Abstract
The lumen of the endoplasmic reticulum (ER) differs from the cytosol in its content of ions and other small molecules, but it is unclear whether the ER membrane is as impermeable as other membranes in the cell. Here, we have tested the permeability of the ER membrane to small, nonphysiological molecules. We report that isolated ER vesicles allow different chemical modification reagents to pass from the outside into the lumen with little hindrance. In permeabilized cells, the ER membrane allows the passage of a small, charged modification reagent that is unable to cross the plasma membrane or the lysosomal and trans-Golgi membranes. A larger polar reagent of ∼5 kDa is unable to pass through the ER membrane. Permeation of the small molecules is passive because it occurs at low temperature in the absence of energy. These data indicate that the ER membrane is significantly more leaky than other cellular membranes, a property that may be required for protein folding and other functions of the ER.
INTRODUCTION
Cellular membranes generate compartments in which the concentration of ions and other small molecules can differ dramatically from that of the surroundings. The imbalance of these small molecules across the membrane is the result of both the barrier that the phospholipid bilayer represents and of pumps that generate the gradients. Cellular membranes generally show some leakiness, caused by the imperfect nature of the bilayer, particularly by kinks in unsaturated hydrocarbon chains of phospholipids and by the presence of proteins that disturb its organization (de Gier, 1992). However, leakiness of a membrane is usually not a problem because the pumps that generate the gradients are strong. Nevertheless, the mere existence of gradients does not indicate how tight a membrane is.
Although the plasma and lysosomal membranes are considered to be highly impermeable, the situation with the endoplasmic reticulum (ER) membrane is less clear. The ER lumen differs from the cytosol in its concentration of calcium ions (Meldolesi and Pozzan, 1998) and probably of oxidized glutathione (Hwang et al., 1992), but whether other small molecules are concentrated or depleted in the ER is unclear. For example, there is no difference in pH between the ER lumen and the cytosol, in contrast to later compartments in the secretory pathway, which are more acidic (Kim et al., 1998; Kneen et al., 1998; Wu et al., 2000). The concentration of calcium ions in the ER lumen may be a very special case. They are bound to many luminal proteins, so that the free concentration is much lower than the total, and they are transported into the ER by powerful pumps. Reduced glutathione seems to be transported into the ER in an energy-independent manner and causes a burden on the oxidizing system in the ER (Banhegyi et al., 1999; Cuozzo and Kaiser, 1999). These considerations raise the possibility that for many small molecules, there may be no steep gradient across the ER membrane, a situation that could be caused by a higher permeability of the ER membrane compared with other cellular membranes.
Experimental evidence for the permeability of the ER is controversial. Fluorescence quenching experiments have shown that iodide and NAD+ ions do not cross isolated ER membranes (Crowley et al., 1994; Hamman et al., 1997). On the other hand, when a small polar substrate, 4-methylum-belliferyl α-d-glucopyranoside (4MαG), of the ER luminal enzyme α-glucosidase was added to broken cells, it was able to cross the ER membrane (Heritage and Wonderlin, 2001; Roy and Wonderlin, 2003). Rough microsomes from rat liver also had a high permeability to small solutes (Meissner and Allen, 1981). Some permeability may be caused by protein-conducting channels that are kept open by the association with nontranslating ribosomes (Simon and Blobel, 1991). This is suggested by the observation that the flow of ions, the passage of 4MαG, and the efflux of calcium ions out of the ER are all stimulated by puromycin, a drug that releases nascent chains from ribosomes but leaves them bound to the membrane (Simon and Blobel, 1991; Heritage and Wonderlin, 2001; Lomax et al., 2002; Roy and Wonderlin, 2003; Wirth et al., 2003). However, significant permeability was seen after ribosomes had been stripped from the membrane, leading to the closure of the protein-conducting channels (Simon et al., 1989; Simon and Blobel, 1991; Heritage and Wonderlin, 2001; Roy and Wonderlin, 2003), suggesting that these channels may not be the only reason for permeability of ER membranes. The high protein content (Fujiki et al., 1982) and the low level of cholesterol, known to fill “gaps” in lipid bilayers (Papahadjopoulos et al., 1972; de Gier, 1992; Allan, 1996; Haines, 2001), may contribute to the leakiness of ER membranes.
To test the permeability of the ER membrane it is advantageous to use small molecules that the cell would not normally encounter. When physiological molecules, such as 4MαG or Ca2+ are used, permeability may be caused by specific transport systems, rather than by nonspecific leakage. Here, we have used chemical modification reagents to test the permeability of the ER membrane with both isolated microsomes and permeabilized cells. Our results show that the ER membrane is significantly more leaky than the plasma membrane, lysosomes or the trans-Golgi, supporting the idea that the ER membrane may have unique properties required for its specialized functions.
MATERIALS AND METHODS
Reagents
Streptolysin O (SLO) was kindly provided by S. Bhadki (Mainz, Germany). Maleimide biotin (EZ-Link PEO maleimide biotin; (+)-biotinyl-3-maleimidopropionamidyl-3,6-dioxaoctanediamine, MB), sulfo-NHS-biotin (N-hydroxysulfosuccinimidobiotin), EZ-Link sulfo-NHS-LC-biotin [sulfosuccinimidyl-6-(biotinamido) hexanoate], and streptavidin beads were purchased from Pierce Chemical (Rockford, IL). Maleimide propionyl biotin (MPB) was purchased from Molecular Probes (Eugene, OR). Maleimide-polyethyleneglycol (mPEG) was from Shearwater (Huntsville, AL). Rabbit anti-protein disulfide isomerase (PDI) serum was from Stressgen (San Diego, CA). Goat sera against GRP94, BiP, calnexin, calreticulin, Hsp90, cathepsin D, and β-1,4-galactosyl transferase 1 were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit antibodies to Sec61β and translocon-associated protein complex α (TRAPα) were described previously (Neuhof et al., 1998). A goat serum against human hexosaminidase B was kindly provided by R. Proia (National Institutes of Health, Bethesda, MD). Secondary antibodies coupled to peroxidase were obtained from Sigma-Aldrich (St. Louis, MO).
In Vitro Translation and Translocation
Translation and translocation assays and the isolation of ribosome-nascent chain complexes, rough microsomes, and puromycin-stripped microsomes were carried out as described previously (Neuhof et al., 1998). In vitro mutagenesis was used to generate a plasmid coding for prepro-α-factor with a cysteine at position 56.
