Abstract
Recently a novel cell division system comprised of homologues of eukaryotic ESCRT-III (endosomal sorting complex required for transport III) proteins was discovered in the hyperthermophilic crenarchaeote Sulfolobus acidocaldarius. On the basis of this discovery, we undertook a comparative genomic analysis of the machineries for cell division and vesicle formation in Archaea. Archaea possess at least three distinct membrane remodelling systems: the FtsZ-based bacterial-type system, the ESCRT-III-based eukaryote-like system and a putative novel system that uses an archaeal actin-related protein. Many archaeal genomes encode assortments of components from different systems. Evolutionary reconstruction from these findings suggests that the last common ancestor of the extant Archaea possessed a complex membrane remodelling apparatus, different components of which were lost during subsequent evolution of archaeal lineages. By contrast, eukaryotes seem to have inherited all three ancestral systems.
Cell division ensuring equal distribution of the genetic material between daughter cells is a key mechanism in all cellular life forms. However, the processes of division in eukaryotes and bacteria have fundamental mechanistic differences. Cell division in bacteria is coupled to replication, and a septum-associated, ATP-dependent DNA pump, FtsK, is responsible for the final stages of DNA segregation1. By contrast, in eukaryotes replication and division are separated by a gap phase (G2), and chromosomes replicated in the S phase of the cell cycle are subsequently partitioned into the daughter cells by spindle motors that depend on several ATPases and GTPases, including tubulins, actins, kinesins, and dyneins2,3. Despite these differences, sequence and structural analysis of bacterial proteins involved in cell division led to the unexpected discovery that the bacterial proteins FtsA, FtsZ and MreB are highly diverged homologues of the eukaryotic cytoskeleton proteins tubulin (FtsZ) and actin (FtsA and MreB)4–6. FtsZ, a GTPase, is an important protein in bacterial cell division1,7,8 (BOX 1).
Box 1. Bacterial cell division.
Cell division in bacteria is a well-studied process. In the Gram-negative bacterium Escherichia coli, which contains an outer and an inner membrane, the outer membrane constricts concomitantly with the generation of the division septum (see the figure). By contrast, in the Gram-positive bacterium Bacillus subtilis constriction of the membrane precedes formation of the crosswall of peptidoglycan (see the figure). In both bacteria, FtsZ forms the so-called Z ring that drives the formation of the septum, separating the daughter cells. The membrane-tethered actin homologue FtsA, which is present in many bacteria, interacts with FtsZ and facilitates the assembly of the Z ring. MreB, another actin-like protein, is essential for division in rod-shaped bacilli, in which it forms filaments governing cell wall elongation, but not in cocci50,51. Like FtsA, ZapA is broadly conserved throughout the Bacteria and acts as a positive regulator of Z ring assembly. SepF is found in a broad range of bacteria, including the phyla Firmicutes, Actinobacteria and Cyanobacteria, and it seems to contribute to both formation of the Z ring and establishment of the correct septum morphology. EzrA has been detected in only Gram-positive bacteria with low GC content, to date. This membrane-associated protein is a negative regulator of FtsZ and contributes to the dynamics of FtsZ polymerization at the division site7.
FtsZ is essential for bacterial survival, although deletion of ftsZ in B. subtilis results in a wall-less (L) form of the bacterium that is capable of anomalous, budding-like division52. The ftsZ gene is present in most bacteria and many archaea, although there are notable exceptions, such as the bacterial class Mollicutes30. The bacteria of the superphylum Verrucomicrobia–Chlamydiae and the phylum Planctomycetales possess altered division machineries, with no FtsZ in most planctomycetes and most chlamydiae, and accelerated evolution of FtsZ in verrucomicrobia (some of which also possess tubulins of probable eukaryotic origin)53,54.
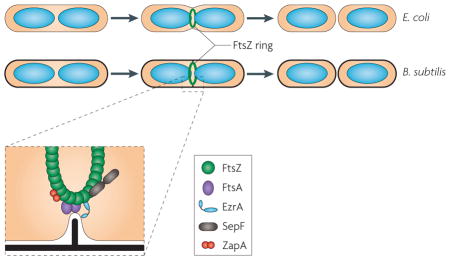
Figure is modified, with permission, from REF. 7 © (2009) Macmillan Publishers Ltd. All rights reserved.
The mechanisms of cell division in Archaea display remarkable diversity and can differ from those in bacteria in many aspects, including DNA replication and membrane organization9,10. Five archaeal phyla have been identified to date: the Crenarchaeota, the Euryarchaeota, the Thaumarchaeota, the Nanoarchaeota and the Korarchaeota, of which the Crenarchaeota and Euryarchaeota are the best characterized. Almost all members of the Euryarchaeota encode FtsZ and, thus, are thought to possess a bacterial-type division mechanism11,12. By contrast, FtsZ is missing in those crenarchaeotes that live at high temperature, which nevertheless undergo division by binary fission. In contrast to bacteria, which segregate their genomes concomitantly with genome replication, the crenarchaeote Sulfolobus solfataricus shows sister chromatid cohesion for a considerable period of the post-replicative stage of the cell cycle, preceding cell division13. In this respect, cell division in the model hyperthermophilic crenarchaeotes of the genus Sulfolobus parallels eukaryotic cytokinesis11,12,14,15. Following the period of cohesion, nucleoids are segregated before the invagination of the cell membrane.
Unexpectedly, it was shown that Sulfolobus acido−caldarius uses an alternative cell division apparatus that is homologous to the eukaryotic ESCRT (endosomal sorting complex required for transport)16,17. The ESCRT apparatus has key roles in diverse manipulations of intra-cellular membranes in eukaryotes, including the formation of multivesicular bodies (MVBs) in particular18–20 (BOX 2).
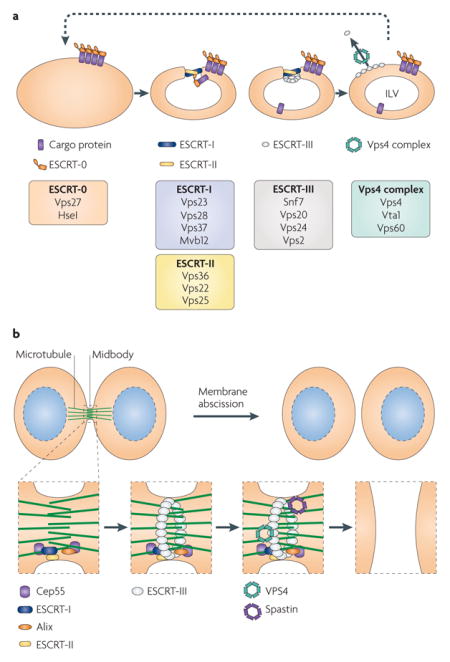
Box 2. The endosomal sorting complex required for export.
