Abstract
Protein post-translational modifications (PTMs) represent important regulatory states that when combined have been hypothesized to act as molecular codes and to generate a functional diversity beyond genome and transcriptome. We systematically investigate the interplay of protein phosphorylation with other post-transcriptional regulatory mechanisms in the genome-reduced bacterium Mycoplasma pneumoniae. Systematic perturbations by deletion of its only two protein kinases and its unique protein phosphatase identified not only the protein-specific effect on the phosphorylation network, but also a modulation of proteome abundance and lysine acetylation patterns, mostly in the absence of transcriptional changes. Reciprocally, deletion of the two putative N-acetyltransferases affects protein phosphorylation, confirming cross-talk between the two PTMs. The measured M. pneumoniae phosphoproteome and lysine acetylome revealed that both PTMs are very common, that (as in Eukaryotes) they often co-occur within the same protein and that they are frequently observed at interaction interfaces and in multifunctional proteins. The results imply previously unreported hidden layers of post-transcriptional regulation intertwining phosphorylation with lysine acetylation and other mechanisms that define the functional state of a cell.
Keywords: kinase, N-acetyltransferase, network, phosphatase, post-translational modification
Introduction
Cells constantly need to adapt their endogenous biochemical activities to a changing environment. An important level of adaptation is achieved by series of post-translational modifications (PTMs) that affect the chemical properties of proteins, conferring molecular diversity beyond the amino-acid sequence. More than 200 different PTMs have been described, and these are known to affect many aspects of protein function, such as activity, stability and interaction (Singh et al, 2007; Li et al, 2009; Deribe et al, 2010; Wang et al, 2010; Zhao et al, 2010). Among all PTMs, reversible protein phosphorylation and lysine acetylation represent prominent and ubiquitous regulatory mechanisms that are conserved from bacteria (Yu et al, 2008; Zhang et al, 2009a; Wang et al, 2010) to humans (Kim et al, 2006; Choudhary et al, 2009; Huttlin et al, 2010; Zhao et al, 2010). Protein phosphorylation is regulated by a variety of kinases and phosphatases, which are themselves regulated by phosphorylation within complex networks (Bodenmiller et al, 2010; Zorina et al, 2011). A series of mass spectrometry (MS)-based methods are currently available that allow the characterization of phosphorylation and lysine acetylation at unprecedented scales (Kim et al, 2006). Most recent analyses have captured 5000–10 000 phosphorylated (Van Hoof et al, 2009; Huttlin et al, 2010; Rigbolt et al, 2011) and 1700 lysine-acetylated (Choudhary et al, 2009) proteins, and large inventories of phosphorylation and acetylation sites are currently available (e.g., PHOSIDA (Gnad et al, 2011), Phospho.ELM (Dinkel et al, 2011) or PhosphoSite (Hornbeck et al, 2004)).
In eukaryotes, many proteins have been observed to be modified on multiple sites, and some nuclear transcription factors (Yang and Seto, 2008b), cytoskeletal proteins (Reed et al, 2006; Zhang et al, 2009b) and protein chaperones (Scroggins et al, 2007) even carry several different PTMs, reminiscent of a molecular barcode. Inspired by histones, for which phosphorylation or lysine acetylation at specific positions influence further modifications and form complex regulatory circuits (Strahl and Allis, 2000; Jenuwein and Allis, 2001), the hypothesis of a protein modification code has been proposed, whereby dynamic patterns of protein modification would encode for alternative protein functions (Yang and Seto, 2008a). However, large-scale studies that consistently investigated direct modulation of one PTM by another are sparse (Yao et al, 2011). It is also unclear when such a code has evolved as prokaryotes are poorly studied in this respect with only a few instances of multiple PTMs in individual proteins having been previously identified (Soufi et al, 2008; Prisic et al, 2010).
Accordingly, here we exhaustively studied two important PTM events—serine/threonine/tyrosine phosphorylation and lysine acetylation in the bacterium Mycoplasma pneumoniae, a human pathogen that causes atypical pneumonia (Waites and Talkington, 2004). This organism is established as a suitable model organism for large-scale systems-wide analyses on proteome, transcriptome, metabolic and protein networks (Guell et al, 2009; Kuhner et al, 2009; Yus et al, 2009; Maier et al, 2011). This self-replicating organism has one of the smallest known genomes (691 protein encoding genes) (Dandekar et al, 2000; Guell et al, 2009). It encodes a reduced PTM machinery, perturbation of which would reveal many of the regulatory cascades. Since it contains only one protein phosphatase (PrpC∣Mpn247), two known serine/threonine protein kinases (HprK∣Mpn223 and PknB∣PrkC∣Mpn248) and two putative N-acetyltransferases (Mpn027 and Mpn114), M. pneumoniae represents an ideal model organism in which to study system-wide impact of phosphorylation on other PTMs. We combined genetics and high-resolution quantitative MS to measure the global effect of kinase and phosphatase deletions on proteome abundance, phosphorylation and lysine acetylation. The study provides a first unbiased and quantifying view on cross-talk between phosphorylation and lysine acetylation and also suggests that these regulatory circuits are a fundamental principle of regulation that might have evolved before the divergence of prokaryotes and eukaryotes.
Results
Quantifying the M. pneumoniae proteome, phosphoproteome and lysine acetylome
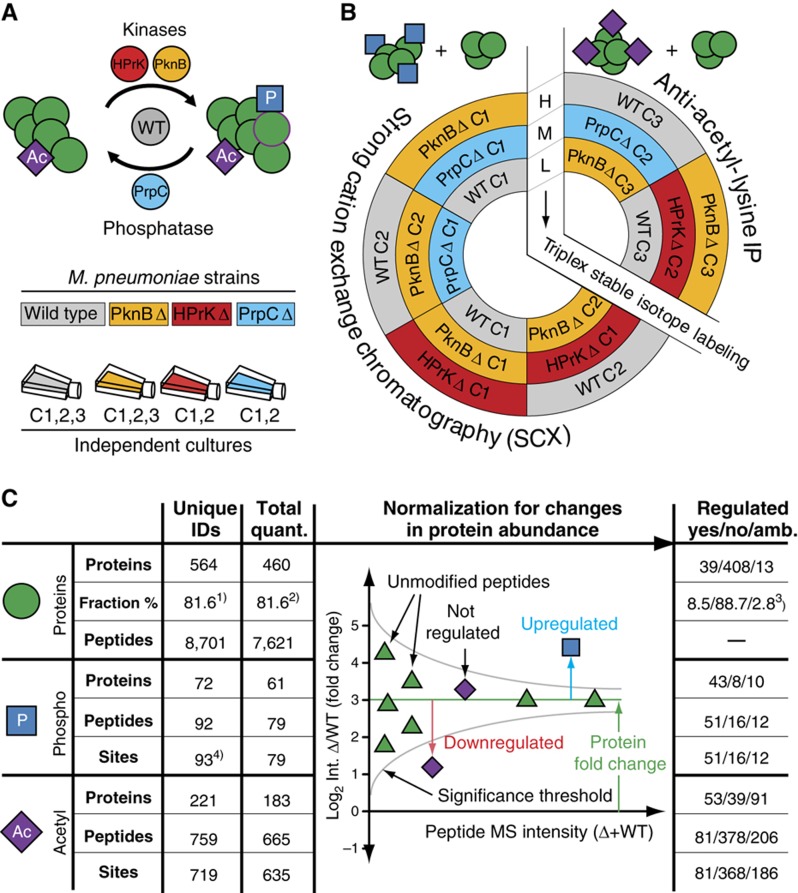
To gather insights into the mechanism of prokaryotic phosphorylation, and to systematically chart impacts of protein phosphorylation on lysine acetylation, we profiled both modifications in wild-type strains of M. pneumoniae and three isogenic mutants deficient in either one of the two protein kinases, HprK and PknB, or the phosphatase, PrpC (Halbedel et al, 2006) (Figure 1A). We applied a quantitative proteomics approach based on chemical, differential labeling with three isotopic dimethyl forms (Boersema et al, 2009). The chemically encoded digested proteomes (originating from the four strains) were combined according to a scheme that includes both technical and biological replicates to ensure that each proteome is chemically labeled with at least two different stable isotopes (Figure 1B; see Materials and methods). To reduce the complexity of the samples and increase sensitivity, peptides were subjected to fractionation: non-phosphorylated and phosphorylated peptides were separated by strong cation exchange (SCX) chromatography (Mohammed and Heck, 2010), whereas lysine-acetylated peptides were enriched using a specific antibody (Choudhary et al, 2009). All fractions were analyzed using a nano LC-LTQ-Orbitrap (Thermo, San Jose, CA) (see Materials and methods). Unmodified, phosphorylated and lysine-acetylated peptides were identified with the Mascot search engine using the M. pneumoniae sequence (UniProt) and corresponding decoy databases: peptide thresholds were set at false discovery rates (FDRs) of 1%. The majority (75%) of phosphorylation and all lysine acetylation sites could be localized to a single amino acid (see Materials and methods). Modified and unmodified peptides were quantified using the software MSQuant (Mortensen et al, 2010). Importantly, to prevent possible biases due to variation in protein expression, the relative intensities of modified peptides were normalized for changes in protein abundance (Figure 1C) (Wu et al, 2011). For each peptide, the statistical significance of the observed change in abundance was computed with the software OutlierD (Cho et al, 2008). The test provides a P-value based on the variation in the normalized ratios observed for all peptides with similar intensities (see Materials and methods). The thresholds were set stringently. Only changes that were statistically significant and that could be further confirmed by visual inspection were considered for further analysis. Additionally, changes in intensities lower than ∼2.8 × (log2<1.5, for proteins and phosphopeptides) or 4 × (log2<2, for lysine-acetylated peptides) were disregarded. We also used the reproducibility between the different technical and biological duplicates to further assess the reliability of the experimental and computational approaches (Supplementary Figure S1C–F). The biological reproducibility for the identification of proteins and phosphopeptides (between the four analyzed mixtures) were 86% and 98%, respectively. The reproducibility for the identification of lysine-acetylated peptides (two mixtures) is 70%. We also measured the reproducibility for the quantification as follows: (1) technical replicates with reverse labeling were included for all phosphorylation measurements (all four strains), whereas for lysine acetylation measurements, technical duplicates were present in the wild-type and PknB mutants and (2) biological replicates were included for the quantification of proteins and phosphopeptides in the wild-type and the PknB mutant (Figure 1). Technical reproducibility of detecting upregulation or downregulation reaches 93%, whereas biological reproducibility varies from 63 to 90%. However, for the final data sets (Supplementary Tables SI–SIII), all changes in abundances that were not reproducible between the technical duplicates were excluded.
