Abstract
Background
Vibrio cholerae causes the diarrheal disease cholera and utilizes different survival strategies in aquatic environments. V. cholerae can survive as free-living or in association with zooplankton and can build biofilm and rugose colonies. The bacterium expresses cholera toxin (CT) and toxin-coregulated pilus (TCP) as the main virulence factors. These factors are co-regulated by a transcriptional regulator ToxR, which modulates expression of outer membrane proteins (OmpU) and (OmpT). The aims of this study were to disclose the role of ToxR in expression of OmpU and OmpT, biofilm and rugose colony formation as well as in association with the free-living amoeba Acanthamoeba castellanii at different temperatures.
Results
The toxR mutant V. cholerae produced OmpT, significant biofilm and rugose colonies compared to the wild type that produced OmpU, decreased biofilm and did not form rugoes colonies at 30°C. Interestingly, neither the wild type nor toxR mutant strain could form rugose colonies in association with the amoebae. However, during the association with the amoebae it was observed that A. castellanii enhanced survival of V. cholerae wild type compared to toxR mutant strain at 37°C.
Conclusions
ToxR does seem to play some regulatory role in the OmpT/OmpU expression shift, the changes in biofilm, rugosity and survival with A. castellanii, suggesting a new role for this regulatory protein in the environments.
Keywords: V. cholerae, Outer membrane proteins, Rugose colonies, Biofilm, Association with amoebae
Background
Vibrio cholerae is a gram-negative bacterium causing severe diarrhoeal disease cholera in Asia, Africa, and America. Many millions of cholera cases occurred endemically and pandemically worldwide [1].
V. cholerae adopts several survival strategies in aquatic environments. The bacterium can survive as free-living or in association with phytoplankton/zooplankton, crustaceans, and molluscs in coastal and estuarine environments [2]. V. cholerae forms biofilms on biotic and abiotic surfaces, thereby protecting themselves with this exopolymer barrier [3], biofilm can provide protection from toxic compounds, such as antibiotics, thermal stress, and predation [4]. The bacterium has been described to switch between the smooth and rugose colony morphotypes contributing to its environmental survival. The exopolysaccharide (EPS) materials of rugose colony-forming V. cholerae strains were recognized as a heavy, fibrous, electron-dense, ferretin-stained layer surrounding the cells, but smooth colony V. cholerae did not appear to have this EPS layer surrounding it [5]. The cell surface EPS materials confer a rugose colony morphology and resistance to osmotic and oxidative stresses. The regulation of EPS synthesis in bacteria is complex and involves multiple systems utilizing both positive and negative regulation [5].
Recent studies have shown that V. cholerae has developed a new survival strategy to grow and survive inside free-living amoeba Acanthamoeba castellanii [6-10].
V. cholerae expresses cholera toxin and toxin-coregulated pilus as the main virulence factors, which are co-regulated by a transcriptional regulator ToxR. ToxR, however, independently of the transcriptional activators TcpP and ToxT, modulates expression of two outer membrane proteins OmpU and OmpT. Transcription of ompU is induced by ToxR, whereas transcription of ompT is repressed by ToxR [11,12].
In this paper we study the effect of ToxR regulatory protein in expression of OmpU and OmpT, biofilm and rugosity as well as survival of V. cholerae O1 with A. castellanii by constructing a toxR deletion mutant.
