Abstract
Post-translational modification of proteins is a universal form of cellular regulation. Phosphorylation on serine, threonine, tyrosine or histidine residues by protein kinases is the most widespread and versatile form of covalent modification. Resultant changes in activity, localization or stability of phosphoproteins drives cellular events. MS and bioinformatic analyses estimate that ~30 % of intracellular proteins are phosphorylated at any given time. Multiple approaches have been developed to systematically define targets of protein kinases; however, it is likely that we have yet to catalogue the full complement of the phosphoproteome. The amino acids that surround a phosphoacceptor site are substrate determinants for protein kinases. For example, basophilic enzymes such as PKA (protein kinase A), protein kinase C and calmodulin-dependent kinases recognize basic side chains preceding the target serine or threonine residues. In the present paper we describe a strategy using peptide arrays and motif-specific antibodies to identify and characterize previously unrecognized substrate sequences for protein kinase A. We found that the protein kinases PKD (protein kinase D) and MARK3 [MAP (microtubule-associated protein)-regulating kinase 3] can both be phosphorylated by PKA. Furthermore, we show that the adapter protein RIL [a product of PDLIM4 (PDZ and LIM domain protein 4)] is a PKA substrate that is phosphorylated on Ser119 inside cells and that this mode of regulation may control its ability to affect cell growth.
Keywords: kinase, peptide array, phosphoproteome, protein kinase A (PKA)
INTRODUCTION
Protein kinases regulate all aspects of cellular behaviour. These key signal transduction enzymes act by phosphorylating target sequences in substrates. This results in modulation of protein function or stability, alterations in association with other proteins, or the movement from one cellular compartment to another [1]. The evolutionary advantage of protein phosphorylation as a versatile and simple form of post-translational modification is supported by evidence that protein kinase genes represent approximately 2 % of most vertebrate genomes. The human ‘kinome’ contains nearly 500 protein kinases, most of which target serine/threonine or tyrosine residues [2]. Historically, one of the first protein kinases to be investigated at the molecular level was PKA (cAMP-dependent protein kinase/protein kinase A). Peptide-based elucidation of the substrate specificity and X-ray crystallography of the catalytic subunit of PKA have provided a framework for understanding the molecular and cellular roles of this abundant enzyme [3].
Although some enzymes such as RAF and MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase] exhibit exquisite substrate specificities and generally phosphorylate single substrates, most protein kinases are promiscuous and modify a broad spectrum of proteins. Furthermore, many kinases share overlapping substrate specificity. Thus the same residues can be targeted by multiple kinases in vivo. For example, PKA phosphorylates sites that conform loosely to serine or threonine residues found just distal to basic residues. However, there are a number of other basophilic kinases that target similar sites, including Akt, PAK (p21-activated kinase) and RSK (ribosomal S6 kinase). Therefore a major challenge in deciphering the kinome is determining substrate specificity for particular kinases and identifying targets that may mediate the physiological effects of these enzymes.
Several technologies are currently available to facilitate the identification of phosphorylation sites in proteins [4]. In addition to advances in MS that have allowed optimized proteome-level characterization of phosphorylation events, peptide and protein arrays have become popular for determining substrate preferences and finding new potential target proteins [5,6]. Finally, phospho-specific antibodies directed against phosphorylation site motifs or individual phosphorylation sites are powerful tools for examining phosphorylation events [7,8].
In the present paper we describe a strategy for protein kinase substrate identification that combines solid-phase phosphorylation and blotting with phospho-specific motif-selective antibodies. These studies led us to further characterize phosphorylation sites in several proteins, including the PDZ-LIM protein RIL [encoded by PDLIM4 (PDZ and LIM domain protein 4)]. RIL is known to be associated with the actin cytoskeleton. RIL is phosphorylated on Ser119, just distal to the PDZ domain, both in vitro and inside cells. Finally, expression of a phosphorylation-site mutant of RIL in PC-3 prostate adenocarcinoma cells increases cell growth as compared with wild-type RIL.
MATERIALS AND METHODS
Reagents and antibodies
Reagents for SPOT synthesis were purchased from Intavis. The polyclonal anti-phospho-PKA substrate antibody was from Cell Signaling Technology. The anti-phospho-RIL antibody was generated in rabbits against the peptide PATSRRS[pS]ISGISLE. Other chemicals and reagents were from Sigma–Aldrich, EMD Biosciences or New England Biolabs.
Database searching
A number of different motif scanners available online were used to identify R-X-X-S/T motifs, including Scansite (http://scansite.mit.edu/) [9], eMotif (http://motif.stanford.edu/distributions/emotif/index.html) [10] and GenomeNet Motif (http://www.genome.jp/tools/motif/). Searches were performed using several different motifs that were variations on the basic R/K-R/K-X-S/T PKA phosphorylation-site motif. Among these were the following: R-X-R-R-X-S-Φ (where Φ is a hydrophobic residue), R-R-X-S-Φ and R-R-X-S/T. When using the Scansite motif scanner, we used both the PKA substrate motif residing in the program as well as the feature for creating new input motifs for uses by the scanner. Typically results from each database largely overlapped, although some hits were found in only a single database. Results from different searches were generally pooled; we were not concerned with categorizing the most reliable motif or program, but instead focused on large pools of candidates that could meet our criteria. The selection process was curated by individually analysing potentially interesting substrates that might reveal previously unrecognized cross-talk between cAMP/PKA and other signalling processes. Sequences of interest were copied into a text file used to program the AutoSpot synthesizer. The main criterion was that the sequence contained a R-X-X-S/T motif that might be phosphorylated by PKA and recognized by the anti-phospho-PKA substrate antibody.