Figure 1.
Synopsis of the systematic quantification of M. pneumoniae proteome. (A) Experimental design for the proteomic comparison of three M. pneumoniae deletion strains: hprK (red), pknB (yellow) and prpC (blue) with wild type (gray). The analyses account for biological variations and include several independent cultures of each strain (C1–C3). (B) Sample labeling and mixing scheme for the relative quantification of the differently perturbed proteomes. The various proteomes are differentially labeled with three stable isotopic forms of dimethyl, light (L), medium (M) and heavy (H). The mixing scheme includes both technical and biological duplicates. (C) Overall number of proteins and peptides identified and quantified. All PTMs data were normalized for changes in protein abundance (illustrated here for one protein). The fraction refers to (1) all ORFs annotated in Uniprot (UniProt Consortium, 2011), (2) the number of identified proteins, (3) the number of quantified proteins and (4) two peptides were identified with two phosphosites and two overlapping peptides cover one single phosphosite.
Overall, we identified 564 proteins, of which 460 (81.6%) were quantified. For 104 proteins, we did not obtain quantitative measurements because the proteins were identified with too few peptides (fewer than three) or the extracted ion chromatograms (XICs) peaks for some peptides were overlapping, precluding unambiguous and reliable quantification. Close to half of all identified proteins (241; 42.7%) were found modified by either phosphorylation or lysine acetylation. In total, 93 phosphorylation and 719 lysine acetylation sites were characterized on 72 and 221 proteins, respectively (Figure 1; Supplementary Tables S1–S3). We observed phosphorylation on serines (58%), threonines (37%) and tyrosines (5%) consistent to previous studies in other bacteria (Mijakovic et al, 2006; Macek et al, 2007, 2008; Soufi et al, 2008). The phosphorylation data set is extensive, and adding an extra enrichment step, using titanium dioxide (TiO2), lead to only two additional phosphopeptides. Similarly, the consideration of previous phosphoproteomics studies in M. pneumoniae (based on two-dimensional gel electrophoresis) (Schmidl et al, 2010b) added only 11 additional phosphosites (11%). The vast majority (98%) of the lysine-acetylated peptides were identified in the anti-acetyl-lysine fraction, which contained a 20-fold enrichment in sites. This represents the largest lysine acetylation data set performed in a prokaryote. To assess quality further, we randomly selected 11 proteins from the lysine acetylome data set and independently confirmed nine lysine acetylations by immunostaining with an anti-acetyl-lysine antibody, two could not be confirmed using this method (Supplementary Figure S2). For example, we found all enzymes in the glycolytic pathway to be phosphorylated or lysine-acetylated, consistent with previous reports in human (Zhao et al, 2010) and other bacteria (Mijakovic et al, 2006; Macek et al, 2007, 2008; Soufi et al, 2008; Wang et al, 2010).
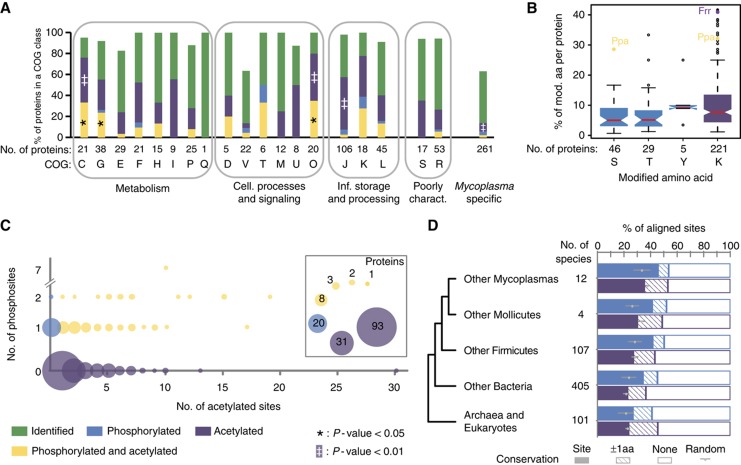
The data set covers 81.6% of all annotated open reading frames (ORFs) or 93% of the previously identified M. pneumoniae proteome (Jaffe et al, 2004; Kuhner et al, 2009) (Supplementary Figure S1A). The sets of protein identified broadly cover all cellular functions (Figure 2A). The median sequence coverage of all identified proteins reaches 43%, a value close to the known upper detection limit inherent to current MS-based protocols (Supplementary Figure S1B) (Swaney et al, 2010). We tested the set for several possible biases, and found that the sets of proteins identified broadly cover all biophysical and biochemical properties (Supplementary Figure S3). Taken together, the data set is among the most comprehensive analyses of both phosphorylation and lysine acetylation in a single prokaryote. The results show that lysine acetylation in M. pneumoniae is very common, being at least as frequent as phosphorylation.
Figure 2.
M. pneumoniae proteome, phosphoproteome and lysine acetylome. (A) Modified proteins are significantly enriched in functions related to metabolism and cellular processes and signaling. (B) Box plots indicating the relative extents of modification for serines, threonines, tyrosines and lysines within all observed modified proteins. (C) Modified proteins show a complex pattern of modification. Bubble plot showing the number of proteins with distinct modification profiles. (D) Phosphorylation is more positionally conserved in bacteria than lysine acetylation. The fraction of conserved modified residues is shown for either precise site conservation (site conservation) or a more plastic conservation within three residues (±1 amino acids conservation).
Evolutionary conservation of phosphorylation and lysine acetylation sites
Both serine/threonine phosphorylation and lysine acetylation are ancient PTMs conserved throughout evolution (Kennelly, 2002; Choudhary et al, 2009). Due to their high fraction in M. pneumoniae proteins, we were able to evaluate the phylogenetic conservation of phosphorylated and lysine-acetylated sites. We observed that for 23% of acetylated-lysines in proteins with a eukaryotic ortholog, the lysine residues were conserved from Mycoplasma species to eukaryotes (Figure 2D). The most conserved PTM sites (>60% conserved in >80 eukaryotes) were frequently found in metabolic enzymes compared with other sites within other evolutionary ubiquitous proteins (P=2.0 × 10−4, Fisher's exact test). When considering exactly the same residues, lysine acetylation sites appear slightly less conserved than phosphorylation sites (P=2.8 × 10−3). However, when the acetylated-lysine was not conserved, an alternative lysine could be frequently found in other species within a window of three amino acids, one upstream or one downstream of the original aligned site (Figure 2D). This suggests that for some lysine acetylation sites, the exact position may not be so critical to maintain function.
We also observed that proteins only occurring in species of the Mycoplasma genus were found to be less frequently modified than other proteins (Figure 2A), indicating that regulation through PTMs can evolve only secondary to proteome differentiation. The sites in these proteins likely represent recently acquired regulatory signals. Interestingly, none of these sites is conserved across all of the 12 sequenced Mycoplasma species, suggesting that they play species-specific regulatory functions.
Dissecting the roles of kinases and phosphatase in M. pneumoniae phosphoproteome
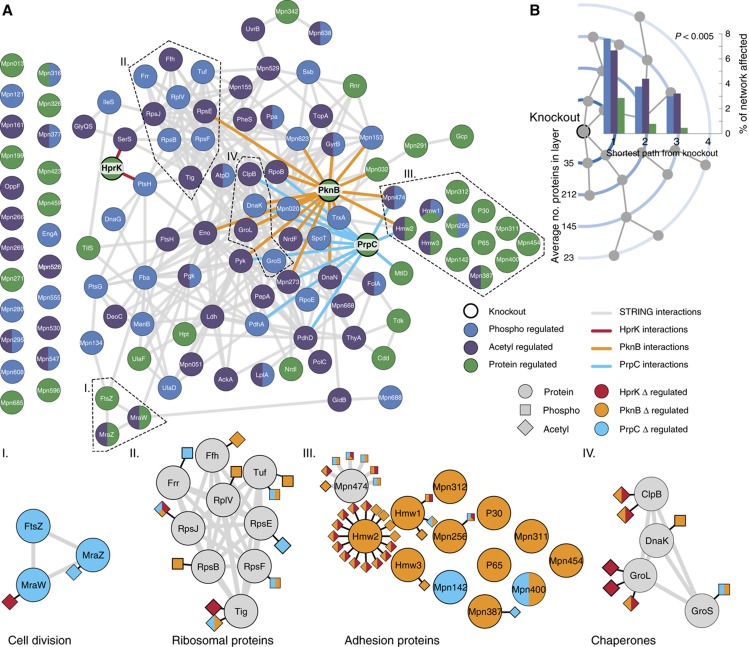
As consideration of putative changes in protein abundance is critical for the proper interpretation of PTM data (Wu et al, 2011), we first quantified the impact of kinase and phosphatase deletion on overall protein abundance. We observed that the levels of 39 of the 447 proteins quantified consistently were significantly affected (Supplementary Figure S4; Supplementary Table S1). We selected 15 for validation by western blot. Of the 45 abundances measured by western blotting in the three knockouts (k.o.), 39 (86.6%) showed upregulation and downregulation consistent with the previous MS data (Supplementary Figure S5). Among the proteins affected by PknB deletion, we found eight of the nine cytadherence proteins previously known to be downregulated upon PknB k.o. (Schmidl et al, 2010a) together with four uncharacterized proteins that could represent new players in the process of cell adhesion (Mpn256, Mpn387, Mpn400 and Mpn454; Supplementary Figure S4 and see Supplementary information). For proteins encoded by the cell-cycle operon: the ribosomal RNA small subunit methyltransferase H (MraW∣Mpn315), the cell-cycle proteins MraZ (Mpn314) and the tubulin-like protein FtsZ (Mpn317) we observed a decreased abundance upon PrpC deletion that correlated with changes in corresponding transcripts (Supplementary Table S4). However, in general, we found that mRNA levels were largely unaffected, indicating the existence of post-transcriptional regulatory mechanisms (Supplementary Table S4) (Schmidl et al, 2010a). These results show that perturbations of the phosphorylation network in M. pneumoniae affect protein abundance and turnover, acting at both transcriptional but also post-transcriptional levels.
