Abstract
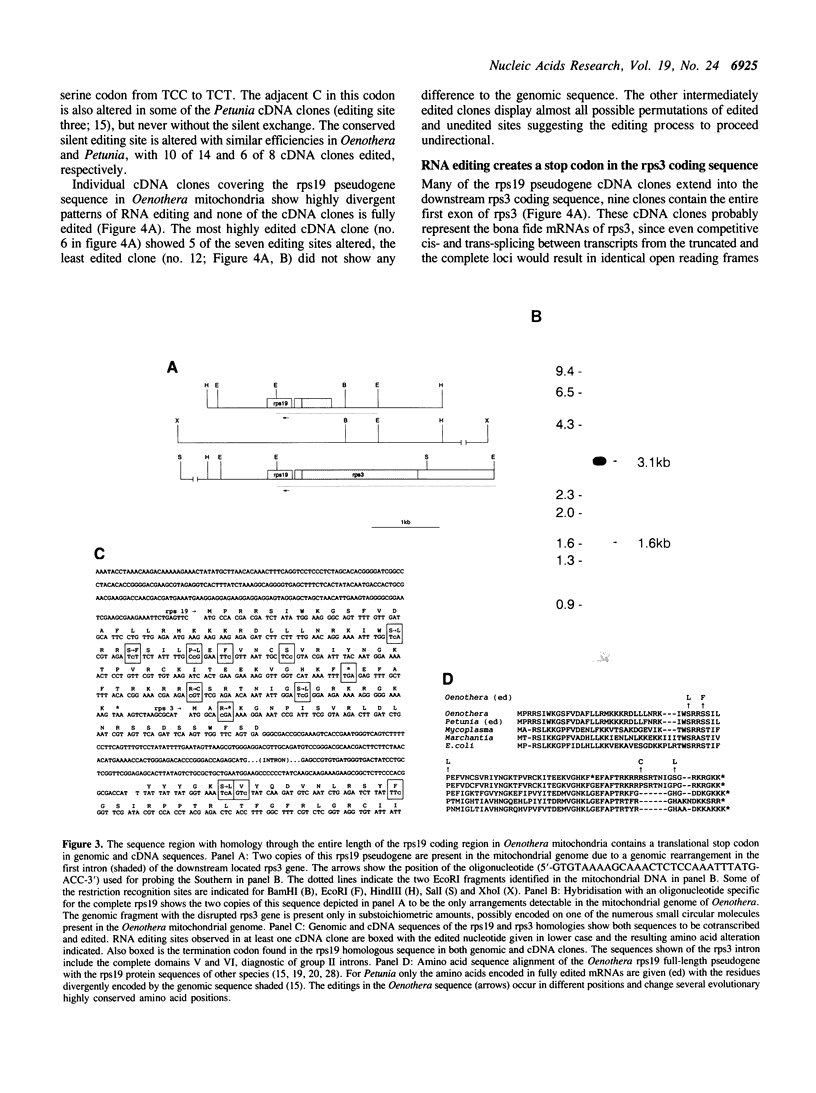
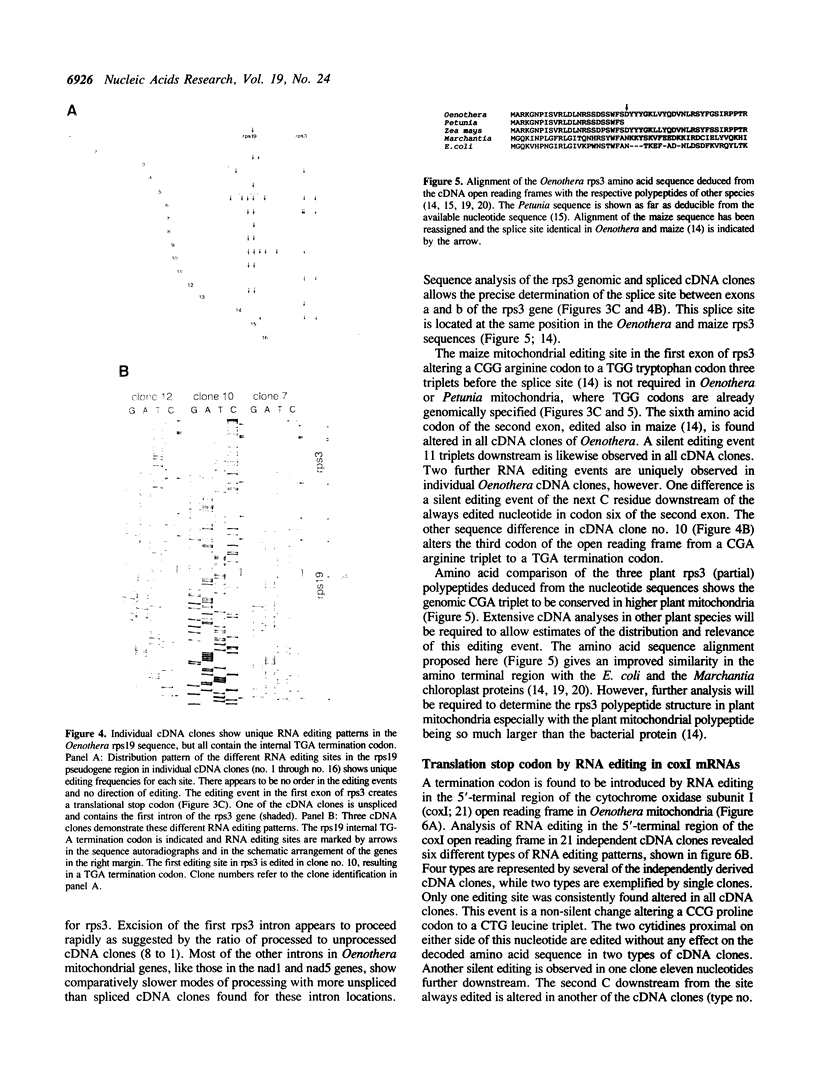
An intact gene for the ribosomal protein S19 (rps19) is absent from Oenothera mitochondria. The conserved rps19 reading frame found in the mitochondrial genome is interrupted by a termination codon. This rps19 pseudogene is cotranscribed with the downstream rps3 gene and is edited on both sides of the translational stop. Editing, however, changes the amino acid sequence at positions that were well conserved before editing. Other strange editings create translational stops in open reading frames coding for functional proteins. In coxI and rps3 mRNAs CGA codons are edited to UGA stop codons only five and three codons, respectively, downstream to the initiation codon. These aberrant editings in essential open reading frames and in the rps19 pseudogene appear to have been shifted to these positions from other editing sites. These observations suggest a requirement for a continuous evolutionary constraint on the editing specificities in plant mitochondria.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bégu D., Graves P. V., Domec C., Arselin G., Litvak S., Araya A. RNA editing of wheat mitochondrial ATP synthase subunit 9: direct protein and cDNA sequencing. Plant Cell. 1990 Dec;2(12):1283–1290. doi: 10.1105/tpc.2.12.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapdelaine Y., Bonen L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a trans-splicing model for this gene-in-pieces. Cell. 1991 May 3;65(3):465–472. doi: 10.1016/0092-8674(91)90464-a. [DOI] [PubMed] [Google Scholar]
- Conklin P. L., Hanson M. R. Ribosomal protein S19 is encoded by the mitochondrial genome in Petunia hybrida. Nucleic Acids Res. 1991 May 25;19(10):2701–2705. doi: 10.1093/nar/19.10.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res. 1990 Sep 11;18(17):5189–5196. doi: 10.1093/nar/18.17.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Weil J. H., Grienenberger J. M. Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res. 1990 Jul 11;18(13):3771–3776. doi: 10.1093/nar/18.13.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Wintz H., Weil J. H., Grienenberger J. M. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988 Dec;215(1):118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Schobel W., Schuster W., Brennicke A. The cytochrome oxidase subunit I and subunit III genes in Oenothera mitochondria are transcribed from identical promoter sequences. EMBO J. 1987 Jan;6(1):29–34. doi: 10.1002/j.1460-2075.1987.tb04714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Brennicke A. Cytochrome oxidase subunit II mRNAs in Oenothera mitochondria are edited at 24 sites. Curr Genet. 