SPOT synthesis
Peptide arrays were synthesized on cellulose membranes using an Auto-Spot Robot ASP 222 (Intavis). After synthesis, the N-termini were acetylated with 2 % acetic acid anhydride in dimethyl formamide. Peptides were then deprotected by a 1 h treatment with dichloromethane/trifluoroacetic acid (1:1), containing 3 %tri-isopropylsilane and 2 %water [11].
Membrane phosphorylation and blotting
Membranes were briefly wetted in ethanol and then placed in pre-incubation buffer [20 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT (dithiothreitol) and 0.2 mg/ml BSA] for 1 h at room temperature (25 °C). Membranes were then blocked overnight in pre-incubation buffer supplemented with 1 mg/ml BSA and 100 μM Mg-ATP. Phosphorylation was performed in pre-incubation buffer containing 100 μM Mg-ATP (or 50 μM Mg-ATP and 10 μCi [32P]ATP) and the PKA C-subunit (catalytic subunit) (1:5000 dilution from bacterially purified stock) for 30 min at 30 °C with rocking. Membranes were then washed three times for 10 min each in 1 M NaCl and 0.2 %Triton X-100, three times for 2 min each in distilled H2O, three times for 10 min each in 1 M NaCl, three times for 2 min each in distilled H2O, and three times for 10 min each in TBST [Tris-buffered saline (50 mM Tris/HCl and 150 mM NaCl, pH 7.5) containing 0.1 % Tween 20]. For detection of [32P]ATP phosphorylation, membranes were rinsed in ethanol, dried and exposed for autoradiography. For Western blotting, membranes were blocked in 5 % non-fat dried milk/1 % BSA for 1 h, rinsed briefly with TBST, and incubated overnight at 4 °C in 5 % BSA containing anti-phospho-PKA substrate antibody (1:500). Membranes were then washed, incubated with the appropriate secondary antisera and developed using ECL (enhanced chemiluminescence).
cDNA cloning
cDNAs for putative PKA substrates were RT (reverse transcriptase)–PCR cloned from total RNA (Clontech). These cDNAs were inserted into the pcDNA3D-V5/HIS-TOPO vector (Invitrogen), which introduces a C-terminal V5/His tag. cDNAs for PKD (protein kinase D) were provided by Dr Vivek Malhotra (University of California San Diego, San Diego, CA, U.S.A.). Point mutations and deletions were introduced using the QuikChange® XL system (Strategene).
Phosphorylation experiments
cDNAs for potential substrates were transiently transfected into HEK (human embryonic kidney)-293 cells using Mirus Transit-LT1. After 24 h, cells were washed in PBS, fed with fresh medium and incubated for a further 20–24 h. Cells were then stimulated with appropriate agonists (see Figure legends for details) or DMSO (vehicle control). After treatment, cells were placed on ice, washed with ice-cold PBS and lysed in lysis buffer [20 mM Hepes (pH 7.4), 150 mM NaCl, 1 % Triton X-100, 50 mM NaF, 100 nM okadaic acid, 1 mM orthovanadate, 1 mM PMSF, 2 mg/ml leupeptin, 2 mg/ml pepstatin and 1 mg/ml aprotinin]. After incubation on ice for 10 min, lysates were centrifuged at 15 000 g for 15 min at 4 °C. Substrate proteins were either immunoprecipitated using tag-specific antibodies or isolated using glutathione–Sepharose. Precipitated proteins were separated by SDS/PAGE, transferred on to nitrocellulose and immunoblotted with the anti-phospho-PKA substrate antibody. Membranes were then stripped and re-probed using tag-specific antibodies to determine total protein. In vitro phosphorylation and dephosphorylation was performed as described in [12].
Confocal microscopy
Cells were plated on glass coverslips and incubated overnight at 37°C under 5 % CO2. Cells were starved for 6 h in serum-free DMEM (Dulbecco’s modified Eagle’s medium). Cells were then treated for 20 min with forskolin/IBMX (isobutylmethylxanthine) as indicated, followed by washing with PBS and fixation in PBS/3.7 % formaldehyde for 20 min. Cells were permeabilized and blocked with 0.1 % Triton X-100 in PBS containing 0.2 % BSA. Coverslips were incubated with specific primary antibodies in blocking buffer overnight at 4 °C. Cells were washed, incubated with Alexa Fluor®-conjugated secondary antibodies (Invitrogen) and Texas Red–phalloidin for 1 h, and washed. Coverslips were mounted using Prolong Antifade reagent (Molecular Probes) and visualized on a Bio-Rad 1024 UV laser-scanning confocal microscope.
