Abstract
The human X and Y chromosomes evolved from an ordinary pair of autosomes during the past 200–300 million years1–3. Due to genetic decay, the human MSY (male-specific region of Y chromosome) retains only three percent of the ancestral autosomes’ genes4,5. This evolutionary decay was driven by a series of five “stratification” events. Each event suppressed X-Y crossing over within a chromosome segment or “stratum”, incorporated that segment into the MSY, and subjected its genes to the erosive forces that attend the absence of crossing over2,6. The last of these events occurred 30 million years ago (mya), or 5 million years before the human and Old World monkey (OWM) lineages diverged. Although speculation abounds regarding ongoing decay and looming extinction of the human Y chromosome7–10, remarkably little is known about how many MSY genes were lost in the human lineage in the 25 million years that have followed its separation from the OWM lineage. To explore this question, we sequenced the MSY of the rhesus macaque, an OWM, and compared it to the human MSY. We discovered that, during the last 25 million years, MSY gene loss in the human lineage was limited to the youngest stratum (stratum 5), which comprises three percent of the human MSY. Within the older strata, which collectively comprise the bulk of the human MSY, gene loss evidently ceased more than 25 mya. Likewise, the rhesus MSY has not lost any older genes (from strata 1–4) during the past 25 million years, despite major structural differences from the human MSY. The rhesus MSY is simpler, with few amplified gene families or palindromes that might enable intrachromosomal recombination and repair. We present an empirical reconstruction of human MSY evolution in which each stratum transitioned from rapid, exponential loss of ancestral genes to strict conservation through purifying selection.
The human Y chromosome no longer engages in crossing over with its once-identical partner, the X chromosome, except in its pseudoautosomal regions. During evolution, X-Y crossing over was suppressed in five different chromosomal regions at five different times, each probably resulting from an inversion within the Y chromosome2,3. Each of these regions of the Y chromosome then began its own individual course of degeneration, experiencing deletions and gene loss. Comparison of the present-day X and Y chromosomes enables identification of these five evolutionary “strata” in the MSY (and X chromosome); their distinctive degrees of X-Y differentiation indicate their evolutionary ages2,3. The oldest stratum (stratum 1) dates back over 240 million years2 and is the most highly differentiated, while the youngest stratum (stratum 5) originated only 30 mya and displays the highest X-Y nucleotide sequence similarity within the MSY3. The five strata and their respective decay processes, playing out over tens to hundreds of millions of years of mammalian evolution, offer replicate experiments of nature from which to reconstruct the trajectories and kinetics of gene loss in the MSY.
The only MSYs sequenced prior to the present study were those of human and chimpanzee, separated by just six million years of evolution. We decided to examine the MSY of a much more distant relative, the rhesus macaque (Macaca mulatta), to enable us to reconstruct gene loss and conservation in the MSY during the past 25 million years. We sequenced the rhesus MSY using bacterial artificial chromosome (BAC) clones and the SHIMS (single-haplotype iterative mapping and sequencing) strategy previously employed in the human and chimpanzee MSYs4,11–13 as well as in the chicken Z chromosome5. The resulting sequence comprises 11.0 megabases (Mb), is complete aside from three small gaps, and has an error rate of about one nucleotide per Mb. We ordered and oriented the finished sequence contigs by fluorescence in situ hybridization and radiation hybrid mapping (Supplementary Figs 1–6, Supplementary Table 1, Supplementary Files 1–2 and Supplementary Note 1).
We then compared the structure of the rhesus Y chromosome to those of human and chimpanzee (Fig. 1). The rhesus Y chromosome has virtually no heterochromatin apart from the centromere, and the euchromatic segment of the MSY is dramatically smaller compared to human and chimpanzee (Fig. 1). The single pseudoautosomal region (PAR) in rhesus corresponds to the short-arm PAR in human and to the single PAR in chimpanzee. The precise boundary between PAR and MSY is identical in the three species (Supplementary Fig. 7), confirming that stratification in all three lineages concluded before the divergence of apes from OWMs.
Figure 1.