Modification Reactions with Isolated ER Membranes
After in vitro translocation of radiolabeled proteins, the membranes were pelleted and resuspended in membrane buffer (50 mM HEPES/KOH pH 7.6, 250 mM sucrose, 150 mM potassium acetate, 8 mM magnesium acetate). MB and MPB were dissolved in dimethyl sulfoxide, whereas sulfo-NHS-biotin was dissolved in water. The final concentration of dimethyl sulfoxide was <0.5% for MB and <1% for MPB. They were added for the indicated time periods and were quenched with either 10 mM dithiothreitol (DTT) (for MB and MPB) or with 10 mM Tris/HCl, pH 7.5 (for sulfo-NHS-biotin) for 5 min on ice. The samples were adjusted to 2% SDS, 67 mM Tris/HCl, pH 7.5, 5 mM DTT and incubated for 10 min at 60°C. They were diluted 1:20 in a buffer containing 1% Triton X-100 (TX-100) and incubated for 3 h with 25 μl of streptavidin beads, pretreated with 1% bovine serum albumin, and pre-washed with buffer. The supernatant fraction was precipitated with trichloroacetic acid (TCA), the beads were washed three times with buffer, and the bound material was eluted with SDS-sample buffer.
Modification of endogenous ER proteins was performed with MB as described above. Immunoprecipitation for PDI and Sec61α was carried out as described previously (Gorlich et al., 1992; Tsai et al., 2001).
Preparation of Permeabilized Cells
HeLa cells were grown at 37°C and 5%CO2 in Dulbecco's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (all from Invitrogen, Carlsbad, CA). The cells were removed with trypsin, rinsed twice with supplemented Dulbecco's medium, followed by washes in cold HCN (50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM CaCl2). For permeabilization in SLO, 2 × 106 cells were gently resuspended in 1.5 ml of cold HCN with or without SLO at 0.5 μg/ml and incubated for 10 min on ice in siliconized Eppendorf tubes. Unbound SLO was removed by two washes in cold HNE (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EGTA). The cells were resuspended in 100 μl of HNE, transferred to a new tube, and incubated for 10 min at 37°C to induce pore formation. Eighty to 90% of cells were permeable to trypan blue. For permeabilization in digitonin, the cells were resuspended in 100 μl of HCN and incubated with 0.04 or 0.1% digitonin (Merck, Whitehouse Station, NJ; purified as in Gorlich et al., 1992) for 10 min on ice. They were washed twice in HNE and transferred to a new tube. Ninety-five to 100% of the cells were permeable to trypan blue.
After permeabilization, 10% of cells were spun down in a Microfuge at 3300 rpm for 10 min at 4°C. The pellet was resuspended in the original volume of HNE buffer. Cell lysates were prepared in 50 mM Tris, pH 8, NaCl 150 mM 1% Triton and proteases inhibitors and clarified at 4°C by centrifugation at full speed in a Microfuge.
Experiments with Metabolically Labeled Cells
A HeLa cell line was generated that stably expressed hemagglutinin (HA)-tagged myosin heavy chain (MHC) class I heavy chains (HLA-A2), in which the three lysines in the cytosolic tail were replaced by arginines (HA-heavy chain [K-R]). The cells were labeled for 10 min with [35S]methionine and -cysteine, permeabilized with 0.04% digitonin, and incubated with 0.5 mM sulfo-NHS-LC-biotin, freshly dissolved at 5 mM in water, for 30 min at 4°C. The samples were immunoprecipitated with HA antibodies, and a portion of the precipitated material (80%) was subjected to incubation with streptavidin-agarose beads to detect biotinylated molecules. Finally, one-half of each sample was incubated with 10 U of endoglycosidase H (Endo H) for 1 h at 37°C. The samples were separated by SDS-PAGE and analyzed by autoradiography.
Hybridoma B cells producing anti-HA tag antibodies (12CA5) were metabolically labeled and processed as described above, except that plasma membrane permeabilization was performed with 0.02% digitonin and that the IgG heavy chains were collected with protein A/G-Sepharose before subjecting them to incubation with streptavidin beads.
Chemical Modification
Sulfo-NHS-LC-biotin (0.5 mM from a 5 mM stock in water) was added to permeabilized cells for 30 min on ice with gentle agitation. Ten percent of the sample was used to analyze the distribution of proteins among the pellet and supernatant fractions, as described above. The biotinylation reaction was stopped with three washes in cold 50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM CaCl2. The cell pellets were resuspended in 100 μl of HNC with 1% Triton X-100 and protease inhibitors and incubated for 20 min at 4°C. The lysates were clarified by centrifugation at full speed in a Microfuge and adjusted to 0.1% Triton X-100, 0.5% IGEPAL CA-630.
Modification with 1.5 mM mPEG, dissolved in 20 mM Tris, pH 7.5, was performed for 30 min at 4°C and the reaction was stopped with 10 mM DTT for 10 min at 4°C.
Binding of Biotinylated Proteins to Streptavidin Beads
Streptavidin agarose beads were washed four times with phosphate-buffered saline (Invitrogen). Slurry (40 μl) was added to each sample. After incubation for 50 min at 4°C, the beads were washed three times with 50 mM sodium-phosphate buffer, pH 7.4. The bound material was eluted with 120 μl of SDS sample buffer (Neuhof et al., 1998) by boiling for 20 min.
