Background: Formin FH1 domains deliver subunits to actin filament barbed ends.
Results: The transfer rate of formin Bni1p depends on FH1 polyproline track sequences and positions.
Conclusion: The two FH1 domains of formin dimers deliver actin independently of one another.
Significance: Sequence features of other FH1 domains are similar to Bni1p, suggesting a common strategy to maximize actin polymerization rates.
Keywords: Actin, ATP, Biophysics, Cytoskeleton, Microscopy, FH1 Domain, Formin, Polymerization
Abstract
Formin-mediated elongation of actin filaments proceeds via association of Formin Homology 2 (FH2) domain dimers with the barbed end of the filament, allowing subunit addition while remaining processively attached to the end. The flexible Formin Homology 1 (FH1) domain, located directly N-terminal to the FH2 domain, contains one or more stretches of polyproline that bind the actin-binding protein profilin. Diffusion of FH1 domains brings associated profilin-actin complexes into contact with the FH2-bound barbed end of the filament, thereby enabling direct transfer of actin. We investigated how the organization of the FH1 domain of budding yeast formin Bni1p determines the rates of profilin-actin transfer onto the end of the filament. Each FH1 domain transfers actin to the barbed end independently of the other and structural evidence suggests a preference for actin delivery from each FH1 domain to the closest long-pitch helix of the filament. The transfer reaction is diffusion-limited and influenced by the affinities of the FH1 polyproline tracks for profilin. Position-specific sequence variations optimize the efficiency of FH1-stimulated polymerization by binding profilin weakly near the FH2 domain and binding profilin more strongly farther away. FH1 domains of many other formins follow this organizational trend. This particular sequence architecture may optimize the efficiency of FH1-stimulated elongation.
Introduction
The formin family of proteins nucleates and promotes elongation of linear actin filaments that form a variety of structures in eukaryotic cells such as cytokinetic contractile rings (1), stress fibers (2), adherens junctions (3), and filopodia (4). Formins are large, multi-domain proteins, but actin polymerization is largely mediated through their Formin Homology 1 and 2 (FH1 and FH2)3 domains (5–8) (Fig. 1B). FH2 domains form stable, head to tail dimers (9) that stabilize filament nuclei consisting of two or three actin monomers (10). FH2 domains remain processively associated with growing actin filament barbed ends (11, 12) by transferring very reliably from the second-to-last actin subunit onto each newly incorporated subunit. Filaments associated with FH2 domains typically elongate slower than filaments with free ends (13–15). This phenomenon is explained as “gating” based on the interpretation that the complex between the FH2 domains and the end of the filament fluctuates rapidly between two conformations, one where the gate is closed and the other where the gate is open (16, 17). The fraction of time that the gate is open varies from <0.1 to >0.8 for the formins that have been tested (13, 14).
FIGURE 1.
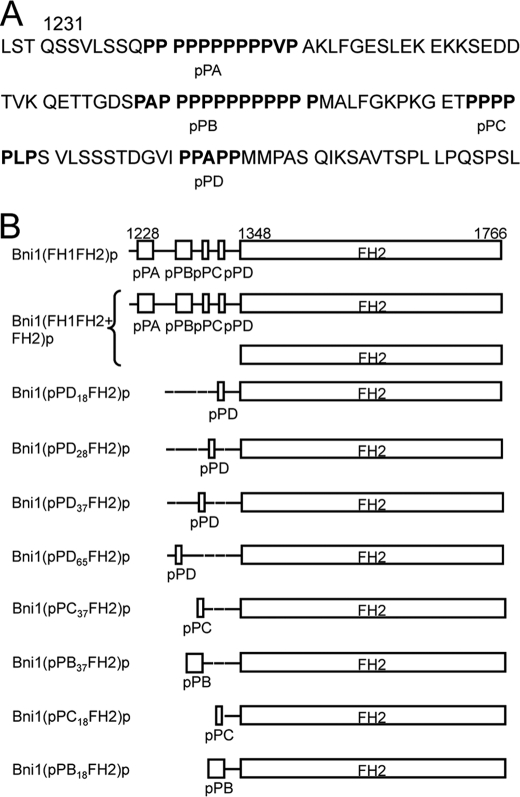
FH1 domain sequence and variants of Bni1(FH1FH2)p. A, sequence of FH1 domain. B, schematic representation of constructs used in this study. Residue numbers indicate domain boundaries. FH1 domains are to scale, but FH2 domains are not. All constructs form homodimers, except for the heterodimer of Bni1(FH1FH2+FH2)p. Both subunits are depicted in this case.
The FH1 domain, located directly N-terminal to the FH2 domain, enables formins to increase the rate of polymerization of FH2-bound filaments in the presence of profilin (17). FH1 domains contain multiple polyproline tracks that bind the actin-binding protein profilin. The poorly conserved sequences between the polyproline tracks are predicted to be disordered (18), thus giving FH1 domains an overall flexible structure. Transfer of actin from the FH1 domain to the barbed end involves four reactions: (1) rate-limiting binding of profilin-actin to a polyproline track in the FH1 domain; (2) loop closure where diffusion of the profilin-actin-polyproline track brings associated profilin-actin complexes into contact with the barbed end of the filament; (3) association of actin with the barbed end; and (4) dissociation of profilin from FH1 and the new terminal subunit at the barbed end of the filament. Reactions 2–4 are very fast, allowing profilin-actin bound to polyproline tracks to transfer at rates >1000 s−1 to the FH2-bound barbed end (13). The ability of profilin-actin to drive elongation increases with the number of profilin binding sites in the FH1 domain, but each additional profilin-binding site is less effective than the last (16).
