating atherosclerosis.71
miR-503 is upregulated in myocardial ECs from GK rats,64 in 3T3-L1 insulin-resistant adipocytes,42 and in the muscles of T2DM and insulin-resistant patients.73 We further examined the role of miR-503 in angiogenesis in diabetes.74 miR-503 was upregulated in HUVECs and human microvascular ECs (HMVECs) cultured in HG/low growth factor conditions (that mimic ischaemia-induced tissue starvation in the context of diabetes). HUVECs infected with a lentiviral vector expressing premiR-503 showed impaired proliferation, migration, and cell networking capacities. Under similar conditions, miR-503 reduced expression of the cell cycle regulators cdc25A and cyclin E1 (CCNE1). Furthermore, miR-503 inhibition in HG/low growth factor conditions restored normal EC proliferation and angiogenesis.74 We went on to examine the role of miR-503 in diabetic limb ischaemia to find that diabetes increased miR-503 expression in the ischaemic muscles and ECs extracted from them in comparison to non-diabetic/non-ischaemic controls. Moreover, in diabetic mice with induced limb ischaemia, local miR-503 inhibition by injection of an adenoviral vector containing a decoy sequence for miR-503 improved capillary and arteriolar density, promoted blood flow recovery, and normalized the expression of cdc25 and CCNE1. Importantly, miR-503 expression is also increased in the limb muscles and plasma of diabetic patients undergoing amputation for critical limb ischaemia compared with calf biopsies of non-diabetic/non-ischaemic controls.74 Our study demonstrates that miR-503 may be a suppressor of post-ischaemic neovascularization in diabetes and thus a potential therapeutic target. The roles of miRNAs in endothelial dysfunction and angiogenesis in diabetes are summarized in Figure 3A.
Figure 3.
(A) MicroRNAs involved in endothelial dysfunction and angiogenesis in the hyperglycaemic environment. Note the conflicting effects of HG on miR-221 from two separate studies (green arrows68 and red arrows70). (B) microRNAs in cardiovascular disease in diabetes.
Atherosclerosis is a major vascular complication of diabetes. Although there is little research on miRNAs in atherosclerosis in the context of diabetes, the role of miRNAs in the pathogenesis of atherosclerosis in general has been the subject of two recent reviews.75,76
5. Cardiac disease
miR-133 is believed to be expressed specifically in cardiac and skeletal muscle, with its function in skeletal muscle being to modulate myoblast proliferation and differentiation.77 Moreover, miR-133 controls cardiac hypertrophy and is downregulated in failing and hypertrophic hearts.78 The GLUT4 glucose transporter is the major mechanism by which glucose uptake into cardiomyocytes can be increased.79 Horie et al.80 found that miR-133 overexpression lowered GLUT4 levels and reduced insulin-induced glucose uptake in cardiomyocytes. Additionally, increased miR-133 also reduces the Krüppel-like transcription factor KLF15, which induces GLUT4 expression. Furthermore, transfection of cardiac myocytes with a miR-133 decoy increased expression of both KLF15 and GLUT4, suggesting a direct role in cardiac glucose transport.