The ESCRT (endosomal sorting complex required for transport) machinery is a versatile and modular apparatus that is involved in many membrane manipulation processes. It contains 4 distinct complexes (ESCRT-0 to ESCRT-III) that are comprised of at least 16 protein subunits (see the figure, part a).
In its role in multivesicular body (MVB) formation (see the figure, part a; protein names refer to the Saccharomyces cerevisiae components), ESCRT-0 recognizes ubiquitylated cargo proteins. This leads to the recruitment of ESCRT-I and ESCRT-II, which jointly promote membrane invagination and the formation of ingressed buds, producing a tubular neck that ESCRT-III acts on to drive membrane scission20. Membrane scission by ESCRT-III alone has been reproduced in vitro with a complex reconstituted from three paralogous coiled-coil subunits, vacuolar protein sorting 20 (Vps20), Vps24 and Snf7; a fourth paralogue, Vps2, joins the complex and recruits the Vps4 complex. The catalytic subunit, Vps4, is an ATPase associated with various cellular activities (AAA+ ATPase) that mediates disassembly of ESCRT-III and, thus, facilitates recycling of the subunits, providing multiple rounds of vesicle formation55,56. The concentration of ESCRT-III proteins required for membrane scission is 40-fold lower in the presence of ESCRT-I and ESCRT-II than in the absence of these two complexes. In addition, ESCRT-I and ESCRT-II facilitate the exclusion of ESCRT-III from the intralumenal vesicles (ILVs).
During the final stages of cytokinesis in mammalian cells, the two daughter cells are joined by the midbody, a membranous tube containing a high density of microtubules. The ESCRT machinery plays a role in the resolution of this structure during membrane abscission (see the figure, part b). Cep55, a component of the midbody, recruits ESCRT-I and another cellular protein, Alix (also known as PDCD6IP); ESCRT-II binds to ESCRT-I, and the complex subsequently mediates ESCRT-III recruitment. It is thought that ESCRT-III then acts to directly drive membrane constriction and scission, as suggested by the observations that overexpression of a defective allele of VPS4 leads to aberrant cell division57,58. In addition, the ESCRT-III assembly directs the recruitment of spastin. Spastin is an AAA+ ATPase that is related to VPS4 and is recruited to ESCRT-III through a microtubule interaction and transport (MIT) domain-mediated interaction59. Spastin severs microtubules and, thus, might clear the way for membrane cleavage by ESCRT-III and its associated ESCRT-III-like factors, Chmp1A, Chmp1B and IST1.
Although no archaeal homologues of ESCRT-0, ESCRT-I or ESCRT-II components have been detected, the sequenced genomes of members of the orders Desulfurococcales and Sulfolobales, representing two of the three branches of the hyperthermophilic crenarchaeotes, encode homologues of ESCRT-III coiled-coil subunits (referred to here as archaeal Snf7-like proteins) as well as homologues of the ATPase vacuolar protein sorting 4 (VPS4). The protein encoded by Saci_1373, one of the four Snf7-like proteins of S. acidocaldarius, is present in a ring-like structure at the site of membrane constriction during cell division16,17; cytokinesis is inhibited by a dominant mutant of this VPS4 homologue16. As this species lacks FtsA, MreB and a typical FtsZ, these data indicate that the counterpart of ESCRT-III constitutes the core of the cell division machinery in S. acidocaldarius and, by implication, in other Sulfolobales and in the Desulfurococcales. In addition, the Snf7-like proteins and VPS4 homologues have been detected in membrane vesicles excreted by S. acidocaldarius as well as in egressing viral particles, suggesting that, as in eukaryotes, these archaeal proteins mediate diverse membrane manipulations16,21. The presence of the archaeal Snf7-like and VPS4-like proteins in secreted vesicles and viruses contrasts with the exclusion of these proteins from MVBs in eukaryotes and may be attributable to the absence of archaeal analogues of ESCRT-I and ESCRT-II components (BOX 2).
Thus, Archaea seem to use at least two unrelated membrane remodelling systems, a bacterial-type system based on FtsZ and a eukaryotic-type system based on ESCRT-III. This diversity and the obvious relevance of archaeal membrane remodelling mechanisms for the origin of the respective systems in eukaryotes, underscored by studies of the ESCRT-III homologues in S. acidocaldarius, prompted us to undertake a comparative genomic analysis of the archaeal proteins known to be involved in, or at least implicated in, cell division (for methods, see Supplementary information S1 (box)).
Archaeal ESCRT-III and associated genes
We started with the three proteins that are encoded in the S. acidocaldarius gene cluster containing a VPS4-like gene (FIG. 1) and that have been shown to participate in cytokinesis: the Snf7-like protein encoded by Saci_1373, the VPS4 orthologue encoded by Saci_1372 and a coiled-coil protein that is apparently unrelated to ESCRT-III subunits, encoded by Saci_1374. The phyletic patterns of these genes were initially extracted from the arCOG (archaeal clusters of orthologous groups) database (BOX 3), and additional database searches and genomic-context analyses were carried out in an attempt to identify other homologous or functionally related genes. However, a search for distant members of the Snf7-like protein family is not trivial and requires a combination of various sequence analysis approaches, secondary-structure prediction and genomic-context examination, as these proteins contain coiled-coil structures and can show spurious sequence similarity to many unrelated proteins.
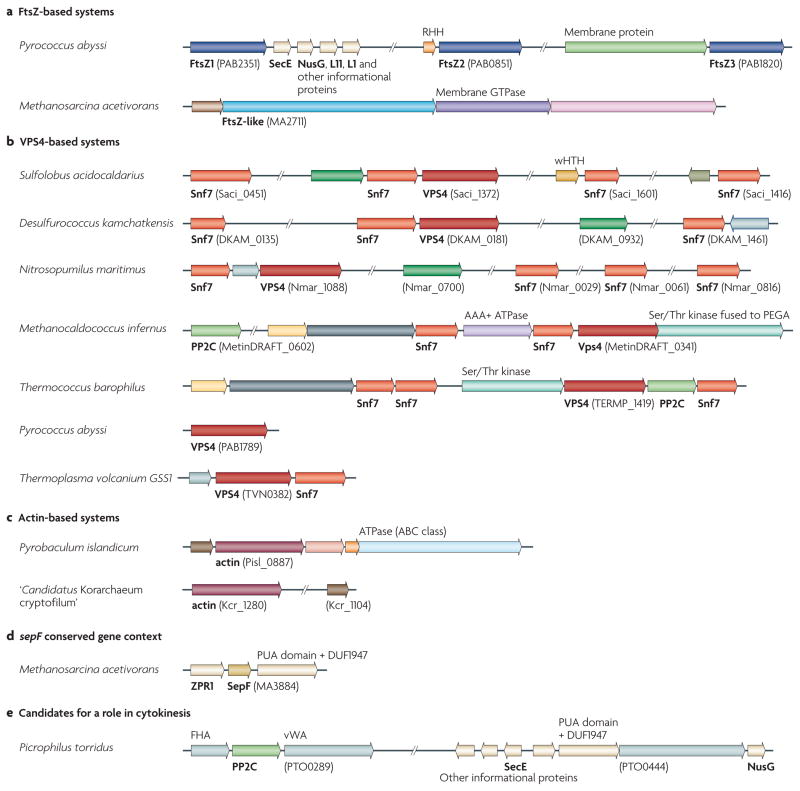
Figure 1. representative gene neighbourhoods of known and predicted membrane remodelling and cell division systems in the Archaea.