RESULTS
Permeability of Isolated Microsomes
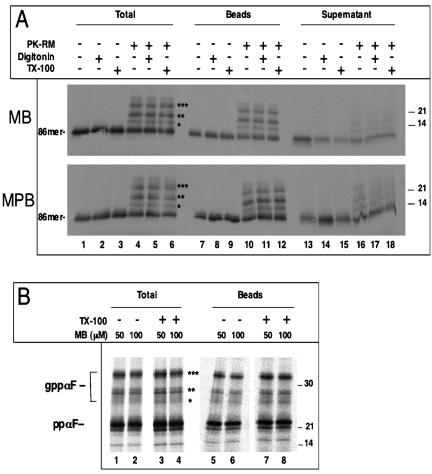
We first tested whether isolated ER membranes are permeable to small molecules. Rough microsomes (RMs) were purified from dog pancreas and stripped of ribosomes by treatment with puromycin and high salt (PK-RM) (Simon and Blobel, 1991; Gorlich et al., 1992). A radiolabeled polypeptide was then imported into these vesicles and its accessibility to chemical modification reagents tested. In initial experiments, we used microsomes in which the protein-conducting channels are expected to be open owing to their association with nontranslating ribosomes. A fragment of prepro-α-factor (ppαF) coding for the first 86 amino acids was synthesized in vitro in the wheat germ system in the presence of [35S]methionine. Because a stop codon is lacking in the mRNA, the C terminus of the nascent chain remains associated with the ribosome. The ribosome-nascent chain complex was isolated and incubated with PK-RM (Neuhof et al., 1998). Although the polypeptide fragment contains three N-glycosylation sites, it cannot be modified by the oligosaccharyl transferase in the ER lumen because the C terminus is tethered to the ribosome. On release of the nascent chain from the ribosome by treatment with puromycin at physiological salt concentration, the polypeptide moves into the ER lumen and becomes glycosylated (Figure 1A, lane 4, and B). As expected, when the membranes are omitted from the reaction, no modification of the nascent chain is seen (lane 1). The puromycin treatment should leave the protein-conducting channel associated with the ribosome and keep it open (Simon and Blobel, 1991).
Figure 1.
Ribosome-stripped ER membranes are permeable to small molecules. (A) Ribosomes carrying a radiolabeled fragment of ppαF (86mer) with a single cysteine at position 56 were incubated with ribosome-stripped microsomes (PK-RM). After treatment with puromycin to release the nascent chains into the ER lumen, the samples were incubated with 0.1 mM MB or 0.075 mM MPB for 10 min on ice, either in the absence of detergent, or in the presence of 0.5% TX-100 or 0.1% digitonin, as indicated. One-half of the samples was analyzed after precipitation with TCA (total), the other was subjected to incubation with streptavidin beads, and the bound (beads) and unbound (supernatant) fractions were analyzed. All samples were analyzed by SDS-PAGE and autoradiography. The stars indicate molecules with one to three carbohydrate chains. (B) Full-length ppαF with a single cysteine at position 56 was imported into untreated RMs. Membranes were pelleted before the modification reaction was carried out. The samples were then treated for 10 min on ice with the indicated concentrations of MB in the absence or presence of 0.5% Triton X-100. One half of the sample was analyzed directly (total), the other was subjected to incubation with streptavidin beads and the bound material was analyzed (beads). All samples were subjected to SDS-PAGE and autoradiography. ppαF, gppαF, pro-α-factor without or with one to three carbohydrate chains, respectively. Molecular masses (in kilodaltons) are indicated on the right side.
To test for permeability of the vesicle membrane, we used two different SH-modifying reagents, MB and MPB, which both attach biotin to cysteines and are reported to poorly permeate the plasma membrane (Loo and Clarke, 1995; Scheel and Pelham, 1998; Spura et al., 2000). Both are hydrophilic reagents with a molecular mass of ∼525 Da. Because wild-type ppαF does not contain cysteines, we introduced a single cysteine at position 56. The modification reagents were added after termination of translation with puromycin, and biotinylated proteins were detected by their association with streptavidin beads. A large percentage of both nonglycosylated and glycosylated ppαF was found on the streptavidin beads (Figure 1A, lane 10 versus 16). Glycosylated ppαF seemed to be modified even more efficiently than nonglycosylated ppαF, perhaps because the cysteine in non-transported ppαF is somewhat shielded by cytosolic chaperones. In any case, the data show that the reagents are able to pass the membrane and reach the glycosylated, luminal protein. The reaction was essentially uninhibited by the presence of a membrane because the same amount of material was modified if the membrane integrity was disrupted by 0.1% digitonin or 0.5% TX-100 (lanes 11 and 12). Thus, two different low-molecular-weight reagents can pass the ER membrane in an essentially unrestricted manner.
Next, we tested the permeability of untreated RMs whose protein-conducting channels were not opened by puromycin. Full-length ppαF with a single cysteine at position 56 was imported into RMs and subjected to modification with MB. The biotinylated proteins were again collected by streptavidin beads (Figure 1B). As before, the reagent was able to cross the ER membrane to the same extent as after Triton X-100 treatment (lanes 5 and 6 versus 7 and 8), suggesting that permeability is an inherent property of the microsomal membranes. Similar results were obtained with the reagent MPB (our unpublished data).
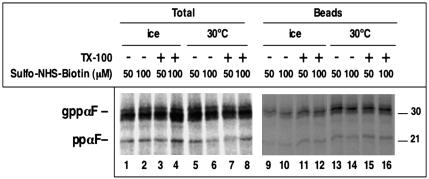
To exclude the possibility that permeability is restricted to a certain class of small reagents, we used sulfo-NHS-biotin, a reagent that modifies amino groups and is considered to be membrane impermeable (Hurley and Finkelstein, 1990). It is a negatively charged reagent with a molecular mass of 443 Da. For these experiments, we used again full-length ppαF and untreated RMs, and the modification reagent was added either on ice or at 30°C. Biotinylated ppαF was separated from nonmodified material by incubation with streptavidin beads (Figure 2). We found that a significant percentage of ppαF was modified even after 10 min on ice (lanes 9 and 10), although the labeling was somewhat stronger at 30°C (lanes 13 and 14). Addition of Triton X-100 before modification increased the labeling efficiency slightly (lanes 11 and 12) or not at all (lanes 15 and 16). A concentration of 50 μM was sufficient to maximally label the protein in the lumen of the vesicles, even when the reaction was performed on ice. At this concentration, passage through the plasma membrane is not observed (Hurley and Finkelstein, 1990). These data show that permeability can be seen with different small molecules and that these permeate even into untreated rough microsomes.
Figure 2.
Rough microsomes are permeable to a modification reagent. Radiolabeled full-length ppαF was translocated into rough microsomes. The membranes were isolated and incubated with different concentrations of sulfo-NHS-biotin, an amino group-modifying reagent, for 10 min on ice or at 30°C. Where indicated, 0.5% TX-100 was present during the modification reaction. One portion of the samples was analyzed directly (total), the other was incubated with streptavidin beads and the bound fraction was analyzed (beads). All samples were subjected to SDS-PAGE and autoradiography. ppαF, gppαF, pro-α-factor without or with three carbohydrate chains, respectively. Molecular masses (in kilodaltons) are indicated on the right side.
