Abstract
Werner Syndrome is a premature aging disorder characterized by genomic instability, elevated recombination, and replication defects. It has been hypothesized that defective processing of certain replication fork structures by WRN may contribute to genomic instability. Fluorescence resonance energy transfer (FRET) analyses show that WRN and Flap Endonuclease-1 (FEN-1) form a complex in vivo that colocalizes in foci associated with arrested replication forks. WRN effectively stimulates FEN-1 cleavage of branch-migrating double-flap structures that are the physiological substrates of FEN-1 during replication. Biochemical analyses demonstrate that WRN helicase unwinds the chicken-foot HJ intermediate associated with a regressed replication fork and stimulates FEN-1 to cleave the unwound product in a structure-dependent manner. These results provide evidence for an interaction between WRN and FEN-1 in vivo and suggest that these proteins function together to process DNA structures associated with the replication fork.
INTRODUCTION
Werner Syndrome (WS) is a premature aging disorder characterized by genomic instability and increased cancer risk (Martin, 1978). The WRN gene product defective in WS belongs to the RecQ family of DNA helicases (Yu et al., 1996). In humans, mutations in RecQ family members BLM and RECQ4 are responsible for two other disorders associated with elevated chromosomal instability and cancer, Bloom and Rothmund-Thomson syndromes, respectively (Ellis et al., 1995; Kitao et al., 1998, 1999). RecQ helicase mutants display defects in DNA replication, recombination, and DNA repair, suggesting a role for RecQ helicases in maintaining genomic integrity (Wu and Hickson, 2002; Cobb et al., 2002).
A DNA processing defect during replication or recombination has been suggested to contribute to the molecular pathology of WS. WS cells have a prolonged S phase (Poot et al., 1992), slower rate of repair associated with DNA damage induced in S-phase, reduced induction of RAD51 foci, and higher level of DNA strand breaks (Pichierri et al., 2001). More recently, it was demonstrated that WS cells initiate recombination at a normal rate but fail to resolve recombination intermediates in a RAD51-dependent pathway (Prince et al., 2001; Saintigny et al., 2002). A DNA substrate for WRN was suggested by the demonstration that RusA, a bacterial enzyme that cleaves four-way junctions, rescued cell survival and restored the ability to generate viable recombinants following exposure of WRN-/- cells to DNA damaging agents (Saintigny et al., 2002).
A stalled replication fork can be converted to a four-way junction resembling a Holliday Junction (HJ) by branch migration and reannealing of nascent DNA strands (McGlynn et al., 2001; Postow et al., 2001). WRN has been shown to unwind the HJ and catalyze branch fork migration on α-structures (Constantinou et al., 2000), suggesting a potential role in processing the recombination intermediate to prevent aberrant recombination events at sites of stalled replication forks. Evidence from studies of Escherichia coli DNA repair and recombination mutants indicates that the DNA damage-induced regressed replication fork intermediate can be processed by the DNA helicase RecQ and the 5′ to 3′ exonuclease RecJ before replication restart once the lesion is removed (Courcelle and Hanawalt, 1999; Courcelle et al., 2003). The mammalian equivalent of the RecQ-RecJ helicasenuclease that participates in regressed fork processing is yet to be identified.