Cell growth assays
WT (wild-type) and S119A RIL were transfected into PC-3 cells and stable lines were generated under neomycin selection. Clones expressing equal levels of WT or mutant RIL were then selected for further analysis. The growth rates of different RIL-expressing cell lines was measured using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega). Cells (1000 from each RIL line) were plated in triplicate on each of two 96-well plates. One plate was read immediately to confirm correct plating of cells. The second plate was read on day 7. Triplicate wells were averaged and day 7 readings for each cell line were normalized to original plating densities on day 1. Means for WT and mutant RIL lines were compared using an unpaired Student’s t test with 0.05 as the level of significance.
RESULTS AND DISCUSSION
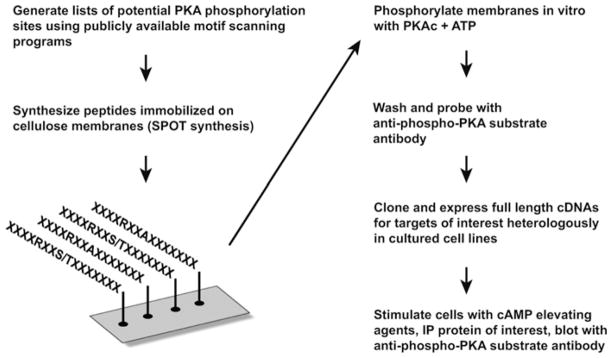
We have developed a strategy that combines peptide array, in vitro phosphorylation and Western blotting with motif-selective antibodies to discover previously unidentified phosphorylation sites in proteins. The basic workflow of this method is outlined in Figure 1. We reasoned that membrane-immobilized phospho-peptides recognized by phospho-motif-specific antibodies might also be detected in the context of the full-length protein by Western blotting. Thus this approach could provide a valuable tool for rapid screening of potential phosphorylation sites without the use of large amounts of radiolabelled ATP.
Figure 1. Strategy for identification of novel PKA substrates.
Our approach involves bioinformatic screening to locate peptide sequences containing consensus PKA phosphorylation site motifs. These peptides were synthesized on a derivatized cellulose membrane using the SPOT technique. Immobilized peptides were then phosphorylated using purified PKA C-subunit, washed and probed with an antibody that specifically recognizes the motif R-X-X-[pS/T]. For candidate substrates, cDNAs are expressed in HEK-293 cells that are treated with either vehicle or forskolin and IBMX to elevate cAMP levels and activate PKA. Proteins of interest are immunoprecipitated (IP) and tested for phosphorylation by Western blot analysis using the anti-phospho-PKA substrate antibody.
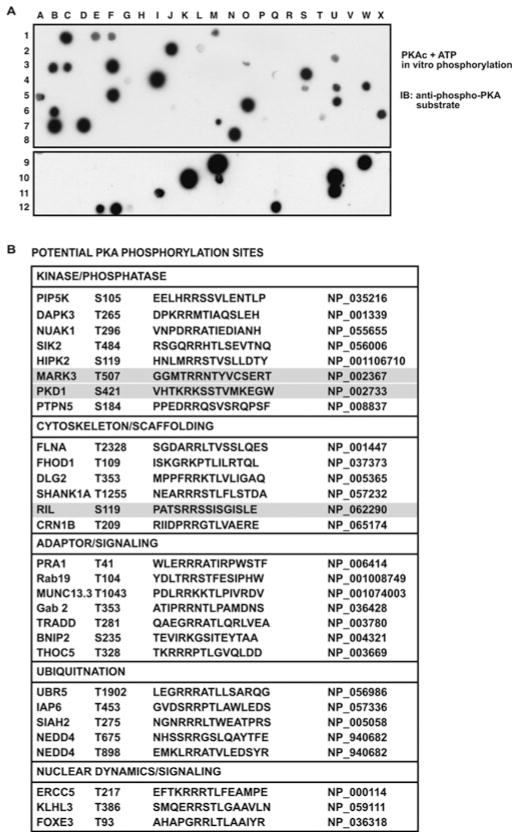
We chose to look for PKA substrates owing to the availability of a well-characterized phospho-specific antibody that recognizes a canonical PKA target motif, R-X-X-pS/T. Several motif-based bioinformatic tools were used to look for sites matching this consensus PKA phosphorylation site. Peptides corresponding to 15 amino acids surrounding these sites were synthesized on cellulose membranes. Peptides containing alanine substitutions in potential phosphoacceptor serine or threonine positions were included as negative controls. Membranes were phosphorylated in vitro with recombinant PKA catalytic subunit and ATP. After extensive washing, membranes were subjected to Western blot analysis with the anti-phospho-PKA substrate antibody (Figure 2A and Supplementary Figure S1 at http://www.BiochemJ.org/bj/438/bj4380103add.htm). Peptides that were positive in the first round were collected and re-synthesized for confirmation in a second round of profiling.
Figure 2. SPOT membrane screening for PKA substrates.