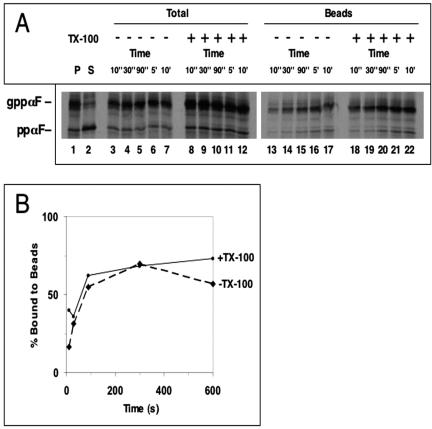
Next, we tested whether the membrane delays the access of a reagent to a luminal protein. The reagent MB was added for different time periods to microsomes containing imported full-length ppαF containing a single cysteine at position 56 (Figure 3A, lanes 13-17). Even after 10 s, there was significant labeling of full-length ppαF (lane 13). Compared with the reaction in the presence of Triton X-100, the labeling was less efficient at the earliest time points, but after 30 s there was no difference (lanes 18-22; see quantification in Figure 3B). Thus, the membrane represents an only small barrier to the passage of the reagent.
Figure 3.
Time course of modification of ppαF. (A) Radiolabeled full-length ppαF with a cysteine at position 56 was translocated into rough microsomes. The samples were centrifuged resulting in P and S fractions. The membranes were incubated with 50 μM MB for different time periods on ice. One-half of the samples was analyzed after TCA precipitation; the other was incubated with streptavidin beads and the bound material was analyzed. ppαF, gppαF, pro-α-factor without or with three carbohydrate chains, respectively. (B) Quantification of the experiment in A.
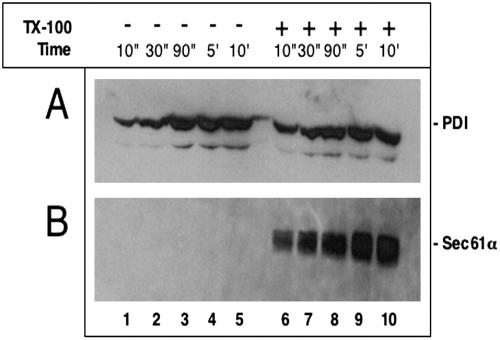
To exclude the possibility that proteins newly imported into the ER lumen may be more accessible to modifying reagents than endogenous proteins, we added the reagent MB to microsomes and tested the modification of PDI by immunoprecipitation and subsequent blotting with streptavidin antibodies (Figure 4A). Even when the reaction was performed on ice, the modification of PDI in intact membranes was as fast as after solubilization with Triton X-100 (lanes 1-5 versus 6-10). As a control, we used the integral membrane protein Sec61α, which contains most of its cysteines inside the membrane (Gorlich et al., 1992). As expected, this protein was only modified in the presence of Triton X-100 (Figure 4B, lanes 1-5 versus 6-10), indicating that the modification reagent does not act on cysteines inside the lipid phase. These results show that even an endogenous luminal protein is accessible to a small molecule added to the outside of the membrane vesicles.
Figure 4.
An endogenous luminal ER protein is accessible to a modification reagent. (A) Ribosome-stripped microsomes (PK-RM) were incubated with 100 μM MB on ice for different time periods in the absence or presence of 0.5% TX-100. The samples were immunoprecipitated with antibodies to PDI, subjected to SDS-PAGE, transferred onto a nitrocellulose filter, and incubated with peroxidase-coupled streptavidin. (B) As in A, but immunoprecipitation with antibodies to Sec61α.
Passage of Small Molecules into the ER of Permeabilized Cells
We next tested the permeability of ER membranes in a more physiological context using permeabilized cells. Tissue culture cells were treated with either the toxin SLO or with a low concentration of the detergent digitonin to generate holes in the plasma membrane, which would allow the addition of chemical modification reagents to the cytosol, so that their permeation into the ER could be tested.
To permeabilize the plasma membrane with SLO, cells were incubated with the toxin at low temperature at which it binds to the plasma membrane but does not form holes in it (Bhakdi et al., 1993). The cells were then washed to remove unbound toxin and incubated at 37°C for 10 min to induce pore formation. The procedure should ensure that holes are only generated in the plasma membrane and not in any internal membrane. To test whether proteins remain in the ER after permeabilization of the plasma membrane, the cells were put back at low temperature and subjected to centrifugation to separate a membrane pellet (P) from a supernatant (S) fraction. The distribution of different proteins was determined by immunoblotting with various antibodies. On treatment with SLO, the cytosolic protein Hsp90 occurred in the supernatant, whereas both the ER membrane protein Sec61β and the ER luminal proteins GRP94, BiP, PDI, and calreticulin all stayed in the membrane pellet, as in nonpermeabilized cells (Figure 5, lanes 3 and 4 versus 1 and 2).
Figure 5.