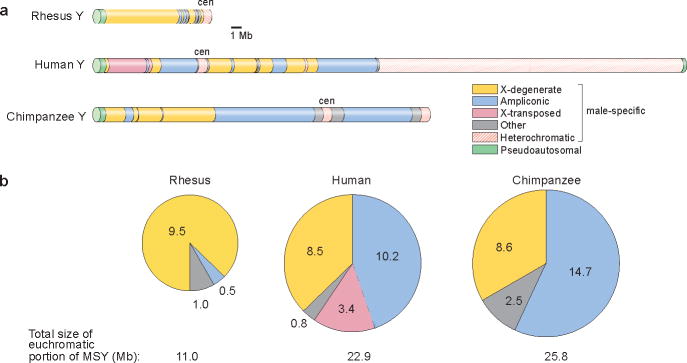
Comparison of rhesus, human and chimpanzee Y chromosomes. (a) Schematic representations, to scale. “Other” denotes single-copy, male-specific sequences that are neither X-degenerate nor X-transposed. (b) Sizes (in Mb) of euchromatic sequence classes in MSYs.
The euchromatic portions of the rhesus, human and chimpanzee MSYs are comprised primarily of two distinct sequence classes: X-degenerate and ampliconic. The X-degenerate regions, relics of shared X-Y ancestry, are dotted with single-copy homologues of X-linked genes. The X-degenerate regions are relatively well conserved among the rhesus, human and chimpanzee MSYs, with large blocks of homology that are readily identifiable (Supplementary Figs 8 and 9). Indeed, the X-degenerate regions are the only portions of the rhesus and human MSYs whose sequences can be aligned over distances > 50 kb. We found rhesus-human nucleotide divergence there to be 9.4% (Supplementary File 3). This is significantly higher than the 6.5% divergence observed when the rhesus and human female genomes are compared14. The difference probably reflects the MSY’s restriction to the male germ line, where base-pair substitutions are more frequent than in the female germ line15. From these data, we calculate the male-to-female mutation rate ratio (αm) to be 2.78 (95% confidence interval 2.74–2.81), in agreement with previous but less precise estimates14,16. The X-degenerate sequences in rhesus, human and chimpanzee are not entirely collinear, as large-scale rearrangements have occurred in each lineage (Supplementary Figs 8–10).
For all three species, the MSY’s ampliconic regions are composed of long, nearly identical repeat units, arrayed in either direct or inverted orientation, that undergo frequent gene conversion – a process thought to slow or prevent the decay of genes that reside there4,17. Ampliconic genes display testis-specific expression patterns, consistent with critical roles in spermatogenesis4,11,18. Only 0.5 Mb of the rhesus MSY euchromatin is ampliconic, compared to 10.2 Mb and 14.7 Mb in human and chimpanzee, respectively (Fig. 1, Supplementary Figs 11 and 12). In human and chimpanzee, the MSY’s ampliconic regions feature large palindromes, each composed of two inverted repeats (arms) separated by a short spacer. The human and chimpanzee MSYs have, respectively, eight and 19 palindromes, and these span, respectively, 5.5 and 7.5 Mb4,13. By contrast, the rhesus MSY has only three palindromes that collectively span 437 kb (Supplementary Table 2, Supplementary Fig. 13). Two of the rhesus MSY palindromes are orthologs of human MSY palindromes, demonstrating that these structures have been maintained for at least 25 million years (Supplementary Fig. 13).
We identified protein-coding genes in the rhesus MSY using three complementary approaches. First, we electronically searched the rhesus MSY for homologs of all known human and chimpanzee MSY genes and pseudogenes. Second, we searched for homologs of all known human X-linked genes, to identify any X-Y shared genes that had been lost in both the human and chimpanzee MSY, but retained in the rhesus MSY. Third, we searched for additional rhesus-specific MSY genes using electronic prediction tools and high-throughput sequencing of rhesus testis cDNA (245 Mb total). We validated each putative gene by verifying transcriptional activity (Supplementary Fig. 14) and, where applicable, by comparing its predicted open reading frame to that of its human ortholog (Supplementary Table 3).