Feng et al.81 looked at the role of miR-133a in cardiomyocytes in the context of streptozotocin (STZ)-induced T1DM in mice. Haemodynamic studies confirmed that the T1DM model resulted in cardiac hypertrophy and poor contractility, and the diabetic hearts displayed increased expression of MEF2A and MEF2C, two transcription factors associated with myocardial hypertrophy. Cardiac tissue from the diabetic mice displayed significant downregulation in miR-133a expression. In addition, culturing rat neonatal cardiomyocytes in HG also resulted in downregulation of miR-133a expression.81 In contrast, transfection of rat neonatal cardiomyocytes with miR-133a mimics prevented HG-induced cardiomyocyte hypertrophy, and miR-133a was shown to mediate its effects by preventing HG-induced upregulation of IGF-1 receptors and SGK1.82
A prolonged QT interval, an adverse cardiac feature of diabetes, can result in arrhythmias and has been suggested as an independent predictor of mortality in DM.83 Zhang et al.84 confirmed a 20% prolongation of the QT interval in diabetic rabbits compared with controls. This occurs as a result of dysfunction of multiple ion currents/channels, predominantly the IK/HERG (human ether-a-go-go) channel. The same group found that levels of miR-133 and miR-1 were significantly upregulated in the hearts of diabetic rabbits compared with controls.85 Furthermore, miR-133 overexpression reduced HERG protein levels, while miR-133 inhibition partially reversed this. This suggests a role for miR133 dysfunction in prolonging the QT interval, and causing the resultant arrhythmias, in diabetic hearts.85 The above two studies suggest that miR-133 has two potential roles in the diabetic heart, depending upon whether expression is increased or decreased. While the significance of the differing findings has not been determined, there may be species- and/or age-specific differences determining the cardiac expression and function of miR-133 under hyperglycaemia.
Hyperglycaemia-induced apoptosis of cardiomyocytes is related to diabetic complications,86 although the mechanisms for this are not well-defined. IGF-1 is an anti-apoptosis factor which is mediated through mitochrondria and the cytochrome-c pathway.87 Yu et al.88 studied the effects of IGF-1 and miRNAs in HG-induced mitochondrial dysfunction. They found that IGF-1 exerted a protective effect towards apoptosis in rat cardiomyocytes by decreasing the cytotoxic effects of glucose. miR-1 is overexpressed in the hearts of diabetic patients.85 HG increased miR-1 expression, and miR-1 overexpression inhibits the antiapoptotic action of IGF-1.88 Heat shock protein (Hsp) 60 prevents apoptotic cardiomyocyte death, but has been shown to be underexpressed in the diabetic heart.89 Shan et al.89 examined miR-1 and miR-206 expression in the hyperglycaemic rat myocardium as well as in rat neonatal cardiomyocytes exposed to HG. They found that HG induced upregulation of miR-1 and miR-206, and that both miRNAs negatively regulated Hsp60 expression. Pim-1 (proviral integration site for Moloney murine leukaemia virus-1) plays a key role in the cardiac response to stressors.90 We found that in STZ-T1DM mice, Pim-1 levels decline during progression of diabetic cardiomyopathy, and this was associated with a rise in miR-1 expression.91 Furthermore, both forced Pim-1 expression and miR-1 inhibition rescued Pim-1 levels in cardiomyocytes under HG conditions, and this resulted in a restoration of prosurvival signalling and reduction in cardiomyocyte apoptosis, suggesting a direct role of miR-1 in inhibiting Pim-1.91
Another study looking into cardiomyocyte glucose metabolism found that miR-223 was upregulated in the left ventricle of T2DM patients. Moreover, miR-223 overexpression induces GLUT4 protein levels in cardiomyocytes,92 and in vivo miR-223 inhibition significantly decreased GLUT4 expression. Given that GLUT4 is downregulated in the diabetic heart, it is possible that increasing miR-223 levels is an adaptive response aimed to restore normal glucose uptake in the insulin-resistant heart.
The miRNAs involved in cardiovascular disease in diabetes are summarized in Figure 3B.