In this study, we used budding yeast formin Bni1p to investigate the determinants of the rates of profilin-actin binding to FH1 and the transfer of actin onto the end of the filament. We found that two FH1 domains are not essential for formins to stimulate actin polymerization beyond the rate mediated by the FH2 dimer alone, and that each FH1 domain functions independently of the other. We also found that the transfer reaction is diffusion-limited and influenced by variations in polyproline track sequences that determine profilin binding. Taken together, position-specific sequence variations within the FH1 domain optimize the efficiency of Bni1p-mediated polymerization.
EXPERIMENTAL PROCEDURES
Plasmid Construction
David Kovar provided the construct encoding the C-terminally His6-tagged protein Bni1(FH1FH2)p (residues 1227–1766) (19). We made other Bni1p constructs by standard cloning methods. The construct Bni1(FH1FH2+FH2)p was made by cloning Bni1(FH1FH2)p (residues 1227–1766) with an N-terminal His6 tag and Bni1(FH2)p (residues 1348–1766) into Multiple Cloning Sites (MCS) 1 and 2 of a pETDuet-1 vector (Novagen). The N-terminal primer used to clone Bni1(FH2)p into MCS2 included an Avitag sequence upstream of the 5′ sequence of Bni1(FH2)p that is biotinylated during protein expression. All other constructs were tagged N-terminally with GST and C-terminally with a His6 tag by cloning into the vector pGV67 (20).
Protein Purification
All formin constructs were expressed in BL21 DE3 RP CodonPlus cells (Stratagene). Bni1p (FH1FH2)p was purified as described (16). Bni1(FH1FH2+FH2)p was purified by tandem avidin and nickel affinity columns. Cells were resuspended in 10 volumes of 500 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1 mm DTT, sonicated, and centrifuged at 16,000 rpm for 30 min to remove insoluble material. The supernatant was incubated with 1 ml of SoftRelease Avidin resin (Promega) by rotation for 1 h at 4 °C. The supernatant and resin were then poured into an empty column, washed with 20 ml of lysis buffer, and eluted with 5 mm biotin in lysis buffer. The eluted protein was then applied to Ni-NTA resin (Qiagen), washed with lysis buffer, and eluted with 250 mm imidazole (pH 8.0) in lysis buffer.
All other formin constructs were purified using Sepharose-glutathione and nickel affinity columns. After lysis and centrifugation, the supernatant was incubated with 2 ml of Sepharose-glutathione resin (GE Healthcare Life Sciences) by rotation for 1 h at 4 °C. The supernatant and resin were then poured into a column, washed with 25 ml of lysis buffer, followed by a wash with 25 ml of low salt buffer (100 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1 mm DTT) and eluted with 100 mm glutathione in low salt buffer. The eluted protein was incubated with ∼2–5 μm TEV protease overnight at 4 °C. Cleaved formin was separated from GST and TEV by a second round of purification on Ni-NTA resin (Qiagen) as described above.
We used ProtParam to calculate extinction coefficients. All formin constructs were concentrated to ∼5–20 μm, dialyzed into KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole (pH 7.0)) with 1 mm DTT, flash-frozen in 10 or 100 μl aliquots, and stored at −80 °C.
Ca2+-ATP-actin was purified from an acetone powder of frozen chicken skeletal muscle (Trader Joe) by one cycle of polymerization and depolymerization (21), and monomers were purified by gel filtration on S-300 resin in G-Buffer (2 mm Tris-HCl (pH 8.0), 0.1 mm CaCl2, 1 mm NaN3, 1 mm DTT). Actin was labeled on cysteine 374 with Oregon Green (OG) 488 iodoacetamide (Invitrogen) (22). Labeled and unlabeled actins were stored at 4 °C. We used an extinction coefficient of 26,000 m−1 cm−1 at λ = 290 nm for unlabeled actin. For OG-labeled actin, we used an extinction coefficient of 78,000 m−1 cm−1 at λ = 491 nm to measure the concentration of OG, and the following relation to calculate the concentration of actin in each fraction: [total Ca2+-actin] = [A290 − (A491 × 0.171)]/26,000 m−1 cm−1.
Saccharomyces cerevisiae profilin was expressed in Escherichia coli BL21 DE3 cells from plasmid pMW172 (16). Purified profilin was dialyzed into KMEI and stored long-term at 4 °C. Before use profilin was centrifuged at high speed (128,000 × g, 30–60 min, 4 °C) to remove any precipitate that developed during storage. We used an extinction coefficient of 19,060 m−1 cm−1 at λ = 280 nm to measure the concentration of profilin.
Circular Dichroism
FH1 domain was dialyzed into 150 mm NaCl, 25 mm Tris-HCl (pH 8.0). Circular dichroism was measured at wavelengths between 195 and 300 nm using an Aviv 62A DS Spectropolarimeter (Aviv Associates, Lakewood, NJ).
Microscopy and Data Analysis
We prepared open-ended glass flow chambers (16) from 22 × 50 mm No. 1 coverslips (Fisherbrand, Pittsburgh, PA), stretched strips of Parafilm spaced 3 mm apart and cleaned glass slides (75 × 25 × 1 mm; Eric Scientific, Portsmouth, NH). The chamber was sealed with moderate pressure and brief flaming. Before microscopy, 8 μl of 0.5% Tween 80 was passed twice through the flow cell by capillary action and allowed to incubate for 1 min. The flow cell was then washed with two changes of 8 μl of HS-TBS (600 mm NaCl, 50 mm Tris-HCl (pH 7.0)), and 0.2–1 μm NEM-inactivated skeletal-muscle myosin (22) was passed twice and allowed adhere to the surface for at least 1 min. The flow cell was washed again with HS-TBS, followed by two passes of 10% (w/v) BSA in HS-TBS, which was allowed to incubate for 1 min.
