Abstract
Dispersed repetitive DNA sequences have been described recently in eubacteria. To assess the distribution and evolutionary conservation of two distinct prokaryotic repetitive elements, consensus oligonucleotides were used in polymerase chain reaction [PCR] amplification and slot blot hybridization experiments with genomic DNA from diverse eubacterial species. Oligonucleotides matching Repetitive Extragenic Palindromic [REP] elements and Enterobacterial Repetitive Intergenic Consensus [ERIC] sequences were synthesized and tested as opposing PCR primers in the amplification of eubacterial genomic DNA. REP and ERIC consensus oligonucleotides produced clearly resolvable bands by agarose gel electrophoresis following PCR amplification. These band patterns provided unambiguous DNA fingerprints of different eubacterial species and strains. Both REP and ERIC probes hybridized preferentially to genomic DNA from Gram-negative enteric bacteria and related species. Widespread distribution of these repetitive DNA elements in the genomes of various microorganisms should enable rapid identification of bacterial species and strains, and be useful for the analysis of prokaryotic genomes.
Full text
PDF




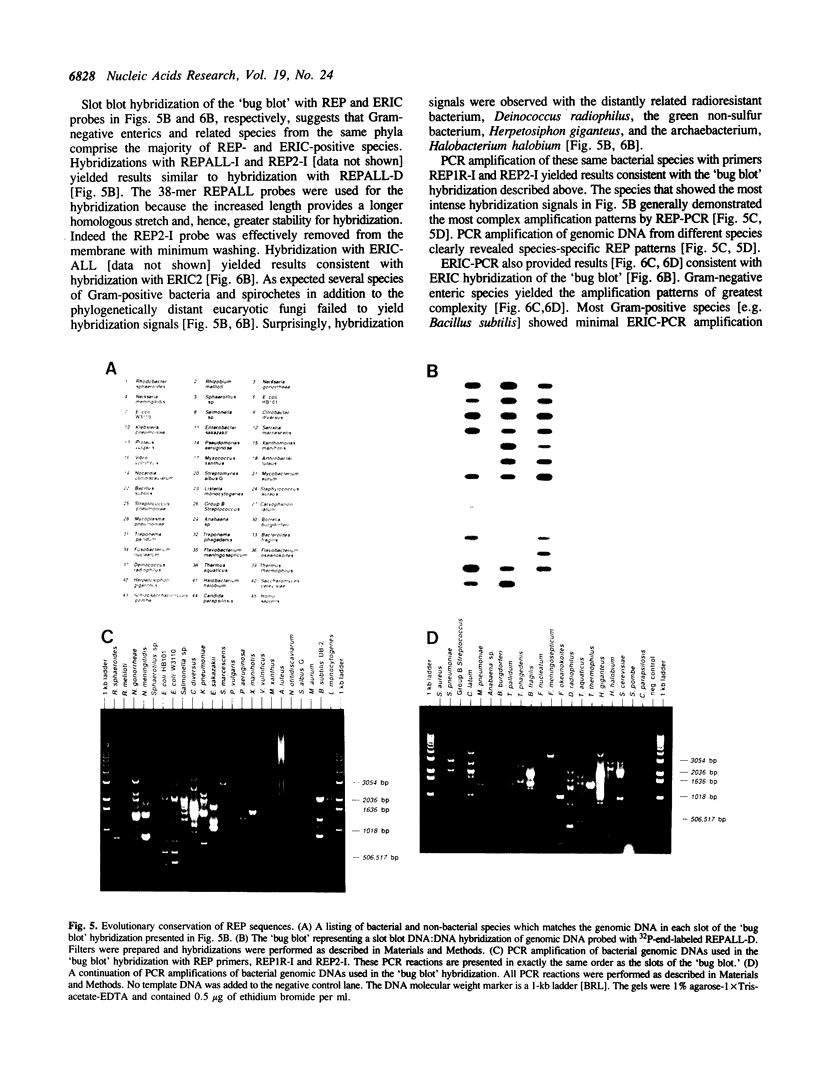
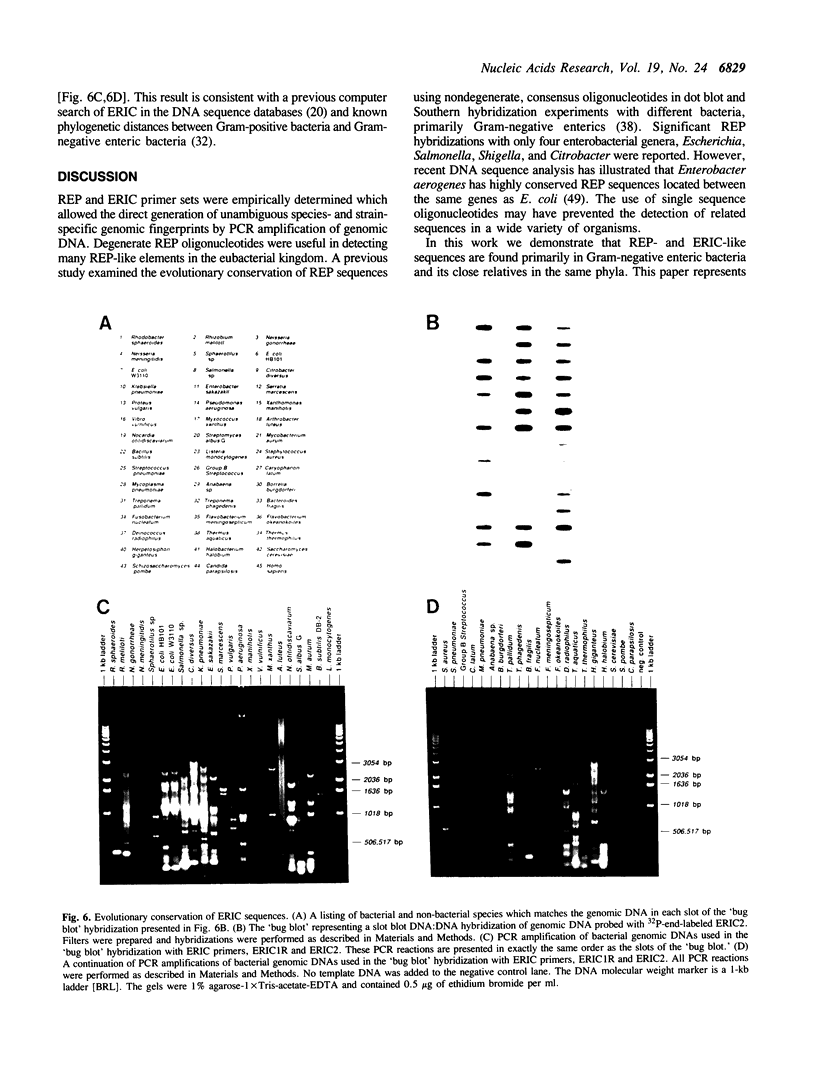


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin D., Larson T. J. Nucleotide sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1991 Jan;173(1):101–107. doi: 10.1128/jb.173.1.101-107.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Retroposons--seeds of evolution. Science. 1991 Feb 15;251(4995):753–753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Carriere C., Allardet-Servent A., Bourg G., Audurier A., Ramuz M. DNA polymorphism in strains of Listeria monocytogenes. J Clin Microbiol. 1991 Jul;29(7):1351–1355. doi: 10.1128/jcm.29.7.1351-1355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., Maley F., Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci U S A. 1984 May;81(10):3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia F. F., Inouye S., Inouye M. A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J Bacteriol. 1986 Sep;167(3):1009–1015. doi: 10.1128/jb.167.3.1009-1015.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl M. K., Francoz E., Saurin W., Boos W., Manson M. D., Hofnung M. Comparison of sequences from the malB regions of Salmonella typhimurium and Enterobacter aerogenes with Escherichia coli K12: a potential new regulatory site in the interoperonic region. Mol Gen Genet. 1989 Aug;218(2):199–207. doi: 10.1007/BF00331269. [DOI] [PubMed] [Google Scholar]
- Eisenach K. D., Cave M. D., Bates J. H., Crawford J. T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990 May;161(5):977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- Gilson E., Bachellier S., Perrin S., Perrin D., Grimont P. A., Grimont F., Hofnung M. Palindromic unit highly repetitive DNA sequences exhibit species specificity within Enterobacteriaceae. Res Microbiol. 1990 Nov-Dec;141(9):1103–1116. doi: 10.1016/0923-2508(90)90084-4. [DOI] [PubMed] [Google Scholar]