(A) Peptides corresponding to potential PKA phosphorylation sites were arrayed on a cellulose membrane using SPOT synthesis. For each peptide, the WT sequence and one or more mutant sequences were tested. The membrane was phosphorylated by PKA in vitro, followed by Western blot (IB) analysis using the anti-phospho-PKA substrate antibody (1:500 dilution). Two representative membranes are presented. (B) A number of potential PKA phosphorylation sites were identified. These sites are listed along with the sequence and GenBank® accession number. We chose PKD (spot I-11 in A, bottom panel), MARK3 (spot S-4 in A, top panel) and RIL (spot M-7 in A, top panel) for further analysis.
Promising candidates were selected for further investigation, including the protein kinases PKD1 and MARK3 [MAP (microtubule-associated protein)-regulating kinase 3]/c-TAK1 (Cdc25C-associated kinase 1), and the adapter protein RIL (Figure 2A, spots I-11, S-4 and M-7 respectively). We were intrigued by the possibility that PKA might regulate signal flow through cascades involving PKD and MARK3. RIL (PDLIM4) was chosen because it is a small-molecular-mass protein that contains a PDZ domain and a LIM domain, both well-studied protein–protein interaction motifs [13].
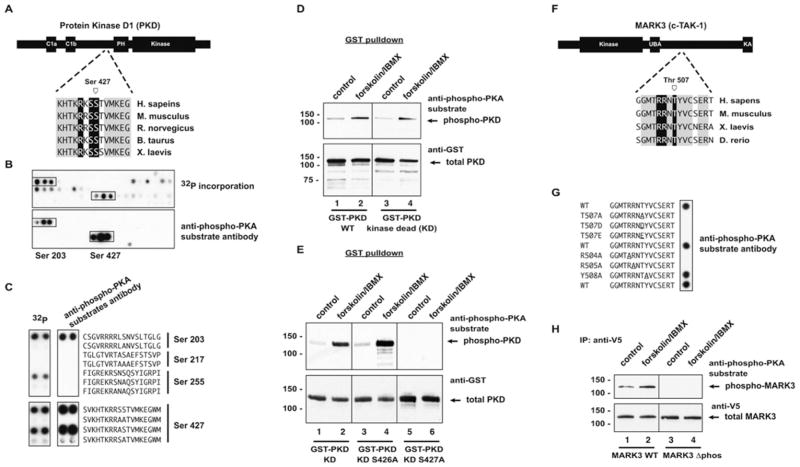
PKD is a multifunctional kinase involved in cardiac remodelling and regulated by anchored PKA [12,14]. BLAST searching confirmed that the site in PKD that was identified in the original screen, Ser427, is conserved in vertebrate species (Figure 3A); however, PKD contains multiple potential PKA phosphorylation sites in addition to Ser427. In order to identify which of these could be phosphorylated and recognized by the anti-phospho-PKA substrate antibody, PKD was arrayed out sequentially as 20-mer peptides, with an overlap of three residues in each subsequent peptide (Figure 3B). Duplicate membranes were phosphorylated in vitro with the PKA C-subunit and [32P]ATP (Figure 3B, top panel), or unlabelled ATP followed by Western blot analysis (Figure 3B, bottom panel). Two distinct regions were phosphorylated and recognized by the antibody. These corresponded to peptides containing Ser203 and Ser427 (Figure 3B). Further analysis of these potential PKA sites showed that the anti-phospho-PKA substrate antibody could recognize both Ser203 and Ser427 when phosphorylated in vitro (Figure 3C). WT or KD (kinase-dead) PKD were expressed as GST (glutathione transferase) fusion proteins in HEK-293 cells. Cells were serum-starved and then stimulated with 20 μM forskolin/75 μM IBMX for 20 min at 37 °C. PKD was collected on glutathione agarose and separated by SDS/PAGE. Western blotting with the anti-phospho-PKA substrate antibody showed a clear increase in phosphorylation of PKD upon cAMP elevation and PKA activation (Figure 3D, top panels). In most cases there was a higher basal level of phosphorylation in WT PKD, indicating that this site may be an auto-phosphorylation site. For this reason, we continued our characterization using the KD mutant of PKD. Loading controls verified that equal amounts of total protein were present in each experiment (Figure 3D, bottom panels).
Figure 3. PKD and MARK3 are substrates for PKA phosphorylation.