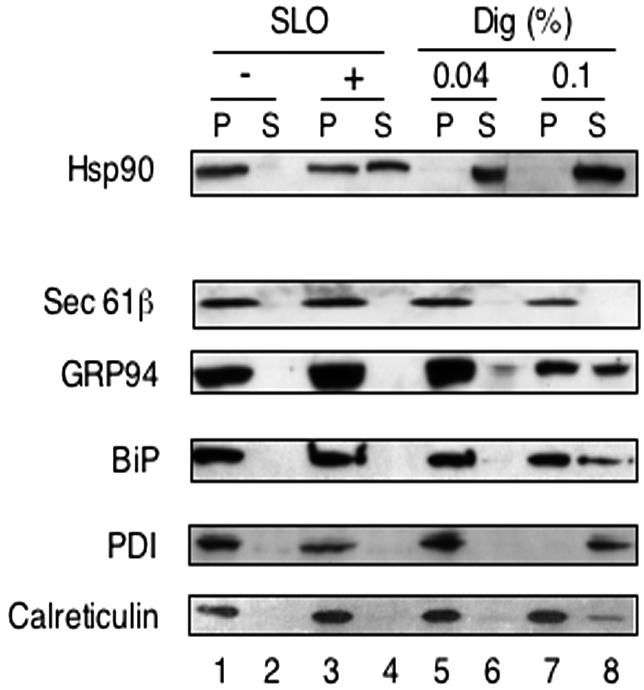
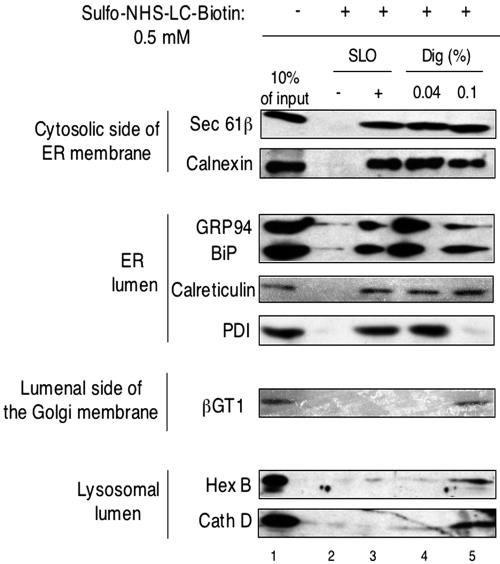
Differential permeabilization of plasma and ER membranes. HeLa cells (2 × 106) were permeabilized with SLO or digitonin (Dig; 0.04 or 0.1%). The cell remnants were pelleted in a Microfuge, and S and P fractions, corresponding to 5 × 104 cells, were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
Similar results were obtained when cells were permeabilized with 0.04% digitonin (lanes 5 and 6), as expected from the fact that at this concentration the cholesterol-interacting detergent remains confined to the plasma membrane. At higher digitonin concentration (0.1%), leakage of ER luminal proteins, in particular of PDI, was observed (lanes 7 and 8). We conclude that, with SLO and 0.04% digitonin, the barrier of the plasma membrane can be broken without allowing proteins to leak out of the ER.
Next, we added the chemical modification reagent sulfo-NHS-LC-biotin (negatively charged, molecular mass 557 Da) to cells that were either untreated or permeabilized with SLO or digitonin. Biotinylation of different proteins was tested by their association with streptavidin beads, followed by blotting with various antibodies (Figure 6). In the absence of SLO or digitonin, only a small percentage of the tested proteins was modified by the reagent (Figure 6, lane 2). These results confirm that the plasma membrane is impermeable to the modification reagent. On treatment of the cells with SLO, the cytosolic domain of Sec61β, which contains essentially all amino groups of this membrane protein; calnexin, which contains amino groups on both sides of the ER membrane; and the ER luminal proteins GRP94, BiP, PDI, and calreticulin were all modified (lane 3). We estimate that 5-10% of the luminal proteins were biotinylated (compare with lane 1). The somewhat lower percentage of modification compared with the results with isolated microsomes (Figure 2) may perhaps be explained by cytosolic proteins that remained in the permeabilized cells and could have partially quenched the reagent. Indeed, some Hsp90 was not washed out during permeabilization (Figure 5), and larger protein complexes, such as proteasomes, or cytoskeletal proteins probably remained completely inside the cells. Selective permeability of the ER membrane for the modification reagent is indicated by the observation that proteins in the lumen of lysosomes (hexosaminidase B and cathepsin B) remained inaccessible (Figure 6). Likewise, β-1,4-galactosyl transferase 1 (βGT1), a trans-Golgi membrane protein containing all amino groups in its luminal domain, could not be modified (Masri et al., 1988; Lo et al., 1998). Similar results were obtained when the cells were treated with 0.04% digitonin before addition of the chemical modification reagent (lane 4). Again, proteins in the ER lumen, but not those inside lysosomes or the trans-Golgi could be modified. When cells were treated with 0.1% digitonin, the labeling of some ER luminal proteins, in particular of PDI, was reduced (lane 5), likely because they were released from the analyzed membrane pellet (Figure 5). Significantly, under these conditions the lysosomal proteins and βGT1 in the trans-Golgi were labeled (Figure 6, lane 5), indicating that the lack of labeling at milder conditions is not due to the absence of reactive amino groups in these proteins. It should be noted that the biotinylation reaction did not cause the release of luminal proteins from the ER or lysosomes, as demonstrated by sedimentation experiments (our unpublished data).
Figure 6.
A small molecule passes into the ER but not into lysosomes or the trans-Golgi. HeLa cells (2 × 106) were permeabilized with SLO, 0.04% digitonin (Dig), or 0.1% digitonin as indicated. Then 0.5 mM sulfo-NHS-LC-biotin was added for 30 min on ice. After solubilization in detergent, cell lysates were incubated with streptavidin agarose beads, and the bound material was analyzed by SDS-PAGE and immunoblotting with antibodies to various proteins. Hex B, hexosaminidase B; Cath D, cathepsin D; βGT1; β-1,4-galactosyl transferase. For βGT1, twice as much sample was used. Cell lysate corresponding to 10% of the input material was loaded as a control (lane 1).
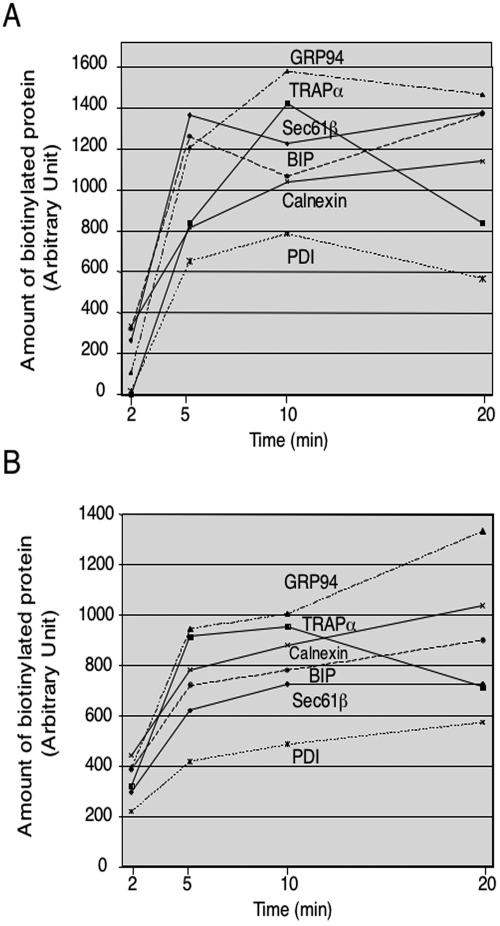
To further test whether the ER membrane represents a barrier for the chemical modification reagent, we compared the time course of modification for different proteins after cells were permeabilized with either 0.04% digitonin or SLO (Figure 7, A and B, respectively). In both cases, all proteins were modified with similar kinetics within the limits of accuracy of the experiments. Sec61β and translocon-associated protein complex α have essentially all their reactive amino groups on the cytosolic side of the ER membrane, and yet they were not modified faster than the ER luminal proteins tested. We therefore conclude that the ER membrane does not constitute a significant barrier to the small chemical reagent.