Homologous genes are the same colour; genes are shown to scale (with the exception of various informational genes shown in cream). Specific domains predicted to be encoded in the genes are listed above the gene, and predicted proteins (or the names of their eukaryotic homologues) are given below the gene in bold. The locus identifier is indicated for one gene in each putative operon (bracketed text). Representative gene neighbourhoods are shown from selected species. Complete information is available in Supplementary information S4,S5 (figure, table). a | The gene organization of the FtsZ-encoding gene and FtsZ-like protein-encoding genes of two representative organisms with an FtsZ or FtsZ-like division machinery. b | Representative organizations of the genes encoding two components of ESCRT (endosomal sorting complex required for transport), vacuolar protein sorting 4 (VPS4) and Snf7. c | An overview of the genetic organization of the genes encoding actin-like proteins. d | The genetic neighbourhood of the gene encoding the SepF homologue in Methanosarcina acetivorans. e | The genetic neighbourhood of the genes encoding proteins that are potentially involved in cytokinesis in Picrophilus torridus, which lacks the FtsZ, ESCRT and actin-based systems. AAA+ ATPase, ATPase associated with various cellular activities; FHA, forkhead-associated domain; l1, ribososmal protein l1; l11, ribosomal protein l11; PP2C, protein phosphatase 2C; RHH, ribbon–helix–helix domain (a predicted transcriptional regulator); vWA, von Willebrand factor type A domain; wHTH, winged helix–turn–helix domain; ZPR1, zinc finger protein 1.
Box 3. Archaeal clusters of orthologous genes.
Comparative genomic analyses and reconstruction of the gene sets of ancestral forms rely on robust identification of orthologous genes in extant genomes. Orthologues are genes that evolved from a single ancestral gene in the last common ancestor of the compared genomes, in contrast to paralogues, which are genes that are related by duplication60. The identification of orthologues in distantly related organisms may be a difficult task, because the complex processes of gene duplication, lineage-specific gene loss and horizontal gene transfer confound orthologous relationships. Originally, clusters of orthologous groups (COGs) were derived for all of the proteins encoded by the genomes of bacteria, archaea and unicellular eukaryotes that were available at the time, and they became a popular platform for functional and evolutionary genomics61,62. With the accumulation of hundreds of genome sequences, it became clear that more reliable, better resolved clusters of orthologues could be obtained by comparisons of limited subsets of genomes. Accordingly, archaeal COGs (arCOGs) were constructed by comparison of 41 archaeal genomes44 and were subsequently updated to include several new genomes. The gene and protein classification and the ancestral genome reconstruction described here uses the 2009 update of the arCOGs.
Sulfolobus spp. encode a total of four Snf7-like proteins. The protein encoded by Saci_1373, mentioned above, belongs to arCOG00453 and is represented by a single orthologue in all sequenced genomes of the Desulfurococcales and Sulfolobales (see Supplementary information S2 (table)). This protein contains a characteristic carboxy-terminal helix–turn–helix (HTH) domain and is encoded in the same operon as the VPS4 homologue in most archaeal genomes17. The Snf7-like proteins encoded by Saci_0451 and Saci_1416 belong to the related arCOG00452. Interestingly, this arCOG also includes seven proteins from two members of the Thaumarchaeota (three from Cenarchaeum symbiosum A and four from Nitrosopumilus maritimus SCM1). The fourth Snf7-like protein from S. acidocaldarius, encoded by Saci_1601, belongs to arCOG00454, which is represented only in the Sulfolobales.
The other two genes in the Saci_1373 gene cluster also show interesting distributions. The phyletic pattern of the coiled-coil-containing protein encoded by Saci_1374 (arCOG04054) mirrors the pattern of arCOG00452 (see Supplementary information S2 (table)). The phyletic pattern of the VPS4 homologue encoded by Saci_1372 unexpectedly reveals the presence of orthologues not only in the Crenarchaeota and the Thaumarchaeota but also in some Euryarchaeota — namely, in the two available genomes from the order Thermoplasmatales and three of the five available genomes from the order Thermococcales (see Supplementary information S2 (table)). Thermococcus gammatolerans and Thermococcus sp. AM4 each encode two VPS4 homologues. The VPS4 homologues in the Thermococcales had not been identified in previous analyses14,15.
The microtubule interaction and transport domain
A diagnostic, functionally critical feature of the VPS4 family proteins is the presence of the amino-terminal microtubule interaction and transport (MIT) domain, a versatile protein–protein interaction module that consists of three antiparallel α-helices and interacts with an MIT-interacting motif (MIM) found in Snf7-like partner proteins16,22,23. As the MIT domain sequence is poorly conserved, identification of this domain requires careful sequence analysis. In particular, we were interested in determining whether the MIT domain is present in the VPS4-like proteins from the Thermoplasmatales and Thermococcales. First, we searched for all archaeal VPS4 homologues containing an N-terminal extension. In addition to the VPS4 homologues that are included in the arCOGs, this search identified VPS4-like ATPases encoded by several recently sequenced species, including three members of the genus Thermococcus (T. gammatolerans, Thermococcus barophilus and Thermococcus sp. AM4) as well as Methanocaldococcus infernus, Methanocaldococcus vulcanius, Ferroplasma acidarmanus fer1 and Halomicrobium mukohataei. We constructed a multiple alignment of the N-terminal portions of these proteins and used the aligned regions of these sequences as queries for the HHpred program. The HHpred search detected statistically significant similarity with the MIT domain for the sequences from the Thermoplasmatales but not for those from other Euryarchaeota. Nevertheless, secondary structure prediction revealed three α-helices characteristic of the MIT domain, as well as a conserved GXXXXA motif (where X is any amino acid) at the beginning of the second α-helix and several conserved hydrophobic positions (see Supplementary information S3 (figure)). These features, together with the N-terminal localization of the domain in the VPS4-like ATPase, suggest that these regions are bona fide MIT domains. Accordingly, we predict that all archaeal VPS4-like ATPases contain N-terminal domains that are homologous to the MIT domain and so are expected to interact with ESCRT-III-like proteins.
