Background: SRP serves as a paradigm for understanding the molecular basis of protein localization.
Results: Varying translation elongation rates changes the stringency of substrate selection by the SRP.
Conclusion: Kinetic competition with ongoing protein synthesis regulates the fidelity of SRP.
Significance: Unraveling mechanisms that govern the fidelity of protein localization is essential for understanding this fundamental cellular process.
Keywords: GTPase, Molecular Cell Biology, Protein Targeting, Protein Translocation, Translation, Signal Recognition Particle, Protein Localization Fidelity
Abstract
The signal recognition particle (SRP) is a universally conserved cellular machinery responsible for delivering membrane and secretory proteins to the proper cellular destination. The precise mechanism by which fidelity is achieved by the SRP pathway within the in vivo environment is yet to be understood. Previous studies have focused on the SRP pathway in isolation. Here we describe another important factor that modulates substrate selection by the SRP pathway: the ongoing synthesis of the nascent polypeptide chain by the ribosome. A slower translation elongation rate rescues the targeting defect of substrate proteins bearing mutant, suboptimal signal sequences both in vitro and in vivo. Consistent with a kinetic origin of this effect, similar rescue of protein targeting was also observed with mutant SRP receptors or SRP RNAs that specifically compromise the kinetics of SRP-receptor interaction during protein targeting. These data are consistent with a model in which ongoing protein translation is in constant kinetic competition with the targeting of the nascent proteins by the SRP and provides an important factor to regulate the fidelity of substrate selection by the SRP.
Introduction
Co-translational protein targeting by the signal recognition particle (SRP)2 is an evolutionarily conserved and essential pathway that mediates the localization of many membrane and secretory proteins to the eukaryotic ER or the bacterial plasma membrane (1, 2). Targeting begins when SRP recognizes an N-terminal signal sequence on nascent polypeptides that emerge from a translating ribosome. The ribosome-nascent chain complex, also termed the cargo, then enables efficient complex assembly between two GTPases in the SRP and the SRP receptor (SR), thus localizing the targeting complex to the membrane. At the membrane, the cargo is transferred from the targeting complex to the sec61p (or secYEG in bacteria) translocation machinery, where the nascent polypeptide is either integrated into the membrane or translocated across the membrane to enter the secretory pathway. Finally, GTP is hydrolyzed from the SRP-SR complex to drive the disassembly and recycling of the targeting factors.
The size and composition of SRP vary among different species. Mammalian SRP is a large complex comprised of six protein subunits and a 7S SRP RNA (2, 3). It contains two structurally and functionally distinct domains: the S domain, comprised of domains II–IV of 7S RNA and the SRP19, 54, and 68/72 protein subunits, and the Alu domain, comprised of domain I of 7S RNA and the SRP9/14 subunits. The most conserved subunit, SRP54, contains two structurally and functionally dissectable domains: a methionine-rich M domain that binds the signal sequences (4–6) and the SRP RNA (6, 7), and a special GTPase, NG domain that interacts with the SR (8, 9). Bacterial SRP is much simpler and is comprised of a SRP54 homologue, Ffh, in complex with a smaller 4.5S SRP RNA, which does not contain domains I and III in the 7S RNA (10, 11). Surprisingly, the much smaller and simpler bacterial SRP can replace its eukaryotic homologues to carry out efficient targeting of mammalian proteins into ER microsomes (10–12); this demonstrates the remarkable evolutionary conservation of SRP and shows that SRP54 and the 4.5S SRP RNA comprise the functional core of SRP.
Extensive studies in the bacterial SRP pathway showed that SRP-dependent protein targeting is, in many respects, a kinetically controlled process. For example, the assembly of a stable SRP-SR complex, which mediates the delivery of cargo to the target membrane, is an intrinsically slow process (kon = ∼102 m−1 s−1) and is accelerated ∼103-fold only when SRP is loaded with a correct cargo (13–17). GTP hydrolysis in the SRP-SR complex would act as a “timer” that aborts the targeting reaction beyond a critical time window; this hydrolysis event is also delayed by the correct but not the incorrect cargos, which can provide the correct cargos an extended time window to complete the targeting reaction (16, 17). Further, the other essential component of the SRP, the SRP RNA, kinetically stimulates both the assembly and activation of the SRP-SR complex without altering its equilibrium stability (13–15, 18). Together, these kinetic regulations allow the binding of correct cargos to be tightly coupled to their membrane delivery and ensure that incorrect cargos are rejected from the SRP pathway despite the highly degenerate nature of signal sequences.
Another intriguing aspect of co-translational protein targeting is that as the nascent polypeptide exceeds a critical length of ∼140 amino acids, the SRP loses the competence to target substrate proteins (19, 20). The molecular basis underlying this phenomenon is still unclear. Nevertheless, at a rate of ∼20–30 amino acids/second for translation elongation in rapidly growing bacterial cells, this length requirement imposes a critical time window of ∼3–5 s for the SRP to complete the targeting reaction. Given the extensive kinetic control that exists in the SRP pathway described above, it is conceivable that the rate of translation elongation could provide another important layer of regulation on the SRP pathway. The interplay between translation elongation and SRP function has been suggested in previous studies: low doses of antibiotics that slow down translation elongation can rescue the growth of cells in which SRP function is compromised (21). Nevertheless, how the ongoing synthesis of nascent proteins affects the efficiency of the SRP pathway and whether this effect is the same or different with correct or incorrect SRP substrates have not been explored.
Compared with the bacterial SRP, the mammalian SRP contains an additional “Alu” domain, which can arrest translation elongation just after the signal sequence emerges from the ribosome (22, 23). Both biochemical work and recent cryo-EM analyses found that the Alu domain of mammalian SRP interacts with the elongation factor binding site of the ribosome (21, 24), suggesting that it blocks the binding of elongation factors and thereby arrests translation. A recent study further showed that although elongation arrest is not a prerequisite for protein targeting in vitro (25), abolishing this function in vivo leads to severe defects in protein targeting and mammalian cell growth (26). Together with the observation that the SRP could not target proteins when the nascent polypeptide exceeds a critical length, these results have led to the proposal that elongation arrest provides a crucial time window that allows the mammalian SRP to find and engage the translocon (25, 26). Thus, the mammalian SRP further demonstrates the intricate interconnection between ongoing protein synthesis and protein targeting by the SRP. It also raised questions as to whether the mammalian SRP, because of this additional elongation arrest activity, have different efficiency or distinct patterns of substrate selection than its bacterial homologue.
In this work, we systematically explored the role of translation elongation on the efficiency and specificity of the SRP pathway. We show that reducing the rate of translation elongation specifically rescues the targeting defect of suboptimal substrate proteins both in vitro and in vivo, resulting in significantly relaxed specificity of the SRP. Thus, rapid ongoing protein synthesis, through kinetic competition with SRP-dependent protein targeting, is a major contributor to the specificity of the SRP pathway. Curiously, the mammalian SRP exhibits a similar pattern of substrate selection and can also be subject to regulation by translation elongation rates, raising new questions as to the extent of elongation arrest by the mammalian SRP and the role of this activity in facilitating protein targeting.