(A) PKD is a serine/threonine kinase that contains two C1 domains and a PH (pleckstrin homology) domain. SPOT array phosphorylation site screening predicted a well-conserved putative PKA phosphorylation site near the N-terminus of the PH domain. (B) PKD was arrayed out as a series of sequential 20-mer peptides offset by three residues with each subsequent peptide. Duplicate membranes were phosphorylated with PKA and [32P]ATP or unlabelled ATP, followed by autoradiography or blotting with the anti-phospho-PKA substrate antibody (1:500 dilution). Several PKA sites were identified by both 32P-labelling and immunoblotting. Ser203 and Ser427 (boxes) were both phosphorylated by PKA in vitro. (C) PKA sites predicted by Scansite were arrayed out on SPOT membranes and analysed as described in (B). Ser203 and Ser427 were detected using both 32P and the phospho-PKA substrate antibody, whereas Ser255 was detected using 32P, but not by immunoblotting. Mutation of the phosphoacceptor serine residues to alanine results in loss of phosphorylation in all cases. (D) GST–PKD or GST–PKD-KD were expressed in HEK-293 cells. Cells were stimulated with vehicle or 20 μM forskolin/75 μM IBMX for 10 min at 37 °C. PKD was isolated by GST pulldown, followed by Western blotting using the phospho-PKA substrate antibody. Both PKD and PKD-KD were phosphorylated in response to forskolin/IBMX treatment. (E) Mutation of Ser427 to alanine in PKD-KD abolishes phosphorylation of PKD in response to forskolin/IBMX treatment. (F) MARK3/c-TAK1 is a MARK family kinase that contains a UBA and a KA (kinase-associated) domain. The predicted PKA phosphorylation site is distal to the UBA domain and conserved in vertebrate species. (G) SPOT analysis of phosphorylation determinants confirms that the phospho-PKA substrate antibody detects Thr507. Replacement of Thr507 with alanine, aspartate or glutamate residues abolishes phosphorylation, as does replacement of Arg504 and Arg505 with alanine. (H) Deletion of a 27-amino-acid peptide containing this phosphorylation site (residues 486–512) results in loss of forskolin/IBMX-stimulated MARK3 phosphorylation. For Western blots, the molecular mass in kDa is indicated on the left-hand side. B. taurus, Bos taurus; H. sapiens, Homo sapiens; M. musculus, Mus musculus; R. norvegicus, Rattus norvegicus; X. laevis, Xenopus laevis; IP, immunoprecipitation.
The sequence surrounding Ser427 contains the consensus PKA motif R-R/K-S-S. Since either serine residue could potentially act as a phosphoacceptor, we mutated either Ser426 or Ser427 to alanine, and performed similar experiments. Mutation of Ser426 had no effect on cAMP-mediated PKD phosphorylation, whereas the S427A mutation abolished phosphorylation, as detected by immunoblotting (Figure 3E, top panels). This result corresponds well with the peptide mapping shown in Figure 3(C).
MARK3 (also known as PAR-1 and c-TAK1) is a member of the microtubule-affinity regulating kinases. MARK3 contains a single kinase domain and a UBA (ubiquitin-associated domain) (Figure 3F). MARK3/PAR-1 homologues are important for establishing cellular polarity in Drosophila and Caenorhabditis elegans [15], whereas mammalian MARK/PAR-1/c-TAK kinases have been implicated in cell polarity [16], Wnt signalling [17], regulation of microtubule function [18] and cell-cycle regulation [19]. Our peptide array screening identified a potential PKA phosphorylation site at Thr507.
Peptides containing Thr507, as well as various mutated versions, were arrayed and probed for PKA phosphorylation by immunobloting. Replacement of the putative phosphoacceptor threonine residue with alanine results in loss of signal (Figure 3C, peptide 2). The signal was also lost following replacement with phosphomimetic amino acids (aspartate and glutamate; Figure 3G, peptides 3 and 4). Phosphorylation and antibody recognition of this peptide was also abolished by replacement of the − 3 and − 2 arginine residues with alanine (Figure 3G, peptides 6 and 7).
A cDNA for MARK3 was amplified from human brain cDNA and cloned for expression as a V5/His-tagged protein in mammalian cells. Cells expressing MARK3 were treated with vehicle or forskolin/IBMX, and MARK3 was immunoprecipitated with anti-V5 antisera. Following separation by SDS/PAGE, Western blot analysis with the anti-phospho-PKA substrate antibody revealed that MARK3 is phosphorylated in response to increases in cAMP concentration and PKA activation (Figure 3H, lanes 1 and 2). We also deleted the region surrounding this putative PKA phosphorylation site. Expression of MARK3Δphos results in loss of phosphorylation by PKA as detected using the anti-phospho-PKA substrate antibody (Figure 3H, lanes 3 and 4). These results further confirm the presence of a PKA site in the region of MARK3/c-TAK1 distal to the UBA.
Despite our strong evidence for PKA phosphorylation of both PKD and MARK3, so far we have not detected changes in kinase activity or localization in phosphorylation site mutants, and are thus unable to ascribe a convincing function to these phosphorylation events. This highlights the fact that a proportion of phosphorylation events in the phosphoproteome is likely to be structural or non-functional [20,21].
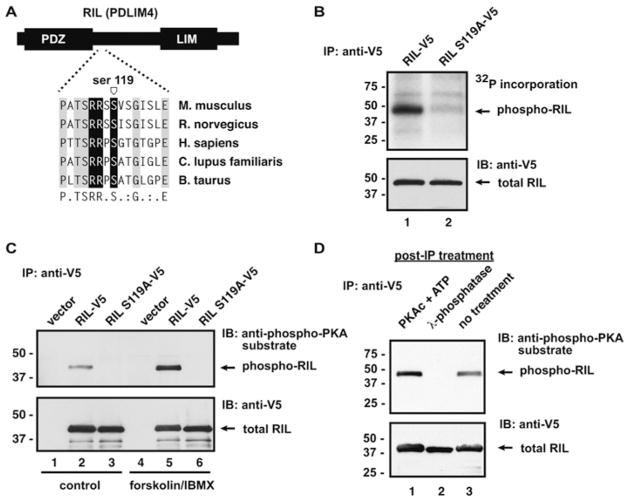
We next chose to focus on the signalling adapter RIL/PDLIM4. RIL is a member of a family of PDZ/LIM proteins that have been implicated in multiple cellular processes, including regulation of the actin cytoskeleton and Src kinase activation [22,23]. Our array analysis predicted a PKA phosphorylation site at Ser119, which is well conserved in mammalian species (but not in avian or amphibian sequences) (Figure 4A).