Genomic neighbourhoods of VPS4-like genes
We next compared the genomic neighbourhoods of all identified archaeal genes encoding VPS4-like ATPases in an attempt to predict additional functional links using the ‘guilt-by-association’ approach. In most genomes of the Desulfurococcales and Sulfolobales, the VPS4 homologue is encoded in the three-gene cluster described above14 (see Supplementary information S4 (figure) and Supplementary information S5 (table)). Although the three-gene cluster was initially thought to be an operon, it has been shown recently that the first gene in the cluster, SSO0911 (orthologue of Saci_1374), is transcribed separately from the Snf7-like and VPS4-like genes in S. solfataricus24. The remaining three Snf7-like paralogues are located elsewhere in the genome and are differentially expressed15. In Aeropyrum pernix, Desulfurococcus kamchatkensis and the Thaumarchaeota, the cluster containing the VPS4-like gene is disrupted and the Saci_1374 paralogues are at different locations (see Supplementary information S4,S5 (figure, table)). A detailed analysis of the genomic neighbourhoods revealed several potential lineage-specific components of the ESCRT system (see Supplementary information S4,S5 (figure, table) and Supplementary information S6 (table)). Among these proteins, the predicted winged HTH (wHTH) domain-containing transcriptional regulator (in arCOG00731) that is associated with SSO0619-like genes is of particular interest, because it is possibly involved in differential regulation of the ESCRT-III-related genes.
In Thermoplasma volcanium, the VPS4-like gene (TVN0382) belongs to a predicted three-gene operon (containing TVN0381, TVN0382 and TVN0383). Using HHpred, a Conserved Domain Database search, PSI-BLAST (position-specific iterative basic local alignment search tool), secondary-structure prediction and multiple alignment (see Supplementary information S1 (box) and Supplementary information S7 (figure) for details), we showed that TVN0383 and its orthologue from T. acidophilum (Ta1181) encode proteins belonging to the Snf7 family, although they are far diverged from the homologues encoded by the Desulfurococcales and Sulfolobales. A PSI-BLAST search of the archaeal protein sequences using the protein encoded by TVN0383 as a query identified highly similar homologues of this protein in the genomes of three thermococci. Given that these euryarchaeal proteins are encoded in the vicinity of VPS4-like genes (FIG. 1) and contain confidently predicted coiled-coil regions, we conclude that they are previously unrecognized representatives of the Snf7 family related to arCOG07402 (see Supplementary information S7 (figure)). The orthologues of the protein encoded by TVN0381 are encoded in all three of the available genomes from the Thermoplasmatales but do not show any similarity with other proteins.
Other genes associated with VPS4-like genes
Further analysis of the genomic contexts of VPS4-like genes and arCOG07402 revealed an additional suite of genes that shows considerable diversity across genomes but has well-defined components (see Supplementary information S6 (table)). This group of ESCRT-III-associated genes is exemplified by the predicted eight-gene operon in T. barophilus (FIG. 1). Each gene from this operon is present in similar genomic neighbourhoods in three thermococci, two methanocaldococci and H. mukohataei (see Supplementary information S4,S5 (figure, table)). Our analysis using exhaustive PSI-BLAST and HHpred searches led to the identification of homologous relationships between some of these genes. In particular, we detected a link between the Snf7 family genes and those encoding the entries in arCOG09747 and arCOG09749 (see Supplementary information S7 (figure)). Thus, most archaeal genomes possessing VPS4 homologues also encode paralogous Snf7-like proteins, as originally observed in the Desulfurococcales and Sulfolobales (FIGS 1,2; see Supplementary information S4,S5 (figure, table)). Two proteins belonging to arCOG08177 and arCOG09748 (see Supplementary information S6 (table)) are not similar to any known proteins and, so far, are unique to the VPS4-like gene neighbourhoods.
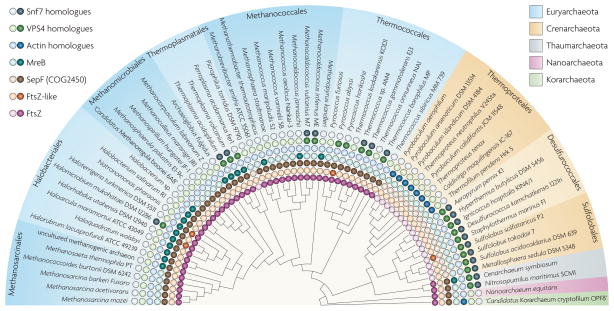
Figure 2. The distribution of the key components of membrane manipulation systems among the Archaea.
The phyletic patterns for indicated proteins are represented by filled circles to show the presence of the proteins and empty circles to show their absence. This is arranged according to the archaeal phylogenetic tree, the topology of which is a consensus based on several recent analyses63–65. Species are coloured by phyla and then subdivided into orders. VPS4, vacuolar protein sorting 4.
The eight-gene cluster also encodes a serine/threonine protein kinase (arCOG03683) and a protein phosphatase 2C (arCOG05302), which probably constitute a distinct regulatory system. Notably, these proteins are present in a limited subset of archaeal lineages (the classes Thermococci, Thermoplasmata, Methanococci and Halobacteria, and the orders Desulfurococcales and Sulfolobales) that is almost identical to the subset of lineages in which at least one genome encodes a VPS4 homologue (see Supplementary information S2 (table)). Taken together with the operon organization that is partially conserved in the Thermococci, the Methanocaldococci and H. mukohataei, this pattern strongly suggests that the kinase and phosphatase (or, alternatively, the FHA domain-containing proteins that are encoded in a predicted operon with the kinase in several other archaea) regulate the activity of the archaeal ESCRT-III system. Archaeal protein kinases remain mostly uncharacterized, but it is notable that the apparent orthologues of the ESCRT-associated serine/threonine protein kinases in Mycobacterium tuberculosis (PknA and PknB) are involved in the regulation of cell division25–27. In particular, PknA phosphorylates FtsZ. Moreover, actinobacteria possess conserved predicted operons that contain both kinase genes and genes involved in cell division, such as FtsW. Interestingly, the ESCRT-associated kinases in different archaea are fused to different domains — for example, to β-propeller and HEAT repeat domains in H. mukohataei — and to the so-called PEGA domains that are typically associated with S-layer proteins28 in members of the class Thermococci, and these kinases are in some cases encoded within the VPS4-like gene neighbourhoods (see Supplementary information S4,S5 (figure, table)). Finally, genes that are predicted to encode AAA+ ATPases (in arCOG03166, arCOG03167 and arCOG03169) are also present in the VPS4-like gene neighbourhoods in members of the Thermococci. These proteins might have additional regulatory functions, given that some of them are fused to wHTH domains similar to those found in transcription regulators.