The physical/functional interactions of WRN with human nuclear proteins implicated in DNA metabolic processes (Brosh and Bohr, 2002) suggest that WRN plays a direct and coordinate role in processing DNA structures that arise during replication or recombination that might interfere with normal DNA transactions. WRN interacts with FEN-1 (Brosh et al., 2001, 2002a), a structure-specific nuclease (Lieber, 1997) implicated in replication (Bambara et al., 1997), DNA repair (Klungland and Lindahl, 1997; Kim et al., 1998; Wu et al., 1999), and recombination (Negritto et al., 2001). A number of studies indicate that FEN-1, like WRN, plays an important role in DNA metabolism. Yeast studies have implicated the FEN-1 homolog RAD27 in genome stability pathways (Johnson et al., 1995; Sommers et al., 1995; Vallen and Cross, 1995; Tishkoff et al., 1997; Freudenreich et al., 1998; Kokoska et al., 1998; Schweitzer and Livingston, 1998), DNA damage response (Reagan et al., 1995), and stabilization of telomeric repeats (Parenteau and Wellinger, 1999). RAD27 mutations result in elevated mitotic crossing over and are lethal in combination with RAD51 and RAD52 mutations (Tishkoff et al., 1997). FEN-1 is directly implicated in Okazaki fragment processing (Bambara et al., 1997), and biochemical and genetic evidence indicates that the double-flap structure that arises during DNA synthesis strand displacement is the physiological substrate of the enzyme (Kao et al., 2002; Xie et al., 2001). WRN and FEN-1 can be coimmunoprecipitated from human nuclear extracts and the physical interaction between WRN and FEN-1 is mediated by a C-terminal region of the WRN protein residing after its helicase domain (Brosh et al., 2001). The domain of WRN responsible for the physical interaction with FEN-1 mediates a functional interaction with FEN-1 whereby the WRN protein domain stimulates FEN-1 endonucleolytic cleavage on fixed 5′ flap or nicked duplex substrates. WRN can stimulate FEN-1 cleavage of very long 5′ flaps (up to 80 nt) by a mechanism distinct from that of the replication/repair factor proliferating cell nuclear antigen (PCNA; Brosh et al., 2002a).
The in vivo evidence implicating WRN in the recovery of DNA synthesis after replication arrest posed the question of whether WRN functions with FEN-1 to resolve a key replication or recombinational intermediate that arises from fork stalling or collapse. In this study, evidence is presented that WRN and FEN-1 form a complex upon replication arrest and together process a double-flap substrate or a regressed replication fork structure. Replication and recombination defects and genomic instability due to fork demise in WRN-/- cells may be a consequence of abnormal DNA structures that persist at arrested replication forks that fail to be processed faithfully.
MATERIALS AND METHODS
Cloning of Fusion Proteins
ECFP-FEN-1, constructed by cloning a cDNA encoding FEN-1 into the XhoI site of the pECFP-C1 vector (CLONTECH, Palo Alto, CA), was a generous gift from Dr. Emma Warbrick (Department of Surgery and Molecular Oncology, University of Dundee, Dundee, Scotland). The ECFP-PCNA, EYFP-WRN and UNG2-EYFP constructs are previously described (Baynton et al., 2003).
Cell Culture, Confocal Microscopy, and FRET Measurements
HeLa cells were grown in Dulbecco's minimal essential media supplemented with 10% fetal bovine serum (FBS) and antibiotics. For exposure to the DNA-damaging agent mitomycin C (MMC), cells were incubated with MMC (0.5 μg/ml) for 16 h in media supplemented with 10% FBS. For exposure to the chemical carcinogen 4-nitroquinoline-1-oxide (4-NQO), cells were incubated with 4-NQO (0.1 μg/ml) in serum-free media for 1 h followed by a PBS wash and additional incubation for 16 h in media supplemented with 10% FBS. A Zeiss LSM 510 laser scanning microscope (Thornwood, NY) equipped with a Plan-Apochromate 63×/1.4 oil immersion objective was used to examine images of 1-μm-thick slices of living HeLa cells. Fluorescence energy transfer (FRET) was determined by modifying the general equations given by Matyus (1992) as described in (Baynton et al., 2003). FRET occurs if I2 - I1[ID2/ID1] - I3[IA2/IA3] >0, where I represents intensities in three channels given in arbitrary units between 0 and 250. Normalized FRET is defined as NFRET = FRET/(I1 × I3)1/2 (Xia and Liu, 2001). Intensities were measured as follows: channel 1: I1, A1, D1 = excitation (ex.) at λ = 458, detection (det.) at 475 nm <λ>525 nm (ECFP); channel 2: I2, D2, A2 = ex. at λ = 458 nm, det. at λ>560 nm; channel 3: I3, D3, A3 = ex. at λ = 514 nm, det. at λ>560 nm (EYFP). ID1, D2, D3 and IA1, A2, A3 are determined separately for cells transfected with only ECFP- and EYFP-fusion proteins, respectively, under the same settings and at the same levels of fluorescence intensities (I1 and I3) as cotransfected cells.