Figure 7.
The ER membrane does not constitute a significant barrier to small molecules. HeLa cells were permeabilized with 0.04% digitonin (A) or SLO (B) and incubated with 0.5 mM sulfo-NHS-LC-biotin for different periods of time. Biotinylated proteins were recovered after incubation with streptavidin beads and analyzed by SDS-PAGE and immunoblotting with antibodies to different proteins. The bands were quantitated using a Fujix PhosporIimager and the software Image Gauge 3.0.
It should be noted that all experiments were performed at 4°C in the absence of ATP or any other energy source. Addition of ATP did not change the modification of ER luminal proteins (our unpublished data). Thus, the passage of the chemical reagent into the ER is likely a passive process.
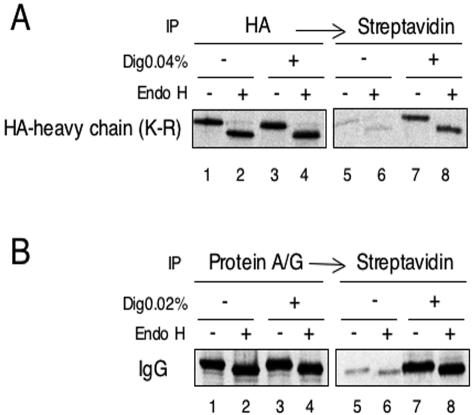
Further experiments confirmed that ER luminal proteins can be readily modified with a small reagent once the plasma membrane has been permeabilized (Figure 8). To test the modification of a luminal domain of a membrane protein, we expressed in HeLa cells MHC class I heavy chains, in which all lysine residues in the cytoplasmic tail were replaced by arginines. Thus, modification of this protein by sulfo-NHS-biotin can occur only on lysines in the luminal domain. The heavy chains were metabolically labeled with [35S]methionine and -cysteine. They were located in the ER, as demonstrated by their sensitivity to treatment with Endo H, an enzyme that cleaves ER-type carbohydrate chains (Figure 8A, lanes 2 versus 1, and 4 versus 3). Without permeabilization of the cells, only a small portion of the heavy chains could be biotinylated (lanes 5 and 6). However, upon treatment with digitonin, a much higher percentage of heavy chains was modified, as demonstrated by their association with streptavidin beads (lanes 7 and 8; 10-15% of the total population). Similar results were obtained with metabolically labeled IgG heavy chains in the ER lumen of hybridoma cells. Again, the labeled protein was localized to the ER, as shown by endoglycosidase H-sensitivity (Figure 8B, lanes 1-4), and it was biotinylated to a much higher extent upon permeabilization of the cells with digitonin (lanes 7 and 8 versus 5 and 6). Thus, in two different cell types, the ER membrane is significantly more permeable to the small modification reagent than the plasma membrane.
Figure 8.
Modification of metabolically labeled ER proteins occurs only upon cell permeabilization. (A) HeLa cells expressing HA-tagged MHC class I heavy chains with lysine residues only in the luminal domain (HA-heavy chain; K-R) were labeled with [35S]methionine and -cysteine. The cells were permeabilized with digitonin (Dig), as indicated, and treated with the amino group modifying reagent sulfo-NHS-biotin. The heavy chains were immunoprecipitated with HA antibodies. 20% of the immunoprecipitated material was analyzed directly by SDS-PAGE (lanes 1-4), and 80% was incubated with streptavidin beads to detect biotinylated material (lanes 5-8). Where indicated, one-half of each sample was treated with Endo H. All samples were subjected to SDS-PAGE followed by autoradiography. (B) Hybridoma B cells expressing IgG were analyzed as in A, except that the labeled IgG was collected with protein A/G-Sepharose.
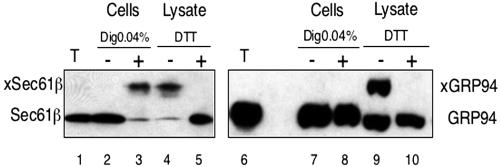
Finally, we tested whether a larger chemical modification reagent would also be able to cross the ER membrane. We used mPEG, a hydrophilic reagent of ∼5 kDa that reacts with SH groups. In nonpermeabilized cells, neither the cysteine in the cytosolic tail of Sec61β, nor the nondisulfide bonded cysteine in the ER luminal protein GRP94 were accessible to the reagent (Figure 9, lanes 2 and 7). After treatment of the cells with 0.04% digitonin, the reagent could reach the cytosolic side of the ER as demonstrated by the size shift of Sec61β (lane 3), but not the ER lumen (lane 8). When the membranes were completely solubilized (lysate), both Sec61β and GRP94 could be modified (lanes 4 and 9) and in both cases the reaction could be blocked by the SH reagent DTT (lanes 5 and 10). Thus, the 5-kDa reagent was unable to cross the ER membrane.
Figure 9.
A reagent of about 5 kDa cannot pass the ER membrane. HeLa cells (2 × 106) were permeabilized with 0.04% digitonin or lysed in 1% Triton X-100 as indicated. Modification was carried out for 30 min at 4°C with 1.5 mM mPEG, an SH-reagent of ∼5 kDa. Where indicated, DTT was present during the reaction. Samples corresponding to 5 × 104 untreated (T) or mPEG-modified cells were analyzed by SDS-PAGE on a 7.5-17.5% urea gel, followed by immunoblotting with antibodies to GRP94 or Sec61β. xSec 61β and xGRP94 indicate the positions of the modified proteins.