Figure 4. RIL is a novel PKA substrate.
(A) RIL is a small adapter protein containing a PDZ domain and a LIM domain. A potential PKA phosphorylation site just downstream of the PDZ domain was identified by peptide array phosphorylation. B. taurus, Bos taurus; C. lupus familiaris, Canis lupus familiaris; H. sapiens, Homo sapiens; M. musculus, Mus musculus; R. norvegicus, Rattus norvegicus. (B) In vitro phosphorylation of immunopurified RIL using [32P]ATP and purified PKA C-subunit shows that RIL is phosphorylated by PKA and mutation of Ser119 abolishes PKA phosphorylation. (C) RIL or mutant RIL (S119A) were expressed as C-terminally tagged proteins in HEK-293 cells. Cells were stimulated with vehicle (DMSO, control) or with 20 μM forskolin/75 μM IBMX for 20 min at 37 °C. Cells were lysed in the presence of phosphatase inhibitors and RIL was immunoprecipitated using the anti-V5 antibody. Phosphorylated RIL was detected by Western blotting with the anti-phospho-PKA substrate antibody. Total RIL in immunoprecipitations was detected using a horseradish peroxidase-conjugated anti-V5 antibody (1:5000 dilution). (D) RIL was expressed in HEK-293 cells and immunopurified as described above. RIL was either phosphorylated in vitro with PKA or was dephosphorylated with λ-phosphatase. Phosphorylated RIL was detected by Western blot analysis with the anti-phospho-PKA substrate antibody. Total RIL was detected using a horseradish peroxidase-conjugated anti-V5 antibody. For Western blots, the molecular mass in kDa is indicated on the left-hand side. IB, immunoblot; IP, immunoprecipitation.
In vitro phosphorylation assays confirmed that RIL could serve as a PKA substrate. Mouse RIL was PCR cloned from a cDNA library. Next, Ser119 was mutated to alanine to abolish this putative phosphorylation site. HEK-293 cells were then transfected with vectors encoding epitope (V5-His)-tagged wild-type RIL or RIL S119A. Cells were lysed and RIL was immunoprecipitated with anti-V5 antisera. After washing, immunoprecipitations were incubated with purified PKA C-subunit and [32P]ATP. RIL was robustly phosphorylated by PKA (Figure 4B, lane 1), whereas RIL S119A was not labelled (Figure 4B, lane 2).
WT RIL or RIL S119A was expressed in HEK-293 cells. Cells were serum-starved and then treated with forskolin and IBMX to stimulate cAMP production and PKA activation. After cell lysis, RIL was immunoprecipitated as before. Precipitated protein was separated by SDS/PAGE followed by Western blot analysis with the anti-phospho-PKA substrate antibody. RIL was weakly detected in vehicle-treated samples, but was strongly phosphorylated upon PKA activation (Figure 4C, lane 5). Mutation of Ser119 abolished the signal, indicating that this site was solely responsible for detection of phosphorylated RIL by the anti-phospho-PKA substrate antibody (Figure 4C, lane 6).
In order to confirm that the antibody detection of RIL was dependent on phosphorylation, recombinant RIL was immunoprecipitated from HEK-293 cells and incubated with PKA C-subunit and unlabelled ATP or with λ-phosphatase. Untreated RIL gave a basal signal on Western blotting with the anti-phospho-PKA substrate antibody (Figure 4D, lane 1). Phosphatase treatment completely abolished the signal, indicating that the antibody is specifically detecting phospho-RIL (Figure 4D, lane 2). Furthermore, the signal was augmented by in vitro phosphorylation with PKA (Figure 4D, lane 3), suggesting that there is basal, but non-stoichiometric, phosphorylation inside cells.