Just before introduction of the polymerization reaction into the sample chamber, the BSA/HS-TBS solution was replaced with microscopy buffer (9.6 mm imidazole (pH 7.0), 48 mm KCl, 0.96 mm MgCl2, 0.96 mm EGTA, 96 mm DTT, 0.19 mm ATP, 14.4 mm glucose, 19.2 μg/ml catalase, 96 μg/ml glucose oxidase, 0.48% methylcellulose (4000 cP at 2%)). Mixtures of unlabeled and OG-labeled Ca2+-ATP actin were converted to Mg2+-ATP actin by addition of 0.2 volume of 1 mm EGTA and 0.25 mm MgCl2. After a 5-min incubation, polymerization was initiated by addition of 2× microscopy buffer with formin and profilin. Samples were immediately introduced into the flow chambers and imaging was begun.
We generated time-lapse movies of growing filaments using prism-style total internal reflection fluorescence microscopy (TIRFM) on an Olympus IX-70 inverted microscope by collecting images every 10 s with a Hamamatsu C4742–95 CCD (Orca-ER) camera and MetaMorph software (Molecular Devices, Union City, CA). Time-lapse movies of growing filaments were processed with ImageJ software. For each sample, we measured the rates of barbed end elongation of ∼10–20 filaments, typically from at least 15 frames of imaging over a span of at least 300 s. To calculate the contribution of FH1-mediated polymerization, we subtracted the polymerization rate measured in the absence of profilin (FH2-mediated) from the rate measured at each profilin concentration.
Simulations of Formin-mediated Actin Polymerization
We adapted a previously described model using Virtual Cell software (National Resource for Cell Analysis and Modeling and the National Center for Research Resources) to model formin-mediated polymerization (11, 23). This model treated free profilin, free actin, profilin-actin, FH2-associated barbed ends, and individual polyproline tracks as distinct species and the minimal components required for polymerization. Supplemental Table S1 describes the rate constants for the formation of intermediate species (FH1-profilin, FH1-profilin-actin, FH1-profilin-actin-barbed end, barbed end-profilin, and filamentous actin).
RESULTS
To investigate how the structure of the FH1 domain influences formin-mediated actin polymerization, we designed a number of variants of the budding yeast formin Bni1p consisting of the FH1 and FH2 domains (called Bni1(FH1FH2)p) with mutations in the FH1 domain. We selected this particular formin as a background for making mutations, because its gating factor (the ratio of elongation rates with and without a formin on the barbed end) is 0.5, which allows it to slow barbed end elongation in the absence of profilin while still incorporating actin subunits at a measurable rate. Its FH1 domain also contains four polyproline tracks that vary in length and sequence, a convenient feature that lends itself to track deletion and swapping. A far-UV circular dichroism (CD) spectrum (supplemental Fig. S1) of the FH1 domain of Bni1p (Bni1(FH1)p; Fig. 1A) had a major peak at 200 nm and was devoid of other features, consistent with a natively disordered polypeptide chain with stretches of type II proline helices (24). The absence of overall structure gave us confidence that mutations in the FH1 domain would not alter the folding and stability of our variants.
We observed actin filament elongation in the presence of formin constructs by time-lapse TIRFM with 1.5 μm actin 33% labeled with Oregon Green on cysteine 374 (Fig. 2). These samples of growing filaments contained free and formin-bound actin filaments, which we distinguished by two criteria. (a) Filaments growing in association with a Bni1p construct having an FH1 domain had a lower fluorescence intensity in the presence of profilin than free filaments, because much of the actin is delivered to the barbed end from the FH1 in association with profilin, and profilin has a higher affinity for unlabeled actin than OG-actin (25). (b) In samples where the fluorescence of filaments with and without formins was similar, two populations could usually be distinguished based on two different polymerization rates.
FIGURE 2.
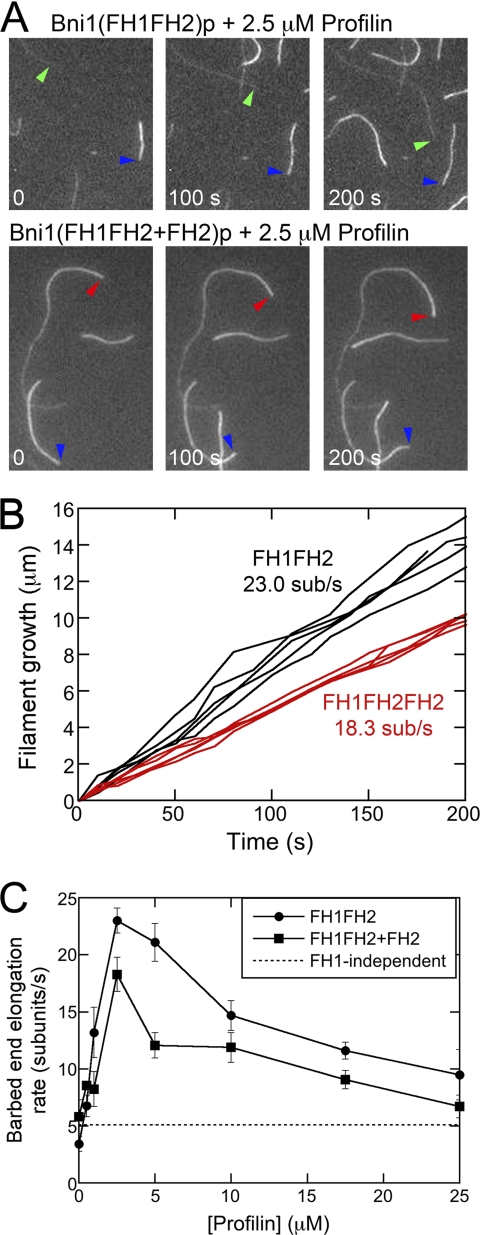
Effect of profilin on actin filament barbed end elongation mediated by Bni1p with a single FH1 domain. Conditions: 1.5 μm actin (33% Oregon Green) in microscopy buffer (9.6 mm imidazole (pH 7.0), 48 mm KCl, 0.96 mm MgCl2, 0.96 mm EGTA, 96 mm DTT, 1.92 mm ATP, 50 mm CaCl2, 14.4 mm glucose, 19.2 μg/ml catalase, 96 μg/ml glucose oxidase, 0.48% methylcellulose (4000 cP at 2%), 0.19% BSA). Data were collected with TIRFM. A and B, time series of images of formin-bound actin filaments growing in the presence of 20 nm wild-type Bni1(FH1FH2)p or 20 nm Bni1(FH1FH2 + FH2)p and 2.5 μm profilin. Colored arrows denote composition of barbed ends: blue marks free ends; green marks ends with Bni1(FH1FH2)p bound; and red marks ends with Bni1(FH1FH2+FH2)p bound. B, time courses of the growth of five filament barbed ends associated with Bni1(FH1FH2)p- (black data) or Bni1(FH1FH2+FH2)p- (red data) in the presence of 2.5 μm profilin. C, profilin concentration dependence of the elongation rates of barbed ends stimulated by wild-type Bni1(FH1FH2)p with two FH1 domains (circles) and one-armed Bni1(FH1FH2+FH2)p with one FH1 domain (squares). The dotted line represents the average contribution of FH1-independent polymerization, mediated through direct binding of actin subunits from the bulk phase, to the overall measured polymerization rates for wild-type and one-armed Bni1p. The error bars are S.E.