6. Retinopathy
Diabetic retinopathy (DR), one of the leading causes of blindness in developed countries,93 is a microvascular complication of diabetes with multiple underlying pathogenic mechanisms. The altered expression of growth factors in sustained hyperglycaemia results in both structural and functional changes in the retina.63 Elevated VEGF-A levels, increasing both vessel permeability and neovascularization, have long been associated with diabetic retinopathy.94 Only two published studies have specifically looked at the role of miRNAs in DR. McArthur et al.95 investigated whether miRNA alterations are involved in DR in a STZ-T1DM rat model, which is known to result in early changes of DR. miR-200b, which targets VEGF-A, was found downregulated in the retinas of diabetic rats. Moreover, in vitro exposure of HUVECs and bovine retinal capillary ECs (BRECs) to HG resulted in a downregulation of miR-200b and an upregulation in VEGF-A mRNA.95 Subsequent transfection of a miR-200b inhibitor in HUVECs cultured under normal conditions resulted in a gluco-mimetic effect of upregulating VEGF-A.95 VEGF-A-mediated vascular permeability and tube formation in HUVECs exposed to HG were reduced upon miR-200b mimic transfection.95 Additional experimentation was performed in animal models: miR-200b mimic injection into the vitreous humour of one eye of diabetic mice resulted in local decreased VEGF-A expression and vascular. Conversely, intravitreal injection of miR-200b antagomir increased VEGF-A expression, thus suggesting a role for miR-200b in the pathogenesis of diabetic retinopathy and raising support for intravitreal injections as a possible therapeutic route for this condition.95
Kovacs et al.96 performed miRNA expression profiling in the retina and retinal ECs of STZ-T1DM rats. NF-kB is a regulator of the immune response and is known to play an important role in the early pathogenesis of DR by triggering a pro-apoptotic program in retinal pericytes.97 miRNAs which are thought to be transcriptionally regulated by NF-kB (miR-146, miR-155, and miR- 21)98–100 were also demonstrated to be upregulated in the retinal ECs of diabetic rats.96 The authors confirmed that NF-kB was able to directly activate miR-146 expression. Furthermore, miR-146 overexpression inhibited interleukin-1β-induced NF-kB activation in retinal ECs, representing a regulatory negative feedback loop to control NF-kB and miR-146 expression.96 Thus, overexpression of miR-146 could be exploited therapeutically through inhibition of NF-kB activation in DR.
7. Biomarkers
There are currently no reliable plasma biomarkers for vascular disease and endothelial dysfunction. Furthermore, current imaging methods can visualize the latter stages of macrovascular disease, e.g. atherosclerotic plaque, but not endothelial dysfunction, particularly at the level of microcirculation. Levels of miRNAs in the serum of humans have been shown to be stable, reproducible, and consistent amongst healthy individuals, allowing them to be of potential value as biomarkers of disease.101 The stability of circulating miRNAs may be explained by their being carried in membrane-bound vesicles,102 complexed with Argonaute2,103,104 or transported by high-density lipoproteins.104 The potential role of miRNAs as biomarkers in wider cardiovascular disease is the subject of another review in this issue. The discovery of potential-specific biomarkers may help predict or detect the development and progression of diabetes and/or diabetes complications at an early stage, and therefore allow timely intervention to delay severe complications.
Zampetaki et al.105 aimed to understand if there is a plasma miRNA signature for T2DM using a prospective population-based cohort. They studied 80 patients with T2DM and a further 80 age- and gender-matched controls.105 In T2DM, miR-28-3p was overexpressed, and a further 12 miRNAs were underexpressed. These findings were confirmed in T2DM ob/ob mice. A decrease in circulating miR-126 in non-diabetic people was found to be a significant predictor of DM. In fact, the authors detected a gradual decline in plasma levels of miR-126 from normal glucose tolerance, through IGT, to diabetes. Some additional miRNAs were predictive of the future development of diabetes: miR-15a, miR-29b, miR-126, and miR-223 were lower in those subjects who went on the develop DM.105 Using expression profiles of the five most significantly different miRNAs (miR-15a, miR-126, miR-320, miR-223, and miR-28-3p), controls and diabetics were correctly identified in 92 and 70% of cases, respectively.105 Of note, the diabetic patients who were not detected by the profiling method had lower fasting glucose levels or had well-controlled diabetes. This research suggests that the use of a miRNAs signature as a biomarker may be a useful predictive tool in diabetes, although this theory will require confirmation in larger prospective populations and in groups with T1DM. miR-126 is known to be highly expressed in ECs and to regulate angiogenesis.106,107 It facilitates VEGF-A signalling by inhibiting two of the negative regulators of the VEGF pathway, the Sprouty-related protein SPRED1 and phosphoinositol-3 kinase regulatory subunit 2 (PIK3R2).106 miR-126 is also the most abundant miRNA in endothelial apoptotic bodies, which are thought to be a novel form of communication between cells.108 Zampetaki et al. showed that HG conditions reduced the miR-126 content in endothelial apoptotic bodies produced by HUVECs in vitro. The authors speculate that, as a consequence of the aforementioned, low delivery of miR-126 to monocytes in diabetes may result in the previously described reduced VEGF-A and subsequent endothelial dysfunction, resulting in defective collateral vessel development.109 Furthermore, decreasing miR-126 levels in plasma were associated with a low ankle brachial pressure index and subsequent new-onset peripheral vascular disease.