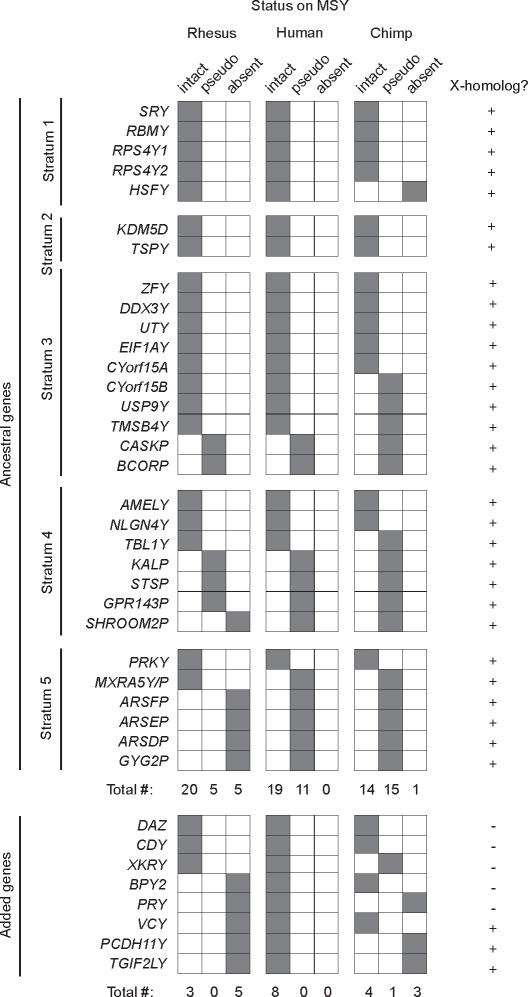
We then compared the catalogs of MSY genes in rhesus, human4 and chimpanzee13 to infer gene loss and conservation during the past 25 million years. To root this analysis in a deep evolutionary context, we first reconstructed which of the modern rhesus MSY genes were present on the common autosomal ancestor of X and Y (Fig. 2, Supplementary Table 4 and Supplementary Note 2). Most “ancestral” MSY genes would be expected to have a homolog both on the human X chromosome and on the chicken autosomes (chromosomes 1 and 4) that share common ancestry with mammalian X and Y chromosomes3,5. Indeed, 33 genes and pseudogenes in the rhesus, human, or chimpanzee MSY have their most closely related human homologs on the X chromosome (Fig. 2), and 29 of these also have homologs within syntenic regions of chicken chromosome 1 or 4. Analyses of a more distant outgroup, Xenopus tropicalis, revealed that two of the four rhesus MSY genes lacking homologs on chicken chromosome 1 and 4 (TSPY and AMELY) are X-Y ancestral; they were lost in the chicken lineage after divergence from mammals (Supplementary Note 2). A few human MSY genes with X homologs are recent additions to the MSY rather than remnants of the ancestral autosome pair – PCDH11Y and TGIF2LY are located in the human-specific X-transposed region4, and the X-linked homolog of VCY is found only in simian primates19. In the end, we arrived at a total of 30 ancestral MSY genes and pseudogenes in rhesus, human or chimpanzee (Fig. 2).
Figure 2.
Inventories of genes, both ancestral and added, in rhesus, human, and chimpanzee MSYs. At top, ancestral genes grouped by stratum, 1 through 5; added genes listed below. In rhesus, human and chimpanzee, current status of each MSY gene is indicated by shading in one of three columns: i. present and intact, ii. inactivated pseudogene or iii. absent/deleted. Total numbers of MSY-intact genes, pseudogenes, and absent/deleted genes – both ancestral and added – are tallied for each species. At far right is indicated, for each MSY gene, whether the most closely related human homolog is located on the X chromosome.
Within strata 1 to 4, which collectively comprise the bulk of the human MSY, the rhesus and human MSYs possess precisely the same 18 ancestral genes (Figure 2). This striking and unanticipated identity leads us to conclude that, 25 million years ago, in the last common ancestor of rhesus and human, MSY strata 1 to 4 also carried these 18 ancestral genes (Table 1, Supplementary Table 5), and that no loss of ancestral genes occurred subsequently in either lineage (Supplementary Note 3). We note that, within strata 3 and 4, the rhesus and human MSYs carry a total of six ancestral pseudogenes, which appear to have lost their function more than 25 mya (Supplementary Fig. 15).
Table 1.
