Abstract
Interindividual variations of microRNA expression are likely to influence the expression of microRNA target genes and, therefore, contribute to phenotypic differences in humans, including cancer susceptibility. Whether microRNA expression variation has any role in ovarian cancer development is still unknown. Here, we evaluated microRNA expression profiles in lymphoblastoid cell lines from 74 women with familial ovarian cancer and 47 unrelated controls matched on gender and race. We found that the cases and unrelated controls can be clustered using 95 differentially expressed microRNAs with 91% accuracy. To assess the potential implications of microRNAs in ovarian cancer, we investigated the associations between microRNA expression and seven ovarian cancer risk variants discovered from genome-wide association studies (GWAS), namely, rs3814113 on 9p22.2, rs2072590 on 2q31, rs2665390 on 3q25, rs10088218, rs1516982, rs10098821 on 8q24.21 and rs2363956 on 19p13. We observed 130 significant associations at a permutation level of 0.01. Compared with other risk variants, rs3814113 and rs2072590 had the greatest number of significant associations (68 and 37, respectively). Interestingly, 14 microRNAs that were associated with ovarian cancer risk alleles belong to five microRNA clusters. The most notable cluster is the tumorigenic miR-17-92 cluster with five microRNAs, all of which are significantly associated with rs3814113. Using pathway analysis, several key biological pathways were significantly overrepresented, such as cellular response to stress (P = 2.87 × 10−06), etc. Further characterization of significant associations between microRNAs and risk alleles could facilitate the understanding of the functions of these GWAS discovered risk alleles in the genetic etiology of ovarian cancer.
Introduction
Epithelial carcinoma of the ovary is one of the most common gynecological malignancies in women (1). Family history is the strongest risk factor for ovarian cancer. Compared with a 1.6% lifetime risk of developing ovarian cancer in the general population, women with one first-degree relative with ovarian cancer have a 5% risk. Familial clustering with an autosomal dominant pattern of inheritance (hereditary ovarian cancer) results from germline mutations in putative tumor suppressor genes (TSGs), such as the BRCA1/2 and MLH1/MSH2 genes (2–5). However, known mutations in BRCA1/2 and MMR genes can only explain a small part of the familial aggregation of ovarian cancer (5–13%). This suggests that other genetic events may contribute to familial ovarian cancers. Recently, genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs), which confer risk to ovarian cancer (6–8). However, most of the ovarian cancer risk variants identified from GWAS reside in non-protein-encoding regions, including intergenic, intronic and untranslated regions (9). Therefore, the observed associations have yet to be translated into a full understanding of the genes and genetic elements mediating disease susceptibility.
Intriguingly, a significant number of microRNAs, which are emerging as key players in the regulation of gene expression, often reside in the non-protein-encoding regions, too (10). MicroRNAs are small non-coding RNAs that regulate >60% of protein-coding transcripts (11). Each microRNA has multiple target genes that are regulated at the posttranscriptional level. They have been implicated in various diseases and may influence tumorigenesis by acting as oncogenes and tumor suppressors (12,13). For example, microRNAs have been linked to ovarian tumor initiation and progression (14–16). Germline variations in microRNAs, messenger RNA transcripts of their target genes, and processing genes have been reported to have an effect not only on tumor progression but also on an individual's risk of developing cancer, including ovarian cancer (17,18). Hence, microRNAs are related to diverse cellular processes and are regarded as important components of the gene regulatory network, which contribute to ovarian carcinogenesis.
It has become clear that gene expression levels vary among individuals and can be analyzed like other quantitative phenotypes, such as height or serum glucose levels (19–21). However, the extent to which microRNA levels are genetically controlled is largely unknown. In a recent expression quantitative traits loci analysis, Borel et al. (22) identified a number of significant expression quantitative traits loci in primary fibroblasts, suggesting that at least part of the microRNA expression variation is regulated by common genetic variants. In human cancer, variations in microRNA expression can be extremely important because microRNAs can act as either TSGs or oncogenes. Reduced expression of TSG like microRNAs and increased expression of oncogene like microRNAs might potentially increase genetic susceptibility to human cancer. Therefore, investigation into microRNA expression variation may provide immediate insight into a probable basis for the disease associations. In addition, it offers valuable tools that may complement the knowledge from GWAS to elucidate the biological functions of SNPs identified from GWAS. In the case of ovarian cancer, studying the associations between microRNAs and ovarian cancer risk alleles will help uncover the potential microRNAs, target genes and biological pathways which these GWAS discovered risk alleles may interact with.
To study microRNA expression variations in lymphoblastoid cell lines (LCLs) and their potential contributions to the development of familial ovarian cancer, we first analyzed the expression profiles of 1145 microRNAs in 121 non-redundant LCLs derived from 74 familial ovarian cancer patients who are non-carriers of known BRCA1/2 and MMR gene mutations, as well as 47 unrelated controls. Then, we studied the associations between microRNA expression variations and seven ovarian cancer risk variants discovered from GWAS (6–8). To our knowledge, this is the first study to examine the roles of microRNA expression variations in LCLs in familial ovarian cancer and evaluate the associations between microRNA expression variations and ovarian cancer risk variants. Since genetic susceptibility of ovarian cancer is far from being fully understood, our study may facilitate candidate gene discovery and lead to better understanding of the genetic susceptibility of ovarian cancer in this post-GWAS era.
Materials and methods
Study population
The study has been approved by the Institutional Research Board of Roswell Park Cancer Institute. Data and samples from women with ovarian cancer and their relatives who were cancer free were obtained from the Gilda Radner Familial Ovarian Cancer Registry (GRFOCR). Seventy-four non-related women with familial ovarian cancer were included in this study as the cases. They were identified from families with inherited ovarian cancer in which at least two first- or second-degree relatives had epithelial ovarian cancer diagnosed at any age. All of the women were non-carriers of BRCA1/2 or MLH1/MSH2 mutations. Over time, different methods have been used to determine the mutation status of BRCA1/2 in GRFOCR samples. For samples collected before 2002, mutation status was determined by screening all exons and intron/exon splice junctions of BRCA 1/2 by a combination of single-strand conformation polymorphism and heteroduplex analysis. Additionally, exon 11 of BRCA1 was assayed by the protein truncation test for stop codon generating mutations. If alterations were found, the altered fragment was sequenced. Since 2002, sequencing of exons and splice junctions was used. In the last 5 years, all samples (old and new) not showing a mutation were assayed for BRCA1 large-scale rearrangements. The cancer-free controls of GRFOCR were family relatives of the cases, including mothers, sisters, nieces, etc. However, in this study, we chose to use unrelated controls. Unrelated controls are women who are not relatives of any cases used in this study. Forty-seven unrelated controls were included. The cases and controls were matched on gender and race. All of the cases and controls were white women. The median age at cancer diagnosis for the 74 cases was 47 (ranging from 21 to 85), whereas the median age for the 47 controls at enrollment in GRFOCR was 58 (ranging from 26 to 89). All study subjects donated blood samples when they were enrolled in the GRFOCR. LCLs were established by Epstein-Barr virus (EBV) transformation using the isolated lymphocytes from the blood samples. The study was approved by the institutional research board.