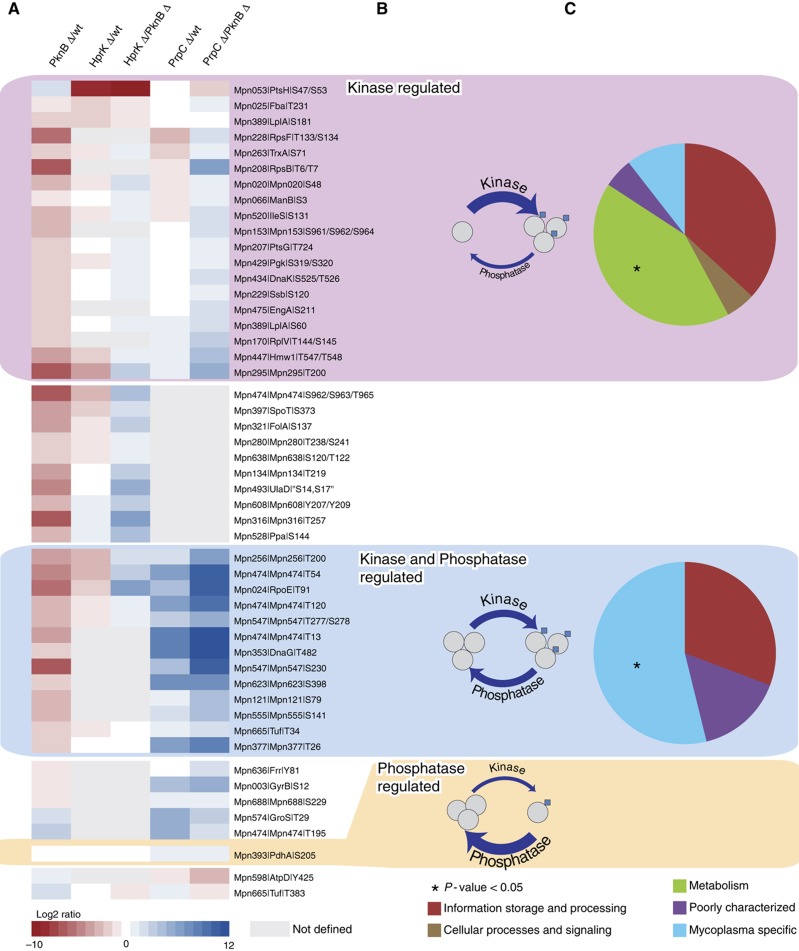
We also determined the impact of systematic kinase and phosphatase perturbation on the M. pneumoniae phosphoproteome (see Materials and methods). Of the 67 phosphosites unambiguously quantified, only 16 (23.9%) were never found to be affected (Supplementary Table S2). They might represent compensatory mechanisms, whereby deletion of one kinase may cause the other kinase to compensate. Alternatively, they might account for HprK-, PknB- and PrpC-independent phosphorylation events, including autophosphorylation of some metabolic enzymes (Jolly et al, 2000) or metabolic intermediates observed at catalytically active sites. For example, the constitutive phosphoserine (S64) in the ATP-binding site of the guanylate kinase (Gmk∣Mpn246) might represent a metabolic intermediate, since in the Escherichia coli structure the equivalent serine lies very close, though not obviously bound, to both phosphate and sulfate groups (Hible et al, 2005). However, the majority (76.1%) of the phosphosites were found regulated in at least one of the k.o., implying that the three enzymes are indeed the major modifiers of this network. The impact of protein kinase and phosphatase deletion largely depends on the extent of their substrate phosphorylation before perturbation, that is, in the wild-type cells. We thought to use the abundance profiles across the different k.o. strains to derive information on the phosphorylation stoichiometries of the different substrates (Figure 3). Phosphosites for which degrees of modification were only affected by deletion of the phosphatase PrpC were rare, possibly because such scenarios are energetically costly, implying futile cycles of phosphorylation and dephosphorylation, whereby the phosphatase activity largely overcomes that of the kinases in the wild type. Instead, for the majority of the sites, phosphorylation levels were affected by deletion of either one of the two kinases PknB and/or HprK. Phosphosites with degrees of modification exclusively downregulated in the kinase mutants were frequent in metabolic enzymes. These include the previously known HprK substrate, the phosphocarrier protein HPr (PtsH∣Mpn053). In contrast, sites with phosphorylation levels regulated in both the kinase and phosphatase k.o. were enriched for proteins specifically conserved among members of the Mycoplasma genus, suggesting recently acquired regulatory mechanisms. Previously known substrates of PknB (Schmidl et al, 2010a, 2010b): Mpn256, RpoE (Mpn024), Hmw1 (Mpn447) and Mpn474 are found in this second category. These results suggest that different cellular processes could require a discrete balance of substrate modification. Phosphorylation dynamics apparently bear functional relevance and could represent a way to extend diversity and complexity beyond the set of available enzymes and their respective specificities.
Figure 3.
Impact of systematic kinase and phosphatase k.o. on M. pneumoniae phosphoproteome. (A) Regulated phosphosites are ordered according to whether they are (1) kinase regulated (PknB or HprK) (pink), (2) kinase and phosphatase regulated (blue) or (3) phosphatase regulated (orange). Heatmaps represent the log2 ratios of the phosphopeptides in the different strain, normalized for protein ratios. Phosphosites outside the box represent some kinase-regulated that phosphosites could not be measured in the phosphatase k.o.: they could be assigned to either class (1) or (2). Similarly, a few phosphatase-regulated phosphosites were not measured in both kinase k.o.. Strains and could belong to either class (2) or (3). (B) Representation of the three different phosphorylation steady states captured by the analysis. For the sites exclusively downregulated in the kinase k.o., the kinase activity largely overcomes that of the phosphatase, whereas for those also upregulated in the phosphatase k.o., kinases and phosphatase have balanced activity. Phosphosites only downregulated in the phosphatase k.o. are rare. They represent cases where the phosphatase activity largely overcomes that of the kinases. (C) Functional classification according to COG. The kinase-regulated substrates are enriched for metabolic functions, whereas the kinase and phosphatase-regulated substrates are enriched for Mycoplasma-specific functions.
We explored if phosphorylation in M. pneumoniae is organized as a network by measuring indirect, downstream responses. For example, we identified a number of phosphosites (13.7% of all regulated sites) that responded with inverted directionality; increased phosphorylation in kinase deletions and decreased phosphorylation in phosphatase deletion that might be indicative of complex regulatory events whereby kinases and phosphatases might directly or indirectly regulate each other. A series of phosphosites were also found to be affected by both PknB and HprK k.o. (17.6% of all regulated sites) (Figure 3; Supplementary Table S2), suggesting that the two kinases act sequentially. Alternatively, for some of the substrates, the two kinases might have redundant specificities. In yeast, such indirect responses have been interpreted as an indication of interconnected signaling networks and they account for more than half of all regulatory events (Bodenmiller et al, 2010). Even though the frequency of indirect phosphorylation we report here is lower, our results support the existence of regulatory networks in M. pneumoniae with principles similar to those described in yeast.
The phosphorylation network modulates protein lysine acetylation states
Because our data set on two different PTMs covers a very significant fraction of an organismal proteome, it offers the opportunity to measure the intertwining of phosphorylation and lysine acetylation globally. Vastly extending early proteomic analyses in other bacteria (Soufi et al, 2008; Prisic et al, 2010), we now show that an important fraction of modified proteins (57%) contain multiple sites, a third being modified on four or more residues (four-fold enrichment, P≪10−4) and 5.8% carrying even 10 or more modifications (29-fold enrichment, P≪10−4) (Figure 2C; Supplementary Table SI). While on average <10% of all serines, threonines, tyrosines and lysines are modified (Figure 2B), some proteins exhibit an unusually high level of modification. These include the ribosome-recycling factor (Frr∣Mpn636), with 41% of all lysines being acetylated and an inorganic pyrophosphatase (Ppa∣Mpn528) for which 29% of all serines and 33% of all lysines were found modified. Remarkably, a significant fraction (37.7%) of multiply modified proteins contains both lysine acetylation and phosphorylation sites that appear tightly coupled: the vast majority of phosphorylated proteins were also found lysine-acetylated (72%; P≪10−6, Fisher's exact test) and, reciprocally, an over-represented fraction of all lysine-acetylated proteins were also phosphorylated (24%; P≪10−6, Fisher's exact test). In contrast to lipid modifications, only one of the 40 lipidated proteins predicted from UniProt (UniProt Consortium, 2011) and contained in the data set was lysine-acetylated (P≪10−6, Fisher's exact test). For example, eight of the nine protein chaperones (UniProt (UniProt Consortium, 2011)) in M. pneumoniae were found lysine-acetylated and four carried additional phosphorylation sites (Figure 2A; COG class O and Supplementary information), suggesting that PTMs exert pleiotropic effects on chaperonin function, reminiscent of the complex regulation of the eukaryotic chaperone Hsp90 (Scroggins et al, 2007).