Recombinant Proteins
Recombinant His-tagged WRN protein (wild-type, WRN-K577M, WRNE84A) was overexpressed using a baculovirus/Sf9 insect system as previously described (Orren et al., 1999). Sf9 cell pellet (∼5 g) containing overexpressed recombinant WRN protein was suspended in 15 ml Buffer A (150 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.5% NP-40, 10% glycerol, 5 mM β-mercaptoethanol) with protease inhibitors (1 mM phenylmethylsulfonylfluoride, 2 μg/ml chymostatin, 2 μg/ml pepstatin A, 2 μg/ml aprotinin, 2 μg/ml leupeptin) and incubated at 4°C for 45 min. In all successive steps, protease inhibitors were present in buffers. After brief sonication, the suspensions were centrifuged at 20,000 × g twice for 15 min at 4°C. The supernatant was passed through a 0.4-μm filter and loaded onto a 20 ml DEAE Sepharose column (Amersham Biosciences, Piscataway, NJ) using an AKTA FPLC system (Amersham Biosciences). After successive washes with Buffer B (Buffer A with NP-40 omitted) containing 10 and 80 mM NaCl, respectively, WRN protein was eluted with Buffer B containing 180 mM NaCl. Peak fractions were pooled, diluted twofold with Buffer B with no NaCl, and loaded onto a 20 ml Q Sepharose column (Amersham Biosciences) using the AKTA FPLC system. After successive washes with Buffer B containing 90, 100, and 200 mM NaCl, respectively, WRN was eluted with Buffer B containing 400 mM NaCl. Peak fractions were pooled and manually loaded onto a 1 ml HisTrap column (Amersham Biosciences) charged with Ni2+ ions. The column was washed successively with Ni2+ Wash Buffer 1 (50 mM Tris-HCl, pH 8.0, 500 mM LiCl, 10% glycerol, 5 mM β-mercaptoethanol), Ni2+ Wash Buffer 2 (10 mM PIPES, pH 7.0, 50 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol), Ni2+ Wash Buffer 2 containing 10 mM imidazole, and Ni2+ Wash Buffer 2 containing 25 mM imidazole. WRN protein was eluted with Ni2+ Wash Buffer 2 containing 300 mM imidazole. Peak fractions were pooled and loaded manually onto a 1 ml SP Sepharose column (Amersham Biosciences) and washed successively with Buffer B containing 50 mM NaCl and 100 mM NaCl, respectively. WRN was eluted with Buffer B containing 400 mM NaCl. Protein fractions were analyzed by SDS PAGE, and those fractions containing WRN were pooled. Bovine serum albumin (BSA) was added to a concentration of 100 μg/ml to the pooled WRN for stability during storage at -80°C. The purified WRN recombinant protein was judged to be 98% pure from analysis on Coomassie-stained SDS polyacrylamide gels (see Figure 1). WRN protein concentration was determined by the Bradford assay (Bio-Rad Protein Assay, Richmond, CA) using BSA as a standard.
Figure 1.
SDS polyacrylamide gel analysis of the purified recombinant proteins. Purified full-length recombinant WRN (∼400 ng) and FEN-1 (∼1.1 μg) were resolved on 8-16% polyacrylamide SDS gel along with molecular weight markers and stained with Coomassie Brilliant Blue. Lane 1, Molecular weight marker; lane 2, purified recombinant WRN; lane 3, purified recombinant human FEN-1.
Two human recombinant wild-type FEN-1 proteins were used in this study: 1) FEN-1 with a hexa-histidine tag at the amino terminus, designated FEN-1N-HIS; and 2) FEN-1 with hexa-histidine tag at the carboxy terminus, designated FEN-1C-HIS. In addition, FEN-1C-HIS with a site-directed mutation D181A in the nuclease domain, designated FEN-1C-HIS-D181A, was also used (Shen et al., 1996). FEN-1C-HIS and FEN-1C-HIS-D181A purified as previously described (Nolan et al., 1996) were provided by Dr. D. Wilson III (NIA-NIH).