Examples of isolated VPS4-like genes
In Pyrococcus horikoshii, Pyrococcus abyssi and Thermococcus kodaka−raensis, VPS4-like genes do not belong to operons. Furthermore, we failed to identify members of the Snf7 family or most of the other typical components of VPS4-like gene operons (see above) in these genomes. However, in Pyrococcus furiosus we detected an apparent remnant of this operon that includes a gene encoding a protein which belongs to arCOG08177, a kinase-encoding gene and an ATPase-encoding gene (arCOG03167) but not VPS4-like or Snf7-like genes (see Supplementary information S4,S5 (figure, table)). These observations suggest that most members of the Thermococcales encode the ancestral version of the ESCRT-III system, whereas Pyrococcus spp. and a few Thermococcus spp. have lost it completely or partially. The monocistronic VPS4-like proteins encoded in some Pyrococcus spp. are predicted to be active ATPases, given the conservation of all characteristic motifs in the AAA+ domain (data not shown). Even more conspicuously, these proteins contain the MIT domain, suggesting that they interact with Snf7-like proteins despite our inability to detect these proteins in Pyrococcus spp. However, it is also conceivable that the MIT domain interacts with non-Snf7-like, MIM-containing partner proteins. Given that the Snf7 family proteins are small (150–250 amino acids) and form coiled-coil structures, we identified all proteins with such characteristics in the pyrococcal genomes and analysed their phyletic patterns and gene neighbourhoods. This search yielded several proteins with compatible characteristics, including a small operon that encodes two coiled-coil proteins (arCOG05733 and arCOG05734) and is reminiscent of the arCOG09749–arCOG09747 Snf7-like pair, and a coiled-coil protein (arCOG05820) encoded in the same operon as a V4R domain-containing ATPase (data not shown), but no solid candidates were revealed. Recently, however, it has been shown that the N-terminal MIT domain of the VPS4-like ATPase katanin interacts with tubulin29, suggesting an interaction between this domain and FtsZ in Pyrococcus spp.
New divergent FtsZ homologues in the Archaea
FtsZ proteins are encoded in all sequenced euryarchaeal genomes, with the sole exception of Picrophilus torridus DSM 9790, as well as in the genomes of the Thaumarchaeota, in Nanoarchaeum equitans and in the one available korarchaeal genome (FIG. 2). Most euryarchaeotes encode multiple paralogues of FtsZ; phylogenetic analysis revealed three branches that are thought to have evolved from three FtsZ paralogues in the last common ancestor of the Euryarchaeota30. In most archaea, the genes from the three branches (ftsZ1, ftsZ2 and ftsZ3) are located in three distinct conserved neighbourhoods; ftsZ1 is located near highly conserved informational genes and can be predicted to function as the key cell division protein (FIG. 1). The FtsZ–tubulin superfamily is characterized by a diagnostic motif in loop T4 — GGGTG(S/T)G — that is involved in GTP binding. We used this motif to search the protein sequences from the arCOG database and, along with the known FtsZ family proteins, we identified a diverged family that includes two membrane-associated proteins (encoded by SSO1376 and SSO1378) from the closely related S. solfataricus strains P2 and 98/2. The genes encoding these proteins are located in a region of the chromosome that has a high density of insertion sequences, and they are disrupted by transposon insertions. Combined with the absence of these genes from the genomes of other Sulfolobales, including the closely related Sulfolobus islandicus, this observation suggests that the genes for membrane-associated, FtsZ-like proteins were acquired in a recent horizontal gene transfer event. To our knowledge, this is the first finding of FtsZ–tubulin superfamily proteins in the Crenarchaeota. In addition, this diverged family of FtsZ-like proteins includes members from several of the Euryarchaeota and from numerous bacteria31. These FtsZ-like proteins contain a long C-terminal extension with a predicted coiled-coil structure. In most archaeal genomes, proteins of this novel FtsZ-like family are encoded in a predicted four-gene operon that also encodes a diverged membrane-associated GTPase (MA2712, in arCOG06794), as well as a large α/β-protein (MA2713, in arCOG07816) and a smaller protein belonging to arCOG07815 (MA2710) that both contain predicted membrane-spanning segments. Interestingly, the corresponding operon described in Halorhabdus utahensis is next to the gene encoding a kinase in arCOG03683 (see above). The genomic context of the genes that encode these archaeal FtsZ-like proteins is unrelated to the context of their bacterial homologues, suggesting that this archaeal operon is an ancient feature rather than a result of recent horizontal gene transfer from bacteria. Therefore, it seems likely that this operon encodes a distinct membrane remodelling system.
SepF orthologues in FtsZ-containing archaea
Although most archaea use the FtsZ-based mechanism of cytokinesis, few archaeal homologues of the bacterial functional partners of FtsZ have been detected. In particular, the actin-like ATPase FtsA, which is essential in bacteria, is present in only Methanopyrus kandleri, whereas another actin-like ATPase, MreB, which determines cell shape in rod-shaped bacteria, is found in only a subset of the archaea that encode FtsZ (FIG. 2). In an attempt to predict potential partners of FtsZ, we searched for arCOGs with the same phyletic pattern as the arCOG containing the FtsZ homologues. Only two such arCOGs were found: arCOG00872, containing an ERCC4-like helicase, and arCOG02263, containing an uncharacterized, conserved protein. The PSI-BLAST and HHpred searches using the Archaeoglobus fulgidus protein from arCOG02263, encoded by AF0782, showed a highly significant similarity (for example, an E value of 10−25 for HHpred) between arCOG02263 and COG1799, which includes the recently characterized bacterial SepF proteins. Thus, arCOG02263 seems to consist of the archaeal orthologues of SepF, which has been shown to interact with FtsZ and has an overlapping function with FtsA32,33. Given that, with a single exception, the available archaeal genomes do not encode FtsA, and taking into account the identical phyletic patterns of arCOG02263 and archaeal FtsZ, it seems highly likely that the archaeal SepF functionally substitutes for FtsA and is essential for FtsZ -based cytokinesis in archaea.