LCLs culture and RNA extraction
LCLs were maintained in RPMI 1640 (GIBCO BRL) media supplemented with 15% fetal calf serum and antibiotics at 37°C, 5% CO2 atmospheric condition and 95% humidity. Total cellular RNAs were isolated from LCLs using TRIzol reagent according to the protocols provided by the manufacturer (Invitrogen Corp., Carlsbad, CA). Purified RNAs were further processed to remove any contaminating DNA (DNA-free kit; Ambion Inc., Austin, TX). The quality and quantity of the RNA were evaluated by 260/280 ratio using NanoDrop spectrophotometry (ND-1000; NanoDrop Technologies Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies).
MicroRNA microarray analysis
Two hundred ng of total RNA from each sample were labeled and hybridized on Human v2 MicroRNA Expression BeadChips (Cat. no. MI-102–1024; Illumina) according to the manufacturer's recommendations (Illumina MicroRNA Expression Profiling Assay Guide). The expression profiles have been deposited in NCBI's Gene Expression Omnibus with accession number GSE31801.
Data assembly.
The raw intensity of the Illumina Human V2 microRNA expression array was scanned and extracted using BeadScan, with the data corrected by background subtraction in the GenomeStudio module. The lumi package in the R-based Bioconductor package was used to normalize the log2 transformed intensity data by using the quantile normalization algorithm. For data quality control, we excluded the probes with detection P value >0.05 (the P values were generated in BeadStudio software) across 75% of the samples. A total of 587 microRNAs passed the quality control step and were used for downstream analysis.
Differential expression testing.
We used the Limma program in the R-based Bioconductor package to calculate the statistical significance for the level of differential expression (23). Briefly, a linear model was fit to the data, with cell means corresponding to the different conditions, age adjusted as a continuous covariate and a random effect for array. The Benjamini and Hochberg method was used to control the false discovery rate (FDR) for the multiple testing (24).
Hierarchical clustering.
Hierarchical clustering algorithm based on the average linkage and Pearson correlation metric was employed to cluster residuals of gene expression corrected for age (25). First, unsupervised hierarchical clustering analysis was performed based on gene expression profiles from the top 50% of the most variable (i.e. largest coefficient of variance) microRNAs across 121 samples, 75% of the most variable microRNAs across 121 samples or all 587 microRNAs. Second, following single microRNA-based significance testing, we used the expression value of selected microRNAs (FDR < 0.01 and at least 1.5-fold expression) to perform hierarchical clustering of the samples. The purpose here was to explore and visualize the performance of identified microRNAs signature, as a whole, at differentiating the participants into their corresponding case versus control group.
Machine learning classification.
The performance of microRNA expression profiles to differentiate the case from control group was further assessed by leave-one-out cross validation. The R package classification for microarrays was used to build classifiers with the support vector machines (SVMs) approach (26). SVM approaches the task of classification by constructing a hyperplane or set of hyperplanes in a high or infinite dimensional space. We utilized residuals of gene expression corrected for age in classification for microarrays. For leave-one-out cross validation, we literately split the samples into training sets and testing sets. Each training set, consisting of all but a single test sample, was used to identify a predictor consisting of a panel of microRNAs, the number of which remained constant between sets. The case/control status of the single test sample was then predicted by the resulting predictor. The classification accuracy rate is the percentage of time the SVM prediction is correct. We varied the number of microRNAs composing the predictor from 6 to 20 and found the classification results are not significantly changed (data not shown). As recommended by (26), we used a nested cross-validation loop to choose the optimal value for the cost parameter in the SVM model kernel parameter from 0.1, 0.2, 0.5, 1, 2, 5, 10 and 20.
Association of microRNA expression and SNP genotypes.
The association of SNP genotype with residuals of expression level adjusted for age and case–control status was calculated using linear regression model as described before (27). Ten thousand permutations of the expression phenotypes relative to SNP genotypes were performed (28–30). To derive P values adjusted for multiple testing, we determined the percentage of times out of 10 000 permutations that the observed P value was exceeded in the permuted data analysis. Only microRNAs with expression above the background in at least 25% of the samples (n = 121) and with a known status in the latest miRBase database (version 17) were included in the association analysis.
Real-time quantitative PCR analysis
The expression levels of microRNA were confirmed with a Taqman-based real-time quantitative PCR using individual microRNA-specific primers and probes as described by the manufacturer (Applied Biosystems). The first-strand microRNA-complementary DNA PCR template was generated from 50 ng of total RNA according to the manufacturer's instructions. Approximately 2.5 ng of complementary DNA was then used in the PCR on a StepOnePlus Real-Time PCR System from Applied Biosystems. Triplicate samples, validated endogenous controls and inter-assay controls were used throughout. The real-time quantitative PCR results were analyzed by SDS 2.2.2. A total of four microRNAs were included in the analysis. They were miR-92b, miR-629*, miR-216a and miR-588. According to manufacturer's suggestion, in this study, we chose RNU-24 as the endogenous control. Real-time quantitative PCR data were the normalized expression values in which the endogenous control RNU-24 was used as the reference gene. For each assay, the Ct (cycle threshold) of microRNA of interest in the TaqMan qPCR assay was subtracted from the average RNU-24 Ct value to obtain a ΔCt value (RNU-24—microRNA of interest). A higher ΔCt value indicates a higher expression level of the microRNA of interest.
Genotyping analysis for ovarian cancer risk alleles
Seven SNPs, which are identified from three ovarian cancer GWAS, were included in the genotyping analysis. They are rs3814113 on 9p22.2, rs2072590 on 2q31, rs2665390 on 3q25 in the intron of TCDD-inducible poly(ADP-ribose) polymerase (TIPARP) gene, rs10088218, rs1516982, rs10098821 on 8q24.21 and rs2363956 on 19p13 in the ankyrin repeat and LEM domain containing 1 (ANKLE1) gene. rs2363956 is a nonsynomous SNP which leads to a Leu to Trp amino acid change. Genotyping analysis was carried out using StepOnePlus™ Real-Time PCR system and Assays-on-Demand SNP Genotyping products for fluorogenic PCR allelic discrimination (Applied Biosystems). Each PCR reaction plate included negative controls, positive controls and unknown samples. The minor allele frequencies for each SNP in the cases and unrelated controls were 0.346/0.298 (P = 0.65) for rs3814113, 0.3/0.368 (P = 0.84) for rs2072590, 0.081/0.060 (P = 0.60) for rs2665390, 0.149/0.107 (P = 0.13) for rs10088218, 0.167/0.119 (P = 0.06) for rs1516982, 0.127/0.071 (P = 0.07) for rs10098821 and 0.432/0.488 (P = 0.20) for rs2363956.