FEN-1N-HIS encoded by a plasmid kindly provided by Dr. M. Lieber (University of Southern California) was overexpressed in E. coli by IPTG (0.5 mM) induction of midlog phase cells for 4 h at 37°C. Bacterial cell pellet (10 g) containing overexpressed FEN-1 protein was resuspended in 50 ml FEN-1 purification buffer (25 mM HEPES-KOH, pH 7.9, 100 mM KCl, 10% glycerol) containing the same protease inhibitors as described above. Lysozyme was added to a concentration of 100 μg/ml, and the lysate was further incubated on ice for 20 min. The suspension was then sonicated briefly, quickly frozen on dry ice, and allowed to thaw on ice. Suspension was clarified by centrifugation at 40,000 × g for 15 min at 4°C. Imidazole (5 mM) was added to the supernatant and the material was loaded onto a 5 ml HisTrap column (Amersham Biosciences) using an AKTA FPLC system (Amersham Biosciences). The column was washed successively with FEN-1 purification buffer containing 40 and 100 mM imidazole, respectively. FEN-1 was eluted with FEN-1 purification buffer containing 250 mM imidazole. Fractions collected were analyzed by SDS PAGE, and those fractions containing FEN-1 were pooled and dialyzed against 25 mM HEPES-KOH, pH 7.9, 100 mM KCl, 10% glycerol, and 1 mM dithiothreitol. Aliquots of recombinant FEN-1 were frozen in liquid nitrogen and stored at -80°C. The purified FEN-1 recombinant protein was judged to be 98% pure from analysis on Coomassie-stained SDS polyacrylamide gels (see Figure 1). FEN-1 protein concentration was determined by the Bradford assay (Bio-Rad Protein Assay) using BSA as a standard.
Oligonucleotide Substrates
PAGE-purified oligonucleotides (Midland Certified Reagent Co., Midland, TX) were used for preparation of DNA substrates. Nicked flap, fixed double flap with 1-nt 3′ noncomplementary tail or 3′ complementary tail, and branch migrating double-flap substrate with equilibrating 12-nt 3′ tail were constructed as previously described using D1:T1:U1, D1:T1:U3, D1:T1:U2, and D2:T1:U8, respectively (Kao et al., 2002). Synthetic HJ(X12) was made by annealing four 50-mer oligonucleotides (X12-1, X12-2, X12-3, X12-4) as described previously (Mohaghegh et al., 2001). HJ substrates with a 5′-32P label on oligonucleotides X12-1, X12-2, X12-3, or X12-4 are designated HJ(X12-1), HJ(X12-2), HJ(X12-3), and HJ(X12-4), respectively. Synthetic HJ(X1) and HJ(X2) were constructed by annealing four 66-mer oligonucleotides as described previously (van Gool et al., 1999). The synthetic “chicken foot” HJ structure was constructed by annealing oligonucleotides X1-1 + 25, X1-2 + 25, X1-3, and X1-4. The sequence of X1-1 + 25 was 5′-TATCGAATCCGTCTAGTCAACGCTGCCGAATTCTACCAGTGAGGATGGACTCCTCACCTGCACGTACATGAGCGTTTAGCGAGTCACGACC-3′; the sequence of X1-2 + 25 was 5′-GGTCGTGACTCGCTAAACGCTCATGTACGTGCAGGTGAGGAGTCCATGGTCTTCCGTCAAGCTCGATGCCGGTTGTATGCCCACGTTGACC-3′. The sequences of X1-3 and X1-4 are as previously described (van Gool et al., 1999).
Cleavage Assays
FEN-1 HJ cleavage reactions (20 μl) contained 20 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, 1 mM dithiothreitol, 2.5 fmol HJ(X12-1), WRN (12 nM), and FEN-1N-HIS (abbreviated FEN-1; 29 nM) unless otherwise indicated. Reactions were incubated at 37°C for 15 min, stopped by the addition of 20 μl of 50 mM EDTA, 40% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanole, and electrophoresed on native 10% polyacrylamide gels. Radiolabeled DNA species were visualized using a PhosphorImager. To map the resolution sites, incision reactions were terminated with the addition of 10 μl formamide dye (80% formamide [vol/vol], 0.1% bromophenol blue, and 0.1% xylene cyanole), heated to 95°C, and products were resolved on 15% polyacrylamide 7 M urea denaturing gels with Maxam-Gilbert piperidine sequencing reactions for each oligonucleotide run in parallel (Maxam and Gilbert, 1980). For FEN-1 nick flap and double-flap cleavage reactions, 20 μl reactions contained 30 mM HEPES-KOH, pH 7.6, 5% glycerol, 40 mM KCl, 0.1 mg/ml BSA, 8 mM MgCl2, 10 fmol of the indicated flap substrate, and the specified amounts of WRN and/or FEN-1 in the presence or absence of ATP (2 mM) as noted. Incision reactions were incubated at 37°C for 15 min, and the products were resolved on native 12% polyacrylamide gels or 20% polyacrylamide 7 M urea denaturing gels.
