Abstract
Transcription is coupled with pre-mRNA splicing in metazoans. In the current issue of Molecular Cell, Huang et al. shows that Med23 interacts with the RNA binding protein hnRNP L to regulate alternative splicing, thus expanding mediator’s function beyond transcriptional control.
Pre-mRNA splicing is largely coupled with transcription, which permits sequential recognition of emerging splicing signals by the splicing machinery (Oesterreich et al., 2011). Such coupling not only facilitates efficient gene expression, but also institutes various mechanisms for the regulation of RNA processing (Han et al., 2011). A curious observation made sometime ago by Kornblihtt and colleagues indicated that different gene promoters have varying influence on splice site selection (Cramer et al., 1997). This observation led to two models for coupling between transcription and splicing: The “recruitment model” suggests that the C-terminal domain (CTD) of the largest Pol II subunit plays a central role in recruiting specific splicing regulators to different gene promoters to mediate co-transcriptional regulation of alternative splicing. Alternatively, the “kinetic model” proposes that differentially recruited transcription factors may affect Pol II transcription during the elongation phase, thereby influencing the splice site selection process. Little is currently known about which and how specific transcription or splicing regulators are loaded onto the Pol II complex at different gene promoters, and the mechanisms have remained largely elusive. In the current issue of Molecular Cell, Wang and colleagues report a direct interaction between the specific Mediator component Med23 and the RNA binding splicing regulator hnRNP L (Huang et al., 2012). This finding joins an early discovery of CHD1-mediated recruitment of a U2 snRNP component via H3K4me3 to gene promoters, which may enhances the recognition of weak splice sites in transcribed pre-mRNA (Sims III et al., 2007). Together, these findings provide critical mechanistic insights into the recruitment model.
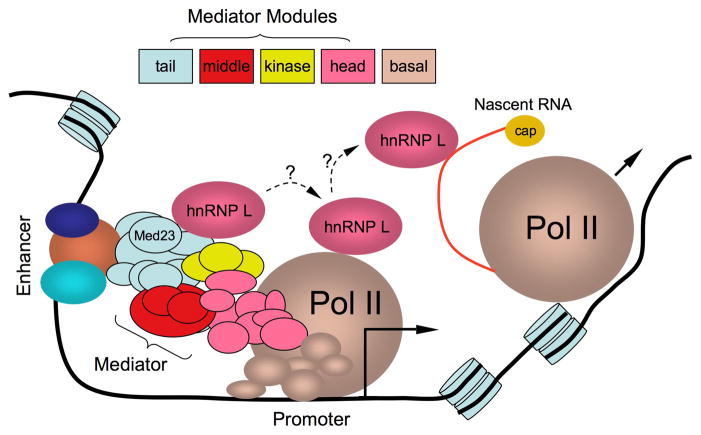
Mediator is a multi-subunit complex that is part of the core transcription machinery. Many years of research have established Mediator as a signaling hub for transcriptional output. Mediator is generally organized into “head, middle, tail, and kinase” modules (Malik and Roeder, 2010; see Figure 1). Specific subunits in the tail module interact with transcription activators or repressors, whereas the head module interacts with the Pol II complex. By connecting Pol II to activators and repressors, Mediator helps to establish the pre-initiation complex at gene promoters. Although Mediator has been purified and characterized as a core complex, recent evidence suggests that different components of Mediator can each mediate specific interactions with other transcription factors and undergo dynamic exchanges during transcription activation (Takahashi et al., 2011). The diverse spectrum of molecular interactions is also evident from the current work, which shows that Mediator components appear to associate with largely distinct groups of proteins (Huang et al., 2012). These findings re-enforce the idea of Mediator as an integrative hub for regulated gene expression in the cell.
Figure 1.