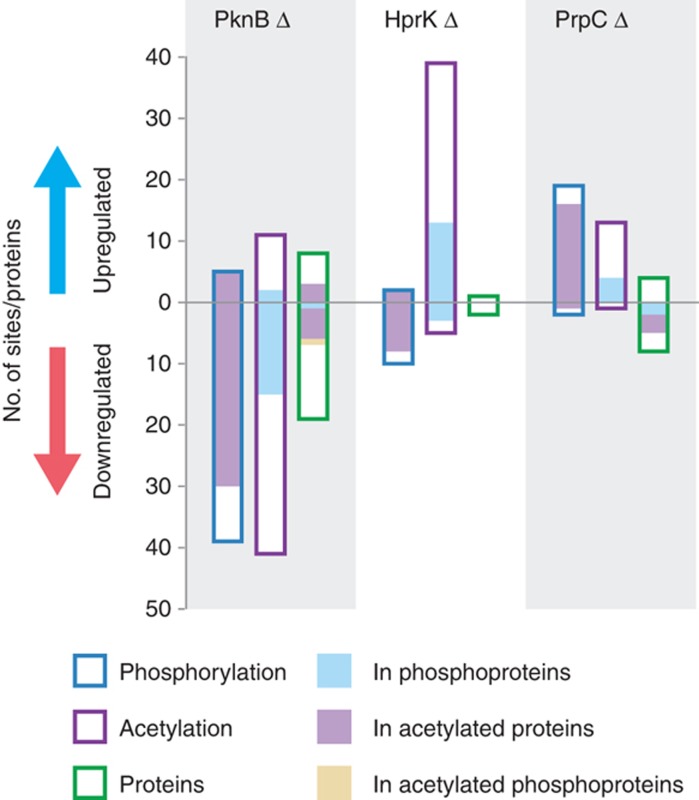
The k.o. permitted us to measure and quantify for the first time the impact of phosphorylation on lysine acetylation patterns on an organismal scale. We observed that the degree of modification of 81 acetylated-lysines (out of 449 unambiguously quantified) was significantly affected in the different kinase or phosphate k.o. (Supplementary Figure S6; Supplementary Table S3). Corresponding unmodified peptides were 1.4-fold more frequently (13%) identified as outliers compared with other non-modified peptides for the same protein (9.8%) (P=8.0 × 10−5) (Supplementary Figure S7). The two kinases have dramatically different global impacts on lysine acetylation. While PknB k.o. leads to a decrease in the overall level of lysine acetylation, HprK mutation induces a corresponding increase, suggesting that the two kinases have antagonistic effects (Figure 4). The individual perturbations caused complex changes in the patterns of lysine acetylation, inducing both increased and decreased degree of modification, sometimes even within the same protein. For example, 16 of the 30 acetylated-lysines within the coiled-coil regions of the cytoskeletal protein Hmw2 (Mpn310) were significantly regulated. Among the sites affected by PknB deletion, 13 showed decreased and three increased acetylation levels (Supplementary Table S3). Four additional cytoskeletal proteins (Mpn474, Hmw1∣Mpn447, Hmw3∣Mpn452 and Mpn387) were found similarly regulated (Figure 5). This suggests that in M. pneumoniae, the regulatory control exerted by the phosphorylation networks on cellular processes such as cytadherence imply cross-talk with lysine acetylation. Supporting this view, previous genetic data showed that M. pneumoniae strains deficient for one of the two putative N-acetyltransferase (Mpn114) had, similarly to PknB deletion, defects in attachment organelle and gliding motility (Hasselbring et al, 2006).
Figure 4.
Phosphorylation induces complex changes in protein abundance and patterns of lysine acetylation. The histogram represents the number of proteins or sites that show changing levels of phosphorylation (open blue bars), lysine acetylation (open purple bars) and protein abundance (open green bars) in the different deletion strains. The filled bars represent the fraction in phosphorylated proteins (blue fill), lysine-acetylated proteins (purple fill) or phosphorylated and lysine-acetylated proteins (yellow fill).
Figure 5.
Multilevel impact of systematic kinases and phosphatase deletion on M. pneumoniae proteome. (A) STRING interactions (>0.7) of proteins regulated at the levels of protein abundance (green), phosphorylation (blue) and lysine acetylation (purple). Direct STRING interactions with HprK (red), PknB (orange) and PrpC (blue) are highlighted. These affected proteins are more frequently interacting with each other than expected from a random set of proteins. Bottom panels: examples of subnetworks. (B) Proteins were ordered by their shortest path to the k.o. proteins in the STRING network (>0.7). The average percentage, over the three k.o., of affected proteins decreases with increasing shortest path length.
We further explored if reciprocally, perturbation of lysine acetylation could affect protein phosphorylation. We applied the same quantitative MS approaches to profile both modifications in two mutant strains of M. pneumoniae deficient in either one of the two putative N-acetyltransferases, Mpn027 and Mpn114. The deletions led to a general decrease in overall levels of lysine acetylation and 64 lysine acetylation sites (out of 600 unambiguously quantified) were significantly regulated (10.7%) (Supplementary Figure S8). Importantly, a significant fraction of the acetylated-lysines though was found unregulated, suggesting that additional N-acetyltransferases might exist. In prokaryotes, these enzymes have not been systematically characterized and the repertoires are still incomplete: for example, no bacterial members of the non-GNAT families of acetyltransferases have been reported to date (Hu et al, 2010). The two k.o. had only marginal impact on overall protein abundance (four proteins affected out of 353 quantified (1.1%). Finally, we found that some of the quantified phosphorylation sites (19%) were affected (Supplementary Tables S1 and S2), consistent with the view that the interplay between phosphorylation and lysine acetylation is bi-directional.
Multilevel impact of kinases and phosphatase perturbation
We observe that phosphorylation exerts pleiotropic effects on cellular proteomes: about a quarter (115) of the quantified proteins showed perturbed abundances or modified phosphorylation and lysine acetylation patterns in the different k.o. Integration of interaction data from the STRING database (version 8.3, COG mode) (Jensen et al, 2009) reveals that these affected proteins are 2.4 more frequently interacting with each other than expected from a random set of proteins (P≪10−6) (Figure 5A), illustrating that they form a highly connected regulatory network. We then determined the impact of phosphoproteome perturbation at the level of the entire interaction network built from STRING. As expected, we observe direct interactors of the perturbed enzymes, PknB, HprK or PrpC are affected most (Figure 5B): phosphorylation and lysine acetylation states and protein abundance levels changed considerably. The fraction of perturbed nodes rapidly decreases with network distance from the source of perturbation, suggesting that the perturbations are spread by direct interactions.
PTMs target interaction interfaces, altering protein oligomerization states
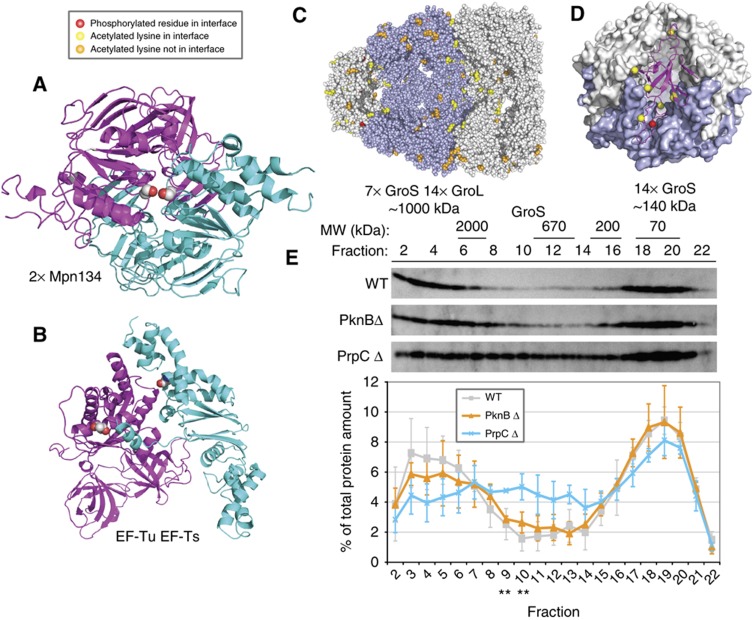
To gain mechanistic insights into the observed dynamic PTM patterns and how they might cause different functional states of the respective proteins, we integrated data on protein–protein interactions and structure (Choudhary et al, 2009). This revealed that multiply modified proteins are found more frequently than expected to be associated with more than one protein complex (P≪10−6, Fisher's exact test), a property that has been proposed to account for protein multifunctionality (de Lichtenberg et al, 2005; Kuhner et al, 2009). The integration of three-dimensional structures (Berman et al, 2000) revealed that compared with non-modified amino acids, the phosphorylated (23% versus 13%; P=0.001, χ2 test) and the acetylated (27% versus 14%; P<0.0001) sites are more frequently located at interaction interfaces, suggesting that modifications could alter the oligomeric state of proteins (Figure 6; Supplementary Figure S9). In some instances, the modification would be predicted to prevent an interaction. For example, serine 392 (S392) in Mpn134, a putative ABC transporter, is predicted to face itself in a homodimeric interface (Figure 6A); phosphorylation would place two negative charges adjacent to each other that would likely lead to a repulsion. For the elongation factor Tu (Tuf∣Mpn665) interacting with the elongation factor Ts (Tsf∣Mpn631), we saw multiple modifications, T34 on Tuf and K133 on Tsf, on both sides of the interaction interface (Kawashima et al, 1996) (Figure 6B). Another example, is threonine 29 (T29) (red in Figure 6C) of GroS (Mpn574) that lies on the outside of the GroS heptamer at an interface that contacts with either the GroL (Mpn573) tetradecamer (Shimamura et al, 2004) (Figure 6C) or a second GroS heptamer (Roberts et al, 2003) (Figure 6D), suggesting that phosphorylation could affect these assemblies. Consistent with this, we found that the massive increase in T29 phosphorylation in the PrpC k.o. (Supplementary Table S2) coincides with changes in GroS sedimentation profiles on sucrose gradients (Figure 6E; Supplementary Figure S9) and elution patterns upon gel filtration (GF) (data not shown). Overall, the results suggest the existence of combinatorial patterns of PTMs in prokaryotes that are part of a global regulatory network. They also suggest how PTMs could be exploited by nature to modulate the functions of individual proteins affecting the way they interact, reminiscent of the molecular barcode proposed in eukaryotes.