DISCUSSION
Our results indicate that the ER membrane is very permeable to small molecules. Several small chemical modification reagents that the cell does not normally encounter were able to pass the ER membrane both in isolated microsomes and in permeabilized cells. Passage seemed to be passive because it occurred in the absence of ATP at low temperatures. In contrast to several reagents of ∼400-500 Da, a reagent of ∼5 kDa was unable to permeate the ER membrane, suggesting that there may be a size limitation. However, it is possible that other features of the larger reagent prevented its passage through the membrane. For the smaller reagents we have not seen a long delay in the modification of luminal proteins compared with cytosolic proteins or cytosolic domains of membrane proteins, indicating that, on the time scale of minutes, the ER membrane does not present a significant hindrance to their passage. We cannot completely exclude the possibility that microsomes are permeable because they are damaged during their preparation, and that both protocols of permeabilization (SLO and digitonin) not only permeabilized the plasma membrane but also generated holes in the ER. However, the latter seems at least unlikely for SLO because excess of the toxin was removed before pores were formed. Moreover, both the lysosomal and trans-Golgi membranes remained intact with both SLO and digitonin permeabilization. Thus, it seems that the ER membrane is more permeable than the other cellular membranes tested.
Our results are in agreement with other studies showing permeability of ER membranes to small molecules (Meissner and Allen, 1981; Meissner, 1988; Heritage and Wonderlin, 2001; Roy and Wonderlin, 2003). The ER is also permeable to protons (Kim et al., 1998; Kneen et al., 1998; Jayaraman et al., 2000) and has numerous ion channels of considerable size (Simon et al., 1989; Simon and Blobel, 1991). On the other hand, fluorescence quenching experiments have shown that the ER membrane is impermeable to iodide and NAD+ ions (Crowley et al., 1994; Hamman et al., 1997). In these studies, high salt-treated microsomes were used, which likely removed nontranslating ribosomes and closed all protein-translocating channels. However, others have found that even after removal of all ribosomes from the ER membrane, there is still sizable permeability to 4MαG, a small polar molecule (Heritage and Wonderlin, 2001; Roy and Wonderlin, 2003). The reasons for this discrepancy are currently unclear.
Because we have used small molecules that likely have no transporters in the ER membrane, it seems that permeation occurs through nonspecific sites in the ER membrane. One possibility is that the small molecules passed through the pores of protein-conducting channels. The untreated rough microsomes used by us contain a large number of nontranslating ribosomes, which may have kept the channels open (Roy and Wonderlin, 2003). However, it is also conceivable that some passage occurred directly through the lipid bilayer. The ER membrane may have a more distorted lipid bilayer than other cellular membranes because it has an exceptionally high protein content and a low cholesterol concentration.
So far, it was assumed that all membranes are designed to provide a strict barrier for small molecules. Although this view may be correct for most membranes, the ER clearly does not entirely fit this paradigm. Rather, it seems that the ER membrane is designed to be tight for protein, and more leaky for small molecules. This would ensure that the lumen of the ER is similar to the cytosol in its composition of ions and other small molecules. For example, ATP might simply move in a passive manner in and out of the ER without a need for any specific transporter. It is indeed remarkable that, besides the calcium pumps, only few other transporters for small molecules have been identified in the ER, although many pumps have been found in the Golgi, the lysosome, or the plasma membrane (Hirschberg et al., 1998). The existence of an ATP transporter has been postulated but has remained elusive (Clairmont et al., 1992; Mayinger et al., 1995).
Obviously, there are gradients across the ER membrane, the most obvious one being for calcium ions (Meldolesi and Pozzan, 1998). Particularly on the time scale of seconds or below, there may be a significant hindrance for the free flow of ions and other small molecules. Together with powerful pumps counteracting the leakage of these molecules, a gradient between the ER and the cytosol can be maintained. Other molecules, such as protons, nucleotides, glutathione, glucose, potassium ions, and phosphate ions may be equilibrated between the ER lumen and the cytosol, and this may be important for slow reactions with a turnover in the range of minutes or longer. The ER lumen differs from other extracytoplasmic space, such as in lysosomes or the Golgi, in that it houses ATP-dependent and other complex reactions. Like in the cytosol, de novo protein folding must occur in the ER lumen, and it seems reasonable to assume that most conditions for the folding process, such as salt concentrations and pH, have to be similar in both compartments. We therefore propose that the unusual permeability properties of the ER membrane have evolved to cope with its specialized functions.
Acknowledgments
We are grateful to R. Proia for providing an antibody to human hexosaminidase B. We thank G. Voeltz, K. Cannon, and S. Heinrich for critical reading of the manuscript. T.A.R. is a Howard Hughes Medical Institute Investigator.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-05-0325. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0325.