Methods
Bacterial strains and plasmids used in this study
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in LB medium at 37°C. Strains were stored frozen in LB medium with 15% glycerol at -80°C. Antibiotics were used at the following concentrations: carbencillin 50 μg/mL, streptomycin 50 μg/ml, kanamycin 50 μg/mL.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| V. cholerae O1 E1Tor Inaba | Strain A1552, Wild type | [13] |
| V. cholerae ΔtoxR | Transcriptional activator deletion mutant of Strain A1552 | This study |
| E. coli DH5α | Φ80dlacZΔM15 recA1gyrA96 thi-1 hsdR17 (rk- mk+) supE44 | [14] |
| Sm10λpir | relA1deoRΔ(lacZYA-argF)U169 | [15] |
| thi-1, thr, leu, tonA, lacY, supE, | ||
| recA::Rp4-2-Tc::Mu,KmR,(λpir) | ||
| Plasmid Puc 18 | ApR lacZaovi colE1 | [16] |
| Plasmid PCVD442 | oriR6K mobRP4 sacB Apr | this study |
Growth conditions
Luria-Bertani (LB) media, All strains were grown in LB medium (1% Bacto Tryptone, 0.5% yeast extract, 0.5% NaCl) with appropriate selection. For antibiotic selection, 50 μg/mL carbenicillin or 50 μg/mL streptomycin was used.
Construction of the internal in-frame toxR deletion mutant
To introduce a deletion at the chromosomal toxR locus we used a double cross over procedure. A plasmid pCVD442 was used as a suicidal vector. In the first step two different asymmetric polymerase chain reactions (PCR) amplified with primers toxR-A (5'CCC ATC CAC TAA ACG GTA CCA TCC GAA CAT CTA ATG TCC CAG-3') toxR-B (5'-GCG TCT AGA ACT CTT TAC CTT CTT CAC GCAG-3') toxR-C (5'-GCG TCT AGA ATC TTC GCT GAA GGT ATG CAGT-3') and toxR-D(5'TGG TAC CGT TTA GTG GAT GGG GCC ATC AAA GTG TGT GAG TAG-3') were used to generate fragments upstream and downstream of the sequences targeted for deletion. In the second step, the upstream and downstream fragments were annealed at their overlapping region and amplified by PCR as a single fragment, using the outer primers (toxR-A and toxR-D). The PCR products were phenol-chloroform extracted, ethanol precipitated, washed with 70% ethanol, vacuum dried, resuspended in 50 mL of Xba-I restriction buffer containing 40 U of XbaI restriction enzyme, and digested overnight at 37°C. The DNA fragments were gel purified, ligated into XbaI digested and alkaline phosphatase-treated pCVD442 and then, electroporated into E. coli SM10λpir. The mutant constructs were checked by PCR and sequencing. The confirmed deletion constructs were introduced into V. cholerae by conjugation, screened on 15% sucrose containing plates, and the carbenicillin sensitive clones were assessed for double cross-over recombination. The recombinant colonies were checked by PCR.
Expression of outer membrane proteins
Bacterial cultures were grown overnight in 2 ml LB media at 37°C. The entire contents were centrifuged at 18,000 × g for 15 min. The supernatants were collected in new tubes and the pellet was suspended in 100 μl sample buffer and boiled for 10 min. The supernatant samples were precipitated with 10% TCA for 30 min on ice. The samples were centrifuged at 18,000 × g for 30 min and washed with 80% acetone. The pellet was resuspended in standard SDS-PAGE sample buffer. Samples were analysed by 13% SDS-PAGE.
Biofilm analysis
V. cholerae wild type and toxR mutant strains were grown on LB agar plates overnight at 37°C. Few colonies from each strain were suspended in 20 mL LB broth and incubated in shaking incubator until the absorbance reached 0.6 units at 600 nm. 5 mL of 1:100 dilution of this suspension was transferred into triplicated glass tubes (13 by 100 mm) and incubated for 18 h at 30°C. Representative biofilm formed by wild type and toxR mutant strains was visualised photographically and the tubes were rinsed with distilled water then filled with 1% crystal violet stain. After 15 min, the tubes were rinsed and the biofilm-associated crystal violet in the tubes was suspended with 95% ethanol. The absorbance of the resulting suspension was measured by spectrophotometry at 570 nm to quantify the biofilm formation.
Colony morphology
For colony morphology assay, V. cholerae wild type and toxR mutant strains were grown on LB agar plates for 1 day at 30°C and left at room temperature until appearance of rugose colonies after 6 days. Percentage of rugose colonies was estimated and representative smooth as well as rugose colonies were visualized photographically.