Figure 6.
PTMs are enriched at interaction interfaces, affecting protein–protein interactions. (A) Homodimerization of Mpn134 modeled on a homodimeric E. coli maltose transporter structure ((Oldham and Chen, 2011), PDB: 3PUY). Cyan and magenta ribbons show the interaction of two copies of Mpn134, spacefilled atoms colored by atom type show the side chains of S392 from both copies. Phosphorylation of S392 is predicted to lead to repulsion of negatively charged phosphates and likely prevent homodimerization. (B) Example of multiple modifications in a single interaction interface: EF-Tu (magenta) interacting with EF-Ts (cyan) modeled on E. coli EF-Tu–EF-Ts ((Kawashima et al, 1996), PDB: 1EFU). EF-Tu phosphorylation site T34 and EF-Ts acetylation site K133 are show as spacefilled atoms colored by atom type. Both are in the interface. (C) GoS/GroL chaperonin modeled on the structure from Thermus thermophilus ((Shimamura et al, 2004), PDB: 1WE3). Phosphorylation sites (T29) are in red spacefill and those of the acetylated-lysines in yellow spacefill. Those sites not in an interface are shown in orange (acetylation) or pink (GroL phosphorylation). One face has been removed to show the surface of the internal cavity. (D) A dimer of GroS heptamer modeled on the structure of a Chaperonin-10 tetradecamer from Mycobacterium tuberculosis ((Roberts et al, 2003), PDB: 1P3H). The overall structure is shown as a surface with one heptamer in blue to emphasize heptamer–heptamer interface. A single monomer is shown with magenta ribbons, with the C-α atom of the lone phosphorylated site (T29) in red spacefill and those of the acetylated-lysines in yellow spacefill. In this model, in at least one monomer, all these sites are in an interface (within 4 Å of a different monomer). Most occur on flexible loops whose conformations are expected to change between the template structure and different models. (C, D) Together show the possibility for some modifications to allow switching between different multimeric states and for those on the inner surface of the cavity to interact with substrate proteins. (E) Sedimentation of GroS on sucrose gradient. In the strain deficient in PrpC, the sedimentation profile of GroS (12 kDa) was found significantly affected; **P<0.01. This correlated with an increase in the level of T29 phosphorylation. For comparison, the sedimentation profile of RplA (Mpn220) remains largely unaffected (Supplementary Figure S9). The results of three independent experiments are represented.
Discussion
Our extensive analysis of protein abundance, phosphorylation and lysine acetylation, simultaneously measured in a small bacterium, gave first insights into the specificity of the key enzymes that modulate phosphorylation networks at an organismal scale and quantifies the penetration of signals through the phosphorylation networks and beyond: it revealed that phosphorylation exerts an unanticipated broad impact on other layers of post-transcriptional regulations in M. pneumoniae. Many components of the translational machinery, including ribosomal proteins, tRNA synthetases, translation initiation and elongation factors, and chaperones were affected by the perturbation of the phosphorylation network, which might account for observed changes in protein abundance that are not obviously the result of transcriptional regulation. Conservation of sites beyond this organism implies that these features can be generalized to other prokaryotes. However, there are a variety of specialized eukaryotic domains recognizing phosphorylated or acetylated residues (e.g., bromodomains, SH2, PTB) that lack known prokaryotic counterparts, suggesting that we have detected earlier, more fundamental mechanisms of regulation by PTM. In contrast to what is currently available in eukaryotes, the number of acetylation sites in M. pneumoniae appears to be nearly an order of magnitude higher than that for phosphorylation, and three times as high as previously observed values for bacteria with larger genomes such as E. coli or Salmonella enterica (Yu et al, 2008; Zhang et al, 2009a; Wang et al, 2010). Extensive modulation of cellular proteomes by lysine acetylation or phosphorylation affects virtually all cellular processes and many sites appear to be conserved in eukaryotes.
Both the extensive degree of phosphorylation and acetylation in this bacterium, and the observed cross-talk between the two PTMs argues for the early evolution of these post-translational events as important regulatory mechanisms in biological systems. A protein modification code might represent an ancient mechanism to diversify protein function outside of the transcription and translation paradigm.
Materials and methods
Cell culture
The k.o. strains of hprK kinase (mpn223), prpC (mpn247) phosphatase and pknB (mpn248) kinase were generated by transposon-mediated insertion of a gentamycin resistance cassette in wild-type M. pneumoniae M129 (ATTC29342 broth passage No. 31) (Halbedel et al, 2006; Schmidl et al, 2010b). Three 100-ml cultures were inoculated with either wild-type M. pneumoniae M129 or one of the three k.o. strains, and the cultures were grown in modified Hayflick medium without antibiotics for 96 h until late exponential phase. Wild-type M. pneumoniae, hprK (mpn223) kinase k.o. strain and prpC (mpn247) phosphatase k.o. strain were then washed three times with ice-cold phosphate-buffered saline (PBS), scraped from the bottom of the flask and centrifuged at 9860 g. As the pknB (mpn248) k.o. strain does not grow adherent, the cells were centrifuged at 9860 g and then washed three times with ice-cold PBS. Of wild-type and pknB (mpn248) k.o., three such pellets were generated and two pellets were generated for hprK (mpn223) k.o. and prpC (mpn247) k.o. to serve as biological replicates.
Cell lysis
Cell pellets were resuspended in urea buffer (8 M urea (Merck), 50 mM ammonium bicarbonate (Fluka), 1 mM sodium vanadate (Merck), 1 mM potassium fluoride (Fluka), 5 mM sodium phosphate (Sigma), supplemented with protease and phosphatase inhibitors (Roche)) and homogenized using a glass douncer. Cells were then lysed by sonication (6 × 20 s, 40 s pause, 80% output level, 50% duty cycle, using an Ultrasonic processor UIS250v and a VialTweeter of Hielscher Ultrasound Technology) and insoluble debris was pelleted at 10 000 g in a table-top centrifuge (Eppendorf 5415D) at 4°C. Protein concentrations were determined for all lysates by Bradford assay (Bio-Rad) and adjusted to 2.5 mg/ml using urea buffer. Cell lysates were snap frozen at –80°C until further processing.
Proteome digestion
For the proteomic and PTM analysis, lysates for three biological replicates of wild-type M. pneumoniae, two hprK (mpn223) k.o., two prpC (mpn247) k.o. and three pknB (mpn248) k.o. (indicated as C1–3 in Figure 1) were produced as described above. From each lysate, two equivalents of 500 μg protein, each, were further processed as technical duplicates. Cysteines were reduced with 5 mM dithiothreitol (DTT) for 15 min at 56°C and subsequently alkylated with 10 mM iodacetamide (Sigma) for 30 min at 25°C in the dark. Proteomes were then digested with 4 μg endoprotease LysC for 4 h at 37°C and the solutions were then diluted with 50 mM ammonium bicarbonate (Sigma) to a final urea concentration of 1 M. The proteomes were further digested by incubation with 8 μg trypsin protease for additional 18 h at 37°C, followed by an additional incubation with 8 μg trypsin at 37°C for another 5 h.
Differential dimethyl labeling of peptides and combining of proteomes
The resulting peptides were bound to a C18 SepPak column and differentially modified with a dimethyl label on the column following the protocol of Boersema et al (2009). ‘Light’-, ‘medium’- and ‘heavy’-labeled peptide solutions were then combined according to the scheme in Figure 1 to give a total of six proteome combinations.
SCX chromatography for peptide fractionation
Each of the six proteome combinations was fractionated using SCX chromatography to separate phosphorylated from unmodified peptides, monitored by UV absorbance.
Peptides from each digest corresponding to 1.5 mg of protein material were loaded onto two C18 cartridges using an Agilent 1100 HPLC system. The flow rate applied was 100 μl/min using water, 0.05% formic acid (FA), pH 2.7, as solvent. Subsequently, peptides were eluted from the trapping cartridges with 80% acetonitrile, 0.05% FA, pH 2.7, onto a PolySULFOETHYL A column 200 × 2.1 mm2 (PolyLCinc.) for 10 min at the same flow rate. Separation was performed using a non-linear 65 min gradient, 0–10 min 100% solvent A (5 mM KH2PO4, 30% Acetonitrile, 0.05% FA, pH 2.7), 10–15 min up to 26% solvent B (5 mM KH2PO4, 30% acetonitrile, 350 mM KCl, 0.05% FA, pH 2.7), 15–40 min to 35% solvent B and from 40 to 45 min to 60% solvent B. At 49 min, the concentration of solvent B was 100%. The column was subsequently washed for 6 min with high salt concentration and finally equilibrated with 100% solvent A for 9 min. The flow rate applied during the SCX gradient was 200 μl/min.
Fractions were collected at 1-min intervals for 40 min. After evaporation of the solvents, fractionated peptides were resuspended in 10% FA. Of each of the fractions 11–22 20% and of the fractions 26–30 0.4% were then analyzed by reversed phase LC-MS/MS.
Enrichment of lysine-acetylated peptides
For the investigation of the acetylome of M. pneumoniae, one lysate of each strain was analyzed. A cocktail of deacetylase inhibitors (Trichostatin A (10 μM), nicotinamid (10 mM) and butyric acid (50 mM)) was added to the urea lysis buffer mentioned for the phosphoproteomic analysis. Lysis, proteome digestion and peptide labeling were essentially performed as described for the phosphoproteomic analysis above. Acetylated peptides were immunoprecipitated following the protocol published by Choudhary et al (2009). In brief, after digestion, dimethyl labeling and proteome combination according to Figure 1, the equivalent of 5 mg of peptides was lyophilized overnight and subsequently dissolved in acetyl-lysine affinity purification buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM sodium chloride). After incubation with anti-acetyl-lysine affinity resins (ImmuneChem, Canada) for 14 h at 4°C on a rotating wheel, the resins were then washed four times with acetylated-lysine affinity purification buffer and two additional times with distilled water. Acetylated peptides were eluted with 0.1% trifluoroacetic acid (TFA). The eluates and flow throughs were desalted using StageTips as described in Rappsilber et al (2007) prior to LC-MS/MS analysis.