References
- Allan, D. (1996). Mapping the lipid distribution in the membranes of BHK cells. Mol. Membr. Biol. 13, 81-84. [DOI] [PubMed] [Google Scholar]
- Banhegyi, G., Lusini, L., Puskas, F., Rossi, R., Fulceri, R., Braun, L., Mile, V., di Simplicio, P., Mandl, J., and Bendetti, A. (1999). Preferential transport of glutathione versus glutathione disulfide in rat liver microsomal vesicles. J. Biol. Chem. 274, 12213-12216. [DOI] [PubMed] [Google Scholar]
- Bhakdi, S., Weller, U., Walev, I., Martin, E., Jonas, D., and Palmer, M. (1993). A guide to the use of pore-forming toxins for controlled permeabilization of cell membranes. Med. Microbiol. Immunol. 182, 167-175. [DOI] [PubMed] [Google Scholar]
- Clairmont, C.A., De Maio, A., and Hirschberg, C.B. (1992). Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP78) and GRP94. J. Biol. Chem. 267, 3983-3990. [PubMed] [Google Scholar]
- Crowley, K.S., Liao, S., Worrell, V.E., Reinhart, G.D., and Johnson, A.E. (1994). Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461-471. [DOI] [PubMed] [Google Scholar]
- Cuozzo, J.W., and Kaiser, C.A. (1999). Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1, 130-135. [DOI] [PubMed] [Google Scholar]
- de Gier, J. (1992). Permeability formed by membrane lipids. Bioelectrochem. Bioenerg. 27, 1-10. [Google Scholar]
- Fujiki, Y., Fowler, S., Shio, H., Hubbard, A.L., and Lazarow, P.B. (1982). Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 93, 103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., Hartmann, E., Prehn, S., and Rapoport, T.A. (1992). A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71, 489-503. [DOI] [PubMed] [Google Scholar]
- Haines, T.H. (2001). Do sterols reduce proton and sodium leaks through lipid bilayers? Prog. Lipid Res. 40, 299-324. [DOI] [PubMed] [Google Scholar]
- Hamman, B.D., Chen, J.C., Johnson, E.E., and Johnson, A.E. (1997). The aqueous pore through the translocon has a diameter of 40-60 A during cotranslational protein translocation at the ER membrane. The aqueous pore through the translocon has a diameter of 40-60 A during cotranslational protein translocation at the ER membrane. Cell 89, 535-544. [DOI] [PubMed] [Google Scholar]
- Heritage, D., and Wonderlin, W.F. (2001). Translocon pores in the endoplasmic reticulum are permeable to a neutral, polar molecule. J. Biol. Chem. 276, 22655-22662. [DOI] [PubMed] [Google Scholar]
- Hirschberg, C.B., Robbins, P.W., and Abeijon, C. (1998). Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49-69. [DOI] [PubMed] [Google Scholar]
- Hurley, J.W., and Finkelstein, E. (1990). Identification of leukocyte surface proteins. Methods Enzymol. 184, 429-433. [DOI] [PubMed] [Google Scholar]
- Hwang, C., Sinskey, A.J., and Lodish, H.F. (1992). Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496-1502. [DOI] [PubMed] [Google Scholar]
- Jayaraman, S., Haggie, P., Wachter, R.M., Remington, S.J., and Verkman, A.S. (2000). Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J. Biol. Chem. 275, 6047-6050. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Johannes, L., Goud, B., Antony, C., Lingwood, C.A., Daneman, R., and Grinstein, S. (1998). Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. USA 17, 2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen, M., Farinas, J., and Verkman, A.S. (1998). Green fluorescent protein as a non invasive intracellular pH indicator. Biophys. J. 74, 1591-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, N.W., Shaper, J.H., Pevser, J., and Shaper, N.L. (1998). The expanding beta 4-galactosyltransferase gene family: messages from the databanks. Glycobiology 8, 517-526. [DOI] [PubMed] [Google Scholar]
- Lomax, R.B., Camello, C., Van Coppenolle, F., Petersen, O.H., and Tepikin, A.V. (2002). Basal and physiological Ca(2+) leak from the endoplasmic reticulum of pancreatic acinar cells. Second messenger-activated channels and translocons. J. Biol. Chem. 277, 26479-26485. [DOI] [PubMed] [Google Scholar]
- Loo, T.W., and Clarke, D.M. (1995). Covalent modification of human P-glycoprotein mutants containing a single cysteine in either nucleotide-binding fold abolishes drug-stimulated ATPase activity. J. Biol. Chem. 270, 22957-22961. [DOI] [PubMed] [Google Scholar]
- Masri, K.A., Appert, H.E., and Fukuda, M.N. (1988). Identification of the full-length coding sequence for human galactosyltransferase (beta-N-acetylglucosaminide: beta 1,4-galactosyltransferase). Biochem. Biophys. Res. Commun. 157, 657-663. [DOI] [PubMed] [Google Scholar]
- Mayinger, P., Bankaitis, V.A., and Meyer, D.I. (1995). Sac1p mediates the adenosine triphosphate transport into yeast endoplasmic reticulum that is required for protein translocation. J. Cell Biol. 131, 1377-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, G. (1988). Ionic permeability of isolated muscle sarcoplasmic reticulum and liver endoplasmic reticulum vesicles. Methods Enzymol. 157, 417-437. [DOI] [PubMed] [Google Scholar]
- Meissner, G., and Allen, R. (1981). Evidence for two types of rat liver microsomes with differing permeability to glucose and other small molecules. J. Biol. Chem. 256, 6413-6422. [PubMed] [Google Scholar]
- Meldolesi, J., and Pozzan, T. (1998). The endoplasmic reticulum Ca2+ store: a view from the lumen. Trends Biochem. Sci. 23, 10-14. [DOI] [PubMed] [Google Scholar]
- Neuhof, A., Rolls, M.M., Jungnickel, B., Kalies, K.U., and Rapoport, T.A. (1998). Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell 9, 103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papahadjopoulos, D., Nir, S., and Oki, S. (1972). Permeability properties of phospholipid membranes: effect of cholesterol and temperature. Biochim. Biophys. Acta 266, 561-583. [DOI] [PubMed] [Google Scholar]
- Roy, A., and Wonderlin, W.F. (2003). The permeability of the endoplasmic reticulum is dynamically coupled to protein synthesis. J. Biol. Chem. 278, 4397-4403. [DOI] [PubMed] [Google Scholar]
- Scheel, A.A., and Pelham, H.R. (1998). Identification of amino acids in the binding pocket of the human KDEL receptor. J. Biol. Chem. 273, 2467-2472. [DOI] [PubMed] [Google Scholar]
- Simon, S.M., and Blobel, G. (1991). A protein-conducting channel in the endoplasmic reticulum. Cell 65, 371-380. [DOI] [PubMed] [Google Scholar]
- Simon, S.M., Blobel, G., and Zimmerberg, J. (1989). Large aqueous channels in membrane vesicles derived from the rough endoplasmic reticulum of canine pancreas or the plasma membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 86, 6176-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spura, A., Riel, R.U., Freedman, N.D., Agrawal, S., Seto, C., and Hawrot, E. (2000). Biotinylation of substituted cysteines in the nicotinic acetylcholine receptor reveals distinct binding modes for alpha-bungarotoxin and erabutoxin a. J. Biol. Chem. 275, 22452-22460. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Rodighiero, C., Lencer, W.I., and Rapoport, T.A. (2001). Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937-948. [DOI] [PubMed] [Google Scholar]
- Wirth, A., Jung, M., Bies, C., Frien, M., Tyedmers, J., Zimmermann, R., and Wagner, R. (2003). The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 12, 261-268. [DOI] [PubMed] [Google Scholar]
- Wu, M.M., Llopis, J., Adams, S., McCaffery, J.M., Kulomaa, M.S., Machen, T.E., Moore, H.P., and Tsien, R.Y. (2000). Organelle pH studies using targeted avidin and fluorescein-biotin. Chem. Biol. 7, 197-209. [DOI] [PubMed] [Google Scholar]