Other potential membrane remodelling systems
The discovery of the archaeal ESCRT-III system revealed the existence of two complementary cell division machineries in the Archaea: the bacterial-like, FtsZ-centred apparatus in the Euryarchaeota and the ESCRT-III-based apparatus in the hyperthermophilic crenarchaeotes. Some archaeal groups, such as the Thaumarchaeota and, among the Euryarchaeota, members of the orders Thermococcales and Thermoplasmatales, encode components of both systems (FIG. 2), generating the intriguing possibility that they may cooperate. However, a glaring gap remains in our knowledge of the cell division systems in archaea: the Thermoproteales, a distinct branch of the hyperthermophilic crenarchaeotes, encode neither the FtsZ-based nor the ESCRT-III-like system (FIG. 2). Members of the Thermoproteales have been reported to undergo a peculiar version of rapid, ‘snapping’ division that is distinct from the gradual membrane invagination seen in other archaea, suggesting that these organisms possess a separate, uncharacterized cell division machinery34,35. With several diverse genomes of the Thermoproteales now available, we identified the arCOGs with phyletic patterns that are specific to this group and exclude other Crenarchaeota. Most conspicuously, members of the Thermoproteales possess a conserved operon containing a gene encoding an actin-like protein (for example, Pisl_0887 from Pyrobaculum islan−dicum) that is also found in ‘Candidatus Korarchaeum cryptofilum’ (FIG. 1) and is unambiguously monophyletic with eukaryotic actins and actin-related proteins (ARPs)36. Actin and ARPs are key components of the eukaryotic cytoskeleton and are essential for, among other functions, mitotic spindle activity and cytokinesis37. The other three genes in the operon that encodes the actin-like protein in Thermoproteales are PAE2275 (the protein encoded by which is in arCOG05582), a gene encoding a globular protein that is also present in ‘Candidatus Korarchaeum cryptophilum’ and is encoded by all available genomes of the Thaumarchaeota, and two genes encoding additional uncharacterized proteins (belonging to arCOG05584 and arCOG07432) that are unique, so far, to the genus Pyrobaculum. Given the complementarity of the phyletic patterns (FIG. 2) and the analogy with the eukaryotic actin family, we predict that the actin-like protein in the Thermoproteales is a key component of a novel membrane remodelling system that is responsible for ‘snapping’ division. The only other protein family that is specific to the Thermoproteales to the exclusion of the other Crenarchaeota and that might be involved in cell division is a distinct ParA–Soj family ATPase (in arCOG00599 and encoded by PAE1775) that is known to be involved in chromosome and plasmid partitioning in bacteria38.
Additional mechanisms of division
Recent experimental breakthroughs (in particular, the discovery of the role of archaeal ESCRT-III homologues in S. acidocaldarius cell division), together with comparative genomic analyses, show that different lineages of the Archaea use at least three distinct systems for cell division (FIGS 2,3). The first system, used by the Euryarchaeota, is similar to that used by the Bacteria and is based on FtsZ (of the tubulin family) and SepF, which probably substitutes for FtsA. The archaeal orthologue of MreB is present in only a subset of organisms that possess FtsZ (FIG. 2), suggesting that, similarly to bacterial cocci, some archaea do not require the MreB function, whereas others might relegate this function to a different protein. The second system, used by two branches of the Crenarchaeota (the Sulfolobales and the Desulfurococcales), is the ESCRT-III (VPS4–Snf7) system that is responsible for vesicle formation and is also involved in cytokinesis in eukaryotes. The third system, found in the crenarchaeal branch Thermoproteales, remains enigmatic, as these organisms lack both FtsZ and ESCRT-III. However, the presence of a protein similar to eukaryotic ARPs that is conserved in Thermoproteales but otherwise seen in only ‘Candidatus Korarchaeum cryptophilum’ (FIGS 1,2) suggests that the third system is actin based.
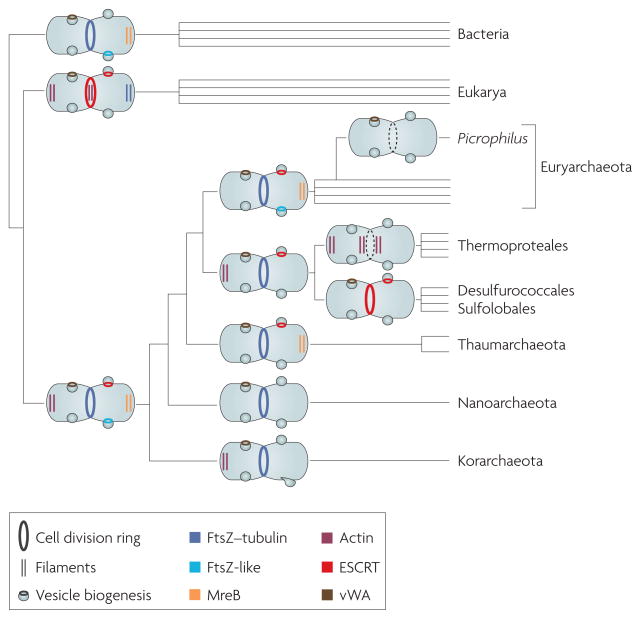
Figure 3. A hypothetical evolutionary scenario for archaeal and eukaryotic membrane remodelling systems.
Using the scenario of evolution of Eukarya from a deep archaeal branch, the last universal common ancestor of the Archaea and the Eukarya probably possessed a full complement of vesicle biogenesis and division proteins (including the ESCRT (endosomal sorting complex required for transport) system, a member of the FtsZ–tubulin family, an FtsZ-like protein and MreB, as well as actin or an actin-like protein and proteins with homology to von Willebrand factor A (vWA)). These were probably dedicated to specific membrane invagination functions within the cell. During the evolution of the Archaea, some of these functions were lost, whereas certain systems acquired new functions. In addition, a new system of cell division may have arisen in Picrophilus torridus and Thermoproteales lineages (dotted cell division rings).
The remarkable absence of any of the identified components of these three systems in P. torridus suggests that there may be additional cell division mechanisms in archaea. It is interesting to note that P. torridus encodes two kinases and a protein phosphatase 2C related to those found in the eight-gene operon described above, and the gene encoding the phosphatase, PTO1092, is colocalized with a gene that encodes a von Willebrand factor type A (vWA) domain-containing protein, which is often encoded in the same operon as FtsZ in bacteria31. In eukaryotes, vWA proteins are commonly involved in basal membrane formation functions39,40. Another uncharacterized protein that is currently unique to the class Thermoplasmata (for example, the protein encoded by PTO0444 of P. torridus) is encoded in a genomic context that is almost identical to the context of FtsZ1 in other archaea and is also likely to have a major role in cell division in this archaeal lineage (FIG. 1). Finally, a role in cell division seems likely for the only ParA–Soj family ATPase of P. torridus (encoded by PTO1106).