Stratification of X-Y ancestral gene loss in primate MSYs
| Age of stratum (millions of years)1 | Number of ancestral genes on human X chromosome2 | Number of ancestral genes on MSY | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Last common ancestor3 | Rhesus | Human | Chimp | |||
| Stratum 1 | 240–320 | 414 | 5 | 5 | 5 | 4 |
| Stratum 2 | 130–170 | 88 | 2 | 2 | 2 | 2 |
| Stratum 3 | 80–130 | 143 | 8 | 8 | 8 | 5 |
| Stratum 4 | 38–44 | 12 | 3 | 3 | 3 | 2 |
| Stratum 5 | 29–32 | 7 | 2–7 | 2 | 1 | 1 |
Gene numbers from ref. 5, Supplementary Table 4 and Supplementary Note 2.
Gene counts in MSY of hypothetical rhesus-human-chimpanzee ancestor deduced from observed gene counts in extant species.
The evolutionary stability of ancestral genes in strata 1–4 could be explained by purifying selection, which, in the absence of sexual recombination, would have preserved critical ancestral genes for tens or even hundreds of millions of years. We previously demonstrated that purifying selection preserved MSY genes during the past 100,000 years of human population expansion and migration20. Here, comparing human and rhesus, we find that most ancestral genes display a ratio of nonsynonymous substitution rate (dN) to synonymous substitution rate (dS) that is significantly less than one (Supplementary Note 4, Supplementary Table 3, Supplementary Fig. 16), demonstrating purifying selection during the past 25 million years.
The pattern of gene loss and conservation in stratum 5, formed only 5 million years before the rhesus and human lineages split, is remarkably different from the pattern in the four older strata. Within the past 30 million years, four ancestral genes have been inactivated or deleted from stratum 5 of the MSY in both rhesus and human (Figure 2; Supplementary Note 5). A fifth ancestral gene, MXRA5Y, remains active in rhesus (Supplementary Fig. 11) but was inactivated by an intragenic deletion in the human lineage (Supplementary Fig. 17). Apart from MXRA5Y, all differences in MSY gene content between rhesus and human involve genes added to the human MSY subsequent to the ape-OWM split (Fig. 2, Supplementary Table 5).
Returning to strata 1 to 4, we note that five ancestral genes have been inactivated or lost from the chimpanzee MSY during the past 6 million years12,13, in sharp contrast to the strict conservation of ancestral gene content in rhesus and human (Fig. 2). We previously speculated that, in the chimpanzee lineage, promiscuous mating behavior21, sperm competition and intense sexual selection that focused on the MSY drove rapid evolution and amplification of MSY sequences associated with spermatogenesis12,13. We further speculated that, in the chimpanzee lineage, inactivated alleles of some ancestral genes became fixed in the population through “genetic hitchhiking” – casualties of positive but indiscriminate selection operating in the absence of sexual recombination in the MSY12,13,22. Among primate species, chimpanzees have a high testis-weight/body-weight ratio, a useful indicator of the degree of sperm competition23,24. Although the rhesus is similarly promiscuous and has an even higher testis-weight/body-weight ratio, the rhesus MSY shows little evidence of intense sexual selection. We speculate that in the rhesus lineage, such selection was focused on spermatogenesis factors encoded elsewhere in the genome. This would also account for the virtual absence in rhesus of the MSY sequence amplification that is prominent in human and even more pronounced in chimpanzee (Fig. 1).