Another study explored seven diabetes-related serum miRNAs (miR-9, miR-29a, miR-34a, miR-30d, miR-124a, miR-146a, and miR-375) in patients with diabetes, IGT, and in people susceptible to T2DM with a normal oral glucose tolerance test.110 While all seven of the tested miRNAs were significantly upregulated in T2DM cases, there was no difference in the expressed levels between IGT cases and normal susceptible controls.110 However, the use of controls that were susceptible to T2DM, rather than those without this susceptibility, may have had a bearing on this result.
8. Therapeutic applications
The deregulation of miRNA function has been linked to diabetes, although it is not yet fully certain whether this is a cause or effect of the pathology. If miRNAs are indeed active in the pathogenesis of diabetes and its related complications, the restoration of normal function by modifying the expression of specific miRNAs may be a therapeutic target for managing this disease. Chemically modified siRNA-like oligonucleotides have been used to decrease miRNA expression (antagomirs) in vivo.111,112 However, due to the hypothetical transient nature of their effects, it is likely that frequent doses may be required to sustain benefit. Given the chronic nature of diabetes, this would require the need for repeated injections with their associated costs. However, a recent study treating chimpanzees with locked nucleic acid-modified oligonucleotides complimentary to miR-122 found that serum cholesterol remained low for over 10 weeks after cessation of treatment, suggesting that longer-lasting effects may be possible.113 Adeno-associated virus (AAV) vectors containing miRNA mimics have been found to promote miRNA expression in vivo.114 For example, Kota et al.115 injected mice with AAV.miR-26a in the context of hepatic cancer. The rise in miR-26a resulted in protection from cancer progression without signs of toxicity. Delivery of these agents to specific tissue targets poses a further problem. Different AAV serotypes have been shown to favour specific tissues, for example serotypes 6, 8, and 9 are predominantly directed to skeletal muscle, liver, and the heart, respectively.116 These results suggest the promises of AAV-mediated miRNA therapeutics. We have also successfully used an adenoviral vector to convey a decoy to inhibit miR-503, which is pathogenic in the setting of diabetic limb ischaemia.74 However, for their short transgene expression, adenovirus may not be the best approach to treat diabetes and its chronic complications in the clinical arena. Another therapeutic strategy involves ‘miRNA sponges’. These artificial miRNA decoys bind native miRNA to create a loss of function of a particular miRNA. These sponges contain multiple binding sites directed against a particular miRNA or against an miRNA seed sequence family.117 miRNA sponges have already been used in vivo to decrease the activity of miR-31 and its role in cancer development.118 The use of miRNA sponges in an AAV vector delivery system maybe a potential novel strategy for miRNA therapeutics. While the first reports on miRNA therapeutics are encouraging, the fact that a typical miRNA targets several genes suggests that clinical intervention may be very complex.
9. Conclusions
miRNAs belong to a class of non-coding RNAs which are involved in the pathogenesis of several diseases. Although many miRNAs have already been identified, their predicted target genes need to be fully researched and functionally characterized. The complexities in the pathogenesis of diabetes make this more challenging. Emerging evidence suggests that miRNAs are differentially expressed, and indeed have a potential causative role, in diabetes and its related cardiovascular complications. In future, these mechanisms may be exploited to help define specific clinical biomarkers and allow appropriate therapeutic intervention in the management of diabetes.