Evolution of cell division systems
Diverged functions of the membrane remodelling systems
The comparative genomic analysis described here substantially extends previous observations14,17 that several groups of archaea encode assortments of cell division-related proteins: some members of the Thermococci and Thermoplasmata and both available genomes of the Thaumarchaeota encode both ESCRT-III subunits and FtsZ, whereas ‘Candidatus Korarchaeum cryptophilum’ encodes components of the FtsZ-based division system (with multiple FtsZ paralogues) and the putative actin-based system (FIG. 2). Snf7-like proteins were identified in only a subset of the archaeal genomes that encode VPS4 homologues. However, given the difficulties of detecting diverged homologues of these coiled-coil proteins and the presence of the MIT domain in all archaeal VPS4-like proteins, it seems likely that VPS4 homologues are reliable markers of the ESCRT-III-like system. The functional relationships between the proteins that constitute alternative cell division machineries in other archaea are interesting targets for further experimental studies. For organisms in which both machineries are present, the simplest hypothesis is that the FtsZ-based system is responsible for cytokinesis, whereas the ESCRT-III system is involved in vesicle biogenesis. Indeed, the formation of exosome-like vesicles during the stress response and during virus egress from infected cells has been reported in the Sulfolobales41,42 as well as in some members of the Thermococci43. The similar topologies of the membrane manipulations involved in cell division and vesicle formation, and the involvement of the eukaryotic ESCRT-III in vesicle formation and cytokinesis, are compatible with this possibility. However, the possibility also remains that the FtsZ-based system and the archaeal ESCRT-III cooperate during cell division in those archaea that possess both systems.
Implications for membrane remodelling systems in the last common ancestor of the Archaea
We cannot formally rule out the possibility that horizontal gene transfers have sculpted the repertoire of cell division proteins in the Archaea. However, considering the phyletic patterns of the analysed components of the membrane remodelling machineries, the most parsimonious explanation is that the last archaeal common ancestor (LACA) possessed a complex division and vesicle biogenesis system that included both FtsZ and ESCRT-III. Indeed, maximum likelihood evolutionary reconstruction gives a probability of >0.99 for the presence of an FtsZ homologue in the LACA and 0.84 for the presence of a VPS4 homologue. According to this reconstruction, the subsequent evolution of the Archaea involved primarily differential, lineage-specific loss of the ancestral components, such as the loss of FtsZ in the Desulfurococcales and independent losses of ESCRT-III in several euryarchaeal lineages, along with some horizontal gene transfers (FIG. 3). The evolution of the archaeal actin-like proteins is less clear. From the phyletic pattern in the extant archaea (FIG. 2), the probability for the presence of MreB in the LACA is <0.01, whereas the probability for the presence of an actin-like protein is ~0.2. However, assuming that the archaeal actin-like proteins of the Thermoproteales and ‘Candidatus Korachaeum cryptophilum’ are orthologues of MreB and, accordingly, considering these proteins jointly, the ancestral actin family protein can be tentatively mapped to the LACA with a probability of ~0.5. Moreover, given the presence of actin-like proteins in two diverged lineages (the Thermoproteales and the Korarchaeota), it cannot be ruled out that the duplication that led to the emergence of the actin-like family was already present in the LACA (FIG. 3). Circumstantial evidence compatible with this possibility comes from the presence, in the Thaumarchaeota, of orthologues of the uncharacterized genes that are encoded near the genes for actin-like proteins in the Thermoproteales (arCOG05582; see Supplementary information S5,S6 (tables)). Thus, the tentative reconstruction of the evolution of archaeal membrane remodelling machinery implies a complex state in the LACA, with subsequent differential reductive evolution resulting in distinct, lineage-specific solutions for the same functional task.
These conclusions are compatible with genome-wide reconstructions that suggest a complex LACA44,45, and they have implications for the evolution of the respective systems in eukaryotes. It is well established that the genetic complement of eukaryotes is a chimaera of genes with bacterial and archaeal origins, but the nature of the archaeal ancestor remains an open question46. Recent large-scale phylogenetic studies suggest at least three alternative candidates: the Thermoplasmatales47, the Crenarchaeota48 and an unidentified deep archaeal branch49. The present analysis of the evolution of membrane remodelling systems seems to be best compatible with the deep archaeal branch possibility. Specifically, we propose that the archaeal ancestor of eukaryotes belonged to a deep, probably extinct — or unidentified — branch of the Archaea (or Archaea-related organisms) that possessed a highly complex membrane remodelling machinery, including FtsZ, an ESCRT-III system with a VPS4 homologue, and an actin-like protein, a combination that is not found in any available extant genome. Moreover, it seems plausible that the hypothetical archaeal ancestor of eukaryotes already possessed the eukaryotic-type system of cell division regulation that uses reversible phosphorylation catalysed by serine/threonine kinase and protein phosphatase 2C.
Phylogenetic analysis of the VPS4-like ATPases reveals three major archaeal branches, one including the Desulfurococcales and the Thaumarchaeota, a second consisting of the euryarchaeal representatives and a third that is limited to the Thermoplasmatales (see Supplementary information S8 (figure)). In the phylogenetic tree, eukaryotic VPS4 clusters with the euryarchaeal orthologues. However, the topology of the tree was not strongly supported by bootstrap analysis, so the origin of the eukaryotic VPS4 proteins could not be inferred with confidence. In contrast, Snf7-like proteins from the Desulfurococcales and the Sulfolobales show greater similarity to eukaryotic homologues than to Snf7-like proteins from the Euryarchaeota (see Supplementary information S7 (figure)). Moreover, the Snf7-like protein encoded in the operon containing the VPS4 homologue in the Desulfurococcales and the Sulfolobales contains a C-terminal wHTH domain15. Given that the eukaryotic ESCRT-II complex consists mostly of wHTH domain-containing subunits16, it seems likely that both eukaryotic ESCRT-II and ESCRT-III have an archaeal origin. Genes encoding Snf7-like proteins apparently duplicated independently in most archaeal lineages14 (see Supplementary information S7 (figure)) and in eukaryotes, making it difficult to infer the number of paralogues in the LACA or the archaeal ancestor of the Eukarya. Despite these uncertainties in the evolutionary reconstructions, it seems likely that the Eukarya inherited a highly complex, diversified membrane remodelling machinery from the Archaea.
Supplementary Material
Acknowledgments
The authors thank Y. Wolf for useful discussions and help with the preparation of figure 3. The authors’ research is supported by the Intramural Research Program of the US National Institutes of Health, National Library of Medicine (K.S.M., N.Y. and E.V.K.) and by the Edward Penley Abraham Trust (S.D.B.).