FH1 Domains Deliver Profilin-Actin Independently to the End of the Filament
FH2 domains must dimerize to nucleate polymerization and bind processively to the barbed end. A consequence of dimerization is also the presence of two FH1 domains near a formin-associated end, both of which can bind and deliver profilin-actin directly to the barbed end. Each FH1 domain of Bni1p contributes four putative profilin binding sites for a total of eight sites per formin-bound filament. Although doubling the number of polyproline tracks within a single FH1 domain increases the rate of formin-mediated polymerization (16), the coordination of profilin-actin delivery by the two FH1 domains of a formin dimer had not been addressed. It was therefore unclear if both FH1 domains are necessary for formins to stimulate the polymerization rate beyond that mediated by FH2 dimers alone, and whether the two FH1 domains interact in a cooperative manner.
To address this question, we designed a “one-armed formin” construct with two FH2 domains and a single FH1 domain (Fig. 1B, Bni1(FH1FH2+FH2)p). After co-expressing His-tagged Bni1(FH1FH2)p and biotinylated Bni1(FH2)p constructs, we isolated heterodimers (denoted Bni1(FH1FH2+FH2)p) by tandem affinity purification (see “Experimental Procedures” and supplemental Fig. S2). The extremely slow dissociation rate of FH2 dimers (10) predicts that, within the timeframe of our experiments, our heterodimeric construct was unlikely to dissociate and allow the subunits to reassociate as homodimers. This prediction was verified by the presence of only two populations of filaments in our experiments with profilin (see below).
In the absence of profilin, filaments bound by either wild-type Bni1(FH1FH2)p or Bni1(FH1FH2+FH2)p elongated slower than free filaments (4.6 and 5.8 subunits/s for wild-type and one-armed formin-bound filaments, compared with 13.6 subunits/s for free filaments), consistent with gating by the FH2 domain (13, 16). These filaments had the same fluorescence intensity as control filaments without bound FH2 (supplemental Fig. S3).
Profilin stimulated elongation of filaments bound by one-armed formin Bni1(FH1FH2+FH2)p. These samples contained just two types of filaments: bright filaments that elongated at the same rate as free barbed ends; and less fluorescent filaments that elongated at rates higher than those mediated by Bni1(FH2)p but lower than wild-type two-armed Bni1(FH1FH2)p (Fig. 2C). At every profilin concentration filaments associated with Bni1(FH1FH2+FH2)p were more fluorescent than filaments associated with Bni1(FH1FH2)p (supplemental Fig. S3) indicating that a larger fraction of subunits came from the bulk phase with Bni1(FH1FH2+FH2)p. For filaments associated with either Bni1(FH1FH2)p or Bni1(FH1FH2+FH2)p elongation rates peaked at 2.5 μm profilin and decreased at profilin concentrations exceeding 5 μm, consistent with free profilin competing with profilin-actin for the polyproline tracks of the FH1 domain. At most profilin concentrations, the contribution of the FH1 domains to elongation (calculated by subtracting the rate of FH2-mediated polymerization, measured in the absence of profilin, from that measured at each profilin concentration) was about 2-fold higher by the two-armed formin than the one-armed formin. Two interesting exceptions were at 0.5 and 2.5 μm profilin where the FH1-mediated rates with the one-armed formin were 82 and 63% of the wild-type formin. As developed in the discussion, these data suggest that individual FH1 domains transfer profilin-actin independently of one another.
FH1-mediated Delivery of Profilin-Actin to the Barbed End Is Diffusion-limited
Each of the four polyproline tracks in the FH1 domain of Bni1p binds and delivers profilin-actin to the filament barbed end (16, 23), but polymerization rates are not proportional to the number of profilin binding sites. Rather, each additional polyproline track has a smaller impact on the overall polymerization rate (16, 23). These results suggest that the efficiency of transferring profilin-actin from the FH1 domain falls with the distance of the profilin-binding site from the FH2 domain, because the reaction rate depends on the volume explored by the flexible FH1 domain.