Results
For the 1145 microRNAs assayed, 74% (n = 849) were detected above the background levels. Of these 849 expressed microRNAs, we included 578 in further analysis because their expression was above the background in at least 30 samples. Using microRNA microarray data, we selected the top 50% of the most variable microRNAs across 121 samples, 75% of the most variable microRNAs across 121 samples or all 587 microRNAs in an unsupervised hierarchical clustering analysis. We found that the age-corrected expression profiles reasonably separated cases from unrelated controls (Supplementary Figure 1, available at Carcinogenesis Online), indicating that the familial ovarian cancer cases and cancer-free unrelated controls have intrinsically different microRNA expression patterns.
Incorporating age as a covariate for adjustment in case versus control comparison, we identified a total of 95 microRNAs with at least 1.5-fold differential expression at the significance level of FDR < 0.01, with 54 microRNAs upregulated in cases and 41 microRNAs downregulated. The detailed list of differentially expressed microRNAs is summarized in Table I. Among these 95 differentially expressed microRNAs, 14 microRNAs had at least two-fold increase in cases and 9 had at least two-fold decrease. These upregulated microRNAs include hsa-miR-216a, hsa-miR-588, HS_33, hsa-miR-521, hsa-miR-675, solexa-2683-338, hsa-miR-1255b, HS_109, hsa-miR-610, HS_169, solexa-15-44487, hsa-miR-1225-3p, hsa-miR-140-5p, hsa-miR-801 and those downregulated microRNAs include HS_228.1, HS_108.1, hsa-miR-92b, hsa-miR-374b, HS_31.1, HS_257, hsa-miR-10b, hsa-miR-629*, hsa-miR-574-3p. We noted that 26 of the 95 (27%) differentially expressed microRNAs belong to unknown microRNAs (i.e. not cataloged by miRBase database). For example, the expression level of HS_228.1 and HS_108.1is the first and second most depleted in case versus control, and the expression level of HS_33 is the first most elevated in case versus control. Our data together indicate a global perturbation in the microRNA expression pattern in familial ovarian cancer cases over controls.
Table I.
Differentially expressed microRNAs between case and control groups
| Upregulated |
Downregulated |
||||||
| ID | Fold change | P value | FDR | ID | Fold change | P value | FDR |
| hsa-miR-216a | 3.58 | 1.77 × 10−25 | 1.78 × 10−23 | HS_228.1 | −3.66 | 1.38 × 10−30 | 2.70 × 10−28 |
| hsa-miR-588 | 3.14 | 6.28 × 10−25 | 4.09 × 10−23 | HS_108.1 | −3.27 | 2.64 × 10−17 | 5.17 × 10−16 |
| HS_33 | 2.99 | 4.76 × 10−32 | 1.40 × 10−29 | hsa-miR-92b | −2.46 | 1.88 × 10−35 | 1.10 × 10−32 |
| hsa-miR-521 | 2.81 | 3.88 × 10−17 | 7.12 × 10−16 | hsa-miR-374b | −2.35 | 1.84 × 10−21 | 7.22 × 10−20 |
| hsa-miR-675 | 2.62 | 2.22 × 10−25 | 1.87 × 10−23 | HS_31.1 | −2.27 | 4.95 × 10−11 | 3.02 × 10−10 |
| solexa-2683-338 | 2.57 | 1.56 × 10−24 | 9.14 × 10−23 | HS_257 | −2.11 | 4.29 × 10−15 | 5.24 × 10−14 |
| hsa-miR-1255b | 2.45 | 9.35 × 10−16 | 1.28 × 10−14 | hsa-miR-10b | −2.1 | 1.18 × 10−10 | 6.66 × 10−10 |
| HS_109 | 2.14 | 4.88 × 10−19 | 1.43 × 10−17 | hsa-miR-629* | −2.04 | 1.05 × 10−18 | 2.71 × 10−17 |
| hsa-miR-610 | 2.13 | 4.83 × 10−14 | 5.06 × 10−13 | hsa-miR-574-3p | −2.04 | 1.34 × 10−16 | 2.07 × 10−15 |
| HS_169 | 2.11 | 3.47 × 10−24 | 1.85 × 10−22 | hsa-let-7b* | −1.97 | 2.53 × 10−15 | 3.16 × 10−14 |
| solexa-15-44487 | 2.1 | 2.05 × 10−17 | 4.16 × 10−16 | hsa-miR-183 | −1.96 | 4.28 × 10−13 | 3.81 × 10−12 |
| hsa-miR-1225-3p | 2.06 | 8.59 × 10−14 | 8.41 × 10−13 | hsa-miR-182 | −1.91 | 4.50 × 10−11 | 2.78 × 10−10 |
| hsa-miR-140-5p | 2.04 | 4.58 × 10−06 | 1.30 × 10−05 | hsa-let-7e | −1.89 | 2.52 × 10−15 | 3.16 × 10−14 |
| hsa-miR-801:9.1 | 2.03 | 1.85 × 10−22 | 8.36 × 10−21 | hsa-miR-505* | −1.84 | 4.78 × 10−12 | 3.30 × 10−11 |
| hsa-miR-891a | 1.93 | 2.33 × 10−12 | 1.78 × 10−11 | hsa-miR-550 | −1.8 | 2.20 × 10−09 | 9.64 × 10−09 |
| solexa-8048-104 | 1.87 | 1.24 × 10−17 | 2.69 × 10−16 | hsa-miR-942 | −1.78 | 6.18 × 10−17 | 1.10 × 10−15 |
| hsa-miR-600 | 1.8 | 1.16 × 10−06 | 3.54 × 10−06 | HS_91.1 | −1.77 | 1.12 × 10−06 | 3.45 × 10−06 |
| HS_45.1 | 1.8 | 4.14 × 10−19 | 1.28 × 10−17 | hsa-miR-598 | −1.74 | 4.26 × 10−12 | 3.04 × 10−11 |
| hsa-miR-490-5p | 1.78 | 4.64 × 10−09 | 1.95 × 10−08 | hsa-miR-7-1* | −1.74 | 1.06 × 10−18 | 2.71 × 10−17 |
| solexa-603-1846 | 1.75 | 6.51 × 10−15 | 7.64 × 10−14 | hsa-miR-130b* | −1.73 | 1.98 × 10−18 | 4.85 × 10−17 |