EXPERIMENTAL PROCEDURES
Materials
Wheat germ translation extract was from Promega. Microsomal membranes (RM) from dog pancreas were prepared by J. Miller (University of California, San Francisco, San Francisco, CA) according to published procedures (27) and were treated with high salt wash and partial trypsin digestion to generate TKRM as described (27). Rabbit reticulocyte lysate (RRL) was a kind gift from R. Hegde. The in vitro transcription plasmid for pPL was from E. Powers (University of California, Davis, Davis, CA) (11). Ffh, FtsY, and 4.5S RNA were expressed and purified as described previously (11, 14). Construction of the mutant FtsY and 4.5S RNA have been described (8, 28, 29). Mutant FtsY and RNAs were purified using the same procedures as those for wild-type protein and RNA. [35S]Methionine was from GE Healthcare. β-OH-Leu was from Sigma.
Escherichia coli strain HDB52 (WAM113 secB::Tn5 zic-4901::Tn10) was a kind gift from Dr. H. Bernstein (30). In this strain, the expression of Ffh is under the control of an arabinose-inducible promoter, and the secB gene was deleted (30). Antibiotics were used at the following final concentrations unless otherwise specified: 200 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 0.1 μg/ml tetracycline where applicable.
The coding region of the biotinylatable domain from Propionibacterium shermania transcarboxylase (PSBT) was amplified by PCR using pHP42 (31, 32) as template. A FLAG tag (DYKDDDDK) was also encoded in the PCR primers. The resulting PCR fragment was cloned into pJH29 (30) using the NdeI and SacI restriction sites to generate pPSBT. The coding sequences of FtsQ, phoA, and EspP were PCR-amplified and cloned into pPSBT using the NdeI and XbaI sites to generate pFtsQ-PSBT, pPhoA-PSBT, and pEspP-PSBT, respectively. Because the phoA-PSBT fusion protein did not express in E. coli, the PSBT fragment was replaced by the biotinylatable Avi tag (SGLNDIFEAQKIEWHE) using QuikChange mutagenesis to yield pPhoA-Avi.
Co-translational Protein Targeting Assay in Vitro
SRP-dependent protein targeting by the bacterial SRP and FtsY was measured using a heterologous co-translational targeting assay described previously (11, 12). Briefly, wheat germ extract was used to translate a mammalian SRP substrate, pPL, at 26 °C. Shortly (1–2 min) after translation is initiated, a cap analogue, 7-methyl-GTP, was added to inhibit additional rounds of translation initiation, such that translocation of only the first round of translation product is followed. E. coli SRP (the Ffh protein bound to the 4.5S RNA), FtsY, and TKRM were added 1 min later to initiate targeting and translocation of pPL. Translation is continued for 30 min to allow completion of pPL synthesis, at which time the reaction is stopped and analyzed by SDS-PAGE and quantified by autoradiography using the ImageQuant software. For reactions in the presence of CHX, the translation was allowed to continue for 45–60 min. Co-translational protein targeting by the mammalian SRP and SR was carried out similarly, except that translation was carried out at 32 °C and that RRL and unwashed ER microsomes were used, providing the source for mammalian SRP and SR (33).
In Vivo Detection of SRP-dependent Protein Targeting
Overnight culture of HDB52 harboring pFtsQ-PSBT, pPhoA-Avi, or pEspP-PSBT was grown in LB medium containing 0.1% arabinose at 37 °C and was washed and diluted 1:100 in fresh LB medium with or without arabinose to generate SRP+ and SRP− cells, respectively. The cells were cultured for 2 h; depletion of SRP is complete over this time window, as established in a previous work (30, 34). Expression of FtsQ-PSBT, PhoA-Avi, or EspP-PSBT was induced with 0.2 mm isopropyl thiogalactopyranoside. To attenuate translation elongation, 0.1 μg/ml tetracyclin was added into the culture for 2 h as described previously (35). The effect of tetracyclin in attenuating translation was also corroborated by a moderate reduction in cell growth rate upon the addition of the drug (supplemental Fig. S1). The samples were harvested and analyzed by SDS-PAGE and Western blotting using the ECL protocol (GE Healthcare). Biotinylated fusion proteins were detected using the streptavidin-horseradish peroxidase conjugate (streptavidin-HRP; GE Healthcare). The total amount of fusion protein was detected using anti-FLAG antibody (Cell Signaling).
RESULTS
β-OH-Leucine as Probe for Fidelity of Protein Targeting
As a simple and convenient means to change the property of signal sequence and probe substrate selection by the SRP, we took advantage of the ability of β-OH-leucine to compete with leucine and incorporate into the nascent polypeptide during translation (11, 36). The additional hydroxyl group in β-OH-leucine allows the generation of a less hydrophobic signal sequence in a model SRP substrate, pPL. To quantitatively analyze the efficiency of protein targeting by the SRP, we used a heterologous assay described previously, in which purified E. coli SRP and SRP receptor (FtsY) was used to target and translocate pPL across microsomal membranes depleted of endogenous SRP and SRP receptor (11, 12). Successful translocation of pPL results in efficient cleavage of the signal sequence, allowing the efficiency of targeting and translocation to be quantified. As expected, incorporation of β-OH-leucine into pPL resulted in dose-dependent reductions in the efficiency of protein targeting and translocation (Fig. 1A), from ∼80% with wild-type pPL to ∼20% at saturating concentrations of the leucine analogue. The efficiency of protein translation was unaffected by the presence of β-OH-leucine (supplemental Fig. S2). In addition, the targeting and translocation of a mutant pPL, in which all the leucines in the signal sequence were replaced with valine or isoleucine (Table 1, pPL-VI), was independent of β-OH-leucine (supplemental Fig. S3). Thus, β-OH-leucine specifically affects the SRP pathway by altering the hydrophobicity of the signal sequence.
FIGURE 1.