We designed four constructs to investigate the relationship between transfer efficiency and the distance separating an individual polyproline track from the FH2-bound filament barbed end. Each construct contained a single polyproline track located at a different distance from the FH2 domain (Fig. 1B). We selected the sequence of the pPD binding site (located closest to the FH2 domain in wild-type Bni1p), because an N-terminally truncated variant of Bni1(FH1FH2)p consisting of the pPD track followed by the FH2 domain stimulates barbed end elongation robustly (16). We constructed our variants by replacing one of the native polyproline tracks in the FH1 domain with the pPD track and deleting the others. We named the constructs Bni1(pPDxxFH2)p, where xx refers to the number of residues separating the pPD sequence from the FH2 domain.
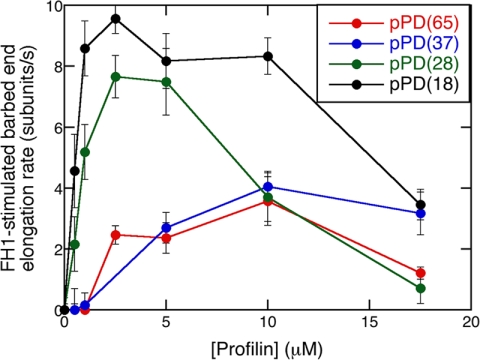
The position of the profilin binding site in the FH1 domain of the Bni1(pPDxxFH2)p constructs strongly influenced the elongation rates over a range of profilin concentrations (Fig. 3). In the absence of profilin, all four constructs slowed polymerization to similar extents, so the mutations in the FH1 domain did not affect the activity of the FH2 domain. To isolate the contribution of the pPD sequence in each construct, we subtracted the rate of FH2-mediated polymerization from that measured at each profilin concentration. Profilin stimulated FH1-mediated polymerization for constructs with the pPD track located at four different sites along the FH1 domain, but the rate stimulation declined with the distance separating the pPD track and the FH2 domain. For the two constructs with the polyproline track separated from the FH2 domain by more than 28 residues low concentrations of profilin had no impact on elongation, and the peak elongation rates shifted to 10 μm profilin. These results are consistent with a diffusion-limited model for FH1-mediated polymerization (23) and a clue for a connection (developed below) between the affinity of the polyproline track for actin and the position of the track in the FH1 domain.
FIGURE 3.
The effect of the distance between a single FH1 polyproline track pPD and the FH2 domain on Bni1p-mediated polymerization. Conditions: 1.5 μm actin (33% Oregon Green) in microscopy buffer with a range of profilin concentrations. Data were collected by TIRFM. Dependence of the FH1-stimulated barbed-end elongation rate of Bni1(pPD18FH2)p- (black), Bni1(pPD28FH2)p- (green), Bni1(pPD37FH2)p- (blue), and Bni1(pPD65FH2)p- (red) associated filaments on the concentration of profilin. The rate of FH2-mediated polymerization without profilin was subtracted from the polymerization rate measured at each profilin concentration to give the contribution of the pPD track to polymerization. The error bars are S.E.
Polyproline Track Sequences Stimulate Elongation in a Position-dependent Manner
The polyproline tracks in the FH1 domain of Bni1p vary in length and sequence. The particular pPD track used in our position-dependence experiments is the shortest and least contiguous of the four polyproline tracks in the FH1 domain. The pPD track stimulated filament elongation robustly when located close to the FH2 domain, but less well at longer distances (Fig. 3).
To explore the relationship between polyproline track sequence and profilin-actin transfer rate, we designed constructs with a single polyproline track located either 18 or 37 residues from the FH2 domain. At each of these sites, we inserted one of three polyproline tracks: either the pPB track with 13 prolines out of 14 residues; the pPC track with 6 prolines out of 7 residues; or the pPD track with 5 prolines out of 8 residues (Fig. 1B). We named these constructs Bni1(pPX18FH2)p and Bni1(pPX37FH2)p, where X is B, C, or D. Because the affinity of profilin for polyproline depends on the length and composition of the polyproline track (26), we assume that pPB has the highest affinity for profilin and pPD has the lowest affinity.
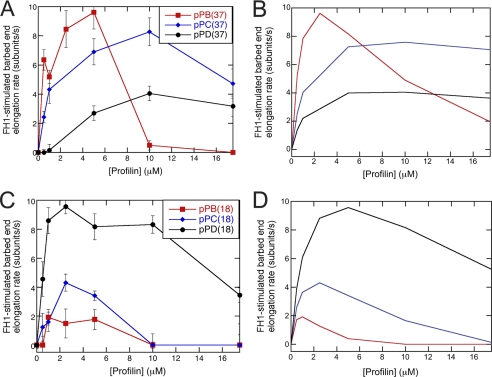
Elongation experiments over a range of profilin concentrations revealed that the relative activities of these polyproline tracks were opposite when they were located close to or far from the FH2 domain (Fig. 4). When located 37 residues from the FH2 domain, the native pPB sequence stimulated elongation much more strongly than pPC, followed by pPD (Fig. 4A). Low profilin concentrations (4–6 μm) gave maximal elongation rates with the long native pPB sequence, while 10 μm profilin gave maximal elongation rates with the shorter pPC and pPD tracks. To the contrary when located 18 residues from the FH2 domain, the short, native pPD sequence produced the largest rate stimulation, whereas elongation rates were lower with the longer pPB and more proline-rich pPC tracks (Fig. 4C).
FIGURE 4.