| hsa-miR-20a* | 1.75 | 1.95 × 10−05 | 5.19 × 10−05 | hsa-miR-1301 | −1.73 | 1.93 × 10−12 | 1.49 × 10−11 |
| HS_260 | 1.74 | 6.04 × 10−27 | 8.86 × 10−25 | HS_10 | −1.72 | 3.44 × 10−10 | 1.77 × 10−09 |
| hsa-miR-330-5p | 1.74 | 3.96 × 10−10 | 1.99 × 10−09 | hsa-miR-193b* | −1.71 | 6.48 × 10−09 | 2.68 × 10−08 |
| hsa-miR-579 | 1.74 | 4.16 × 10−03 | 7.62 × 10−03 | hsa-miR-27a* | −1.71 | 5.76 × 10−08 | 2.10 × 10−07 |
| HS_128 | 1.72 | 2.77 × 10−14 | 3.01 × 10−13 | hsa-miR-29b-1* | −1.71 | 3.38 × 10−16 | 4.73 × 10−15 |
| hsa-miR-33a | 1.69 | 3.47 × 10−05 | 8.91 × 10−05 | hsa-miR-629 | −1.69 | 5.71 × 10−14 | 5.88 × 10−13 |
| hsa-miR-885-5p | 1.68 | 3.27 × 10−05 | 8.43 × 10−05 | hsa-miR-126* | −1.64 | 7.29 × 10−06 | 2.02 × 10−05 |
| hsa-miR-591 | 1.68 | 4.25 × 10−07 | 1.40 × 10−06 | hsa-miR-150 | −1.61 | 6.53 × 10−17 | 1.13 × 10−15 |
| HS_116 | 1.67 | 3.77 × 10−17 | 7.12 × 10−16 | hsa-miR-105* | −1.61 | 2.34 × 10−20 | 8.08 × 10−19 |
| hsa-miR-1268 | 1.67 | 2.99 × 10−09 | 1.27 × 10−08 | hsa-miR-30a* | −1.59 | 8.74 × 10−17 | 1.43 × 10−15 |
| HS_32 | 1.62 | 9.03 × 10−09 | 3.58 × 10−08 | hsa-miR-200c | −1.58 | 9.61 × 10−11 | 5.48 × 10−10 |
| hsa-miR-1322 | 1.62 | 1.74 × 10−09 | 7.84 × 10−09 | hsa-miR-628-3p | −1.57 | 3.60 × 10−11 | 2.25 × 10−10 |
| HS_175 | 1.61 | 6.48 × 10−18 | 1.52 × 10−16 | HS_67 | −1.57 | 1.94 × 10−13 | 1.84 × 10−12 |
| HS_112 | 1.59 | 6.53 × 10−07 | 2.10 × 10−06 | hsa-miR-200b | −1.55 | 1.98 × 10−13 | 1.85 × 10−12 |
| hsa-miR-335 | 1.58 | 1.80 × 10−14 | 1.99 × 10−13 | hsa-miR-342-5p | −1.55 | 2.60 × 10−08 | 9.78 × 10−08 |
| hsa-miR-612 | 1.58 | 2.24 × 10−08 | 8.49 × 10−08 | hsa-miR-1238 | −1.54 | 2.39 × 10−05 | 6.33 × 10−05 |
| hsa-miR-554 | 1.58 | 1.19 × 10−09 | 5.55 × 10−09 | hsa-miR-199a-3p, | −1.54 | 2.78 × 10−05 | 7.32 × 10−05 |
| hsa-miR-615-5p | 1.58 | 6.66 × 10−11 | 3.95 × 10−10 | hsa-miR-454* | −1.54 | 2.43 × 10−09 | 1.05 × 10−08 |
| HS_126 | 1.58 | 2.06 × 10−11 | 1.33 × 10−10 | hsa-miR-1287 | −1.52 | 2.70 × 10−11 | 1.71 × 10−10 |
| hsa-miR-1303 | 1.58 | 4.94 × 10−08 | 1.82 × 10−07 | hsa-miR-628-5p | −1.52 | 5.54 × 10−06 | 1.55 × 10−05 |
| hsa-miR-647 | 1.56 | 1.16 × 10−07 | 4.03 × 10−07 | hsa-miR-525-5p | −1.51 | 1.51 × 10−16 | 2.28 × 10−15 |
| solexa-9124-90 | 1.55 | 1.82 × 10−25 | 1.78 × 10−23 | ||||
| hsa-miR-21* | 1.54 | 2.07 × 10−04 | 4.79 × 10−04 | ||||
| hsa-miR-18b | 1.54 | 3.13 × 10−03 | 5.91 × 10−03 | ||||
| hsa-miR-326 | 1.54 | 9.12 × 10−07 | 2.86 × 10−06 | ||||
| HS_287 | 1.53 | 9.66 × 10−17 | 1.53 × 10−15 | ||||
| hsa-miR-296-5p | 1.53 | 1.88 × 10−06 | 5.56 × 10−06 | ||||
| hsa-miR-548g | 1.52 | 5.91 × 10−25 | 4.09 × 10−23 | ||||
| HS_282 | 1.52 | 6.95 × 10−17 | 1.17 × 10−15 | ||||
| hsa-miR-1281 | 1.52 | 7.22 × 10−11 | 4.21 × 10−10 | ||||
| hsa-miR-1226* | 1.52 | 9.54 × 10−10 | 4.52 × 10−09 | ||||
| hsa-miR-101 | 1.51 | 1.44 × 10−03 | 2.87 × 10−03 | ||||
| HS_147 | 1.51 | 5.25 × 10−15 | 6.29 × 10−14 | ||||
| hsa-miR-603 | 1.51 | 2.08 × 10−06 | 6.12 × 10−06 | ||||
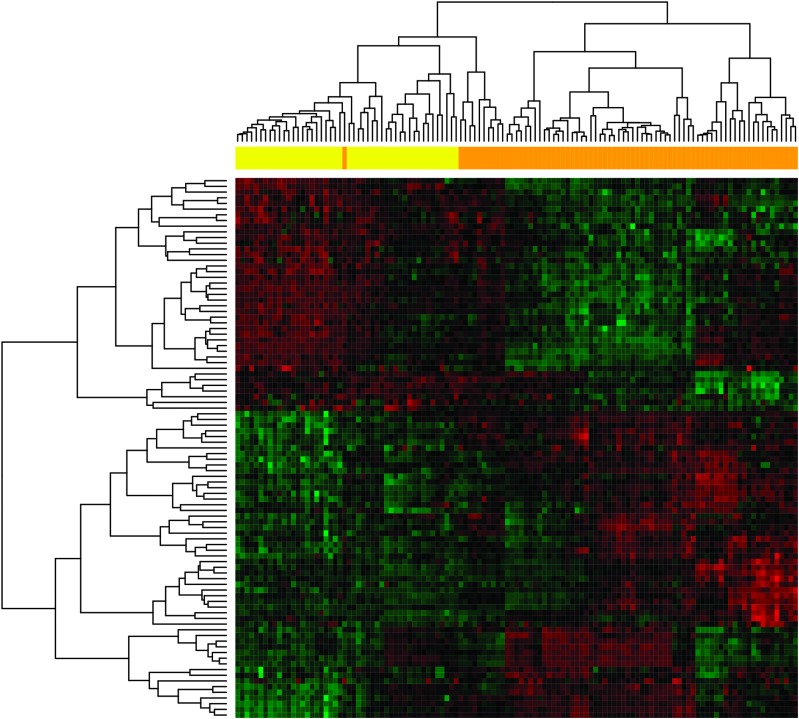
We next used hierarchical clustering to explore the differentiation of case from control samples based on the age-corrected expression signature of the 95 differentially expressed microRNAs described above. As shown in Figure 1, the 74 familial ovarian cancer cases and 47 unrelated controls were separately grouped, except for a small group of 11 cases that were grouped with the unrelated controls. The overall accuracy or classification rate is 91% (110/121). To further explore the potential of a microRNA signature for discriminating ovarian cancer cases and unrelated controls, we employed the classification for microarrays package to automatically perform microRNA selection (from the initial 587 microRNAs), parameter tuning, classifier construction and unbiased evaluation of the constructed classifiers. Specifically, a SVM classifier composed of eight microRNAs (varying the number did not significantly change the accuracy) was built using a training set consisting of 120 samples and tested on the remaining single test sample. This procedure was repeated 121 times until all the samples had been left out (i.e. leave-one-out cross validation), and the resulting classification accuracy for the 121 classifiers was 95.9%. Seven microRNAs, hsa-miR-548g, hsa-miR-588, hsa-miR-216a, solexa-2683-338, hsa-miR-92b, HS-260 and HS-228.1, were present in all 121 classifiers, and HS-169 was present in 103 (85.1%) classifiers.