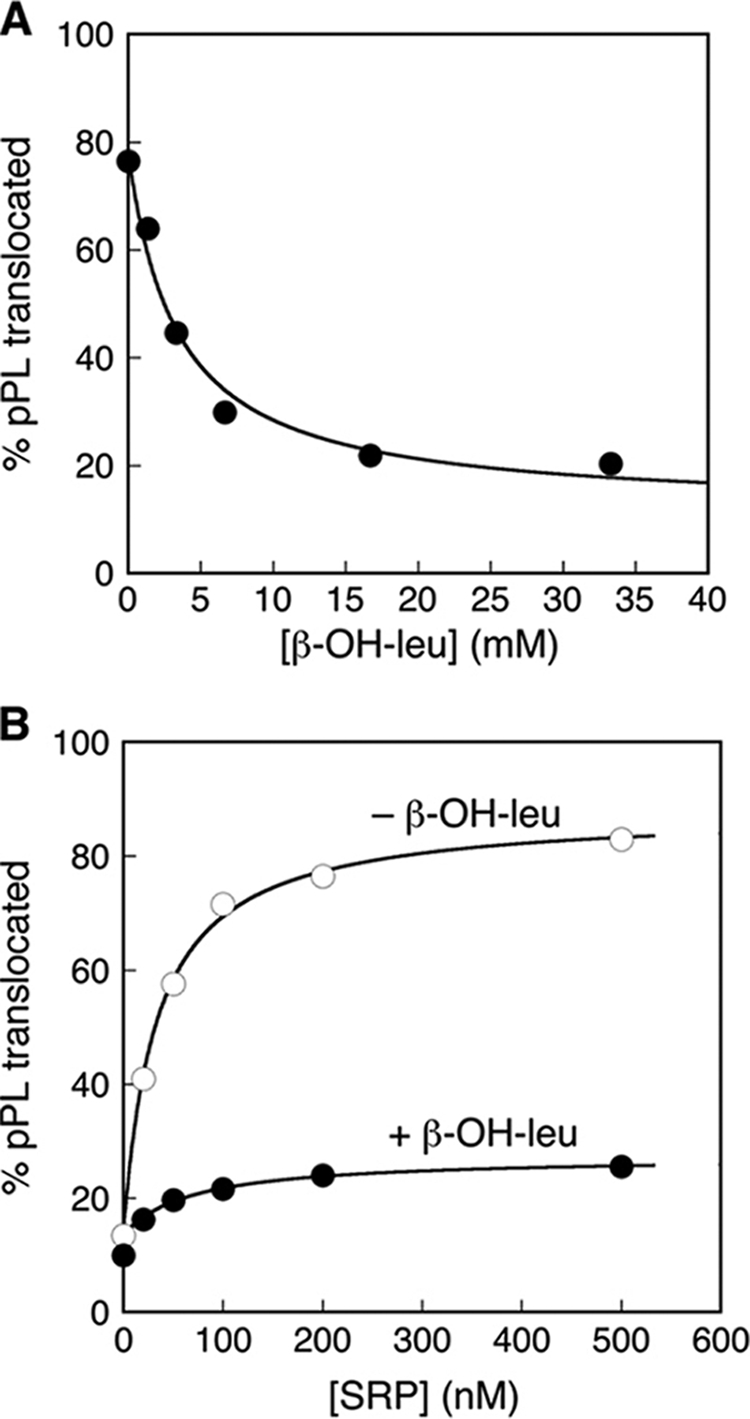
Use of β-OH-Leu to probe the fidelity of protein translocation. A, incorporation of β-OH-Leu into nascent polypeptide reduces the efficiency of pPL translocation. B, the translocation defect of proteins containing β-OH-Leu cannot be rescued by increasing SRP concentration.
TABLE 1.
Signal sequence variants used in this study
| Signal sequence mutationsa | PLb | |
|---|---|---|
| pPL | MNIKGSPWKGSLLLLLVSNLLLCQSVAP | LPICP … |
| pPL-VI | MNIKGSPWKGSVIVVVVSNIIVCQSVAP | LPICP … |
| 2A8L | MNIKGSPWKGSLALLLLLLLACQSVAP | LPICP … |
| 3A7L | MNIKGSPWKGSLALLLLLALACQSVAP | LPICP … |
| 4A6L | MNIKGSPWKGSLALALLLALACQSVAP | LPICP … |
| 5A5L | MNIKGSPWKGSLALALALALACQSVAP | LPICP … |
| phoA | MNIKGSPWKGSIALALLPLLFCQSVAP | LPICP … |
| 8A2L | MNIKGSPWKGSLAAAAAAALACQSVAPCQSVAP | LPICP … |
a The chimeric pPL constructs used for the co-translational protein targeting assay (see “Experimental Procedures”). Bold type highlights the hydrophobic core.
b PL indicates mature protein after cleavage by signal peptidase.
The observed defect in the targeting of mutant pPL bearing β-OH-leucine could arise from a reduced binding affinity of SRP to ribosome-nascent chain complexes bearing a defective signal sequence, or from defects in subsequent steps of the SRP pathway. If defective cargo binding by the SRP were responsible, then increasing the concentration of SRP would be expected to rescue the targeting and translocation of mutant pPL bearing β-OH-leucine. Such a rescue was not observed (Fig. 1B). Instead, saturation in translocation efficiency was reached at similar SRP concentrations with the wild-type and mutant pPL, and the translocation efficiency of pPL bearing β-OH-leucine remained at ∼22% at saturating SRP concentrations (Fig. 1B). This suggests that the defect in the targeting of mutant pPL arises from steps in the pathway other than SRP binding, consistent with previous work showing that the affinity of cargo binding is not the sole determinant of substrate selection by the SRP (17).
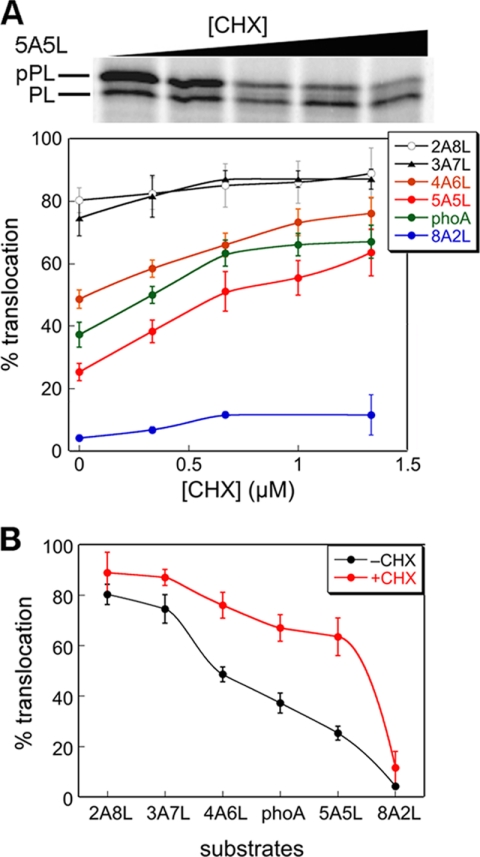
In the search for factors that modulate substrate selection by the SRP, we noticed that the rate of protein translation plays a major role: moderate reductions in the rate of translation elongation in the presence of low doses of cyclohexamide (CHX), a translation elongation inhibitor (Fig. 2A), rescued the targeting and translocation defect of mutant pPL bearing β-OH-leucine (Fig. 2B, lower gel and closed circles). In contrast, the targeting efficiency of wild-type pPL was largely unaffected by CHX (Fig. 2B, upper gel and open circles). At 1 μm CHX, where translation elongation was slowed ∼4-fold (Fig. 2A), the targeting efficiency of mutant pPL bearing β-OH-leucine was ∼65%, approaching that of wild-type pPL (Fig. 2B). Thus, the co-translational protein targeting reaction exhibits significantly reduced discrimination against suboptimal signal sequences under conditions where translation elongation was attenuated. This phenomenon could arise from an effect of the translation rates on the action of SRP or on that of the Sec translocation machinery.