Effect of sequence variations on FH1-stimulated polymerization. Conditions: 1.5 μm actin (33% Oregon Green) in microscopy buffer with varying concentrations of profilin. Data were collected by TIRFM. A, experimental and B, simulated dependence of FH1-stimulated barbed-end elongation on profilin concentration for variants of Bni1p with a single polyproline track (pPB, pPC, or pPD) located 37 residues (pPB position) from the FH2 domain. The error bars are S.E. C, experimental and D, simulated dependence of FH1-stimulated barbed-end elongation on profilin concentration for variants of Bni1p with a single polyproline track (pPB, pPC, or pPD) located 18 residues (pPD position) from the FH2 domain. The sequences inserted at these positions are the polyproline track sequences from sites pPB (red squares), pPC (blue diamonds), or pPD (black circles). Dissociation equilibrium constants for profilin:polyproline interactions are 20 μm for pPB, 600 μm for pPC, and 1200 μm for pPD. The loop closure rate for the profilin-bound FH1 domain is 10,000 s−1 at the pPB position and 50,000 s−1 at the pPD position. To account for a loss of FH1 flexibility upon insertion of the pPB sequence at the pPD position, the loop closure rate for free FH1 domain is 5,000 s−1 for pPB18FH2.
DISCUSSION
FH1 domains are natively unfolded polypeptides that share little overall sequence homology aside from their polyproline tracks, which vary in number, composition and length (18). Despite this variability, these domains play critical roles in formin-mediated polymerization by binding and delivering profilin-actin to the FH2-bound filament barbed end (19). We characterized the relationship between FH1 structure and formin-mediated actin polymerization by examining (a) the number of FH1 domains, (b) the position of a polyproline track in the FH1 domain, and (c) polyproline track sequences.
Two FH1 Domains Are Better Than One
Donut-shaped, head-to-tail dimers of FH2 domains encircle the barbed ends of filaments and remain processively attached as elongation progresses. A consequence of FH2 dimerization is the presence of two FH1 domains, which doubles the number of potential profilin-actin binding sites for each formin-bound filament. The number of polyproline tracks within a single FH1 domain influences formin-mediated elongation (16), but the effect of having two separate FH1 domains had not been investigated. Our experiments with a Bni1p construct containing a single FH1 domain showed that this one-armed formin promoted elongation only half as well as the same formin with two FH1 domains (Fig. 2C). Only at low profilin concentrations was the one-armed formin slightly more efficient per arm than a two-armed formin.
The overall fluorescence of filaments bound by Bni1(FH1FH2)p was dramatically lower in the presence of profilin (supplemental Fig. S3), because of the lower affinity of profilin for Oregon Green-labeled actin than unlabeled actin (25). The overall fluorescence of filaments bound by Bni1(FH1FH2+FH2)p was also lower in the presence of profilin. However, the average fluorescence intensity of filaments with the one-armed formin was ∼50% greater than that of Bni1(FH1FH2)p-bound filaments (supplemental Fig. S3), showing that a larger fraction of subunits came from the bulk phase.
Taken together, these data indicate that formins do not require two FH1 domains to accelerate actin polymerization beyond the rate mediated by FH2 dimers. Secondly, because the rate mediated by the single FH1 domain in our one-armed construct is approximately half that mediated by the two FH1 domains in wild-type Bni1(FH1FH2)p, each FH1 domain appears to function independently of the other, arguing against a cooperative binding and delivery mechanism.
Attaching the C terminus of the flexible FH1 domain to the N terminus of the FH2 domain fixes one of its ends in space and constrains the diffusive conformations available for exploration. Although we cannot conclusively determine whether each FH1 domain favors delivery to one actin long-pitch helix over the other, our interpretation of the crystal structure of profilin 2a bound to two contiguous polyproline tracks of formin mDia1 (27) is that binding of profilin-actin to polyproline tracks of the FH1 domain favors delivery of actin to the nearest long-pitch helix. The presence of two FH1 domains therefore facilitates efficient formin-mediated polymerization.
Multiple Interdependent Variables Influence the Elongation Rate with Bni1(FH1FH2)p
Elongation of an actin filament with an FH1FH2 formin on the barbed end depends on more than ten interdependent reactions (23). The rates of these reactions depend on the concentrations of profilin and actin (fixed parameters in our experiments). Here we modified the FH1 domain of Bni1(FH1FH2) to explore how the positions and affinities of the polyproline tracks for profilin influence the elongation rate.
Transfer of Profilin-Actin from FH1 to the End of the Filament Is Diffusion-limited, so the Elongation Rate Depends on the Distance between the Polyproline Track and FH2
Direct transfer of profilin-actin from the FH1 domain to the FH2-bound filament requires a loop closure reaction, to bring the profilin-actin bound to a polyproline track close enough to the barbed end of the filament for actin to bind to the end of the filament. The number of residues between the polyproline track and the FH2 domain determines the volume explored by diffusing profilin-actin bound to a polyproline track before contacting the barbed end and thus the loop closure rate. Numerous experimental and theoretical studies of disordered polypeptides revealed that the rate of end-to-end loop closure is proportional to n−3/2, where n is the number of residues in the polypeptide (28). The numerical values of n−3/2 for polypeptides consisting of the number of residues between the FH2 domain and each polypeptide track in the FH1 domain of Bni1p are 0.013 for pPD (18 residues), 0.0067 for pPC (28 residues), 0.0044 for pPB (37 residues) and 0.0019 for pPA (65 residues). Therefore, the pPD site contacts the barbed end approximately twice as often as the pPC site, three times as often as the pPB site and seven times as often as the pPA site.