Co-cultivation assay
The co-cultivation assay was based on a method presented previously. Axenically maintained amoebae were grown at 30°C to a final concentration of 2 × 105 CFU/mL in ATCC medium as described above. Co-cultivations of V. cholerae with A. castellanii were incubated in NUNC tissue culture flasks (75 cm) purchased from VWR International (Stockholm, Sweden) filled with 50 mL ATCC medium 712 containing A. castellanii at a concentration of 2 × 105 CFU/mL and the particular V. cholerae species at a concentration of 2 × 106 CFU/mL. Control flasks containing bacteria or amoebae only were prepared in the same way and with the same initial concentration as the co-culture flasks. All flasks were triplicates and incubated at 25°C and 37°C. Samples were taken and plated on blood agar plates regularly to study growth and survival of V. cholerae.
Results
Construction of a Δ toxR in V. cholerae and expression of outer membrane proteins
An internal in-frame toxR deletion mutant in V. cholerae was constructed as described in the materials and methods and examined for expression of outer membrane proteins by SDS-PAGE. The result showed that the V. cholerae toxR mutant strain expressed OmpT compared to the wild type strain, which expressed OmpU (Figure 1)
Figure 1.
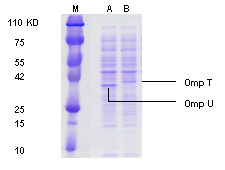
Outer membrane proteins expression by SDS-PAGE gel from whole cell lysates. Lanes M, A and B represent marker, V. cholerae wild and toxR mutant strain. The gel confirmed that the wild V. cholerae expressed OmpU as it is usually activated by ToxR (A) while the constructed toxR mutant expressed OmpT (B).
Biofilm analysis
To study the effect of ToxR in biofilm formation at 30°C in LB broth, the biofilm and biofilm-associated crystal violet formed by the wild type V. cholerae or the toxR mutant strains was visualised photographically and absorbance of the biofilm-associated crystal violet ethanol solution was measured spectrophotometrically.
The photographical analysis showed that the wild type V. cholerae (Figure 2A upper panel) had repressed biofilm formation compared to the toxR mutant strain (Figure 2B upper panel). Moreover, it was found that the absorbance of the biofilm produced by the wild type V. cholerae (Figure 2A lower panel) and the toxR mutant strain (Figure 2B lower panel) were 1.1 ± 0.2 and 1.8 ± 0.35, respectively. However, the spectrophotometry showed that the toxR mutant of V. cholerae had enhanced biofilm formation, which was confirmed by the statistical analysis, since the difference in absorbance was statistically significant (p of t-test was < 0.05).
Figure 2.
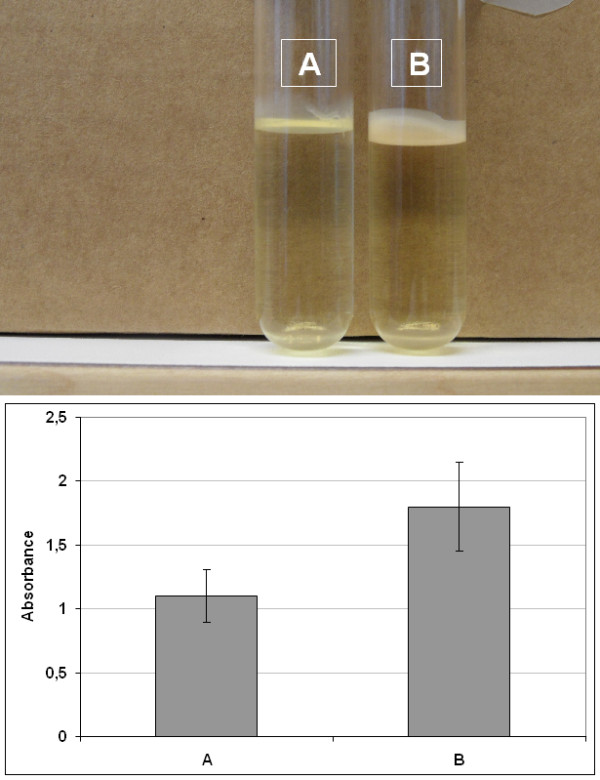
Biofilm formation. Upper panel showed representative photographs of biofilms formed by wild type V. cholerae (A) and toxR mutant strain (B). Lower panel showed representative measurement of the absorbance, which is directly proportional to the concentration of biofilm-associated crystal violet formed by wild type V. cholerae (A) and toxR mutant (B) strains. Data indicates mean ± SD of three independent experiments.