Glossary
- Orthologue
One of two or more homologous genes (or the encoded proteins) that are derived by vertical descent from a common ancestor
- Paralogue
One of two or more homologous genes (or their encoded proteins) that have evolved following duplication of an ancestral gene
- FHA domain
A domain that binds phosphopeptides. Most FHA domains recognize phosphothreonine, with additional specificity contributed by residues that are carboxy-terminal to the phosphothreonine
- AAA+ ATPase
A member of the vast superfamily of ATPases associated with various cellular activities (AAA+). These proteins utilize the energy of ATP binding, hydrolysis and release to remodel macromolecular substrates
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
stephen D. Bell’s homepage:
http://users.path.ox.ac.uk/~sbell/Welcome.html
Eugene V. Koonin’s homepage: http://www.ncbi.nlm.nih.gov/CBBresearch/Koonin
References
- 1.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- 3.Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Lowe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Lowe J, van den Ent F, Amos LA. Molecules of the bacterial cytoskeleton. Annu Rev Biophys Biomol Struct. 2004;33:177–198. doi: 10.1146/annurev.biophys.33.110502.132647. [DOI] [PubMed] [Google Scholar]
- 6.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 7.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 8.Vats P, Yu J, Rothfield L. The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci. 2009;66:3353–3362. doi: 10.1007/s00018-009-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, Koonin EV. Comparative genomics of archaea: how much have we learned in six years, and what’s next? Genome Biol. 2003;4:115. doi: 10.1186/gb-2003-4-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci. 2006;361:1007–1022. doi: 10.1098/rstb.2006.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernander R. The archaeal cell cycle: current issues. Mol Microbiol. 2003;48:599–604. doi: 10.1046/j.1365-2958.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 12.Bernander R, Lundgren M, Ettema TJ. Comparative and functional analysis of the archaeal cell cycle. Cell Cycle. 2010;9:794–806. [PubMed] [Google Scholar]
- 13.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ettema TJ, Bernander R. Cell division and the ESCRT complex: a surprise from the archaea. Commun Integr Biol. 2009;2:86–88. doi: 10.4161/cib.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. This study and that described in reference 16 provide evidence that Sulfolobus spp. homologues of ESCRT-III and VPS4 are involved in cell division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Opin Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelet X, Djeddi A, Legouis R. Developmental and cellular functions of the ESCRT machinery in pluricellular organisms. Biol Cell. 2010;102:191–202. doi: 10.1042/BC20090145. [DOI] [PubMed] [Google Scholar]
- 20.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. This article describes the in vitro reconstitution of the eukaryotic ESCRT system with purified proteins and model giant unilamellar vesicles. The work shows that ESCRT-I and ESCRT-II form membrane buds that are then cleaved at the neck by ESCRT-III. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortmann AC, et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott A, et al. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci USA. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 24.Wurtzel O, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sureka K, et al. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS ONE. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Thakur M, Chakraborti PK. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem. 2006;281:40107–40113. doi: 10.1074/jbc.M607216200. [DOI] [PubMed] [Google Scholar]
- 28.Adindla S, Inampudi KK, Guruprasad K, Guruprasad L. Identification and analysis of novel tandem repeats in the cell surface proteins of archaeal and bacterial genomes using computational tools. Comp Funct Genomics. 2004;5:2–16. doi: 10.1002/cfg.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwaya N, et al. A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J Biol Chem. 2010;285:16822–16829. doi: 10.1074/jbc.M110.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of Archaea, Bacteria, and Eukaryota. J Mol Evol. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 31.Makarova KS, Koonin EV. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct. 2010;5:33. doi: 10.1186/1745-6150-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 33.Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F. Characterization of the FtsZ-interacting septal proteins SepF and Ftn6 in the spherical-celled cyanobacterium Synechocystis strain PCC 6803. J Bacteriol. 2009;191:6178–6185. doi: 10.1128/JB.00723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horn C, Paulmann B, Kerlen G, Junker N, Huber H. In vivo observation of cell division of anaerobic hyperthermophiles by using a high-intensity dark-field microscope. J Bacteriol. 1999;181:5114–5118. doi: 10.1128/jb.181.16.5114-5118.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundgren M, Malandrin L, Eriksson S, Huber H, Bernander R. Cell cycle characteristics of Crenarchaeota: unity among diversity. J Bacteriol. 2008;190:5362–5367. doi: 10.1128/JB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. This study describes archaeal actin-like proteins and suggests a hypothetical scenario of eukaryogenesis. In this scenario, the archaeal ancestor of eukaryotes possessed an actin-based cytoskeleton, including branched filaments, that allowed this organism to produce actin-supported membrane protrusions, and these protrusions facilitated engulfment of other bacteria and archaea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Easter J, Jr, Gober J. W ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell. 2002;10:427–434. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- 39.Springer TA. Complement and the multifaceted functions of VWA and integrin I domains. Structure. 2006;14:1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prangishvili D, et al. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J Bacteriol. 2000;182:2985–2988. doi: 10.1128/jb.182.10.2985-2988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellen AF, et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles. 2009;13:67–79. doi: 10.1007/s00792-008-0199-x. This work demonstrates that Sulfolobus spp. ESCRT-III and VPS4 homologues are found in secreted vesicles, suggesting that they may play a part in the biogenesis of these vesicles. [DOI] [PubMed] [Google Scholar]
- 43.Soler N, Marguet E, Verbavatz JM, Forterre P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res Microbiol. 2008;159:390–399. doi: 10.1016/j.resmic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 44.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Csuros M, Miklos I. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 2009;26:2087–2095. doi: 10.1093/molbev/msp123. A sophisticated maximum-likelihood reconstruction of archaeal genome evolution that infers highly complex ancestors of the Archaea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 47.Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 48.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. This and references 47 and 48 provide detailed analyses of the contributions of different groups of archaea to the evolution of eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carballido-Lopez R, Formstone A. Shape determination in Bacillus subtilis. Curr Opin Microbiol. 2007;10:611–616. doi: 10.1016/j.mib.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Graumann PL. Dynamics of bacterial cytoskeletal elements. Cell Motil Cytoskeleton. 2009;66:909–914. doi: 10.1002/cm.20381. [DOI] [PubMed] [Google Scholar]
- 52.Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. An intriguing study showing that, under highly defined conditions, FtsZ can be dispensible for viability in B. subtilis. The cells lacking FtsZ and cell walls divide by a bizarre budding–extrusion mechanism. [DOI] [PubMed] [Google Scholar]
- 53.Jenkins C, et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci USA. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilhofer M, Rosati G, Ludwig W, Schleifer KH, Petroni G. Coexistence of tubulins and ftsZ in different Prosthecobacter species. Mol Biol Evol. 2007;24:1439–1442. doi: 10.1093/molbev/msm069. [DOI] [PubMed] [Google Scholar]
- 55.McDonald B, Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shestakova A, et al. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 58.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. This work, along with that described in reference 57, provides the first evidence that the ESCRT machinery localizes to the midbody and is required for membrane abscission in human cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nature Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. This investigation shows that an ESCRT-III protein, Chmp1b, recruits the microtubule-severing ATPase, spastin, to the midbody through a MIT domain–MIM3 interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koonin EV. Orthologs, paralogs and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 61.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 63.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 64.Elkins JG, et al. A korarchaeal genome reveals insights into the evolution of the archaea. Proc Natl Acad Sci USA. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yarza P, et al. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–50. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.