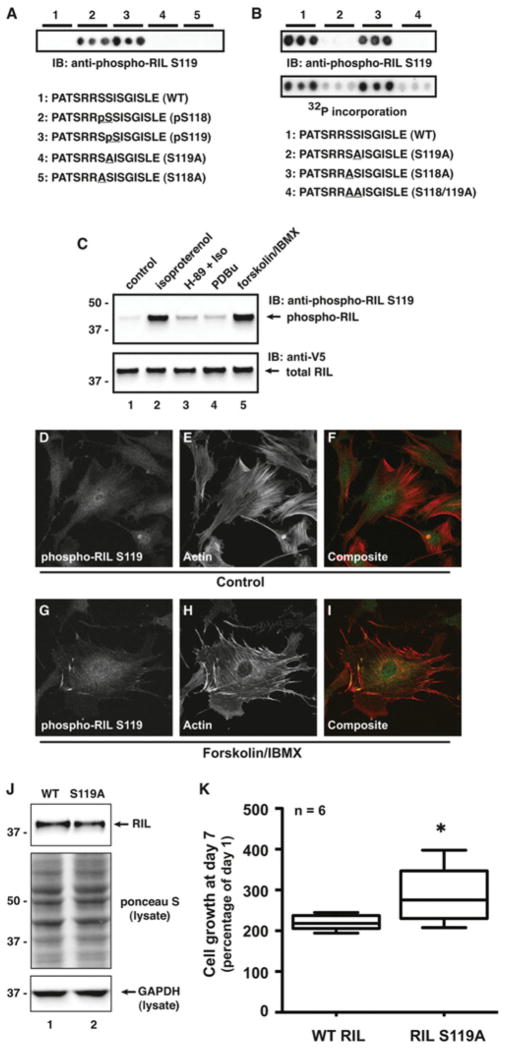
Because the anti-phospho-PKA substrate antibody recognizes a general consensus motif and is not specific to RIL, we developed a phospho-specific antibody against phospho-Ser119. Initial testing of this antibody was performed on peptide arrays containing different variations on the WT sequence.
Peptides corresponding to the region around Ser119 were arrayed out in triplicate. Peptides were synthesized with a phospho-serine residue in either the 118 or 119 position. Membranes were then immunoblotted with the anti-phospho-RIL Ser119 antibody. This antibody recognized both phosphoserine-containing peptides, whereas non-phosphorylated and mutant peptides gave no signal (Figure 5A). Next, we assessed the antibody recognition of in vitro phosphorylated peptides. In addition to the WT peptide, several mutated peptides were also synthesized. First, Ser119 was changed to alanine. As position 118 is also a serine residue (in mouse RIL), this residue was changed to alanine as well. Finally, a peptide with both serine residues replaced with alanine residues was included. Membranes were phosphorylated in vitro with purified PKA C-subunit and either [32P]ATP or unlabelled ATP, followed by either autoradiography or immunoblotting with the anti-phospho-RIL Ser119 antibody. In both cases, strong phosphorylation was detected in WT or S118A peptides, but not in the peptides containing S119A (Figure 5B). Taken together, these results indicate that even though there is a serine residue at position 118, PKA seems to selectively phosphorylate Ser119, a site that is conserved across species.
Figure 5. RIL is phosphorylated in vivo.
(A) A phospho-specific antibody was generated that recognizes phospho-Ser119 in RIL. Epitope mapping by peptide array shows that this antibody can recognize phospho-serine at either position 118 or 119 when incorporated into the peptide during synthesis. IB, immunoblot. (B) Duplicate sets of peptides on membranes were phosphorylated with PKA and unlabelled ATP or [32P]ATP, followed by immunoblotting with the anti-phospho-RIL Ser119 antibody (top panel) or autoradiography (bottom panel). RIL Ser119 is phosphorylated in the WT peptide. Disruption of Ser119, but not Ser118, by alanine substitution blocks phosphorylation. IB, immunoblot. (C) RIL is phosphorylated on Ser119 inside cells. RIL was expressed in HEK-293 cells. Cells were serum-starved and treated with agonists and inhibitors as indicated. Lysates were immunoblotted for phospho-RIL and total RIL levels. Isoproterenol (100 nM, 5 min) induces robust phosphorylation of Ser119, which is blocked by pretreatment with H-89 (10 μM, 30 min). In addition, RIL is not phosphorylated by PKC after stimulation with the phorbol ester PDBu. The molecular mass in kDa is indicated on the left-hand side. IB, immunoblot. (D–I) Immunofluorescence analysis of RIL phosphorylation in cells. REF52 cells were serum-starved and treated with vehicle or forskolin/IBMX. Cells were immunostained with the phospho-RIL Ser119 antibody and Texas Red–phalloidin to visualize F-actin. (J) PC-3 prostate adenocarcinoma cells were stably transfected with WT RIL or RIL S119A and selected for approximately equal expression by immunoblotting. The molecular mass in kDa is indicated on the left-hand side. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (K) Growth analysis of WT RIL and RIL S119A PC-3 cell lines was performed using a MTS assay at day 7 post-plating. Cells expressing WT RIL grew significantly slower than those expressing RIL S119A (n = 6, *P < 0.05, unpaired Student’s t test).
We next tested the antibody on RIL expressed in cells. Treatment of cells expressing RIL–V5/His with 100 nM isoproterenol resulted in robust RIL phosphorylation (Figure 5C, lane 2). Pre-treatment with the PKA inhibitor H-89 blocked β-adrenergic-mediated RIL phosphorylation (Figure 5C, lane 3). Phosphorylation of RIL Ser119 appears to be somewhat selective for PKA, as stimulation of PKC with the phorbol ester PDBu (phorbol 12,13-dibutyrate) had minimal effect on RIL phosphorylation (Figure 5C, lane 4). Finally, as shown in Figure 4, forskolin/IBMX treatment resulted in robust RIL phosphorylation (Figure 5C, lane 5).