Colony morphology
The result showed that all colonies of V. cholerae wild type strain were smooth since this strain did not form any rugose colony (Figure 3 upper panel). However, it was found that 35 ± 10% of toxR mutant strain colonies was rugose (Figure 3 lower panel) and 65 ± 10% was smooth. The ability of each strain to form rugose colonies was significantly differed by χ2 test (p < 0.001).
Figure 3.

Colony morphology of V. cholerae strains. Upper panel represents smooth colonies of wild type strain. Lower panel represents rugose colonies of toxR mutant strain.
Effect of ToxR protein on growth, survival and rugose colony formation of V. cholerae associated with Acanthamoeba castellanii
Rugose colony formation, growth and survival of 2.0 ± × 106 cell/mL V. cholerae wild type and toxR mutant cultivated in ATCC medium were compared with those of the same strains cultivated with 2.0 × 105 cell/mL A. castellanii in the same medium.
The result showed that the V. cholerae wild type and the toxR mutant survived differently at different temperatures. At 37°C the wild type strain grew to 1.3 × 107 ± 5.8 × 106 CFU/mL and survived 1 day in the absence of amoebae compared to 1.4 × 102 ± 5.5 × 10 CFU/mL and survived 3 days in the presence of the amoebae, whereas the toxR mutant strain died on the first day in the absence of amoebae but survived 1 day to 4.7 × 105 ± 4.0 × 105 CFU/mL in the presence of amoebae (Figure 4 upper panel).
Figure 4.
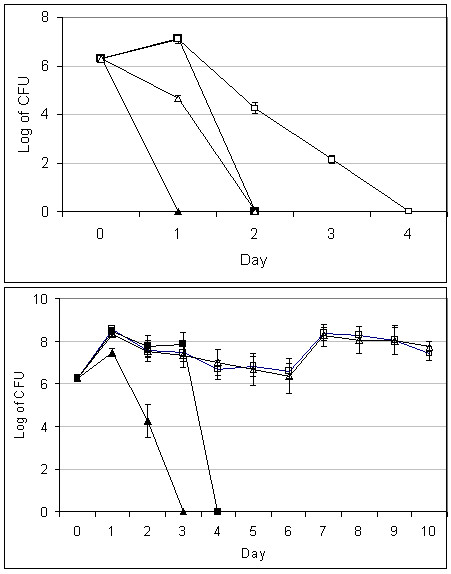
Growth and survival of V. cholerae strains in absence and presence of A. castellanii at 37°C upper panel and at 25°C lower panel. Alone cultivated wild type V. choleare (■) and cultivated with A. castellanii (□). Alone cultivated toxR mutant strain (▲) and cultivated with A. castellanii (Δ). Data indicates mean ± SD of three independent experiments.
At 25°C the wild type strain grew to 7.5 × 107 ± 1.9 × 107 CFU/mL and survived 3 days, in the absence of amoebae compared to the toxR mutant strain, which decreased to 2.0 × 104 ± 1.0 × 104 CFU/mL and survived 2 days. Whereas in the presence of the amoebae, the wild type and the mutant strains grew to 3.3 × 107 ± 2.6 × 107 CFU/mL and 6.0 × 107 ± 3.0 × 107 CFU/mL and survived 10 days, respectively (Figure 4 lower panel).