In our experiment the rate of elongation was generally inversely related to the distance between the test polyproline track in FH1 and the FH2 domain (Fig. 3), consistent with diffusion of the FH1 track limiting the rate of transfer to the barbed end (23). The construct with the polyproline track farthest from FH2 was an exception. Based on length, we expected the loop closure rate for the construct with the polyproline track 65 residues from FH2 to be almost 2.5-fold lower than the construct with 37 residues separating the polyproline track from FH2. However, FH1-mediated polymerization rates for these two constructs were very similar. One possible explanation is that the intrinsic flexibility of the FH1 domain makes the orientation of the distal polyproline track less constrained than that of the proximal track, compensating for the slower end-to-end closure rate.
The Transfer Rate from a Particular Polyproline Track Position in FH1 Depends on the Affinity of the Polyproline Track for Profilin
The affinity of a polyproline track for profilin influences the elongation rate and the optimal profilin concentration for elongation, because elongation depends on profilin-actin being bound to the FH1 at the time of loop closure. However, our experiments and simulations showed that the optimal affinity for profilin also depends on the distance of the polyproline track from FH2.
To characterize profilin-actin transfer from individual polyproline tracks located at different distances from the filament barbed end, we simulated formin-mediated actin polymerization in the presence of profilin with a Virtual Cell model (11, 23) taking into account interactions among actin, profilin, FH1 domains, FH2 domains and filament ends (supplemental Table S1). Because we were unable to measure the very low affinities of the short Bni1p FH1 polyproline tracks for profilin, we estimated the affinities from model studies of Acanthamoeba profilin (26) where the affinity of a peptide of 6 prolines (like the pPC track) is 140-fold lower than a peptide of 11 prolines (like the pPA and pPB tracks). Non-proline residues in a polyproline peptide diminish the affinity ∼100-fold, so the affinity of pPD for profilin is extremely weak. The estimated Kd values used in the simulations were 20 μm for pPB, 600 μm pPC, and 1200 μm for pPD. In each of our simulations, binding of profilin and profilin-actin to the FH1 domain was the rate-limiting step.
For a polyproline track located 37 residues from the FH2 domain, like the pPB track, the elongation rate increased and the optimal profilin concentration decreased with the affinity for profilin in both experiments (Fig. 4A) and simulations (Fig. 4B). In simulations the optimal profilin concentration was 2.5 μm with the estimated affinity for pPB. With the shorter, lower affinity pPC and pPD tracks polymerization was progressively slower at the higher optimal profilin concentrations of 10 μm.
On the other hand, when only 18 residues separated the polyproline track and the FH2 domain, the short, non-contiguous pPD sequence promoted polymerization much better than the longer pPB track or the more proline-rich pPC track in both experiments (Fig. 4C) and simulations (Fig. 4D). Optimal profilin concentrations were lower for longer polyproline tracks. Strong interaction of profilin with a proximal polyproline track therefore seems to hinder efficient profilin-actin transfer when the loop closure rate is high. For these simulations we adjusted the loop closure rate for unbound FH1 domain in the pPD position from 50,000 s−1 for the pPD to 5000 s−1 for the pPB track to compensate for the effects of polyproline track lengths on chain flexibility of an intrinsically disordered region.
These results suggest that the affinity of each polyproline track in the FH1 domain of Bni1p for profilin-actin is tuned relative to its distance to the FH2 domain to optimize transfer efficiency. The expected differences in binding affinity suggest that, near the FH2 domain, a polyproline track with low affinity for profilin favors rapid actin transfer to the barbed end upon FH1 loop closure, whereas efficient transfer of actin from a position farther from the FH2 domain depends on polyproline tracks with higher affinity for profilin (slower dissociation rates) to compensate for the slower loop closure rates (Fig. 5A).
FIGURE 5.
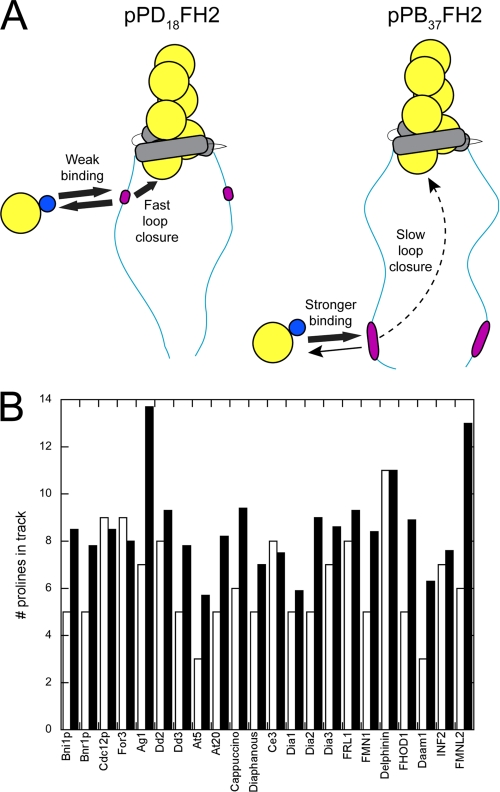
The position and length of polyproline tracks are tuned for efficient FH1-mediated polymerization. A, schematic representations of actin polymerization mediated by the pPD18FH2 versus pPB37FH2 constructs of Bni1(FH1FH2)p. Actin, profilin, FH2 domains, and polyproline tracks (pLp) are shown in yellow, blue, gray, and magenta. B, summary of the length of the polyproline track located closest to the FH2 domain (white bars) and the average length of polyproline tracks within an FH1 domain (black bars) for 22 formins. The formins listed are from S. cerevisiae (Bni1p and Bnr1p), S. pombe (Cdc12p and For3), A. gossypii (Ag1), D. discoideum (Dd2, Dd3), A. thaliana (At5, At20), D. melanogaster (Cappuccino, Diaphanous), C. elegans (Ce3), M. musculus (Dia1, Dia2, Dia3, FRL1, FMN1, Delphinin, FHOD1, Daam1), and H. sapiens (INF2, FMNL2).