Electrophoretic Mobility Shift and FEN-1 Antibody Pull-down Assays
WRN (73 nM) and FEN-1 (83, 166, 332 nM) were incubated in binding buffer (20 mM triethanolamine, pH 7.5, 2 mM MgCl2, 0.1 mg/ml BSA, 1 mM dithiothreitol) with 25 fmol HJ(X12-1) and 1 mM ATPγS in a total volume of 20 μl at 24°C for 20 min. Protein-DNA complexes were fixed in the presence of 0.25% glutaraldehyde for 10 min at 37°C, resolved on native 5% polyacrylamide 0.5× TBE gels, and visualized by phosphorimage analysis. To immunoprecipitate proteins cross-linked to HJ substrate, FEN-1 (332 nM), WRN (73 nM), or both proteins were incubated with radiolabeled HJ(X12-1) (25 fmol) in binding buffer (20 μl) at 24°C for 20 min, and protein-DNA complexes were cross-linked with UV light (1200 ìJ/cm2 [×100]) in a stratalinker (Stratagene). Binding mixtures were then incubated with rabbit anti-FEN-1 polyclonal antibody (1:400; Brosh et al., 2001) on ice for 1 h in 200 μl RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS). Protein-G agarose (20 μl) was added to each reaction, mixed at 4°C for 1 h, and washed three times with 1 ml RIPA buffer. Bound products were eluted with 2× SDS sample buffer, resolved on 8% polyacrylamide SDS gels, and detected by phosphorimage analysis.
WRN-FEN-1 Coimmunoprecipitation Experiments
Purified recombinant WRN (12 nM) and FEN-1 (29 nM) were incubated in HJ resolution buffer (300 μl) with protease inhibitors (0.1 mM PMSF, 1 μg/ml aprotinin, and 5 μg/ml leupeptin) either in the absence or presence of ATP (2 mM) and HJ(X12-1) (38 fmol) for 1 h on ice. Reaction mixtures were then incubated with goat anti-WRN polyclonal antibody (1:40, Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C with gentle mixing. Protein complexes were adsorbed onto protein-G agarose beads by incubating for 1 h at 4°C with gentle mixing. Beads were washed four times with PBS containing 0.1% Tween-20, and proteins were eluted by boiling with SDS sample buffer and resolved on 8-16% gradient polyacrylamide SDS gels, followed by Western blotting. Mouse anti-WRN (1:250, BD PharMingen, San Diego, CA) or rabbit anti-FEN-1 (1:400; Brosh et al., 2001) antibodies were used to detect WRN and FEN-1 on the membranes using ECL Plus (Amersham Biosciences).
RESULTS
WRN and FEN-1 Colocalize and Exhibit FRET In Vivo
WRN has been shown to localize primarily in nucleoli of human cells (Gray et al., 1998) and exit the nucleoli in response to treatment of cells with agents that induce DNA damage (Gray et al., 1998). Similarly, interruption of DNA synthesis by depletion of the nucleotide pool using hydroxyurea results in the transport of WRN from nucleoli to distinct nuclear foci that colocalize with RPA (Constantinou et al., 2000). In a very recent study, WRN-PCNA and WRNRAD52 were shown to colocalize and exhibit FRET in HeLa cells that had been arrested for replication by MMC (Baynton et al., 2003). These studies suggest a role of WRN at stalled replication forks. FEN-1 is localized to replication foci during S phase and repair foci after DNA damage (Qiu et al., 2001). On the basis of these observations and the biochemical interaction between WRN and FEN-1, we examined the localization of WRN and FEN-1 after treatment of human cells with the DNA damaging agents 4-NQO or MMC.