During the cultivation with the amoebae it was observed that the wild type and the toxR mutant strains did not form any rugose colonies at both 25°C and 37°C.
Discussion
V. cholerae adopts several survival strategies in aquatic environments. The bacterium can survive as free-living or in association with zooplankton and can build biofilm and rugose colonies [2,3]. Recent studies have shown that V. cholerae has an enhanced growth in association with the free-living amoeba A. castellanii at 30°C [6-10] and both these microorganisms are detected in same water samples from cholera endemic area [17].
In this study, the internal in-frame toxR deletion mutant V. cholerae strain succeeds to express the ToxR-repressed OmpT instead of the ToxR-activated OmpU. This altered expression of the ToxR-regulated OmpU and OmpT was confirmed by previous study and might affect the viruelence of V. cholerae [11].
Our paper has disclosed the role of ToxR regulatory protein in the environmental survival strategies of V. cholerae such as biofilm formation, switching from smooth to rugose colony morphotypes and association with the free-living amoeba namely A. castellanii.
Our results demonstrate that ToxR clearly affects biofilm and rugose formation since the differences in biofilm and rugose colony formation between V. cholerae wild type and toxR mutant were significantly high by t-test and χ2 test, respectively.
Effect of ToxR on growth and survival of V. cholerae associated with A. castellanii was studied by viable count performed on blood agar. V. cholerae wild type strain survived longer than the toxR mutant strain at 37°C, in the presence or absence of the amoebae. Interestingly, the association with A. castellanii enhanced survival of both bacterial strain but the wild type strain survived longer uncovering a role of ToxR in the survival of V. cholerae associated with protozoa in aquatic environment.
In cultivation with the amoebae at 25°C, it was observed that both V. cholerae wild type and toxR mutant strains survived more than 10 days.
Despite V. cholerae wild type and toxR mutant strains survived more than 10 days in cultivation with the amoebae at 25°C, these strains did not form any rugose colony indicating that the bacteria avoided starvation. However, presence of the amoebae might enrich the cultivation medium and viable count of the bacteria was performed on enriched plates (blood agar plates).
Analogies to rugosity can be found in a number of other bacterial species, including the expression of alginate by mucoid strains of Pseudomonas aeruginosa and the expression of an adhesive EPS by members of the marine genus [5].
Spontaneous and reversible variation in cell-associated and cell-free EPS production represents an optimal adaptive mechanism that facilitates survival in stressful environments [18].
Although, over 20 genes are co-ordinately controlled by the ToxR regulon [19], the mechanism of switching is currently under study and it seemed to be regulated by exopolysaccharide related phase variation. Phase variation can occur via DNA inversion, DNA recombination, and slipped strand mispairing and is known to be involved in controlling the expression of several surface structures of gram-negative bacteria, including fimbriae, flagella, outer membrane proteins, lipopolysaccharide, and capsular polysaccharide [20]. However, role of ToxR was found to be critical for V. cholerae bile resistance, virulence factor expression, and intestinal colonization [11,12]. Surprisingly, ToxR homolog from V. anguillarum was found to regulate its own production, bile resistance, and biofilm formation [21] which might emphasis the regulatory role of ToxR in the expression of virulence factors for Vibrio species.
Conclusions
Our results demonstrate that ToxR does seem to play some regulatory role in the OmpT/OmpU expression shift and in the environmental survival strategies of V. cholerae such as biofilm formation, switching from smooth to rugose colony and association with the free-living amoeba A. castellanii, suggesting a new role for this regulatory protein.
Authors' contributions
SV carried out the construction of deletion, outer membrane detection, the analysis of biofilm-and co-cultivation assay and drafted the manuscript. SW contributed to providing of bacterial strains, guiding and design of deletion, and critical reading. AS participated in writing. GS participated in writing and critical reading. HA conceived the study, designed co-infection assay, performed the statistical analysis and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Soni P Valeru, Email: soni.valeru@ki.se.