FIGURE 2.
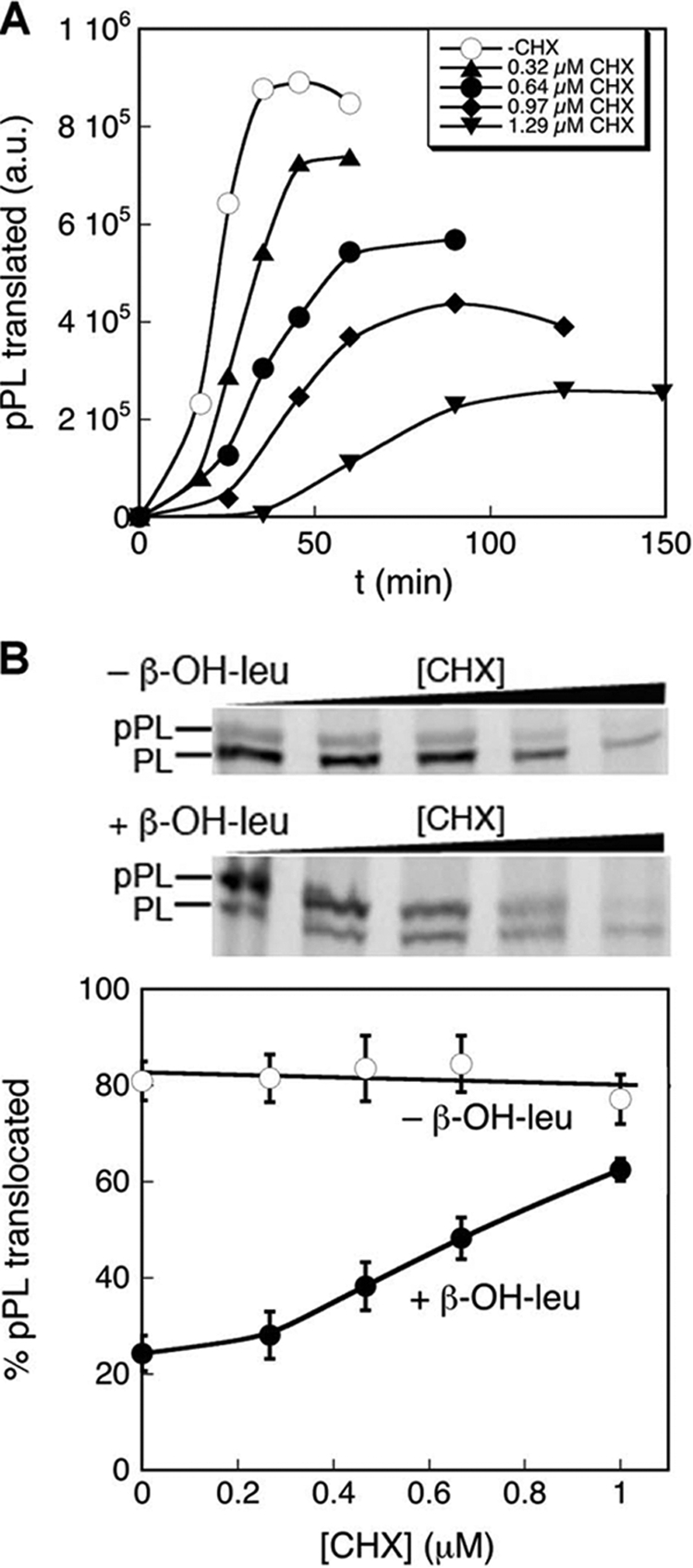
The targeting of pPL bearing β-OH-Leu can be rescued by reducing the rate of translation elongation. A, effect of cycloheximide used in this work on the efficiency of pPL translation. B, CHX rescues the targeting defect of pPL bearing β-OH-Leu.
Slower Translation Elongation Rescues Translocation Defect of Proteins with Suboptimal Signal Sequences
To directly test whether slower translation elongation reduces the stringency of substrate selection by the SRP, we systematically varied the signal sequence. We used a series of signal sequence variants described previously (37–39), in which the hydrophobic core of the pPL signal sequence was replaced by that from phoA, a borderline SRP substrate, or by a combination of leucine and alanine (Table 1). The Leu/Ala ratio was varied to generate signal sequences with different hydrophobicity (Table 1). Analogous to the cases where the signal sequence strength was modulated by incorporation of β-OH-leucine, modest reductions in the rate of translation elongation using CHX rescued the targeting of pPL variants with suboptimal signal sequences (Fig. 3A). The most substantial rescue was observed with substrate proteins whose signal sequences are on the “borderline” of rejection by the SRP, such as phoA and 5A5L (Fig. 3A). In contrast, the targeting and translocation of pPL variants with strong signal sequences, such as 2A8L and 3A7L, were not significantly affected by CHX (Fig. 3A). The pPL variant that contains no signal sequences, 8A2L, could not be efficiently translocated (<11%) regardless of whether CHX was present or not (Fig. 3A). As a result, under conditions of reduced protein synthesis rates, the SRP exhibits a significantly more relaxed threshold of substrate selection, and even substrate proteins with weak signal sequences, such as phoA and 5A5L, could be targeted substantially through the SRP pathway (Fig. 3B).
FIGURE 3.
Targeting of proteins with suboptimal signal sequences can be rescued by reducing translation elongation. A, CHX rescues SRP-dependent targeting of proteins with suboptimal signal sequences but has a much smaller effect on the targeting of strong SRP substrates. B, SRP exhibits a lower threshold of substrate selection in the presence of CHX.
Slower Translation Elongation Rescues Kinetic Defects in SRP-dependent Protein Targeting
Two models could be envisioned to explain the ability of CHX to rescue the targeting of substrate proteins with suboptimal signal sequences. First, as proposed previously, SRP preferentially interacts with the translating ribosome at a distinct stage or conformational state during the translation elongation cycle (21); CHX, by locking the translating ribosome at this stage, could allow better interaction with the SRP. Alternatively or in addition, the effect of CHX could be attributed to kinetic competition between the SRP targeting pathway and translation elongation (40). This is because the SRP loses the ability to target substrate proteins when the nascent polypeptide exceeds a critical length, which imposes a limited time window for its action (11, 19, 20). If the defects in the targeting of substrate proteins with weak signal sequences were primarily kinetic in nature, reducing the rate of translation elongation would provide an extended time window that allows a larger fraction of these substrates to complete their targeting.