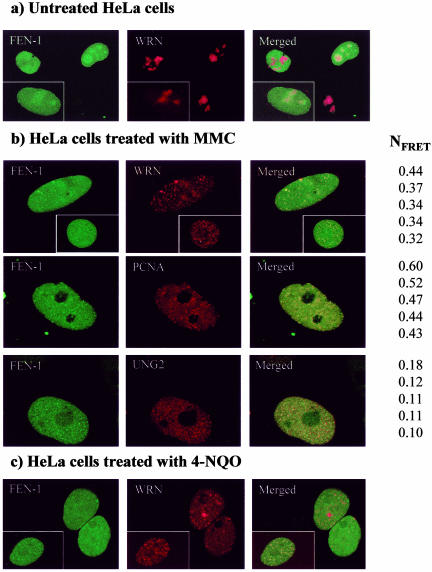
FEN-1 and WRN were expressed as C-terminal fusions to variants of the enhanced green fluorescent protein (ECFP and EYFP, respectively) to determine colocalization and FRET in living cells. Cotransfected FEN-1 and WRN showed only limited colocalization in untreated HeLa cells (Figure 2A). However, in cycling HeLa cells treated with MMC, FEN-1 and WRN foci colocalized as evidenced by the appearance of yellow dots in the merged picture (Figure 2B, top row). Under these same conditions, a substantial proportion of HeLa cells have FEN-1 foci that colocalize with PCNA (Figure 2B, middle panel) and WRN foci that colocalize with PCNA (Baynton et al., 2003), representing arrested replication forks. UNG2 was previously shown to colocalize in replication foci with PCNA (Otterlei et al., 1999) and also colocalizes with FEN-1 in replication foci (Figure 2B, bottom panel). In HeLa cells treated with the chemical carcinogen 4-NQO, FEN-1 and WRN foci also colocalized as evidenced by the appearance of yellow dots in the merged picture (Figure 2C). Apparently, the colocalization of WRN and FEN-1 is induced not only by MMC, but also by other types of DNA damage. We also found the same relocalization of WRN to replication forks after methylmethane sulfate treatment (our unpublished results; Baynton et al., 2003). These results strongly suggest that the observed colocalization between FEN-1 and WRN occurs at arrested replication forks in response to DNA damage.
Figure 2.
Localization of FEN-1 and WRN in live, cycling HeLa cells. Colocalization is indicated in the yellow in the merged views. Cells coexpressing ECFP-FEN-1 and EYFPWRN that were either untreated (A), treated with MMC (0.5 μg/ml) overnight (B), or treated with 4-NQO (0.1 μg/ml) for 1 h and incubated further for 16 h (C) are shown. In B, cells cotransfected with ECFP-FEN-1 and EYFP-WRN (top panel), ECFP-FEN-1 and EYFP-PCNA (middle panel), and EYFPFEN-1 and ECFP-UNG2 (bottom panel) after treatment with 0.5 μg/ml MMC overnight are shown. Five of the representative high FRET values found within the given levels of intensities (donor intensities (I1, ID1) between 85-190, and acceptor intensities (I3, IA3) between 55-155. NFRET represents FRET normalized against protein expression levels. FRET is calculated from the mean of the intensities within one region of interest (ROI) containing more than 25 pixels (i.e., one replication focus). Within ROI, all individual pixels had intensities below 250.
FRET analysis was performed to determine whether fluorescent-tagged FEN-1 and WRN proteins interact in vivo or reside closely together within the same protein complex (i.e., < 100 Å) at the arrested replication forks. The normalized FRET (NFRET) values shown in Figure 2 are calculated from the mean of intensities within a replication focus. FRET values for the FEN-1-PCNA pair and FEN-1-UNG2 pair served as positive and negative controls, respectively, because FEN-1 and PCNA directly interact at replication foci, whereas UNG2 localizes to replication foci but is not known to directly bind to FEN-1. Five positive NFRET values are given for FEN-1 and WRN, FEN-1 and PCNA, and FEN-1 and UNG2, respectively (Figure 2B). The average NFRET value for FEN-1 and WRN was 2.9-fold greater than that of FEN-1 and UNG2, but not quite as great as the 3.9-fold difference between FEN-1-PCNA and FEN-1-UNG2. The greater FRET observed for WRN and FEN-1 compared with UNG2 and FEN-1 indicates an interaction in vivo rather than coresidency of the protein pair in a replication complex. The direct interaction of WRN with FEN-1 at arrested replication foci is supported by our observations that endogenously expressed WRN and FEN-1 colocalize in cells treated with MMC to a significantly greater extent compared with untreated cells (our unpublished results). These results indicate that FEN-1 directly interacts with WRN at stalled replication forks in vivo.