Sun N Wai, Email: sun.nyunt.wai@molbiol.umu.se.
Amir Saeed, Email: amir.saeed@ki.se.
Gunnar Sandström, Email: gunnar.sandstrom@ki.se.
Hadi Abd, Email: hadi.abd@ki.se.
Acknowledgements
This project was supported by the Swedish Civil Contingencies Agency (MSB), Project No. 800/2008.
References
- Tauxe RV, Mintz ED, Quick RE. Epidemic cholera in the new world: translating field epidemiology into new prevention strategies. Emerg Infect Dis. 1995;1(4):141–146. doi: 10.3201/eid0104.950408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64(10):3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Jahid MI, Rahman MM, Rahman MZ, Kabir MS, Sack DA, Schoolnik GK. Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol Immunol. 2007;51(4):369–379. doi: 10.1111/j.1348-0421.2007.tb03924.x. [DOI] [PubMed] [Google Scholar]
- Johnson LR. Microcolony and biofilm formation as a survival strategy for bacteria. J Theor Biol. 2008;251(1):24–34. doi: 10.1016/j.jtbi.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Wrangstadh M, Conway PL, Kjelleberg S. The production and release of an extracellular polysaccharide during starvation of a marine Pseudomonas sp. and the effect thereof on adhesion. Arch Microbiol. 1986;145(3):220–227. doi: 10.1007/BF00443649. [DOI] [PubMed] [Google Scholar]
- Abd H, Saeed A, Weintraub A, Nair GB, Sandstrom G. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol Ecol. 2007;60(1):33–39. doi: 10.1111/j.1574-6941.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- Abd H, Saeed A, Weintraub A, Sandstrom G. Vibrio cholerae O139 requires neither capsule nor LPS O side chain to grow inside Acanthamoeba castellanii. J Med Microbiol. 2009;58(Pt 1):125–131. doi: 10.1099/jmm.0.004721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd H, Weintraub A, Sandstrom G. Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ Microbiol. 2005;7(7):1003–1008. doi: 10.1111/j.1462-2920.2005.00771.x. [DOI] [PubMed] [Google Scholar]
- Amir Saeed AH, Edvinsson B, Sandstrom G. Vibrio cholerae-Acanthamoeba castellanii interaction showing endosymbiont-host relation. Symbiosis. 2007;44:153–158. [Google Scholar]
- Sandstrom G, Saeed A, Abd H. Acanthamoeba polyphaga is a possible host for Vibrio cholerae in aquatic environments. Exp Parasitol. 2010;126(1):65–68. doi: 10.1016/j.exppara.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci USA. 2000;97(18):10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano D, Schuhmacher DA, Barker JL, Klose KE. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun. 2000;68(3):1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. V. cholerae 01 E1 Tor: identification of a gene cluster required for the rugose col type Exopolysaccharide production chlorine resistance and biofilm formation. Proc Natl Acad Sci USA. 1999;96(7):4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequencing of the M13mp18 and pUC9 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanan S, Abd H, Hedenstrom I, Saeed A, Sandstrom G. Detection of Vibrio cholerae and Acanthamoeba species from same natural water samples collected from different cholera endemic areas in Sudan. BMC Res Notes. 2011;4:109. doi: 10.1186/1756-0500-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179(20):6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Merrell DS, Camilli A, Kaper JB. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol Microbiol. 2002;43(6):1577–1589. doi: 10.1046/j.1365-2958.2002.02845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414(6863):555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- Wang SY, Lauritz J, Jass J, Milton DL. A ToxR Homolog from Vibrio anguillaru Serotype O1 regulates its own production, bile resistance, and biofilm formation. J Bacteriol. 2002;184(6):1630–1639. doi: 10.1128/JB.184.6.1630-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