Conflict of interest: none declared.
Funding
S.S. is a British Heart Foundation (BHF) PhD student and C.E. is a BHF Senior Research Fellow. This article was supported by BHF grants (FS/10/001/27959 and FS/10/61/28566 to C.E.). Funding to pay the Open Access publication charges for this article was provided by the BHF.
References
- 1.Adams KF, Schatzkin A, Harris TB, Kipnis V, Mouw T, Ballard-Barbash R, et al. Overweight, obesity, and mortality in a large prospective cohort of persons 50 to 71 years old. N Engl J Med. 2006;355:763–778. doi: 10.1056/NEJMoa055643. [DOI] [PubMed] [Google Scholar]
- 2.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 9999;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. National Diabetes Factsheet: National Estimates and General Information on Diabetes and Prediabetes in the United States. Atlanta, GA: Department of Health and Human Services, Centers for Disease Control and Prevention; 2011. [Google Scholar]
- 4.Ilonen J, Akerblom HK. New technologies and genetics of type 1 diabetes. Diabetes Technol Ther. 1999;1:205–207. doi: 10.1089/152091599317440. [DOI] [PubMed] [Google Scholar]
- 5.Kahn SE. Clinical review 135: the importance of beta-cell failure in the development and progression of type 2 diabetes. J Clin Endocrinol Metab. 2001;86:4047–4058. doi: 10.1210/jcem.86.9.7713. [DOI] [PubMed] [Google Scholar]
- 6.Winer N, Sowers JR. Epidemiology of diabetes. J Clin Pharmacol. 2004;44:397–405. doi: 10.1177/0091270004263017. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods Mol Biol. 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3:140–155. [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolas FE, Lopez-Martinez AF. MicroRNAs in human diseases. Recent Pat DNA Gene Seq. 2010;4:142–154. doi: 10.2174/187221510794751659. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 15.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Xu X, Liang Y, Liu S, Xiao H, Li F, et al. miR-375 enhances palmitate-induced lipoapoptosis in insulin-secreting NIT-1 cells by repressing myotrophin (V1) protein expression. Int J Clin Exp Pathol. 2010;3:254–264. [PMC free article] [PubMed] [Google Scholar]
- 17.Norlin S, Ahlgren U, Edlund H. Nuclear factor-{kappa}B activity in {beta}-cells is required for glucose-stimulated insulin secretion. Diabetes. 2005;54:125–132. doi: 10.2337/diabetes.54.1.125. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Sen S. Myotrophin-kappaB DNA interaction in the initiation process of cardiac hypertrophy. Biochim Biophys Acta. 2002;1589:247–260. doi: 10.1016/s0167-4889(02)00178-7. [DOI] [PubMed] [Google Scholar]
- 19.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3′-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H, Guan J, Lee HM, Sui Y, He L, Siu JJ, et al. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through beta-cell deficit and islet amyloid deposition. Pancreas. 2010;39:843–846. doi: 10.1097/MPA.0b013e3181d12613. [DOI] [PubMed] [Google Scholar]
- 22.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 23.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 24.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 25.Puigserver P, Rodgers JT. Foxa2, a novel transcriptional regulator of insulin sensitivity. Nat Med. 2006;12:38–39. doi: 10.1038/nm0106-38. [DOI] [PubMed] [Google Scholar]
- 26.Guay C, Roggli E, Nesca V, Jacovetti C, Regazzi R. Diabetes mellitus, a microRNA-related disease? Transl Res. 2011;157:253–264. doi: 10.1016/j.trsl.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun LL, Jiang BG, Li WT, Zou JJ, Shi YQ, Liu ZM. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res Clin Pract. 2011;91:94–100. doi: 10.1016/j.diabres.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Chan CB, De Leo D, Joseph JW, McQuaid TS, Ha XF, Xu F, et al. Increased uncoupling protein-2 levels in beta-cells are associated with impaired glucose-stimulated insulin secretion: mechanism of action. Diabetes. 2001;50:1302–1310. doi: 10.2337/diabetes.50.6.1302. [DOI] [PubMed] [Google Scholar]