Mass spectrometry
The analysis of the SCX fractions was performed using a nano LC-LTQ-Orbitrap Classic (Thermo). An Agilent 1200 series LC system was equipped with a 20-mm Aqua C18 (Phenomenex, Torrance, CA) trapping column (packed in-house, i.d., 100 μm; resin, 5 μm) and a 400-mm ReproSil-Pur C18-AQ (Dr Maisch GmbH, Ammerbuch, Germany) analytical column (packed in-house, i.d., 50 μm; resin, 3 μm). Trapping was performed at 5 μl/min for 10 min in solvent A (0.1 M acetic acid in water), and elution was achieved with a gradient of 10–35% B (0.1 M acetic acid in 80/20 acetonitrile/water) in 90 min in a total analysis time of 120 min (fractions 11–22), or in 135 min in a total analysis time of 180 min. The flow rate was passively split to 100 nL/min when performing the elution analysis. Nanospray was achieved using a distally coated fused silica emitter (New Objective, Cambridge, MA) (o.d., 360 μm; i.d., 20 μm, tip i.d. 10 μm) biased to 1.7 kV. A 33-MΩ resistor was introduced between the high voltage supply and the electrospray needle to reduce ion current.
The LTQ-Orbitrap mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full scan MS spectra (300–1500 m/z) were acquired with a resolution of 60 000 at 400 m/z after accumulation to a target value of 500 000. The five (fractions 11–22) or 10 (fractions 26–30) most intense peaks above a threshold of 500 were selected for collision-induced dissociation in the linear ion trap at normalized collision energy of 35% after accumulation to a target value of 30 000.
The acetyl-lysine enriched and depleted peptide mixtures were analyzed by chromatographic separation on a EASY-nLCTM system (Proxeon Biosystems) fitted with a trapping (self-packed Hydro-RP C18 (Phenomenex), 100 μm × 2.5 cm, 4 μm) and an analytical column (self-packed Reprosil C18 (Dr Maisch) 75 μm × 15 cm, 3 μm, 100 Å). The outlet of the analytical column was coupled directly to an LTQ-OrbitrapVelos (Thermo Scientific) using a Thermo Scientific Nanospray Flex Ion Source. Solvent A was water, 0.1% FA and solvent B was acetonitrile, 0.1% FA. The samples (1 μl in 5% acetonitrile, 5% FA) were loaded with a constant flow of solvent A at 20 μl/min onto the trapping column. Trapping time was 1 min. Peptides were eluted via the analytical column at a constant flow of 0.3 μl/min. During the elution step for the acetyl-enriched samples, the percentage of solvent B increased in linear gradients from 5 to 25% B in 40 min, then from 25% B to 80% in 5 min, to a total gradient time of 60 min including a final wash step of 15 min at 80% B. For the elution of the acetyl-lysine-depleted samples, the percentage of solvent B increased in linear gradients from 5 to 25% B in 90 min, then from 25% B to 40% in 10 min and finally from 40 to 80% B in 10 min, to a total gradient time of 120 min including a final wash step of 10 min at 80% B. The peptides were introduced into the mass spectrometer via a Pico-Tip Emitter 360 μm OD × 20 μm ID; 10 μm tip (New Objective), and a spray voltage of 1.9 kV was applied. The capillary temperature was set to 200°C. Full scan MS spectra with a mass range 300–1700 m/z were acquired in profile mode in the FT with a resolution of 30 000. The filling time was set at maximum of 500 ms with limitation of 106 ions. The most intense ions (up to 15) from the full scan MS were selected for sequencing in the LTQ. Normalized collision energy of 40% was used, and the fragmentation was performed after accumulation of 3 × 104 ions or after filling time of 50 ms for each precursor ion (whichever occurred first). MS/MS data were acquired in centroid mode. Only multiply charged (2+ and 3+) precursor ions were selected for MS/MS. The dynamic exclusion list was restricted to 500 entries with maximum retention period of 30 s and relative mass window of 10 p.p.m. In order to improve the mass accuracy, a lock mass correction using a background ion (m/z 445.12003) was applied.
Mascot search results were uploaded into the TRANCHE data repository (https://proteomecommons.org/tranche/) in Scaffold file format, which can be viewed with the free Scaffold viewer available at http://www.proteomesoftware.com/Proteome_software_prod_Scaffold.html. The files can be downloaded using the following Hash:
OejVCeG2v3KJap1OlQbfeYM3KvQwv6EtRRw9+msFLnPpLl/4MBuKKp6hdI/ZgX2JNW1pUoUGAUeMro8FRgIOkLp/tW8AAAAAAAAFNg==. Additionally, the scaffold files and raw data are available at http://vm-lux.embl.de/Docu/VanNoortMSB2012/ and from TRANCHE using the hash 8YQSJO0UiPO2JoOgE2DZ3yXolC5cGOCOhra/0kvrMLRGKagf1fXUJ2w1c/5DdbkS9/k0aIDW0d4+qR/Kpz03zrrvCDAAAAAAAABvYw==. Note that scaffold files contain the raw mascot results loaded into the Scaffold tool with some default filter criteria and were not filtered the same way as described in the Materials and methods section, and also the FDR calculations there will be different than in our final data sets.
Peptide identification
Peak lists in the Mascot generic text file format were extracted from Thermo raw data files using the Quant application in the MaxQuant environment (Cox and Mann, 2008) (version 1.1.1.13). Results from all LC-MS/MS experiments of each proteome combination were combined into a single file and analyzed with the Matrix Science Mascot search engine (version 2.2.03) using a UniProt protein database of M. pneumoniae (downloaded on 18 May 2010 from http://www.uniprot.org) plus previously identified contaminants. Search parameters were chosen as follows: trypsin as the proteolytic enzyme, up to three missed cleavages, cysteine carbamidomethylation as a fixed modification and methionine oxidation as well as serine/threonine/tyrosine phosphorylation as variable modifications. Instead of searching for differential dimethyl labels as variable modifications (see below), dimethyl (‘light’ (12C2 1H6), ‘medium’ (12C2 1H2 2H4) and ‘heavy’ (13C2 2H6)) lysine and peptide N-termini were defined as exclusive modifications in the Mascot-specific ‘Quantitation’ mode. The peptide tolerance was set to 15 p.p.m., and the MS/MS tolerance was set to 0.6 Da. The ‘Decoy’ option was used for subsequent peptide FDR-based filtering (see below). The data of all SCX fractions of a proteome combination were searched together. Only unique peptides in the protein database were taken into account. For this the following rules to identify peptides were applied: (1) the peptide is unique among SwissProt M. pneumoniae proteins or (2) the peptide is unique in UniProt (SwissProt + trEMBL) M. pneumoniae proteins else the peptide identification is discarded.
Modified and unmodified peptide filtering
We used RockerBox (van den Toorn et al, 2011) to filter all peptides (modified and unmodified) to a 1% peptide FDR, using the Mascot Percolator option (Kall et al, 2007) for each proteome combination separately (Supplementary Table S2). For this purpose, we trained RockerBox on the complete set of peptides (including phosphorylated or lysine-acetylated) and then separated the target and decoy peptides into modified and unmodified sets. Two separate score thresholds were set per proteome combination such that both the modified and unmodified peptides had a peptide FDR of 1%. As phosphorylated and lysine-acetylated peptides have more degrees of freedom and therefore there are more options to fit decoy peptides, this procedure results in taking a higher threshold for modified than unmodified peptides. Nevertheless, to reduce the false positive rate, the spectra of modified peptides with a low Mascot score were manually inspected and modified peptides with a Mascot score <10 were removed from the data set.
Automatic peptide quantification
The Mascot result files were then exported to Mascot peptide html file format without any further filtering and loaded into MSQuant version 2.0a81 (Mortensen et al, 2010) together with the respective raw mass spectrometric data files. Peptide abundance ratios were determined automatically by MSQuant using the dimethyl (‘light’ (12C2 1H6), ‘medium’ (12C2 1H2 2H4) and ‘heavy’ (13C2 2H6)) lysine labels in the ‘Quantitation mode’ without any additional peptide or protein filters. Phosphorylation sites were localized using MSQuant's PTM scoring. Selected peptide ratios for which contaminant peptide signal intensities and non-co-eluting peptide pairs were detected were re-calculated by manually adjusting their LC elution time window in MSQuant.
Automatic acetylated peptide identification and quantification
Acetylation as well as the dimethyl labels target lysine residues and protein N-termini. To identify acetylated peptides, both of these modifications have to be chosen as variable modification in MASCOT. However, in contrast to the ‘Quantitation mode,’ the selection of the dimethyl labels as variable modifications includes experimentally impossible combination of these labels at the peptide N-terminus and at the lysine side chain (e.g., light label at the N-terminus and heavy label at the lysine side chain). Together with oxidized methionines, these modifications add up to nine variable modifications, which is the maximum number of variable modifications allowed in Mascot (light, intermediate and heavy dimethyl labels on lysines and peptide N-termini, acetylation of lysines and protein N-termini and oxidized methionines).
The use of a large number of variable modifications in database searches is known to decrease the number of identifications at a fixed FDR. Therefore, we employed a strict filtering of the Mascot search results for peptides with inconsistent labels (i.e., ‘light’ dimethyl N-terminus and ‘intermediate’ lysine) and unlabeled lysines/peptide N-termini as they should be incorrect. As additional control, we also estimated the accuracy of the search to be above 99% by counting the number of peptides with a C-terminally acetylated-lysine (either an in vitro artifact or incorrect identification as trypsin is not expected to cleave C-terminal to acetylated-lysine residues (Choudhary et al, 2009) in the entire acetylation data set. We only found seven such cases among the 759 non-redundant acetylated peptide matches (=0.9% FDR).