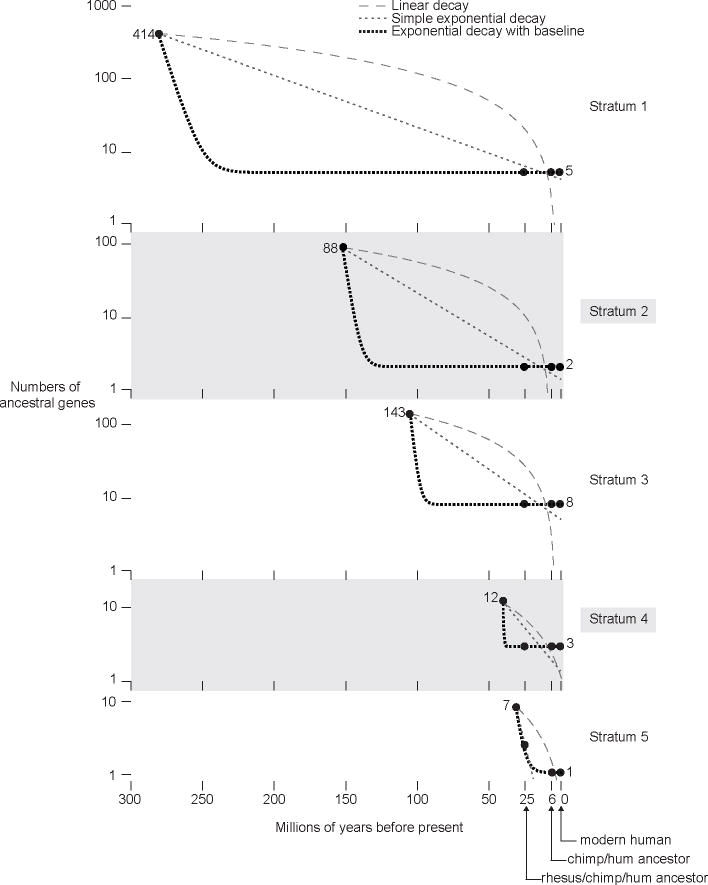
Our knowledge of all five strata of the MSY, gained through our comprehensive comparisons of ancestral gene content in the rhesus, human and chimpanzee MSYs, enabled us to reconstruct the kinetics and trajectory of human MSY evolution. For each of the five MSY strata, we estimated ancestral gene numbers at three points in the human evolutionary lineage: in the last common ancestor of human and chimpanzee (6 mya), in the last common ancestor of human and rhesus macaque (25 mya), and at the time of the stratum’s formation, when X-Y differentiation was initiated (from ~30 to >240 mya; Table 1). For each stratum, we plotted these three estimated numbers against evolutionary time, together with the observed number of ancestral genes in modern human, and fit a curve (Fig. 3, Supplementary Fig. 18). For each of the five strata, a simple two-parameter model, using an exponential decay equation that includes a baseline constant, provides an excellent fit to our data (Fig. 3, Supplementary Table 6). According to this reconstruction, ancestral gene decay within each stratum proceeded rapidly at first – with an ancestral gene half-life < 5 million years (Supplementary Table 6) – but then decelerated markedly, as the ancestral gene count reached a stable level far below its starting point. In our reconstruction, strata 1–4 had already reached a stable level before the human lineage diverged from rhesus; after divergence from rhesus, gene loss in the human lineage was limited to stratum 5, the youngest stratum, which stabilized before the human lineage diverged from chimpanzee.
Figure 3.
Kinetics of ancestral gene loss during evolution of five human MSY strata. Gene numbers are plotted on a log scale on the Y axis, and time (in millions of years before present) is plotted on the X axis. Filled circles represent inferred or observed gene numbers in (from left to right): i. X-Y ancestral chromosome (at time of stratum formation), ii. rhesus-chimpanzee-human ancestral MSY (25 mya), iii. chimpanzee-human ancestral MSY (6 mya), and iv. modern human MSY. Dotted/dashed lines represent best-fit curves to data points using each of three decay models as indicated.
Our empirical reconstruction of MSY evolution is at odds with a linear model7,9,10 and with a simple random decay (exponential) model25, both of which project a continual decline of MSY gene content and cannot account for the stability of gene content that we observe over the past 25 million years (Fig. 3). Our data are better explained by more complex models for MSY gene loss that incorporate a combination of evolutionary forces26. Sequencing additional Y chromosomes from animals representing more divergent mammalian lineages will enable refinement of our reconstruction of MSY gene kinetics in the human lineage.
Methods Summary
BAC selection and sequencing
The SHIMS strategy11 was employed to assemble a path of sequenced clones selected from the CHORI-250 BAC library (bacpac.chori.org) and a custom BAC library (RMAEX) constructed by Amplicon Express (www.genomex.com).
Fluorescence in situ hybridization (FISH) analysis
Assays were performed on rhesus fibroblast cell line PR00112 from Coriell Institute for Medical Research (ccr.coriell.org). Extended metaphase FISH and interphase FISH were performed as previously described27.