- 32.Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of MicroRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS ONE. 2011;6:e18613. doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goossens GH. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol Behav. 2008;94:206–218. doi: 10.1016/j.physbeh.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 35.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 37.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 38.Hotamisligil GS, Murray DL, Choy LN, Spiegelman BM. Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc Natl Acad Sci USA. 1994;91:4854–4858. doi: 10.1073/pnas.91.11.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 41.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH, Chen LX, et al. Changes in microRNA profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin Exp Pharmacol Physiol. 2009;36:e32. doi: 10.1111/j.1440-1681.2009.05207.x. [DOI] [PubMed] [Google Scholar]
- 43.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 44.Krishnaswami A, Ravi-Kumar S, Lewis JM. Thiazolidinediones: a 2010 perspective. Perm J. 2010;14:64–72. doi: 10.7812/tpp/09-052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from PCR mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 47.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White MF. Regulating insulin signaling and beta-cell function through IRS proteins. Can J Physiol Pharmacol. 2006;84:725–737. doi: 10.1139/y06-008. [DOI] [PubMed] [Google Scholar]
- 51.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 54.Huang B, Qin W, Zhao B, Shi Y, Yao C, Li J, et al. MicroRNA expression profiling in diabetic GK rat model. Acta Biochim Biophys Sin (Shanghai) 2009;41:472–477. doi: 10.1093/abbs/gmp035. [DOI] [PubMed] [Google Scholar]
- 55.Igarashi M, Wakasaki H, Takahara N, Ishii H, Jiang ZY, Yamauchi T, et al. Glucose or diabetes activates p38 mitogen-activated protein kinase via different pathways. J Clin Invest. 1999;103:185–195. doi: 10.1172/JCI3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai G, Satoh T, Kumai T, Murao M, Tsuchida H, Shima Y, et al. Hypertension accelerates diabetic nephropathy in Wistar fatty rats, a model of type 2 diabetes mellitus, via mitogen-activated protein kinase cascades and transforming growth factor-beta1. Hypertens Res. 2003;26:339–347. doi: 10.1291/hypres.26.339. [DOI] [PubMed] [Google Scholar]
- 57.Susztak K, Raff AC, Schiffer M, Bottinger EP. Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes. 2006;55:225–233. [PubMed] [Google Scholar]
- 58.Avogaro A, Albiero M, Menegazzo L, de Kreutzenberg S, Fadini GP. Endothelial dysfunction in diabetes: the role of reparatory mechanisms. Diabetes Care. 2011;34(Suppl. 2):S285–S290. doi: 10.2337/dc11-s239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emanueli C, Salis MB, Pinna A, Stacca T, Milia AF, Spano A, et al. Prevention of diabetes-induced microangiopathy by human tissue kallikrein gene transfer. Circulation. 2002;106:993–999. doi: 10.1161/01.cir.0000027104.33206.c8. [DOI] [PubMed] [Google Scholar]
- 60.Waltenberger J. Impaired collateral vessel development in diabetes: potential cellular mechanisms and therapeutic implications. Cardiovasc Res. 2001;49:554–560. doi: 10.1016/s0008-6363(00)00228-5. [DOI] [PubMed] [Google Scholar]
- 61.Rivard A, Silver M, Chen D, Kearney M, Magner M, Annex B, et al. Rescue of diabetes-related impairment of angiogenesis by intramuscular gene therapy with adeno-VEGF. Am J Pathol. 1999;154:355–363. doi: 10.1016/S0002-9440(10)65282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emanueli C, Graiani G, Salis MB, Gadau S, Desortes E, Madeddu P. Prophylactic gene therapy with human tissue kallikrein ameliorates limb ischemia recovery in type 1 diabetic mice. Diabetes. 2004;53:1096–1103. doi: 10.2337/diabetes.53.4.1096. [DOI] [PubMed] [Google Scholar]