Fig. 1.
Hierarchical clustering of 74 familial ovarian cancer cases and 47 cancer-free relatives based on age-corrected microRNA expression levels. In the clustering heat map, red indicates upregulated, whereas green indicates downregulated. In the sample clustering dendrogram, orange indicates samples from cancer patients, whereas yellow indicates samples from cancer-free controls.
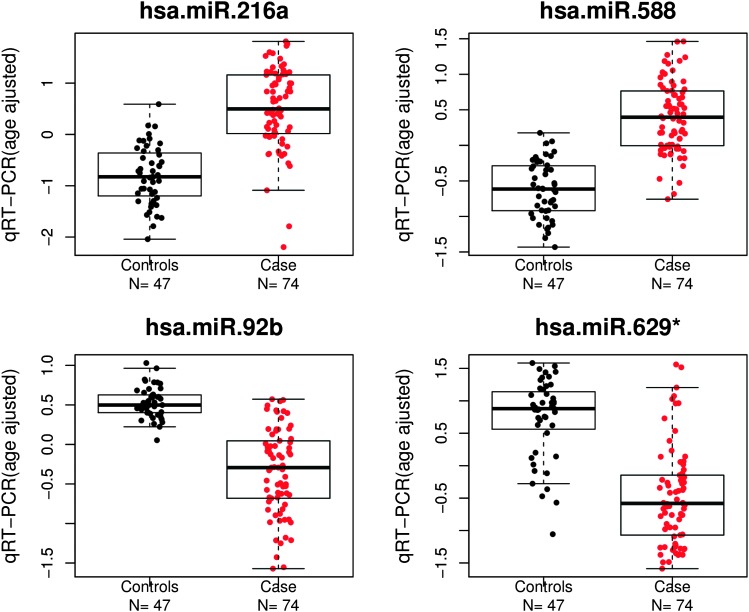
To validate the results from microRNA profiling, Taqman quantitative real-time PCR (qRT–PCR) was performed on these 121 samples. Selected microRNAs included hsa-miR-216a and hsa-miR-588 which were upregulated in cancer cases versus unrelated controls, and hsa-miR-92b and hsa-miR-629* which were downregulated. The qRT–PCR validation results were consistent with those of the initial microarray experiment, with hsa-miR-216a (P = 1.01 × 10−18, fold change = 2.47) and hsa-miR-588 (P = 1.12 × 10−22, fold change = 2.03) significantly upregulated in familial ovarian cancers and hsa-miR-92b (P = 3.34 × 10−22, fold change = −1.82) and hsa-miR-629* (P = 3.05 × 10−16, fold change = −2.30) significantly downregulated in familial ovarian cancers (Figure 2).
Fig. 2.
Validation of microRNA microarray experiments using quantitative reverse transcription real-time PCR. Red point denotes the expression value from samples of cancer patients, whereas black point denotes the expression value from control samples. The residuals of gene expression corrected for age is shown in log2 scale.
Because familial ovarian cancers are more likely to be diagnosed at a younger age, we conducted an exploratory analysis of the relationship between microRNA expression profiles and age of familial ovarian cancer diagnosis. Using Spearman's rank correlation at the significance level of 0.05, 3 of the 95 differentially expressed microRNAs showed an inverse association, with increasing expression value with decreasing age at cancer diagnosis (hsa-miR-92b, ρ = −0.288, P = 0.0129; hsa-miR-629*, ρ = −0.285, P = 0.0139; hsa-miR-548, ρ = −0.248, P = 0.0333), whereas 2 of the 95 differentially expressed microRNAs showed a positive association, with increasing expression value with increasing age at cancer diagnosis (hsa-miR-490-5p, ρ = 0.250, P = 0.0317; hsa-miR-140-5p, ρ = 0.231, P = 0.0481). The significantly inverse associations of hsa-miR-92b and hsa-miR-629* with age at cancer diagnosis in cancer cases were validated by qRT–PCR (hsa-miR-92b, ρ = −0.28, P = 0.015; hsa-miR-629*, ρ = −0.26, P = 0.022).