For automatic quantification of lysine-acetylated peptides in MSQuant, all ‘K’ entries corresponding to acetylated-lysines in the Mascot result (.dat) files were replaced by ‘J’ in the respective peptide sequences, and ‘Acetyl (J)’ was added to the MSQuant parameter file (quantitationModes.xml).
Detection of outlier peptides
The detected unmodified peptides of a particular protein should all exhibit a similar abundance ratio. As the peptide ratios are used to determine the abundance change of the protein, the quality of the determined changes in protein abundance relies on reproducible quantification of the peptides that originate from the same parent protein. We called an unmodified peptide ‘outlier’ in case it displays a significantly (P-value<0.05, corrected for multiple testing according to Benjamini and Hochberg (1995)) different ratio than the remaining peptides originating from the same parent protein. These different abundance changes can arise from co-eluting contaminants in the chromatogram for one of the isotope entities, stoichiometric dynamics in PTM or not considered differences between the combined proteomes (although processed in parallel).
To account for mixing error, the measure signal intensities of each peptide were normalized, such that the sum of signal intensities for each dimethyl label are equal in each proteome combination. Then, the change in protein abundance was estimated by the median of the peptides quantified for each protein and the normalized peptide signal intensities were corrected for this estimated protein abundance change of the parent protein. The corrected peptide signal intensities were analyzed with the R package OutlierD according to the author's recommendations (‘linear’ method and k=1.5 (Cho et al, 2008)) and detected outliers were removed (Supplementary Figure S10).
Protein quantification
The change in protein abundance was calculated as the ratio of the sum of the peptide signal intensities normalized for mixing error. The change in protein abundance was calculated as the weighted average of the peptides identified for this protein. A minimum of two unmodified non-outlier peptides was required and outlier peptides as well as modified peptides were excluded from the protein quantification.
Detection of regulated peptides and regulated proteins
The normalized peptide signal intensities were corrected for protein abundance determined by weighted average. The R package OutlierD (Cho et al, 2008) was used to determine the first and third quartiles of the peptide abundance changes, respectively. This information was used to calculate the z-score and subsequently the P-value for each peptide. The P-values were then corrected for multiple testing according to Benjamini and Hochberg (1995). A modified peptide was called regulated if the corrected P-value was <0.001 or a log2 abundance ratio was >1.5 for phosphorylated peptides and two for acetylated peptides. Essentially the same approach was used for the proteins. As only peptides were measured, but not entire proteins, the sum of peptide intensities was used as ‘protein signal intensity.’ Proteins were considered ‘regulated’ if the log2 abundance change was >1.5 and a corrected P-value <0.01.
Integration of protein quantification from different proteome combinations
The peptide signal intensities from both technical duplicates were added for each protein, as they should represent maximal reproducibility. To integrate the biological duplicates the more significant change was taken. If the P-values were equal, the more severe change was chosen. In case neither of the two abundance changes was significant, the mean of the abundance changes from the biological duplicates was taken and the P-value was set to ‘none significant.’ The regulated proteins were classified according to in which mutant the protein is regulated. Essentially the same method was applied to integrate the abundance change of modified peptides.
Functional enrichment analysis
The classification of the M. pneumoniae proteins in the different clusters of orthologous groups of proteins was extracted from the ‘whog.txt’ file downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/pub/COG/COG/) and the P-value for the enrichment was determined using Fisher's exact test. The P-values were corrected for multiple testing according to Benjamini and Hochberg (1995). Proteins were considered to be enriched in a particular cluster if the corrected P-value was <0.05.
Bias analysis
The molecular weight, the isoelectric point, the hydrophobicity and the instability index were calculated for each M. pneumoniae protein annotated in SwissProt using the protparam tool from ExPASy (http://www.expasy.ch/tools/protparam.html). The distribution of each parameter was then binned into 20 equally spaced bins. Supplementary Figure S3 shows the comparison of the coverage of each of these bins across all identified proteins in this study and all proteins annotated in the SwissProt database.
PTM localization
PTM scores from MSQuant were summed for each possible PTM site within phosphopeptides identified in multiple fractions. The PTM was localized if the highest PTM score was at least 1.25 times the second highest PTM score and the combined score was >4.
Evolutionary conservation of modified residues
PTM sites were localized in alignments of orthologous groups from eggNOG (Muller et al, 2010) version 2. Species were assigned to one phylogenetic group being ‘other Mycoplasmas,’ ‘other Firmicutes,’ ‘other Bacteria’ or ‘Archaea and Eukaryotes’ based on NCBI Taxonomy. If at least one protein form one species has the same amino acid as the PTM site, the site is considered ‘Site conserved’ in this species, otherwise if in a window of three residues, one before to one after the PTM site, the same amino acid is found the site is considered ‘Conserved in window’ in this species. Otherwise, the site is considered ‘Not conserved’ in this species. If there is no protein for the considered species in the orthologous group, it is not counted at all. Each species is counted only once for one PTM site. The conservation level for each phylogenetic group is the number of ‘Site conserved’ divided by the sum of ‘Site conserved,’ ‘Conserved in window’ and ‘Not conserved.’ Random conservation was estimated by taking 10 times the same number of PTM sites with the same amino-acid distribution from the M. pneumoniae proteome and performing the same conservation analysis.
Clustering of modified regulated peptides and proteins
Log2 ratios for regulated proteins and modified peptides in three k.o./wt comparisons and two k.o./k.o. comparisons were used to calculate uncentered correlations between each set of regulated proteins, regulated phosphopeptides and regulated lysine-acetylated peptides. The uncentered correlations were used for hierarchical clustering with hclust as implemented in the R package.
Network generation and analysis
Interactions between M. pneumoniae proteins were derived from STRING (Jensen et al, 2009) version 8.3. Scores between COGs and NOGs were converted to scores between M. pneumoniae proteins by mapping M. pneumoniae proteins to COGs and NOGs as they are in STRING v8.3. A network was generated between proteins regulated on abundance, phosphorylation and acetylation level by taking only interactions with a score >0.7. Random networks were generated by taking 100 times the same number of proteins from all M. pneumoniae proteins and counting the number of interactions between them reveals a random expectation of number of interactions.
Minimum path lengths to HprK, PknB and PrpC were calculated for all M. pneumoniae proteins through the complete network of interactions (>0.7). Number of affected (upregulated or downregulated) proteins at each minimum path length were calculated for each k.o. strain and divided by the total number of proteins at each minimum path length.
Validation of lysine acetylation: TAP purification and quantitative western blot
One-liter cultures were incubated in five 300 cm2 cell culture flasks (Sarstedt) and harvested 96 h after inoculation. Cells were washed twice with ice-cold PBS and centrifuged at 9860 g. Pellets were resuspended in 2 ml lysis buffer (50 mM Tris pH 7.5, 5% glycerol, 1.5 mM MgCl2, 100 mM NaCl, 0.2% NP40, 1 mM DTT, 1 mM AEBSF, 1 mM PMSF, 1 μg/ml pepstatin A, 1 μg/ml antipain, 2 μg/ml aprotinin, 1 μg/ml leupeptin and 16 μg/ml benzamidin) and lysed mechanically using a douncer. TAP purification was done following established protocols (Kuhner et al, 2009) stopped after TEV elusion. The elusion was split into two and each of these samples was analyzed by SDS–PAGE and western blot. A peroxidase anti-peroxidase antibody (Sigma, P1291) was used to detect the TAP-fusion proteins. Acetylated proteins were detected using an anti-acetyl-lysine primary antibody (Immunechem) and a HRP-coupled secondary antibodies (Sigma). For quantification, total band intensity was integrated with Photoshop software (Adobe) and normalized versus the highest detected peak (Supplementary Figure S2).
Validation of protein abundance changes by quantitative western blot
Mycoplasma strains were lysed as for the TAP purification. Total cell lysate of each of the four strains was then analyzed by SDS–PAGE and western blot using polyclonal antibodies raised against endogenous M. pneumoniae proteins. Final detection was done using secondary antibodies coupled to HRP (Sigma). For the quantification, total band intensity was integrated with Photoshop software (Adobe) and normalized versus the highest detected peak (Supplementary Figure S5)
Evaluating the protein complexes by separation on sucrose gradient, GF chromatography and western blot
Lysis was performed as mentioned for the TAP purification supplementing the lysis buffer with the deacetylase inhibitors nicotinamid (10 mM) and butyric acid (50 mM). Volume of 30 μl of samples were layered on a top of 4 ml sucrose gradient (10–35%) and separated by 14 h centrifugation at 130 000 g at 4°C. The gradient was subsequently divided into 22 fractions per 165 μl. GF chromatography was performed at 10°C on a Pharmacia SMART system at a flow rate of 40 μl/min by using a SuperoseTM 6 PC 3.2/30 column, equilibrated with lysis buffer. The chromatographic profile was monitored at 280 nm by using the μPeak monitor (Pharmacia). Volumes of 50 μl of samples were loaded on a column and 60 μl fractions were collected. Fractions from both sucrose gradient and GF were analyzed by SDS–PAGE and western blot (Figure 6). Polyclonal antibodies produced in rabbits have been used to detect the GroS protein (Mpn574) and 50S ribosomal protein RlpA (Mpn220); final detection was done using secondary antibodies coupled to HRP (Sigma) on Image Station 4000 MM Pro (Kodak). For the quantification, the band intensities detected with Photoshop software (Adobe) were first normalized for equal sample loading and subsequently each fraction was represented as a percentage of total protein amounts of particular sample. The values obtained from three independent sucrose gradient separation experiments were used for calculation of average values and standard deviations. To test for a significant difference in the intensities for the three different groups (WT, PrpC, PknB), we performed a one-way ANOVA with a significance threshold of P<0.01. To further test for pairwise differences, we then applied Tukey's Honest significant difference test for the corresponding fractions, with a significance threshold of P<0.05.