- 63.Khan ZA, Chakrabarti S. Growth factors in proliferative diabetic retinopathy. Exp Diabesity Res. 2003;4:287–301. doi: 10.1155/EDR.2003.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36:181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 65.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 66.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabinovsky ED, Draghia-Akli R. Insulin-like growth factor I plasmid therapy promotes in vivo angiogenesis. Mol Ther. 2004;9:46–55. doi: 10.1016/j.ymthe.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Song YH, Li F, Yang T, Lu YW, Geng YJ. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem Biophys Res Commun. 2009;381:81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 70.Togliatto G, Trombetta A, Dentelli P, Rosso A, Brizzi MF. MIR221/MIR222-driven post-transcriptional regulation of P27KIP1 and P57KIP2 is crucial for high-glucose- and AGE-mediated vascular cell damage. Diabetologia. 2011;54:1930–1940. doi: 10.1007/s00125-011-2125-5. [DOI] [PubMed] [Google Scholar]
- 71.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 75.Vickers KC, Remaley AT. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17:150–155. doi: 10.1097/MED.0b013e32833727a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Najafi-Shoushtari SH. MicroRNAs in Cardiometabolic Disease. Curr Atheroscler Rep. 2011;13:202–207. doi: 10.1007/s11883-011-0179-y. [DOI] [PubMed] [Google Scholar]
- 77.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 79.Tian R, Abel ED. Responses of GLUT4-deficient hearts to ischemia underscore the importance of glycolysis. Circulation. 2001;103:2961–2966. doi: 10.1161/01.cir.103.24.2961. [DOI] [PubMed] [Google Scholar]
- 80.Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389:315–320. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 81.Feng B, Chen S, George B, Feng Q, Chakrabarti S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev. 2010;26:40–49. doi: 10.1002/dmrr.1054. [DOI] [PubMed] [Google Scholar]
- 82.Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation. 2005;111:1652–1659. doi: 10.1161/01.CIR.0000160352.58142.06. [DOI] [PubMed] [Google Scholar]
- 83.Rossing P, Breum L, Major-Pedersen A, Sato A, Winding H, Pietersen A, et al. Prolonged QTc interval predicts mortality in patients with type 1 diabetes mellitus. Diabet Med. 2001;18:199–205. doi: 10.1046/j.1464-5491.2001.00446.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Xiao J, Lin H, Luo X, Wang H, Bai Y, et al. Ionic mechanisms underlying abnormal QT prolongation and the associated arrhythmias in diabetic rabbits: a role of rapid delayed rectifier K+ current. Cell Physiol Biochem. 2007;19:225–238. doi: 10.1159/000100642. [DOI] [PubMed] [Google Scholar]
- 85.Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 86.Yoon YS, Uchida S, Masuo O, Cejna M, Park JS, Gwon HC, et al. Progressive attenuation of myocardial vascular endothelial growth factor expression is a seminal event in diabetic cardiomyopathy: restoration of microvascular homeostasis and recovery of cardiac function in diabetic cardiomyopathy after replenishment of local vascular endothelial growth factor. Circulation. 2005;111:2073–2085. doi: 10.1161/01.CIR.0000162472.52990.36. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Higashi Y, Itabe H, Song YH, Du J, Delafontaine P. Insulin-like growth factor-1 receptor activation inhibits oxidized LDL-induced cytochrome C release and apoptosis via the phosphatidylinositol 3 kinase/Akt signaling pathway. Arterioscler Thromb Vasc Biol. 2003;23:2178–2184. doi: 10.1161/01.ATV.0000099788.31333.DB. [DOI] [PubMed] [Google Scholar]