Initiating the coupling between transcription and RNA processing at gene promoter. hnRNP L is initially recruited to gene promoter via direct protein-protein interaction with Med23, a tail component of the large Mediator complex. This recruitment appears to also enhance Pol II binding at gene promoter. The recruited hnRNP L affects downstream splicing events by binding to CA-rich motifs in pre-mRNA, although the mechanism for the RNA binding protein to switch from Mediator to elongating Pol II and then to nascent RNA remains to be defined. This work highlights a new role of Mediator in coupling between transcription and pre-mRNA processing. Specific RNA binding proteins recruited to Mediator may also play critical roles in promoting enhancer-promoter communications via intergenic non-coding RNAs.
Med23 is a “tail” component of Mediator previously shown to interact with several specific transcription activators, including E1A. By using baculovirus-expressed Med23 to pull down cellular proteins from nuclear extracts followed by tandem affinity purification and mass spectrometry, the authors unexpectedly discovered that Med23 specifically interacts with a large number of RNA binding proteins involved in RNA splicing and other RNA metabolic pathways (Figure 1). One of the top hits was hnRNP L, a sequence-specific RNA binding protein previously characterized to recognize CA-rich motifs in pre-mRNA and to function in regulated splicing (Hui et al., 2003; Motta-Mena et al., 2010). Indeed, on both model genes and by using genome-wide approaches, the authors provide evidence that Med23 is involved in the regulation of a large number (but not all) hnRNP L-mediated RNA processing events. Genome-wide ChIP-seq analysis revealed that hnRNP L binds to both gene promoter and gene body, but knockdown of Med23 diminishes hnRNP L binding to gene promoters, but not gene body, suggesting that hnRNP L may be recruited to gene promoters in a Med23-dependent fashion, but recruitment to gene bodies involves additional mechanisms. While it will be interesting to extend the analysis to other Mediator components to further establish specificity, the new work expands the roles of Mediator beyond transcription to RNA processing events.
How does hnRNP L influence alternative splicing after its initial recruitment to gene promoters? A reasonable assumption is that it may be first recruited to gene promoters in a gene-specific fashion, and then travel with the elongating Pol II complex to mediate specific splice site selection during co-transcription splicing. However, because it is generally believed that the Mediator complex does not travel with the elongating Pol II complex, how does the recruited hnRNP L exert its effect on alternative splicing, which may be far downstream from the promoter region? By a yet unknown mechanism, hnRNP L may be “handed” over from Mediator to Pol II during transcription elongation. Alternatively, hnRNP L bound promoters may remain in close proximity to specific downstream transcribed regions, allowing hnRNP L to switch binding sites in pre-mRNA during transcription. In the future, it will be interesting to examine the genome-wide interactions of both Pol II and hnRNP L with DNA and RNA. By looking at how the binding events are perturbed upon depletion of Med23 or hnRNP L, it should be possible to gain some further mechanistic insights.
Interestingly, the authors noticed that Pol II binding diminished on a model gene in response to hnRNP L RNAi, suggesting that hnRNP L may have a role in transcription. While it is currently unclear whether hnRNP L affects transcription initiation or elongation or both, the observation is in line with increasing evidence for functional integration between the transcription and splicing machineries in mammalian cells, which emphasize direct roles of specific components in one machinery in the other process, rather than just the coupling between transcription and splicing in time and space in the nucleus (Pandit et al., 2008). Therefore, specially recruited splicing factors may first benefit the transcription process at the beginning of gene expression and later modulate various RNA processing events. This might be a common feature among a large number of RNA binding proteins expressed in metazoans. In particular, the recruitment of specific RNA binding proteins to the transcription machinery may prove to play a central role in mediating the function of various regulatory non-coding RNAs in regulated gene expression.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD. Trends Cell Biol. 2011;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin Q-J, Yin J-W, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mol Cell. 2012 doi: 10.1016/j.molcel.2011.12.022. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. Nat Struct Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW. Mol Cell. 2010;37:223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterreich FC, Bieberstein N, Neugebauer KM. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Curr Opion Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Millhouse S, Chen FF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Pamely TJ, Sato S, Tomomorl-Sato C, Banks CAS, Kong SE, Szotorisz H, Swanson SK, Martin-Brown S, Washbum MP, Florens L, Seidel CW, Lin C, Smith ER, Shilatifard A, Conaway RC, Conaway JW. Cell. 2010;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]