We also performed immunofluorescence analysis of endogenous RIL phosphorylation in REF52 cells. Cells were starved and treated with forskolin and IBMX. After fixation and permeabilization, cells were stained with the anti-phospho-RIL Ser119 antibody and Texas Red–phalloidin to label F-actin (filamentous actin). Confocal microscopy showed weak basal staining for phospho-RIL Ser119 in untreated cells (Figures 5D–5F). After PKA activation, levels of phospho-RIL Ser119 increased throughout the cells (Figures 5G–5I). Interestingly, phospho-RIL seems to be particularly concentrated at the tips of actin-rich projections that may represent sites of cell-substrate interaction (Figures 5G–5I).
Several recent reports have implicated RIL in control of cell growth and cancer [24,25]. Silencing of RIL expression through promoter methylation occurs in several types of cancers, suggesting that loss of RIL expression may be important for cancer progression and malignancy. Re-expression in several cancer cell types impairs proliferation and anchorage-independent growth through unknown mechanisms that may involve stabilization of F-actin or regulation of cell-cycle progression [24,25]. Vanaja et al. [25] showed that RIL expression in PC-3 prostate adenocarcinoma cells inhibits cell growth. In order to assess the function of Ser119 phosphorylation of RIL, we generated stable PC-3 cell lines expressing either WT RIL or RIL S119A. Cell lines were chosen for matched RIL expression (Figure 5J, top panel). Next, we performed MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assays to examine proliferation of the two cell lines. Cells expressing RIL S119A grew significantly faster than cells expressing WT RIL at 7 days after starting the assay (Figure 5K). Therefore we propose that mutation of Ser119 partially rescues the inhibitory growth effect of expressing RIL in these cells. This suggests that phosphorylation of Ser119 by PKA (or another kinase with similar motif preferences) may be required for suppression of growth by RIL.
The results of the present study show that combining phosphorylation of immobilized peptides and immunoblotting with motif-selective antibodies is an effective way to discover kinase targets. These methods should be easily extendable to a large number of kinase/antibody pairs, particularly as the inventory of phospho-specific antibodies expands and more information is forthcoming on so called ‘orphan kinases’.
Supplementary Material
Acknowledgments
We thank Dr Graeme Carnegie (University of Illinois at Chicago, Illinois, Chicago, U.S.A.) and members of the Scott laboratory for helpful discussions, and Melanie Milnes for administrative support.
FUNDING
This project was supported by the National Institutes of Health [grant number DK54441].
Abbreviations used
- C-subunit
catalytic subunit
- c-TAK1
Cdc25C-associated kinase 1
- F-actin
filamentous actin
- GST
glutathione transferase
- HEK
human embryonic kidney
- IBMX
isobutylmethylxanthine
- KD
kinase dead
- MARK
MAP (microtubule-associated protein)-regulating kinase
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- PDBu
phorbol 12,13-dibutyrate
- PDLIM4
PDZ and LIM domain protein 4
- PKA
cAMP-dependent protein kinase/protein kinase A
- PKD
protein kinase D
- TBST
Tris-buffered saline containing Tween 20
- UBA
ubiquitin-associated domain
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Donelson Smith, Bret Samelson and John Scott designed the experiments. Donelson Smith and Bret Samelson performed the experiments. Donelson Smith and John Scott wrote the paper.
References
- 1.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. PKA: a portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 4.Turk BE. Understanding and exploiting substrate recognition by protein kinases. Curr Opin Chem Biol. 2008;12:4–10. doi: 10.1016/j.cbpa.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 6.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 8.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;13:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang JY, Brutlag DL. The EMOTIF database. Nucleic Acids Res. 2001;29:202–204. doi: 10.1093/nar/29.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217. [Google Scholar]
- 12.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Krcmery J, Camarata T, Kulisz A, Simon HG. Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. BioEssays. 2010;32:100–108. doi: 10.1002/bies.200900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–13208. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- 15.Hurov J, Piwnica-Worms H. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle. 2007;6:1966–1969. doi: 10.4161/cc.6.16.4576. [DOI] [PubMed] [Google Scholar]
- 16.Bohm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr Biol. 1997;7:603–606. doi: 10.1016/s0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- 17.Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 18.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 19.Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, III, Wu Z, Stephenson MT, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 20.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 21.Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Vallenius T, Scharm B, Vesikansa A, Luukko K, Schafer R, Makela TP. The PDZ-LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp Cell Res. 2004;293:117–128. doi: 10.1016/j.yexcr.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Tu Y, Zhao J, Chen K, Wu C. Reversion-induced LIM interaction with Src reveals a novel Src inactivation cycle. J Cell Biol. 2009;184:785–792. doi: 10.1083/jcb.200810155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boumber YA, Kondo Y, Chen X, Shen L, Gharibyan V, Konishi K, Estey E, Kantarjian H, Garcia-Manero G, Issa JP. RIL, a LIM gene on 5q31, is silenced by methylation in cancer and sensitizes cancer cells to apoptosis. Cancer Res. 2007;67:1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- 25.Vanaja DK, Grossmann ME, Cheville JC, Gazi MH, Gong A, Zhang JS, Ajtai K, Burghardt TP, Young CY. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27:264–272. doi: 10.1080/07357900802406319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.