Surface accessibility
Structure models of each sequence, where available, were retrieved from ModBase (Pieper et al, 2009). ModBase gives several models for each UniProt entry, covering different parts of the sequence and with different scores, e-values and sequence identity between the target and the template (template=the known structure on which the target is modeled). For each of the modified residues (Supplementary Tables S2 and S3), and for their unmodified equivalents (unacetylated-lysine, unphosphorylated serine, threonine and tyrosine), the model with the lowest e-value that included the residue was taken. For each of these models, NACCESS (http://www.bioinf.manchester.ac.uk/naccess/) was used to calculate the relative accessible surface area (RSA) of all side chains, and a residue was defined as exposed when RSA>5%.
3D structures of interactions
Structural templates for interactions of any pair of proteins were found by BLAST comparisons (Altschul et al, 1990) of the sequences against sequences of structures in the PDB (E⩽0.001), including biounit assemblies, and looking for pairs of proteins that hit different but interacting parts of the same structure. A particular pair of proteins might have zero, one or more templates. For each template, the residues of one component that are in contact with the other were found and, via the sequence alignments given by BLAST, used to infer the residues in contact in the query pair (with InterPreTS (Aloy and Russell, 2003)). The occurrence of modified residues (Supplementary Tables S2 and S3) in interfaces was compared with that of their unmodified equivalents and significance measured with a χ2 test. Images were produced with PyMol (http://sourceforge.net/projects/pymol/). For selected protein interfaces, we also constructed homology models using MODELLER (Sali and Blundell, 1993).
Microarray analysis of k.o. strains
A custom DNA array was used consisting of 688 70 mers representing 688 ORFs. The M. pneumoniae array consists of oligos (70 bases, amino linker) spotted on the array four times each. The design process was done in cooperation with Operon Biotechnologies, synthesis of the 688 oligos (probes) was performed by Operon Biotechnologies and the spotting was done at the EMBL Genomics Core Facility (Guell et al, 2009). M. pneumoniae M129 was grown in 150 cm2 tissue culture flasks with 100 ml of modified Hayflick medium with the following composition: 18.4 g of PPLO broth, 29.8 g of HEPES, 10 g glucose, 5 ml of 0.5% phenol red and 35 ml of 2 N NaOH per liter. Horse serum and penicillin were included to a final concentration of 20% and 100 U/ml, respectively. In the reference condition, cells were grown for 96 h at 37°C.
After growth, surface-attached cells were washed once with PBS and immediately lysed in the cultivation flask by adding RLT buffer from the QiagenTMRNeasyPlus Mini Kit (Cat. Num. 74134). This isolation method used for RNA extraction removed most RNAs <200 nucleotides, thus preventing the synthesis of cDNA from tRNA. For cell lysis, 2 ml of RLT buffer in the presence of 0.134 M β-mercaptoethanol was used per cultivation flask. The purification was done according to the manufacturer's protocol.
In all, 9 μg of total RNA was used for the reverse transcription polymerase chain reaction (RT–PCR) using SuperScriptTM Indirect cDNA Labeling System from Invitrogen. This kit was used according to the manufacturer's indications, with the exception of two modifications. RT–PCR was carried out at 37°C instead of 46°C and the set of random hexamers (2 μl of 2.5 μg/μl) was used instead of polyT 20 mers. Hybridization and scanning was carried out at the EMBL Genomics Core Facility. Custom microarrays were scanned using an Axon GenePix 4000.
After background subtraction, quantile normalization was done using the bioconductor package marray (http://www.bioconductor.org/packages/release/bioc/html/marray.html).
Supplementary Material
Acknowledgments
We are grateful to Richard Herrmann for fruitful discussion, Christina Besir, the EMBL Proteomics Core Facility and Hinnerk Eilers and Sebastian R Schmidl, Georg-August University Göttingen, for expert help and the sharing of reagents. SM, MO’F and AJRH were supported by the Netherlands Proteomics Centre, which is part of the Netherlands Genomics Initiative. This work was partially founded by Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN) to ACG (BMBF NGNF IG Cellular Systems Genomics, 01GS0865).
Author contributions: VN, JSe, PB and ACG designed the experiments. JSe, SB, SM, MO, AS, EY performed the experiments. IV, SK, TM, VR performed the experimental validation. VN, JSe, SB, MJB, RK analyzed the data. VN, PB and ACG wrote the paper. JSt, LS, RBR and AJRH contributed to the study design, discussed results and commented on the manuscript. Original idea was formulated by PB and ACG.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aloy P, Russell RB (2003) InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics 19: 161–162 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300 [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484–494 [DOI] [PubMed] [Google Scholar]
- Cho H, Kim YJ, Jung HJ, Lee SW, Lee JW (2008) OutlierD: an R package for outlier detection using quantile regression on mass spectrometry data. Bioinformatics 24: 882–884 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res 39: D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, Yuan YP, Herrmann R, Bork P (2000) Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res 28: 3278–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg U, Jensen LJ, Brunak S, Bork P (2005) Dynamic complex formation during the yeast cell cycle. Science 307: 724–727 [DOI] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I (2010) Post-translational modifications in signal integration. Nat Struct Mol Biol 17: 666–672 [DOI] [PubMed] [Google Scholar]
- Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F (2011) Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic Acids Res 39: D261–D267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M (2011) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39: D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L (2009) Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Halbedel S, Busse J, Schmidl SR, Stulke J (2006) Regulatory protein phosphorylation in Mycoplasma pneumoniae. A PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J Biol Chem 281: 26253–26259 [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Page CA, Sheppard ES, Krause DC (2006) Transposon mutagenesis identifies genes associated with Mycoplasma pneumoniae gliding motility. J Bacteriol 188: 6335–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hible G, Renault L, Schaeffer F, Christova P, Zoe Radulescu A, Evrin C, Gilles AM, Cherfils J (2005) Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase. J Mol Biol 352: 1044–1059 [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B (2004) PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4: 1551–1561 [DOI] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ (2010) Bacterial protein acetylation: the dawning of a new age. Mol Microbiol 77: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JD, Berg HC, Church GM (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4: 59–77 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jolly L, Pompeo F, van Heijenoort J, Fassy F, Mengin-Lecreulx D (2000) Autophosphorylation of phosphoglucosamine mutase from Escherichia coli. J Bacteriol 182: 1280–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4: 923–925 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R (1996) The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 A resolution. Nature 379: 511–518 [DOI] [PubMed] [Google Scholar]
- Kennelly PJ (2002) Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett 206: 1–8 [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618 [DOI] [PubMed] [Google Scholar]
- Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castano-Diez D, Chen WH, Devos D, Guell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R et al. (2009) Proteome organization in a genome-reduced bacterium. Science 326: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y (2009) SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol Cell Proteomics 8: 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics 7: 299–307 [DOI] [PubMed] [Google Scholar]
- Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics 6: 697–707 [DOI] [PubMed] [Google Scholar]
- Maier T, Schmidt A, Guell M, Kuhner S, Gavin AC, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Petranovic D, Macek B, Cepo T, Mann M, Davies J, Jensen PR, Vujaklija D (2006) Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res 34: 1588–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed S, Heck AJ (2010) Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr Opin Biotechnol 22: 9–16 [DOI] [PubMed] [Google Scholar]
- Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, Andersen JS, Mann M (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res 9: 393–403 [DOI] [PubMed] [Google Scholar]
- Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, Bork P (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res 38: D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham ML, Chen J (2011) Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A (2009) MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 37: D347–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang CM, Bemis G, Church GM, Steen H, Husson RN (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA 107: 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ (2006) Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol 16: 2166–2172 [DOI] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3. [DOI] [PubMed] [Google Scholar]
- Roberts MM, Coker AR, Fossati G, Mascagni P, Coates AR, Wood SP (2003) Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops. J Bacteriol 185: 4172–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schmidl SR, Gronau K, Hames C, Busse J, Becher D, Hecker M, Stulke J (2010a) The stability of cytadherence proteins in Mycoplasma pneumoniae requires activity of the protein kinase PrkC. Infect Immun 78: 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl SR, Gronau K, Pietack N, Hecker M, Becher D, Stulke J (2010b) The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol Cell Proteomics 9: 1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L (2007) An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell 25: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T, Koike-Takeshita A, Yokoyama K, Masui R, Murai N, Yoshida M, Taguchi H, Iwata S (2004) Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure 12: 1471–1480 [DOI] [PubMed] [Google Scholar]
- Singh KD, Halbedel S, Gorke B, Stulke J (2007) Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC. J Mol Microbiol Biotechnol 13: 165–171 [DOI] [PubMed] [Google Scholar]
- Soufi B, Gnad F, Jensen PR, Petranovic D, Mann M, Mijakovic I, Macek B (2008) The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8: 3486–3493 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Coon JJ (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res 9: 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Toorn HW, Munoz J, Mohammed S, Raijmakers R, Heck AJ, van Breukelen B (2011) RockerBox: analysis and filtering of massive proteomics search results. J Proteome Res 10: 1420–1424 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226 [DOI] [PubMed] [Google Scholar]
- Waites KB, Talkington DF (2004) Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17: 697–728, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP (2011) Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics 10: M111 009654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008a) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008b) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Li H, Liu BQ, Huang XY, Guo L (2011) SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J Biol Chem 286: 27342–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG (2008) The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol 18: 1529–1536 [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, Wodke JA, Guell M, Martinez S, Bourgeois R, Kuhner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutierrez-Gallego R, Russell RB, Gavin AC, Bork P et al. (2009) Impact of genome reduction on bacterial metabolism and its regulation. Science 326: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y (2009a) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 8: 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, Nicosia SV, Zhang X (2009b) Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28: 445–460 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorina A, Stepanchenko N, Novikova GV, Sinetova M, Panichkin VB, Moshkov IE, Zinchenko VV, Shestakov SV, Suzuki I, Murata N, Los DA (2011) Eukaryotic-like Ser/Thr protein kinases SpkC/F/K are involved in phosphorylation of GroES in the Cyanobacterium synechocystis. DNA Res 18: 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.