The FH1 domains of many other formins follow this trend, with the potential affinity of polyproline tracks increasing with the distance from the FH2 domain. In 18 of 22 formin sequences from a wide spectrum of eukaryotes, the polyproline track located nearest to the FH2 domain is shorter than the average length of the polyproline tracks in that FH1 domain (Fig. 5B). In only three cases is the most proximal polyproline track slightly longer than the average track. Similarly, in 18 of the 22 formin sequences examined, either the most distal or second-most distal polyproline track (or both) are longer than the average track length in that FH1 domain (not shown). This particular sequence architecture might therefore maximize the efficiency of FH1-stimulated elongation.
Formins participate in a wide variety of cellular functions, so fast polymerization may not always be optimal. For example, fission yeast formin Cdc12p has the longer of two FH1 domain polyproline tracks next to the FH2 domain. This cytokinesis formin may have been selected for slow polymerization, since it not only has two very similar polyproline tracks in the FH1 domain, but also has an FH2 domain with the smallest known gating factor (13). FH1-mediated polymerization rates also depend on specific profilin isoforms (11, 29, 30), so the sequence variations of FH1 polyproline tracks must also specify interactions with profilin isoforms.
The Contributions of the Four Polyproline Tracks in Bni1p Are Not Additive
A comparison of the polymerization rates mediated by wild-type Bni1(FH1FH2)p with constructs having single polyproline sequences at their native locations shows that the wild-type formin FH1 domain produces a lower elongation rate than the sum of the independent polymerization rates mediated by each of the four sites. For example, in the presence of 2.5 μm profilin, the FH1-stimulated elongation rate produced by wild-type Bni1(FH1FH2)p with four polyproline tracks in each FH1 domain is 20 subunits/s, while the independent contributions of just Bni1(pPB37FH2)p and Bni1(pPD18FH2)p sum to ∼18 subunits/s without considering the additional contributions of sites pPA and pPC. One possible explanation for this interference is that profilin-actin competes for binding polyproline tracks in the FH1 domain, although a crystal structure showed that two profilins can bind to the same long polyproline track (31). A more likely explanation is that profilin-actin bound to the polyproline tracks affects the flexibility of the FH1 domain and impedes diffusion and loop closure. This could affect transfer from sites located far away from the FH2 domain when profilin-actin is bound at pPD.
Supplementary Material
Acknowledgments
We thank Aditya Paul and Julien Berro for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Research Grant GM-026338.

This article contains supplemental Table S1 and Figs. S1–S3.
- FH
- Formin homology
- TIRFM
- total internal reflection fluorescence microscopy.
REFERENCES
- 1. Chang F., Drubin D., Nurse P. (1997) cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143 [DOI] [PubMed] [Google Scholar]
- 3. Kobielak A., Pasolli H. A., Fuchs E. (2004) Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pellegrin S., Mellor H. (2005) The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol.: CB 15, 129–133 [DOI] [PubMed] [Google Scholar]
- 5. Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615 [DOI] [PubMed] [Google Scholar]
- 6. Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., Pellman D., Goode B. L. (2004) A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15, 896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pring M., Evangelista M., Boone C., Yang C., Zigmond S. H. (2003) Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42, 486–496 [DOI] [PubMed] [Google Scholar]
- 8. Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C. J., Blanchoin L. (2005) The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17, 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004) Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 10. Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., Rosen M. K. (2005) Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
- 11. Paul A. S., Pollard T. D. (2009) Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J. Biol. Chem. 284, 12533–12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kovar D. R., Pollard T. D. (2004) Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. U.S.A. 101, 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 14. Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D. (2003) The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161, 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris E. S., Li F., Higgs H. N. (2004) The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 16. Paul A. S., Pollard T. D. (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paul A. S., Pollard T. D. (2009) Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higgs H. N. (2005) Formin proteins: a domain-based approach. Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 19. Kovar D. R., Pollard T. D. (2004) Progressing actin: Formin as a processive elongation machine. Nat. Cell Biol. 6, 1158–1159 [DOI] [PubMed] [Google Scholar]
- 20. Nolen B. J., Pollard T. D. (2008) Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J. Biol. Chem. 283, 26490–26498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 22. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vavylonis D., Kovar D. R., O'Shaughnessy B., Pollard T. D. (2006) Model of formin-associated actin filament elongation. Mol. Cell 21, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ronish E. W., Krimm S. (1974) The calculated circular dichroism of polyproline II in the polarizability approximation. Biopolymers 13, 1635–1651 [DOI] [PubMed] [Google Scholar]
- 25. Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. (1998) Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880 [DOI] [PubMed] [Google Scholar]
- 26. Petrella E. C., Machesky L. M., Kaiser D. A., Pollard T. D. (1996) Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35, 16535–16543 [DOI] [PubMed] [Google Scholar]
- 27. Kursula P., Kursula I., Massimi M., Song Y. H., Downer J., Stanley W. A., Witke W., Wilmanns M. (2008) High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J. Mol. Biol. 375, 270–290 [DOI] [PubMed] [Google Scholar]
- 28. Lapidus L. J., Eaton W. A., Hofrichter J. (2000) Measuring the rate of intramolecular contact formation in polypeptides. Proc. Natl. Acad. Sci. U.S.A. 97, 7220–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neidt E. M., Scott B. J., Kovar D. R. (2009) Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 284, 673–684 [DOI] [PubMed] [Google Scholar]
- 30. Neidt E. M., Skau C. T., Kovar D. R. (2008) The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem. 283, 23872–23883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahoney N. M., Janmey P. A., Almo S. C. (1997) Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol. 4, 953–960 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.