- 88.Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, et al. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun. 2008;376:548–552. doi: 10.1016/j.bbrc.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 89.Shan YX, Liu TJ, Su HF, Samsamshariat A, Mestril R, Wang PH. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J Mol Cell Cardiol. 2003;35:1135–1143. doi: 10.1016/s0022-2828(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 90.Muraski JA, Fischer KM, Wu W, Cottage CT, Quijada P, Mason M, et al. Pim-1 kinase antagonizes aspects of myocardial hypertrophy and compensation to pathological pressure overload. Proc Natl Acad Sci USA. 2008;105:13889–13894. doi: 10.1073/pnas.0709135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katare R, Caporali A, Zentilin L, Avolio E, Sala-Newby G, Oikawa A, et al. Intravenous gene therapy with PIM-1 via a cardiotropic viral vector halts the progression of diabetic cardiomyopathy through promotion of prosurvival signaling. Circ Res. 2011;108:1238–1251. doi: 10.1161/CIRCRESAHA.110.239111. [DOI] [PubMed] [Google Scholar]
- 92.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86:410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 93.Klein R, Klein BE, Moss SE, Davis MD, DeMets DL. The Wisconsin epidemiologic study of diabetic retinopathy. III. Prevalence and risk of diabetic retinopathy when age at diagnosis is 30 or more years. Arch Ophthalmol. 1984;102:527–532. doi: 10.1001/archopht.1984.01040030405011. [DOI] [PubMed] [Google Scholar]
- 94.Shibuya M. Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J Biochem Mol Biol. 2006;39:469–478. doi: 10.5483/bmbrep.2006.39.5.469. [DOI] [PubMed] [Google Scholar]
- 95.McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60:1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kovacs B, Lumayag S, Cowan C, Xu S. microRNAs in Early Diabetic Retinopathy in Streptozotocin-induced Diabetic Rats. Invest Ophthalmol Vis Sci. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 97.Kowluru RA, Koppolu P, Chakrabarti S, Chen S. Diabetes-induced activation of nuclear transcriptional factor in the retina, and its inhibition by antioxidants. Free Radic Res. 2003;37:1169–1180. doi: 10.1080/10715760310001604189. [DOI] [PubMed] [Google Scholar]
- 98.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 101.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vickers AC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2001;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 106.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 109.Waltenberger J, Lange J, Kranz A. Vascular endothelial growth factor-A-induced chemotaxis of monocytes is attenuated in patients with diabetes mellitus: a potential predictor for the individual capacity to develop collaterals. Circulation. 2000;102:185–190. doi: 10.1161/01.cir.102.2.185. [DOI] [PubMed] [Google Scholar]
- 110.Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 111.Kolfschoten IG, Roggli E, Nesca V, Regazzi R. Role and therapeutic potential of microRNAs in diabetes. Diabetes Obes Metab. 2009;11(Suppl. 4):118–129. doi: 10.1111/j.1463-1326.2009.01118.x. [DOI] [PubMed] [Google Scholar]
- 112.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 113.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Snove O, Jr, Rossi JJ. Expressing short hairpin RNAs in vivo. Nat Methods. 2006;3:689–695. doi: 10.1038/nmeth927. [DOI] [PubMed] [Google Scholar]
- 115.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alexander IE, Cunningham SC, Logan GJ, Christodoulou J. Potential of AAV vectors in the treatment of metabolic disease. Gene Ther. 2008;15:831–839. doi: 10.1038/gt.2008.64. [DOI] [PubMed] [Google Scholar]
- 117.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S, et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–1109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herrera BM, Lockstone HE, Taylor JM, Wills QF, Kaisaki PJ, Barrett A, et al. MicroRNA-125a is over-expressed in insulin target tissues in a spontaneous rat model of Type 2 Diabetes. BMC Med Genomics. 2009;2:54. doi: 10.1186/1755-8794-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004699. e4699. [DOI] [PMC free article] [PubMed] [Google Scholar]