Radiation hybrid mapping
Nine STS markers (Supplementary Table 7) were tested on a 10,000-rad panel consisting of 185 hybrid clones28. A genetic map was constructed and analyzed statistically using RHMAPPER 1.2229.
Generation of cDNA for RT-PCR and 454 sequencing
cDNA was generated from total RNA was isolated from male rhesus tissues using the RNeasy kit (Qiagen). For 454 sequencing, cDNA was normalized using the Trimmer kit (Evrogen).
Alignments and dot plots
Rhesus and human Y sequences were aligned using Stretcher (bioweb2.pasteur.fr/docs/EMBOSS/stretcher.html) with gap open penalty of 20 and gap extend penalty of one. Dot plot analyses were performed using custom Perl codes available at jura.wi.mit.edu/page/papers/Hughes_et_al_2005/tables/dot_plot.pl.
Calculation of αm
The male-to-female mutation rate ratio was calculated from the human-rhesus Y divergence rate and the human-rhesus autosomal divergence rate using a previously described method15,30.
Modeling ancestral MSY gene loss
We fit a one-phase exponential decay model (shown below) with a baseline constant to our data (gene numbers shown in Table 1) using nonlinear regression analysis in GraphPad Prism 5.0. Parameters for each stratum are given in Supplementary Table 6.
One-phase exponential decay model:
where
N(t) = number of genes at time t
N0 = number of genes within given stratum in ancestral autosomal/pseudoautosomal portion of genome
K = decay constant
b = baseline (approximated by the number of active ancestral genes within that stratum on human Y chromosome)
Methods
BAC selection and sequencing
The SHIMS strategy11 was employed to assemble a path of sequenced clones selected from the CHORI-250 BAC library (bacpac.chori.org) and a custom BAC library (RMAEX) constructed by Amplicon Express (www.genomex.com). The rate of error in the finished sequence was estimated by counting mismatches in overlapping clones.
Fluorescence in situ hybridization (FISH) analysis
All assays were performed on rhesus fibroblast cell line PR00112 obtained from Coriell Institute for Medical Research (ccr.coriell.org). Extended metaphase FISH and interphase FISH were performed as previously described27.
Radiation hybrid mapping
Nine STS markers (Supplementary Table 7) were tested on a 10,000-rad, male, whole-genome panel consisting of 185 hybrid clones28. The average retention frequency of the markers tested was 16%, ranging from 10–27%. A genetic map was constructed and analyzed statistically using RHMAPPER 1.2229.
RT-PCR
Total RNA was isolated from male rhesus tissues (brain, prostate, liver, lung, spleen testis; Alpha Genesis, Inc.) using the RNeasy kit (Qiagen) and cDNA was generated. RT-PCR primer sequences and product sizes are listed in Supplementary Table 8.
454 sequencing of testis cDNA
Rhesus testis cDNA was generated from total RNA isolated using the RNeasy kit (Qiagen). The cDNA was normalized using the Trimmer kit (Evrogen) and sequenced on a 454 FLX Titanium machine.
Alignments and dot plots
Rhesus and human Y sequences were aligned using Stretcher (bioweb2.pasteur.fr/docs/EMBOSS/stretcher.html) with gap open penalty of 20 and gap extend penalty of one. Dot plot analyses were performed using custom Perl codes available at jura.wi.mit.edu/page/papers/Hughes_et_al_2005/tables/dot_plot.pl.
Calculation of αm
The male-to-female mutation rate ratio was calculated using the human-rhesus Y divergence rate (9.40%, 312,840 substitutions per 3,330,847 sites examined) and the human-rhesus autosomal divergence rate (1.385 × 108 substitutions per 2.248 × 1010 sites examined; hg18-rheMac2 alignments downloaded from genome.ucsc.edu). Miyata’s formula was then used to calculate αm as follows15,30: Y/A = 2αm/(1 + αm). Confidence intervals for ratios of divergence rates were calculated as previously described30.
Modeling ancestral MSY gene loss
We modeled the numbers of ancestral genes within individual MSY strata as a function of time in millions of years before the present by fitting a one-phase exponential decay model with a baseline constant (below) to our data (gene numbers shown in Table 1) using nonlinear regression analysis in GraphPad Prism 5.0. Parameters for each stratum are given in Supplementary Table 6.