To test whether kinetic competition can explain the ability of CHX to rescue the targeting of suboptimal SRP substrates, we reduced the rate of cargo delivery to the target membrane by introducing mutations into the SRP receptor, FtsY, that specifically compromise the kinetics of SRP-FtsY complex assembly (28, 41). All three mutants, FtsY(G455W), FtsY(E475K), and FtsY(D449N), exhibited severe defects in the targeting and translocation of pPL ((12) and Fig. 4A). If CHX rescues the targeting of suboptimal substrate proteins by reducing the kinetic competition between protein translation and SRP-dependent protein targeting, it would be expected to also rescue protein targeting by the mutant SRP receptors. Consistent with this notion, low doses of CHX significantly increased the targeting of pPL in the presence of the mutant receptors, allowing translocation to reach 45–55% completion in the presence of 1.3 μm CHX (Fig. 4A). Similar rescue was observed when the kinetics of protein targeting was reduced by introducing mutations into the tetraloop of the SRP RNA, which also specifically compromise the rate of SRP-FtsY complex assembly (Fig. 4B) (29). In contrast, the targeting of pPL by wild-type SRP and FtsY was largely unaffected by the addition of the translation elongation inhibitor (Fig. 4, A and B, open circles). Together, these results strongly suggest that rescue of suboptimal protein targeting by CHX could be accounted for by kinetic competition between SRP-dependent protein targeting and elongation of the nascent polypeptide.
FIGURE 4.
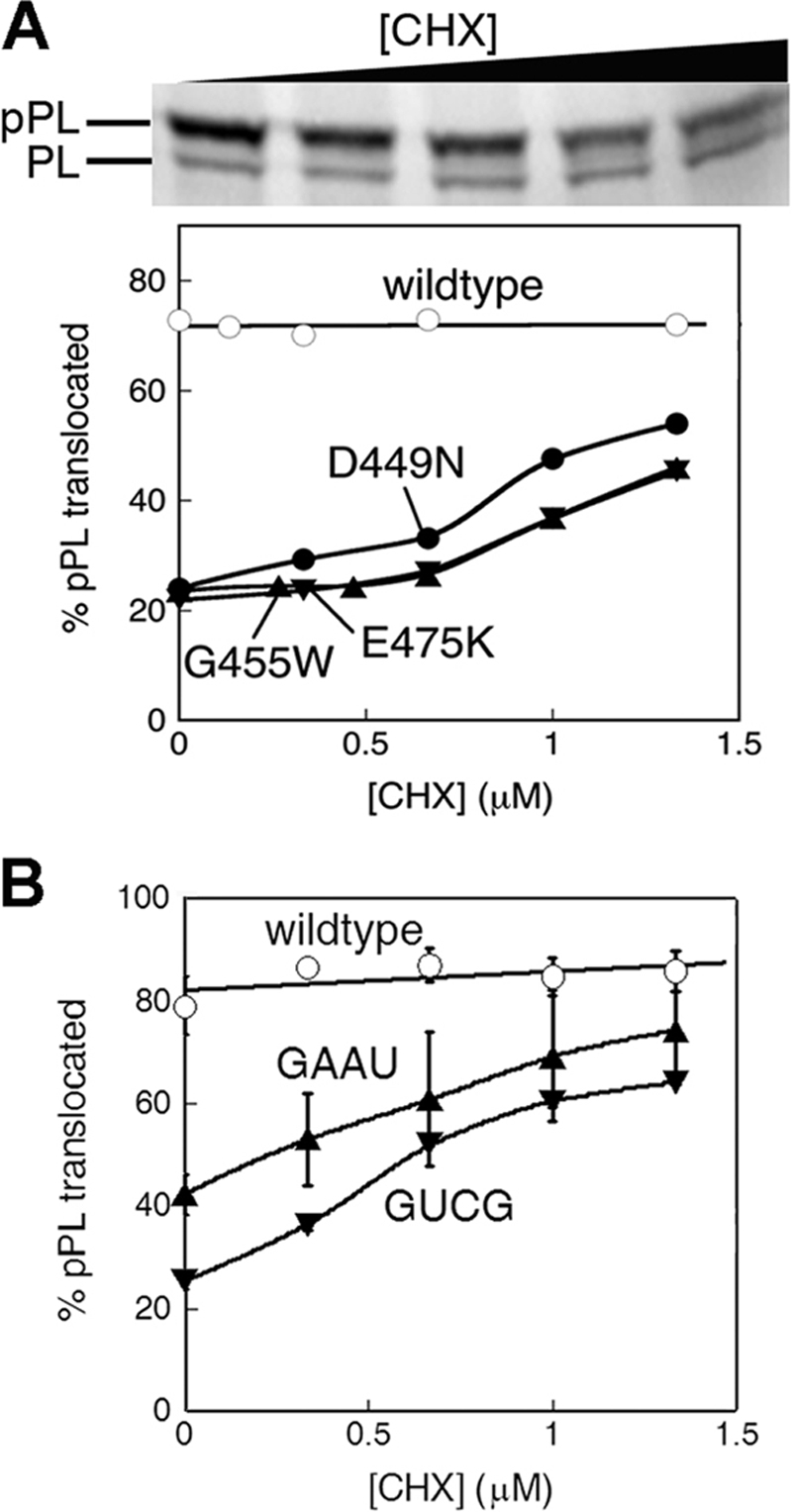
Mutations that compromise the kinetics of SRP-SR interaction can be rescued by reducing the rate of translation elongation. A, CHX rescues the defects of mutant FtsYs in the targeting of pPL. B, CHX rescues the defects of mutant SRP RNAs in the targeting of pPL.
Slower Translation Rescues Targeting of Suboptimal SRP Substrates in Vivo
Thus far, in vitro experiments showed that a reduced rate of translation elongation rescues kinetic defects in SRP-dependent protein targeting, caused by either suboptimal signal sequences or by defective SRP or SRP receptors. To test whether this phenomena occurs in vivo, we adapted a sensitive assay to detect protein targeting in vivo established by Jander et al. (31, 32). In this assay, a small biotinylatable peptide (Avi tag) or the biotinylatable domain from PSBT was fused to the periplasmic domain of the protein substrate of interest. Inefficient targeting of the substrate protein results in its biotinylation because the substrate protein accumulates in the cytoplasm, whereas efficient targeting and translocation of the substrate protein allow it to escape biotinylation. We tested three model substrates: FtsQ, a bona fide SRP substrate; and PhoA and EspP, both of which have weak signal sequences and are targeted preferentially by the Sec pathway (38, 42). An additional FLAG tag on these proteins provided an internal control for their expression levels.