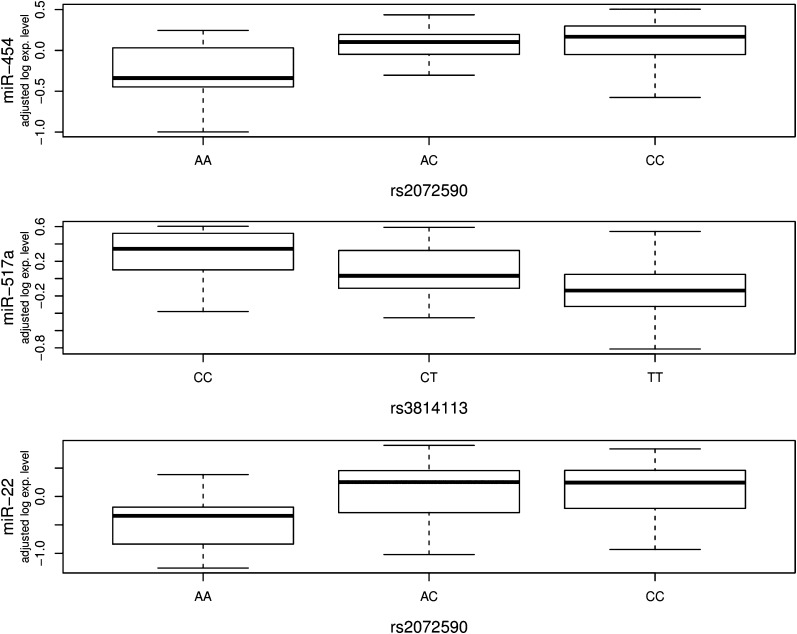
To further assess the potential implications of microRNAs in ovarian cancer, we performed association analysis to analyze the correlations between microRNA expression variations and seven ovarian cancer risk SNPs identified from GWAS. Significant associations were identified by evaluating the relationships between variations of microRNA expression levels (with age and case–control status adjusted) and variant genotypes through 10 000 permutations. Significance was set at permutation level threshold of 0.01. The complete list of 130 identified significant associations is shown in Supplementary Table 1, available at Carcinogenesis Online. As shown in Table II, rs3814113 and rs2072590, the two most significant ovarian cancer risk SNPs, were significantly associated with a large number of microRNA expression variations. At the 0.01 permutation threshold, the number of significant associations with these two variants was 68 and 37. The number was reduced to 14 and 7 at the more stringent permutation threshold of 0.001. On the other hand, the other five variants had a fewer number of significant associations with microRNA expression variations. The most significant associations detected were between rs3814113 and hsa-miR-517a (permutated P < 0.0001; Figure 3, middle), rs2072590 and hsa-miR-454 (permutated P < 10−4; Figure 3, top) and rs2072590 and hsa-miR-22 (permutated P = 10−4; Figure 3, bottom). rs3814113 explained ∼15% of the variation in hsa-miR-517a expression as measured by adjusted r2. rs2072590 explained ∼16% and 12% of the variation in hsa-miR-454 and hsa-miR-22 expression, respectively (Supplementary Table 1, available at Carcinogenesis Online). We examined whether the identified microRNAs were differentially expressed between ovarian cancer cases and unrelated controls. As shown in Table IIB and C, a number of them were differentially expressed at the significance level of FDR < 0.01, and several of them were further characterized by at least 1.5-fold expression change. For example, both hsa-miR-18b and hsa-miR-101 were significantly overexpressed in cancer cases with at least 1.5-fold changes (Table I). As shown in Supplementary Figure 2, available at Carcinogenesis Online, both microRNAs were significantly associated with rs3814113 (permutated P = 4.0 × 10−4 and 8.0 × 10−4, adjusted r2 = 11% and 10%, respectively).
Table II.
Summary of significant associations between SNP genotypes and microRNA expression phenotypes
| A. All discovered eQTLs | |||||||
| rs3814113 | rs2072590 | rs10088218 | rs2363956 | rs2665390 | rs1516982 | rs10098821 | |
| P < 0.01 | 68 | 37 | 6 | 0 | 4 | 7 | 8 |
| P < 0.001 | 14 | 7 | 1 | 0 | 0 | 1 | 1 |
| B. eQTLs involving differentially expressed microRNAs (FDR < 0.01) | |||||||
| rs3814113 | rs2072590 | rs10088218 | rs2363956 | rs2665390 | rs1516982 | rs10098821 | |
| P < 0.01 | 41 | 24 | 3 | 0 | 2 | 2 | 2 |
| P < 0.001 | 8 | 5 | 0 | 0 | 0 | 0 | 0 |
| C. eQTLs involving differentially expressed microRNAs (FDR < 0.01 and FC > 1.5) | |||||||
| rs3814113 | rs2072590 | rs10088218 | rs2363956 | rs2665390 | rs1516982 | rs10098821 | |
| P < 0.01 | 5 | 7 | 1 | 0 | 1 | 0 | 1 |
| P < 0.001 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
Fig. 3.
Examples of significant associations between SNP genotypes and microRNA expression phenotypes. The boxplots shows the relationship between log2 residuals of microRNA expression levels (adjusted for age and case–control status) and genotype of the SNPs. rs3814113 explained ∼15% of the variation in hsa-miR-517a expression as measured by adjusted r2. rs2072590 explained about 16 and 12% of the variation in hsa-miR-454 and hsa-miR-22 expression, respectively.
In further analysis, we investigated the genomic locations of the identified microRNAs, which were significantly associated with ovarian cancer risk SNPs. Among the 109 identified microRNAs, 14 were found to be located in five microRNA clusters (Table III) within the cluster distance of 5000 bp. These five microRNA clusters are on chromosomes X (hsa-miR-18b, hsa-miR-106a and hsa-miR-20b), Chromosome 8 (hsa-miR-30b, hsa-miR-30d), chromosome 13 (hsa-miR-18a, hsa-miR-19a, hsa-miR-20a, hsa-miR-20a*, hsa-miR-17), chromosome 14 (hsa-miR-539 and hsa-miR-377) and chromosome 19 (hsa-miR-517a and hsa-miR-517e). The cluster on chromosome 13 belongs to a well-known microRNA cluster, miR-17-92, one of the first microRNA clusters recognized as a key component of the molecular network that impacts tumorigenesis and tumor maintenance. All five microRNAs belonging to the miR-17-92 cluster are significantly associated with rs3814113, with the strongest association between hsa-miR-20a and rs3814113 (P = 7.0 × 10−4).
Table III.
The microRNA clusters (with cluster distance of 5000 bp) for the microRNAs significantly associated with GWAS discovered SNPs
| SNPs-ID | Associated microRNA | Permulated P value | Chromosome | Cluster distance cutoff (bp) |
| Cluster 1 | ||||
| rs3814113 | miR-18b | 4.00 × 10−4 | Chr. X | 500 |
| rs3814113 | miR-106a | 1.00 × 10−3 | Chr. X | 500 |
| rs3814113 | miR-20b | 2.50 × 10−3 | Chr. X | 500 |
| Cluster 2 | ||||
| rs3814113 | miR-30b | 6.80 × 10−3 | Chr. 8 | 5000 |
| rs1516982 | miR-30d | 9.00 × 10−3 | Chr. 8 | 5000 |
| Cluster 3 | ||||
| rs3814113 | miR-18a | 4.50 × 10−3 | Chr. 13 | 500 |
| rs3814113 | miR-19a | 2.00 × 10−4 | Chr. 13 | 500 |
| rs3814113 | miR-20a | 7.00 × 10−4 | Chr. 13 | 500 |
| rs3814113 | miR-20a* | 2.10 × 10−3 | Chr. 13 | 500 |
| rs3814113 | miR-17 | 9.20 × 10−3 | Chr. 13 | 500 |
| Cluster 4 | ||||
| rs2665390 | miR-539 | 9.50 × 10−3 | Chr. 14 | 5000 |
| rs3814113 | miR-377 | 2.10 × 10−3 | Chr. 14 | 5000 |
| Cluster 5 | ||||
| rs3814113 | miR-517a | <10−5 | Chr. 19 | 5000 |
| rs3814113 | miR-518e | 4.5 × 10−3 | Chr. 19 | 5000 |
We also examined whether the microRNAs that showed significant associations with ovarian cancer risk SNPs might also share similar predicted molecular functions. We downloaded the computational target predictions from the latest miRBase Targets database http://www.mirbase.org/ and obtained the list of genes targeted by at least eight microRNAs whose expression variations were significantly associated with variant genotypes in our study. The list of shared target genes was used for Gene Ontology term enrichment analysis with the NIH DAVID Tools http://david.abcc.ncifcrf.gov/. As shown in Supplementary Table 2, available at Carcinogenesis Online, the list of significantly enriched Gene Ontology terms include ‘cellular response to stress’ (P = 2.87x10−06), ‘cell cycle’ (P = 3.19×10−5), ‘response to DNA damage stimulus’ (P = 1.77 × 10−4), ‘ATP binding’ (P = 2.18 × 10−5) and ‘mitochondrion’ (P = 3.17 × 10−4).