- Gilson E., Clément J. M., Brutlag D., Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984 Jun;3(6):1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Perrin D., Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990 Jul 11;18(13):3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Rousset J. P., Clément J. M., Hofnung M. A subfamily of E. coli palindromic units implicated in transcription termination? Ann Inst Pasteur Microbiol. 1986 Nov-Dec;137B(3):259–270. doi: 10.1016/s0769-2609(86)80116-8. [DOI] [PubMed] [Google Scholar]
- Glare E. M., Paton J. C., Premier R. R., Lawrence A. J., Nisbet I. T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990 Sep;28(9):1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott J. M., Shub D. A., Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986 Oct 10;47(1):81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F., Barnes W. M., Clement J. M., Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982 Aug 19;298(5876):760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Hulton C. S., Higgins C. F., Sharp P. M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991 Apr;5(4):825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye S. Retroelements in bacteria. Trends Biochem Sci. 1991 Jan;16(1):18–21. doi: 10.1016/0968-0004(91)90010-s. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase Y., Iwai S., Inoue H., Miura K., Ohtsuka E. Studies on nucleic acid interactions. I. Stabilities of mini-duplexes (dG2A4XA4G2-dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC) containing deoxyinosine and other mismatched bases. Nucleic Acids Res. 1986 Oct 10;14(19):7727–7736. doi: 10.1093/nar/14.19.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kuhsel M. G., Strickland R., Palmer J. D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990 Dec 14;250(4987):1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon E., Gutman P. D., Yao H. L., Minton K. W. A highly conserved repeated chromosomal sequence in the radioresistant bacterium Deinococcus radiodurans SARK. J Bacteriol. 1991 Mar;173(6):2137–2140. doi: 10.1128/jb.173.6.2137-2140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin C. M., Storm A. L., Chipowsky S., Ryan K. J. Molecular epidemiology of Shigella infections: plasmid profiles, serotype correlation, and restriction endonuclease analysis. J Clin Microbiol. 1991 Jan;29(1):104–108. doi: 10.1128/jcm.29.1.104-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. A., Daniel A. S., Braymer H. D., Murray N. E. Organization and sequence of the hsd genes of Escherichia coli K-12. J Mol Biol. 1987 Nov 20;198(2):159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D., Houmard J., Castets A. M., Tandeau de Marsac N. Highly repetitive DNA sequences in cyanobacterial genomes. J Bacteriol. 