1990 Nov;18(4):371–375. doi: 10.1007/BF00318219. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Hodges P. E., Navaratnam N., Greeve J. C., Scott J. Site-specific creation of uridine from cytidine in apolipoprotein B mRNA editing. Nucleic Acids Res. 1991 Mar 25;19(6):1197–1201. doi: 10.1093/nar/19.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M. D., Newton K. J. The NCS3 mutation: genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 1991 May;10(5):1045–1052. doi: 10.1002/j.1460-2075.1991.tb08043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki K., Suzuki T., Kazama S. A chimeric gene containing the 5' portion of atp6 is associated with cytoplasmic male-sterility of rice. Mol Gen Genet. 1990 Oct;224(1):10–16. doi: 10.1007/BF00259445. [DOI] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Schuster W., Brennicke A. RNA editing of ATPase subunit 9 transcripts in Oenothera mitochondria. FEBS Lett. 1990 Jul 30;268(1):252–256. doi: 10.1016/0014-5793(90)81021-f. [DOI] [PubMed] [Google Scholar]
- Schuster W., Ternes R., Knoop V., Hiesel R., Wissinger B., Brennicke A. Distribution of RNA editing sites in Oenothera mitochondrial mRNAs and rRNAs. Curr Genet. 1991 Nov;20(5):397–404. doi: 10.1007/BF00317068. [DOI] [PubMed] [Google Scholar]
- Schuster W., Unseld M., Wissinger B., Brennicke A. Ribosomal protein S14 transcripts are edited in Oenothera mitochondria. Nucleic Acids Res. 1990 Jan 25;18(2):229–233. doi: 10.1093/nar/18.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Wissinger B., Unseld M., Brennicke A. Transcripts of the NADH-dehydrogenase subunit 3 gene are differentially edited in Oenothera mitochondria. EMBO J. 1990 Jan;9(1):263–269. doi: 10.1002/j.1460-2075.1990.tb08104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I., Suffolk R., Leaver C. J. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell. 1989 Jul 14;58(1):69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- Wissinger B., Schuster W., Brennicke A. Species-specific RNA editing patterns in the mitochondrial rps13 transcripts of Oenothera and Daucus. Mol Gen Genet. 1990 Dec;224(3):389–395. doi: 10.1007/BF00262433. [DOI] [PubMed] [Google Scholar]
- Wissinger B., Schuster W., Brennicke A. Trans splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell. 1991 May 3;65(3):473–482. doi: 10.1016/0092-8674(91)90465-b. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Zurawski S. M. Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1985 Jun 25;13(12):4521–4526. doi: 10.1093/nar/13.12.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]