Discussion
The genetic etiology of familial ovarian cancer is still an enigma. Known mutations in BRCA1/2 and MMR genes can only explain a small part of the familial aggregation of ovarian cancer. The results from recent GWAS studies have identified several common SNPs (6–8). However, most of these SNPs are not in protein-encoding regions, so the functional significance of these variants is largely unknown. The current study presents an approach to dissect the genetic susceptibility of familial ovarian cancer as well as elucidate the potential functional significance of the identified risk SNPs from GWAS. In this study, we applied microarray analysis followed by qRT–PCR validation to assess global microRNA expression in LCLs from patients with familial ovarian cancer and unrelated controls. We found a large number of differentially expressed microRNA between familial ovarian cancer cases and unrelated controls. The differences have been further cross validated by machine learning classification analysis. Two differentially expressed microRNAs, hsa-miR-92b and hsa-miR-629*, are found to be inversely associated with age at cancer diagnosis. The predicted targets of hsa-miR-92b are enriched for metabolism, oncogenesis and transcription, and the predicted targets of hsa-miR-629* appear enriched for transport and apoptosis (Supplementary Table 3 is available at Carcinogenesis Online). In the subsequent association analysis, we observed a significant number of associations between microRNA expression variations and seven ovarian cancer risk SNPs discovered from GWAS (6–8). Among them, rs3814113 (9p22.2) and rs2072590 (2q31) had the highest number of significant correlations (68 and 37, respectively, at the 0.01 permutation threshold). Interestingly, 14 microRNAs that were associated with ovarian cancer risk SNPs are found to belong to five microRNA clusters. The most notable cluster is the miR-17-92 cluster with five microRNAs within 500 bp cluster distance. All of these five microRNAs belonging to the miR-17-92 cluster are significantly associated with rs3814113. In the pathway analysis, using the predicted target genes of microRNAs with expression significantly associated with ovarian cancer risk SNPs, we observed significant enrichment of pathways involved in cell cycle and response to DNA damage.
This study provides the first assessment of the expression level variation of mature human microRNAs in LCLs from familial ovarian cancer patients and healthy unrelated controls. There are several limitations to this study. First, many microRNAs are expressed in a tissue-restricted manner (31). The results from LCLs in this study are probably to represent a small subset of microRNA expression variations. Also, our ability to study the genetics of microRNA expression is limited by the fact that we only include seven SNPs in the analysis, although these seven SNPs have been linked to ovarian cancer in GWAS. Second, there are concerns about using EBV-transformed LCLs and potential cancer treatment effects on microRNA expression patterns in LCLs. EBV-transformed LCLs have been frequently used in genetic studies on gene expression. Several studies suggest that the messenger RNA expression patterns are highly concordant between EBV-transformed LCLs and other cell types, including B cells before transformation (32–34). Therefore, it is possible that microRNA expression pattern before and after EBV immortalization is concordant to a certain degree, although we do not have the answer yet. Third, we do not expect to observe significant effects of cancer treatment on miRNA expression profiles since the analyses were performed using EBV-transformed LCLs. Although we could not exclude the remote chance that certain treatments (e.g. radiation therapy, etc.) might cause inherited changes in the blood cells and thereby affect microRNA expression, it is very unlikely. Last, there is a concern about what the results actually mean when measuring expression in non-tumor tissue at a single point in time. The ultimate goal of our study is to identify the inherited genetic determinants of microRNA expression in normal tissues rather than somatic alterations of microRNA expression in tumor tissues. Studies have been shown that at least part of the messenger RNA/microRNA expression is genetically determined. Therefore, even at a single time point in non-tumor tissue, what we have observed from this study still provides useful information about how microRNA expression is genetically regulated. In the future, we are interested in expanding our analysis to other normal tissues.
One unique feature of our study is the association analysis between microRNA expression and ovarian cancer risk SNPs identified from GWAS. Several GWAS studies in ovarian cancer have successfully identified a few risk SNPs, including a susceptibility cluster on chromosome 9p22.2 containing 12 SNPs, rs2072590 on 2q31, rs2665390 on 3q25, rs10088218, rs1516982, rs10098821 on 8q24.21 and rs2363956 and rs8170 on 19p13 (6–8). All of the SNPs except rs2363956 and rs8170 on 19p13 are located in non-coding regions. Thus, the functional significance for most of these variants is unknown. Our analysis presents the first evidence of potential functional significance of these candidate ovarian cancer risk SNPs in this post-GWAS era. Our goal is to investigate whether microRNAs, and their predicted target genes or pathways, may be associated with these ovarian cancer risk SNPs. In the analysis, we observed 130 different significant associations between microRNAs and variants. rs3814113 (9p22.2) and rs2072590 (2q31), two most significant ovarian cancer risk SNPs from GWAS studies (P = 2.5 × 10−19 and 1.9 × 10−8, respectively), have the greatest number of significant associations (68 and 37, respectively). The number of significant associations does not necessarily positively correlate with the biological functions. However, considering the fact that microRNAs can regulate at least 60% of human protein-encoding genes, they can act as TSGs or/and oncogenes, and they are actively involved in every step of carcinogenesis; our data suggest that these two GWAS discovered variants might be key players involved in a variety of biological pathway and might have important biological functions. On the other hand, our study adds a new level of complexity to cellular gene expression regulation by revealing that common genetic variants can affect the expression of microRNAs that are their own regulatory molecules. Earlier studies have shown that common genetic variants contribute significantly to the individual differences in protein-coding gene expression variation. Whether that is the case for microRNA is not known. Tested genetic variants in this study are not only associated with ovarian cancer risk, but also potential candidates for the involvement in microRNA expression. Differences in the quantity of mature microRNAs have a clear impact on the level of targeted proteins and result in phenotypic differences. The subsequent identification of the functional variation related to each variant may provide important genomic targets for dissecting the molecular basis of susceptibility to familial ovarian cancer.