1990 May;172(5):2755–2761. doi: 10.1128/jb.172.5.2755-2761.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Heath J. D., Sharma B. R., Weinstock G. M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990 Sep;28(9):2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. L., Ledbetter S. A., Corbo L., Victoria M. F., Ramírez-Solis R., Webster T. D., Ledbetter D. H., Caskey C. T. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesin M., Lupski J. R., Svec P., Godson G. N. Possible new genes as revealed by molecular analysis of a 5-kb Escherichia coli chromosomal region 5' to the rpsU-dnaG-rpoD macromolecular-synthesis operon. Gene. 1987;51(2-3):149–161. doi: 10.1016/0378-1119(87)90303-9. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Noda A., Courtright J. B., Denor P. F., Webb G., Kohara Y., Ishihama A. Rapid identification of specific genes in E. coli by hybridization to membranes containing the ordered set of phage clones. Biotechniques. 1991 Apr;10(4):474, 476-7. [PubMed] [Google Scholar]
- Owen R. J. Chromosomal DNA fingerprinting--a new method of species and strain identification applicable to microbial pathogens. J Med Microbiol. 1989 Oct;30(2):89–99. doi: 10.1099/00222615-30-2-89. [DOI] [PubMed] [Google Scholar]
- Rikke B. A., Garvin L. D., Hardies S. C. Systematic identification of LINE-1 repetitive DNA sequence differences having species specificity between Mus spretus and Mus domesticus. J Mol Biol. 1991 Jun 20;219(4):635–643. doi: 10.1016/0022-2836(91)90660-x. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Sharples G. J., Lloyd R. G. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990 Nov 25;18(22):6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Stringer S. L., Hong S. T., Giuntoli D., Stringer J. R. Repeated DNA in Pneumocystis carinii. J Clin Microbiol. 1991 Jun;29(6):1194–1201. doi: 10.1128/jcm.29.6.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Higashitani A., Takanami M., Akiyama K., Kohara Y., Nishimura Y., Nishimura A., Yasuda S., Hirota Y. Construction of an ordered cosmid collection of the Escherichia coli K-12 W3110 chromosome. J Bacteriol. 1989 Feb;171(2):1214–1218. doi: 10.1128/jb.171.2.1214-1218.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuven F. The trouble with PCR machines: fill up the empty spaces! Trends Genet. 1991 May;7(5):142–142. [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., McCabe E. R., Lupski J. R. Use of the polymerase chain reaction for physical mapping of Escherichia coli genes. J Bacteriol. 1991 Sep;173(17):5253–5255. doi: 10.1128/jb.173.17.5253-5255.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991 Feb 25;19(4):861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Q., Kathe S. D., Goodrich-Blair H., Nierzwicki-Bauer S. A., Shub D. A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990 Dec 14;250(4987):1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981 Dec 21;9(24):6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]