One-phase exponential decay model:
where
N(t) = number of genes at time t
N0 = number of genes within given stratum in ancestral autosomal/pseudoautosomal portion of genome
K = decay constant
b = baseline (approximated by the number of active ancestral genes within that stratum on human Y chromosome)
Supplementary Material
Acknowledgments
We thank V. Frazzoni, G. Rogers, and S. Zaghlul for technical assistance; L. Lyons, W. Murphy, and J. Womack for radiation hybrid panels; W. Johnson, E. Curran, S. O’Neil, and A. Kaur for tissue samples; R. Harris for analyses of rhesus-human genome alignments; T. DiCesare for assistance with figures; and D. Bellott, M. Carmell, R. Desgraz, Y. Hu, B. Lesch, J. Mueller, K. Romer and S. Soh for comments on the manuscript. Supported by the National Institutes of Health, the Howard Hughes Medical Institute, and the Charles A. King Trust.
Footnotes
Supplementary Information: is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: J.F.H., H. Skaletsky, W.C.W., S.R., R.G., R.K.W., and D.C.P. planned the project. J.F.H., H.S., L.G.B., T.J.C., and N.K. performed BAC mapping, radiation hybrid mapping and RT-PCR analyses. T.G., R.S.F., S.D., Y.D., C.J.B., C.K., Q.W., H. Shen, M.H., D.V., L.V.N., A.C., L.C., J.V., H.K., and D.M.M. were responsible for finished BAC sequencing. J.F.H. and H.S. performed comparative sequence analyses. T.P. performed FISH analyses. J.F.H. and D.C.P. wrote the paper.
Author Information: cDNA sequences of rhesus Y genes have been deposited in GenBank (www.ncbi.nlm.nih.gov) under accession numbers FJ527009-FJ527028 and FJ648737-FJ648739. 454 testis cDNA sequences have been deposited in GenBank under accession number SRA039857.
LITERATURE CITED
- 1.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 2.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 3.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 5.Bellott DW, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philosophical transactions of the Royal Society of London Series B: Biological sciences. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aitken RJ, Marshall Graves JA. The future of sex. Nature. 2002;415:963. doi: 10.1038/415963a. [DOI] [PubMed] [Google Scholar]
- 8.Sykes B. Adam’s Curse. Norton, W. W. & Company, Inc; 2004. [Google Scholar]
- 9.Graves JA, Koina E, Sankovic N. How the gene content of human sex chromosomes evolved. Current Opinion in Genetics and Development. 2006;16:219–224. doi: 10.1016/j.gde.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Marshall Graves JA. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda-Kawaguchi T, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genetics. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 12.Hughes JF, et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 13.Hughes JF, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 15.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: A model and nucleotide sequence analysis. Cold Spring Harbor Symposia on Quantitative Biology. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 16.Bohossian HB, Skaletsky H, Page DC. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–625. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 18.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 19.Lahn B, Page D. A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum Mol Genet. 2000;9:311–319. doi: 10.1093/hmg/9.2.311. [DOI] [PubMed] [Google Scholar]
- 20.Rozen S, Marszalek JD, Alagappan RK, Skaletsky H, Page DC. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am J Hum Genet. 2009;85:923–928. doi: 10.1016/j.ajhg.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixson AF. Primate Sexuality: Comparative Studies of the Prosimians, Monkeys, Apes and Human Beings. University of Chicago Press; 1998. [Google Scholar]
- 22.Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz AH. The relative weight of the testes in primates. Anatomical Record. 1938;72:387–394. [Google Scholar]
- 24.Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- 25.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 26.Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxena R, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nature Genetics. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 28.Karere GM, Froenicke L, Millon L, Womack JE, Lyons LA. A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly. Genomics. 2008;92:210–218. doi: 10.1016/j.ygeno.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slonim D, Kruglyak L, Stein L, Lander E. Building human genome maps with radiation hybrids. Journal of Computational Biology. 1997;4:487–504. doi: 10.1089/cmb.1997.4.487. [DOI] [PubMed] [Google Scholar]
- 30.Shimmin LC, Chang BH, Li WH. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.