We tested whether the reduction in translation elongation rate could increase the efficiency of SRP-dependent targeting of phoA and EspP and allow them to enter the SRP pathway in vivo. The SRP dependence of the targeting of these model substrate proteins was assessed using the strain HDB52, in which the expression of genomic SRP was placed under the control of the arabinose promoter (30). In the presence and absence of arabinose, the cells exhibit SRP+ and SRP− genotypes, respectively. SecB was also removed from this strain (30), which allows us to focus on SRP-dependent protein targeting. Consistent with FtsQ being a strongly SRP-dependent substrate protein, no biotinylation of FtsQ-PSBT was observed in wild-type cells, whereas depletion of SRP resulted in significant biotinylation of FtsQ despite a slight reduction in its total expression level in SRP− cells (Fig. 5A). The addition of low doses of a translation elongation inhibitor, tetracyclin, did not have a detectable effect on the targeting of FtsQ (Fig. 5A). In contrast, substantial biotinylation of the phoA-avi fusion protein was observed in SRP+ cells in the absence of tetracyclin, consistent with the notion that phoA is a suboptimal substrate for the SRP and is preferentially targeted by the alternative Sec pathway (Fig. 5B). The presence of a translation elongation inhibitor, tetracyclin, completely eliminated the biotinylation of phoA-avi in SRP+ cells but did not affect the degree of its biotinylation in SRP− cells, indicating that phoA is targeted much more efficiently by the SRP when translation elongation was slowed down. Similarly, the addition of tetracyclin removed the biotinylation of EspP in SRP+ cells, allowing it to be efficiently targeted by the SRP (Fig. 5C). Together, these results support conclusions from the in vitro study and strongly suggested that attenuation of translation elongation substantially increases the fraction of suboptimal substrate proteins that can enter the SRP pathway and thus relaxes the stringency of substrate selection by the SRP in bacterial cells.
FIGURE 5.

Slower translation elongation rescues the targeting defect of sub-optimal SRP substrate proteins in vivo. Failures in the efficient SRP-dependent targeting of FtsQ-PSBT (A), PhoA-Avi (B), and EspP-PSBT (C) were detected by their biotinylation in the cytoplasm, as described in the text, in wild-type (arabinose, +) and SRP-depleted (arabinose, −) cells and in the presence and absence of the translation elongation inhibitor tetracyclin.
Protein Targeting by Mammalian SRP Is Also Regulated by Translation Rates
Compared with the bacterial SRP, the mammalian SRP is much more complex and contains an additional Alu domain that can arrest translation elongation by the ribosome (21–24, 26). This raises the question: is regulation of the SRP pathway by translation elongation rates restricted to bacterial cells, or is substrate selection by the eukaryotic SRP subject to similar regulation?
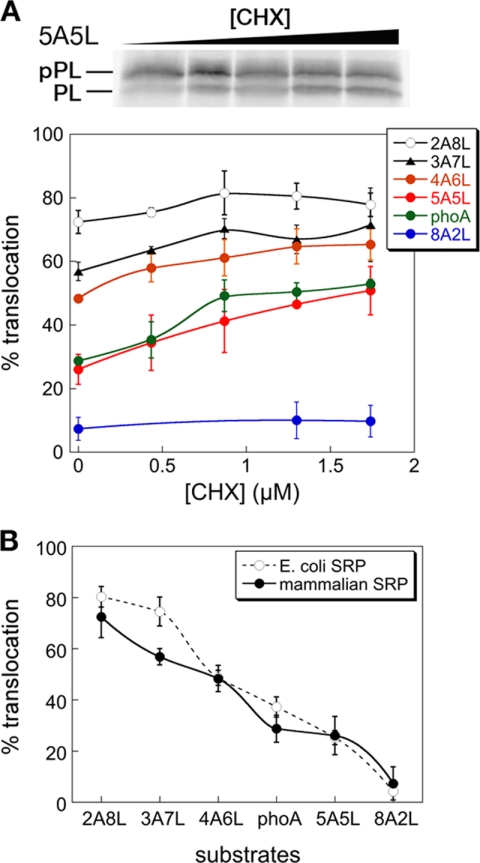
To address this question, we tested the targeting and translocation of the series of pPL variants (Table 1) in RRL, in which the mammalian SRP and SRP receptor mediate the targeting of these proteins. Analogous to observations with bacterial SRP and FtsY, the targeting of suboptimal SRP substrates could be rescued by reducing translation rates using low doses of CHX, whereas the targeting of strong SRP substrates, such as 2A8L, was not significantly affected (Fig. 6A). For example, the targeting and translocation efficiency of two weak SRP substrates, phoA and 5A5L, increased from 26–28% to over 50% (Fig. 6A). The extent of rescue by CHX was ∼10% less than that observed with the bacterial SRP (Fig. 6A versus Fig. 3A) but was substantial nevertheless. We also note that equal amount of CHX was less effective in slowing down translation elongation by the RRL than the wheat germ ribosomes (cf. band intensity in Fig. 6A versus Fig. 3A). Thus, substrate selection by the mammalian SRP could also be modulated by translation elongation rates.
FIGURE 6.
Protein targeting by the mammalian SRP is also subject to regulation by translation elongation rates. A, CHX rescues the targeting of proteins with suboptimal signal sequences by the mammalian SRP and SR. B, comparison of the pattern of substrate selection by the mammalian (solid line) and bacterial (dashed line) SRP/SR systems. The data for the bacterial SRP were from Fig. 3.
It has been suggested that substrate selection by the mammalian SRP is less stringent than that by the bacterial SRP. Our data on the targeting of the same series of substrate proteins by both SRP systems allowed us to directly test this notion. The comparison showed that the bacterial SRP does not exhibit stronger discrimination against suboptimal substrate proteins than the mammalian SRP (Fig. 6B). Indeed, the substrate 3A7L is reproducibly targeted less efficiently by the mammalian SRP (Fig. 6B). Collectively, these results suggest that, despite the complexity of the mammalian SRP compared with its bacterial homologue, the pattern of substrate selection is not substantially different between the two pathways, and both could be subject to kinetic regulation by ongoing translation elongation.
DISCUSSION
Proper localization of proteins to their correct cellular destinations is essential for the order and organization in all cells. How cellular protein targeting machineries select the correct set of substrates based on degenerate signal sequences remains a challenging question. Previous work showed that substrate selection by the SRP is governed by a combination of factors, including the binding affinity between the SRP and translating ribosome (the cargo), the ability of correct cargos to induce rapid SRP-FtsY complex assembly, and kinetic proofreading through GTP hydrolysis in the SRP-FtsY complex (17). The interplay between SRP and other ribosome-associated cellular chaperones, such as trigger factor in bacteria (38, 43–45) and the nascent polypeptide associated complex in eukaryotic cells (46–48), has also been suggested to modulate the fidelity of the SRP pathway, although these models remain to be rigorously tested. The results here demonstrate another important determinant of substrate selection by the SRP: kinetic competition with ongoing translation elongation. Modest reductions in translation elongation rates allowed many suboptimal substrate proteins to be targeted by the SRP, substantially relaxing the stringency of SRP in substrate selection. This phenomenon was observed both in vitro and in vivo and occurred with both the bacterial and mammalian SRP systems.