More intriguingly, five microRNAs (hsa-miR-18a, hsa-miR-19a, hsa-miR-20a, hsa-miR-20a*, hsa-miR-17) on chromosome 13, which are significantly associated with rs3814113, belong to the miR-17-92 microRNA cluster. The miR-17-92 cluster, which is also designated as oncomir-1, is one of the best-characterized oncogenic microRNAs (35). Human major miR-17-92 cluster is located at 13q31.3, a region amplified in several hematopoietic malignancies and solid tumors, including diffuse B-cell lymphomas, follicular lymphomas, Burkitt's lymphomas and lung carcinoma (36). Overexpression of miR-17-92 has been observed in multiple tumor types (37,38). However, studies in the past have revealed that miR-17-92 functions pleiotropically during both normal development and malignant transformation to promote proliferation, inhibit differentiation, augment angiogenesis and sustain cell survival. The relationship between miR-17-92 and ovarian cancer has not been thoroughly investigated (39,40). Loss of heterogeneity at the 13q31.3 locus that harbors human miR-17-92 has been reported in some ovarian and breast cancers and melanomas, suggesting it might act as a tumor suppressor in these cancers (39). The most significant association in this cluster is between hsa-miR-20a and rs3814113. Fan et al. (41) reported that inhibition of hsa-miR-20a in OVCAR3 ovarian cancer cell line could suppress, whereas overexpression of miR-20a could enhance, cell long-term proliferation and invasion. They also confirmed amyloid precursor protein (APP) as a direct target gene of hsa-miR-20a. In addition, Nam et al. (42) reported that higher expression of hsa-miR-18a was associated with poor prognosis of ovarian cancer. The microRNA cluster on chromosome X, including hsa-miR-18b, hsa-miR-106a and hsa-miR-20b, is the paralog of miR-17-92, caused by ancient gene duplication. Along with the other two clusters on chromosome 14 and 19, these three microRNAs clusters have been reported to be regulated through imprinting or epigenetic mechanisms (16). Zhang et al. (16) found the microRNA expression in these three clusters were significantly lower in ovarian tumor tissues compared with normal ovarian surface epithelium cells. The cluster on chromosome 8 includes hsa-miR-30d that has been reported to play a role in the initiation and progression of ovarian cancer. The DNA copy number of the hsa-miR-30d and hsa-miR-30b region has been frequently amplified in multiple types of human cancers, including ovarian cancer (43). Therefore, it seems that all five microRNA clusters that show significant associations with ovarian cancer risk alleles have important functional implications in the etiology of ovarian cancer.
Additionally, using these identified significant associations in the pathway analysis, we have found that the genes predicted to be targeted by multiple SNP-associated microRNAs are enriched in several key biological pathways, such as cell cycle, cellular response to stress/damage, energy metabolism, etc. Interestingly, so far, all known familial ovarian cancer genes (BRCA1/2 and MMR) are key players in these pathways. For example, it has been demonstrated that BRCA1 is the key regulator in sensing DNA stress/damage and subsequently promoting cell cycle arrest (44). Although our association analysis cannot pinpoint the exact functions of these GWAS discovered SNPs, it elucidates the potential biological pathways for which one could focus on in future analysis.
To the best of our knowledge, this is the first report to describe the microRNAs expression profiles in LCLs from familial ovarian cancer cases and unrelated controls. This is also the first report on the significant associations between microRNA expression variations and ovarian cancer risk SNPs. Our discovery not only suggests the microRNA expression in LCLs might be regulated by genetic variants but also proposes the possible functional significance of ovarian cancer risk SNPs identified from GWAS. Further studies are needed to determine the genetic causes of differentially expressed microRNAs and biological consequences and pathways of the identified significant associations between microRNA and ovarian cancer risk SNPs. These studies will facilitate candidate gene discovery and lead to better understanding of the genetic etiology of familial ovarian cancer and development of microRNA-based tools to detect familial ovarian cancer at early age.
Supplementary material
Supplementary Figures 1 and 2 and Tables 1–3 can be found at http://carcin.oxfordjournals.org/.
Funding
National Institutes of Health (5R01CA136483 to H.Z. and P30 CA016056 to Roswell Park Cancer Institute); Ralph Wilson Medical Foundation to H.Z.; Department of Defense Ovarian Cancer Program (OC073116 to H.Z.); and Roswell Park Alliance Foundation to H.Z.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- EBV
Epstein-Barr virus
- FDR
false discovery rate
- GRFOCR
Gilda Radner Familial Ovarian Cancer Registry
- GWAS
genome-wide association studies
- LCL
lymphoblastoid cell line
- SNP
single nucleotide polymorphisms
- SVM
support vector machine
- TSG
tumor suppressor gene
- qRT–PCR
quantitative real-time PCR
References
- 1.Yancik R. Ovarian cancer. Age contrasts in incidence, histology, disease stage at diagnosis, and mortality. Cancer. 1993;71:517–523. doi: 10.1002/cncr.2820710205. [DOI] [PubMed] [Google Scholar]
- 2.Narod SA, et al. An evaluation of genetic heterogeneity in 145 breast-ovarian cancer families. Am. J. Hum. Genet. 1995;156:254–264. [PMC free article] [PubMed] [Google Scholar]
- 3.Ford D, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am. J. Hum. Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Easton DF, et al. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am. J. Hum. Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch HT, et al. Surveillance and management of patients at high genetic risk for ovarian carcinoma. Obstet. Gynecol. 1982;59:589–596. [PubMed] [Google Scholar]
- 6.Bolton KL, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goode EL, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman ML, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, et al. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, et al. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Genet. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Shahab SW, et al. Evidence for the complexity of microRNA-mediated regulation in ovarian cancer: a systems approach. PLoS One. 2011;6:e22508. doi: 10.1371/journal.pone.0022508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Permuth-Wey J, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 19.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 20.Heinzen E, et al. Tissue specific genetic control of gene expression and alternative splicing: implications for the study of human complex traits. PLoS Biol. 2008;6:e1000001. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borel C, et al. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011;21:68–73. doi: 10.1101/gr.109371.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear. models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:R3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, et al. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 25.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slawski M, et al. CMA: a comprehensive Bioconductor package for supervised classification with high dimensional data. BMC Bioinformatics. 2008;9:439. doi: 10.1186/1471-2105-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudoit S, et al. Classification in microarray experiments. In: Speed T, editor. Statistical Analysis of Gene Expression Microarray Data. 1st edn. Chapman and Hall/CRC; 2003. pp. 93–158. [Google Scholar]
- 28.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon AL, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 33.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 34.Duan S, et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olive V, et al. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 37.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrocca F, et al. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 39.Hossain A, et al. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, et al. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 41.Fan X, et al. miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta Biochim. Biophys. Sin. (Shanghai) 2010;42:318–324. doi: 10.1093/abbs/gmq026. [DOI] [PubMed] [Google Scholar]
- 42.Nam EJ, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 43.Lu Y, et al. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22–q24.23 in medulloblastoma. PLoS One. 2009;4:e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, et al. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1:117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.