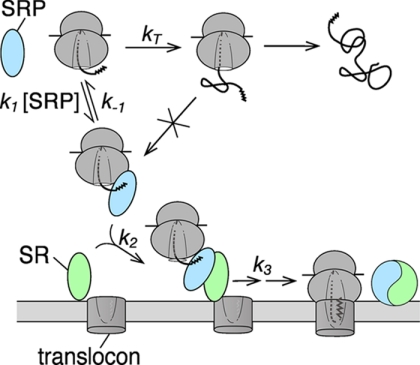
Fig. 7 depicts a model that extends the previous studies and accounts for the effect of translation elongation on substrate selection by the SRP. In this model, the fraction of substrate proteins successfully targeted by the SRP is determined by the relative kinetics of nascent polypeptide elongation (kT) versus the protein targeting reaction, the latter being a collective function of the kinetics of SRP cargo binding (k1 and k-1), recruitment of the SRP receptor (k2), and unloading of cargo to the translocation machinery (k3). Because cargo proteins lose competence to be targeted by the SRP when the nascent polypeptide exceeds a critical length (19, 20), there is a limited time window for the action of SRP that is dictated by the rate of translation elongation (kT). With correct cargos bearing strong signal sequences, targeting occurs more rapidly than translation, allowing most of these proteins to be successfully delivered within this time window. Thus, extending this time window would not have a significant effect on the targeting efficiency of strong SRP substrates. In contrast, cargos with weak signal sequences are much slower in their targeting and are thus out-competed by ongoing translation elongation. Extending the time window for protein targeting by slowing down translation elongation thus allows a larger fraction of these substrates to complete their targeting reaction and relaxes the specificity of SRP.
FIGURE 7.
Model of kinetic competition between protein synthesis and co-translational protein targeting. The ribosome-nascent chain complex can be targeted by the SRP pathway through three major steps, cargo binding (k1 and k−1), SRP-SR assembly (k2), and cargo unloading (k3) within a limited time window, which is dictated by the rate of translation elongation (kT) and the critical nascent chain length beyond which the SRP loses targeting competence.
The concept that translation elongation plays an important role in the SRP pathway has been suggested by several previous studies. For example, the growth of SRP-deficient mutant cells could be partially restored by low doses of translation elongation inhibitor (21); the defect of mutant mammalian SRP lacking the Alu domain in both protein targeting and in supporting cell growth could be rescued by slower translation elongation (26). These results have been interpreted to indicate that translation elongation arrest by the Alu domain provides a longer time window for protein targeting and is essential for the function of the mammalian SRP. The results here describe an important extension, or consequence, of the interplay between translation and the SRP pathway. They demonstrate that, in rapidly growing cells, the competition between translation elongation and co-translational protein targeting provides an important factor that dictates the set of cargo proteins that engage the SRP pathway.
The results here reinforced the notion that, in addition to SRP-cargo binding affinity, the kinetics of subsequent steps in the pathway plays a key role in maintaining the specificity of SRP. The targeting defect of pPL mutant bearing β-OH-leucine in place of leucine in the signal sequence could not be rescued by increasing SRP concentration, suggesting that the targeting defect of this mutant did not arise solely from weaker cargo binding. The observation that slower translation elongation could rescue the targeting of suboptimal SRP substrates also pointed at the kinetic origin of their targeting defects. Although in principle, the kinetics of any of the steps in the targeting pathway, including cargo binding, delivery, and unloading (Fig. 7, k1, k2, and k3, respectively) could be slower with suboptimal signal sequences, SRP-cargo binding is a relatively fast process (k1 = ∼4 × 106 m−1 s−1) (49) and not strongly dependent on the signal sequence.3 In contrast, stable assembly of the SRP-FtsY complex, which mediates the delivery of cargo (k2), differs up to 103-fold between strong and weak signal sequences (15, 17). Thus, downstream steps after the binding of cargo play key roles in determining the efficiency of SRP-dependent protein targeting and in the ability of SRP to discriminate against substrates with suboptimal signal sequences.
Compared with the bacterial SRP, the additional Alu domain in mammalian SRP allows it to arrest translation elongation once the SRP binds a translating ribosome. As proposed previously, this elongation arrest activity extends the time window for protein targeting by the mammalian SRP and, in principle, should exert an effect similar to that of translation elongation inhibitors. Given this, it is curious that modest reductions in the rates of translation elongation also rescued the ability of mammalian SRP to target proteins with weak signal sequences. It is possible that the amount of translation elongation arrest by the mammalian SRP is quite limited (23). Alternatively or in addition, the mammalian SRP does not exert a strong translation elongation arrest for ribosomes bearing weak signal sequences, which could provide an additional mechanism to help the mammalian SRP discriminate against the incorrect cargo. These possibilities remain to be tested and distinguished. Another interesting conclusion from comparison of the mammalian and bacterial SRP in this work is that, despite the much higher complexity of mammalian SRP, the pattern and stringency of substrate selection by the two SRP systems are quite similar. Whether this reflects evolutionarily conserved mechanisms to ensure the fidelity of the SRP pathway or whether distinct mechanisms used by the two systems could reach a similar stringency of substrate selection remains an open question.
The observation that modest reductions in the rate of protein synthesis could significantly rescue the targeting of suboptimal substrate proteins by the SRP raises the intriguing possibility that, in living cells, the spectrum of SRP-dependent substrate proteins is dynamic and subject to changes by environmental or signaling cues. Stress conditions that trigger a reduction in cellular protein synthesis rates would alter the set of substrate proteins targeted by the SRP. In bacterial cells, this could imply that a substantial fraction of proteins that are targeted by the Sec pathway, such as phoA, could become routed through the SRP pathway under these compromised cellular conditions. In mammalian cells, this could lead to increased localization of proteins to the endoplasmic reticulum that are otherwise inefficiently targeted by the SRP. Whether cells could take advantage of this phenomenon to help adapt to stress conditions or whether such relaxed specificity in protein targeting could cause more detriments to cell survival remain intriguing questions for future investigations.
Supplementary Material
Acknowledgments
We thank R. Hegde for RRL and microsomal membranes, H. Bernstein for the strain HDB52, J. Beckwith for antibodies, and X. Zhang and members of the Shan group for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM078024 (to S. S.).

This article contains supplemental Figs. S1–S3.
S. Shan, unpublished results.
- SRP
- signal recognition particle(s)
- SR
- SRP receptor
- pPL
- preprolactin
- RRL
- rabbit reticulocyte lysate
- TKRM
- salt-washed and partial trypsin-digested microsomal membrane
- CHX
- cycloheximide
- PSBT
- Propionibacterium shermania transcarboxylase.
REFERENCES
- 1. Pool M. R. (2005) Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review). Mol. Membr. Biol. 22, 3–15 [DOI] [PubMed] [Google Scholar]
- 2. Walter P., Johnson A. E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10, 87–119 [DOI] [PubMed] [Google Scholar]
- 3. Walter P., Blobel G. (1983) Disassembly and reconstitution of signal recognition particle. Cell 34, 525–533 [DOI] [PubMed] [Google Scholar]
- 4. Janda C. Y., Li J., Oubridge C., Hernández H., Robinson C. V., Nagai K. (2010) Recognition of a signal peptide by the signal recognition particle. Nature 465, 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keenan R. J., Freymann D. M., Walter P., Stroud R. M. (1998) Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94, 181–191 [DOI] [PubMed] [Google Scholar]
- 6. Zopf D., Bernstein H. D., Johnson A. E., Walter P. (1990) The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 9, 4511–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batey R. T., Rambo R. P., Lucast L., Rha B., Doudna J. A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287, 1232–1239 [DOI] [PubMed] [Google Scholar]
- 8. Egea P. F., Shan S. O., Napetschnig J., Savage D. F., Walter P., Stroud R. M. (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427, 215–221 [DOI] [PubMed] [Google Scholar]
- 9. Focia P. J., Shepotinovskaya I. V., Seidler J. A., Freymann D. M. (2004) Heterodimeric GTPase core of the SRP targeting complex. Science 303, 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernstein H. D., Zopf D., Freymann D. M., Walter P. (1993) Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc. Natl. Acad. Sci. U.S.A. 90, 5229–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Powers T., Walter P. (1997) Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16, 4880–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shan S. O., Chandrasekar S., Walter P. (2007) Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J. Cell Biol. 178, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peluso P., Herschlag D., Nock S., Freymann D. M., Johnson A. E., Walter P. (2000) Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 288, 1640–1643 [DOI] [PubMed] [Google Scholar]
- 14. Peluso P., Shan S. O., Nock S., Herschlag D., Walter P. (2001) Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40, 15224–15233 [DOI] [PubMed] [Google Scholar]
- 15. Shen K., Zhang X., Shan S. (2011) Synergistic actions between the SRP RNA and translating ribosome allow efficient delivery of the correct cargos during cotranslational protein targeting. RNA 17, 892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., Schaffitzel C., Ban N., Shan S. (2009) Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc. Natl. Acad. Sci. 106, 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X., Rashid R., Wang K., Shan S. (2010) Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science 328, 757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bradshaw N., Neher S. B., Booth D. S., Walter P. (2009) Signal sequences activate the catalytic switch of SRP RNA. Science 323, 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flanagan J. J., Chen J. C., Miao Y., Shao Y., Lin J., Bock P. E., Johnson A. E. (2003) Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 278, 18628–18637 [DOI] [PubMed] [Google Scholar]
- 20. Siegel V., Walter P. (1988) The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 7, 1769–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogg S. C., Walter P. (1995) SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 22. Walter P., Blobel G. (1981) Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91, 557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolin S. L., Walter P. (1989) Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 109, 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halic M., Becker T., Pool M. R., Spahn C. M., Grassucci R. A., Frank J., Beckmann R. (2004) Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 [DOI] [PubMed] [Google Scholar]
- 25. Siegel V., Walter P. (1985) Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J. Cell Biol. 100, 1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lakkaraju A. K., Mary C., Scherrer A., Johnson A. E., Strub K. (2008) SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walter P., Blobel G. (1983) Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96, 84–93 [DOI] [PubMed] [Google Scholar]
- 28. Shan S. O., Walter P. (2003) Induced nucleotide specificity in a GTPase. Proc. Natl. Acad. Sci. U.S.A. 100, 4480–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X., Kung S., Shan S. (2008) Demonstration of a multistep mechanism for assembly of the SRP × SRP receptor complex. Implications for the catalytic role of SRP RNA. J. Mol. Biol. 381, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee H. C., Bernstein H. D. (2001) The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. U.S.A. 98, 3471–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jander G., Cronan J. E., Jr., Beckwith J. (1996) Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J. Bacteriol. 178, 3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian H., Boyd D., Beckwith J. (2000) A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. 97, 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma A., Mariappan M., Appathurai S., Hegde R. S. (2010) In vitro detection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol. Biol. 619, 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang D., Sweredoski M. J., Graham R. L., Hess S., Shan S. O. (2011) Novel proteomic tools reveal essential roles of SRP and importance of proper membrane protein biogenesis. Mol. Cell Proteomics 11, 10.1074/mcp.M1111.011585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee C. A., Beckwith J. (1986) Suppression of growth and protein secretion defects in Escherichia coli secA mutants by decreasing protein synthesis. J. Bacteriol. 166, 878–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walter P., Ibrahimi I., Blobel G. (1981) Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91, 545–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doud S. K., Chou M. M., Kendall D. A. (1993) Titration of protein transport activity by incremental changes in signal peptide hydrophobicity. Biochemistry 32, 1251–1256 [DOI] [PubMed] [Google Scholar]
- 38. Valent Q. A., Kendall D. A., High S., Kusters R., Oudega B., Luirink J. (1995) Early events in preprotein recognition in E. coli. Interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 14, 5494–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X., Lam V. Q., Mou Y., Kimura T., Chung J., Chandrasekar S., Winkler J. R., Mayo S. L., Shan S. O. (2011) Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 6450–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng N., Gierasch L. M. (1996) Signal sequences. The same yet different. Cell 86, 849–852 [DOI] [PubMed] [Google Scholar]
- 41. Shan S. O., Stroud R. M., Walter P. (2004) Mechanism of association and reciprocal activation of two GTPases. Plos Biol. 2, e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peterson J. H., Szabady R. L., Bernstein H. D. (2006) An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J. Biol. Chem. 281, 9038–9048 [DOI] [PubMed] [Google Scholar]
- 43. Buskiewicz I., Deuerling E., Gu S. Q., Jöckel J., Rodnina M. V., Bukau B., Wintermeyer W. (2004) Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl. Acad. Sci. U.S.A. 101, 7902–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fedyukina D. V., Cavagnero S. (2011) Protein folding at the exit tunnel. Annu. Rev. Biophys. 40, 337–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lakshmipathy S. K., Gupta R., Pinkert S., Etchells S. A., Hartl F. U. (2010) Versatility of trigger factor interactions with ribosome-nascent chain complexes. J. Biol. Chem. 285, 27911–27923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. del Alamo M., Hogan D. J., Pechmann S., Albanese V., Brown P. O., Frydman J. (2011) Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. Plos Biol. 9, e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S., Sakai H., Wiedmann M. (1995) NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 130, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wickner W. (1995) The nascent-polypeptide-associated complex. Having a “NAC” for fidelity in translocation. Proc. Natl. Acad. Sci. U.S.A. 92, 9433–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saraogi I., Zhang D., Chandrasekaran S., Shan S. (2011) Site-specific fluorescent labeling of nascent proteins on the translating ribosome. J. Am. Chem. Soc. 133, 14936–14939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.