WRN Stimulates FEN-1 Cleavage of Its Preferred Double-flap Substrate
The colocalization and interaction of WRN and FEN-1 after DNA damage that blocks cells in S phase raised the possibility that the two proteins act together upon a DNA intermediate found at replicating foci. Recent evidence suggests that the physiological substrate of FEN-1 during DNA replication is a double-flap structure with equilibrating 3′ and 5′ ssDNA tails that arises after strand displacement DNA synthesis catalyzed by a DNA polymerase (Xie et al., 2001; Kao et al., 2002). The double-flap substrate can also potentially arise during DNA repair synthesis which involves many of the same proteins that function at the replication fork. Yeast FEN-1 preferentially cleaves the double-flap substrate with a 1-nt 3′ tail at a precise position 1 nt into the downstream annealed region, allowing the 3′ tail to anneal and generate a nick suitable for ligation (Kao et al., 2002).
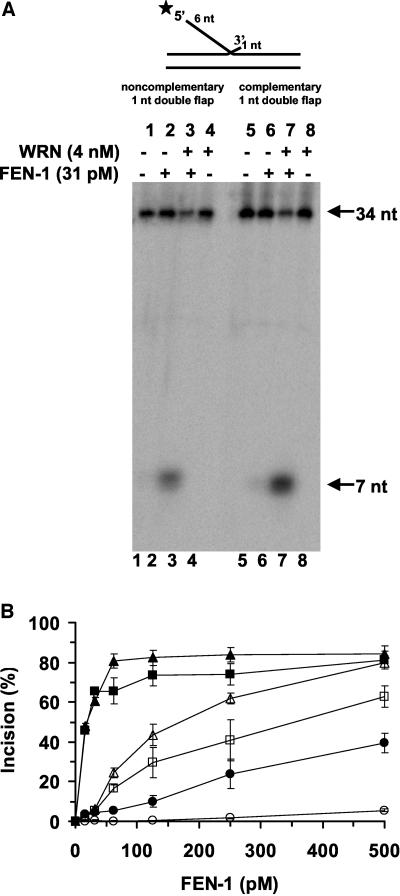
Because the double flap was proposed to be the cellular substrate of yeast FEN-1 and the functions of yeast FEN-1 and human FEN-1 are conserved (Greene et al., 1999), we investigated the effect of WRN on human FEN-1 cleavage of this structure. Initially we tested double-flap substrates with a 1-nt 3′ tail, by far the most active substrate configuration. At a limiting amount of enzyme (31 pM), human FEN-1 cleaved double-flap substrates with either a noncomplementary (Figure 3A, lane 2) or complementary (Figure 3A, lane 6) 3′ tail of 1-nt to yield the predicted 7-nt product. The presence of WRN (4 nM) resulted in a substantial stimulation of the FEN-1 cleavage reaction to yield the 7-nt product for both double-flap substrates (Figure 3A, lanes 3 and 7). In control reactions, WRN did not cleave either substrate (Figure 3A, lanes 4 and 8). A quantitative comparison of the FEN-1 cleavage reactions conducted in the presence or absence of WRN on the double-flap substrates and the nick flap substrate lacking the 3′ 1-nt flap is shown in Figure 3B. As previously observed for yeast FEN-1, human FEN-1 preferentially cleaves the double-flap substrates with a 1-nt 3′ tail compared with the nick flap substrate at all levels of FEN-1 tested (Figure 3B). At 125 pM FEN-1, only a very small amount of the nick flap substrate (<0.5%) was cleaved whereas 28 and 42% of the double-flap substrates with noncomplementary and complementary 3′ 1-nt tails, respectively, were incised. In the presence of WRN and 31 pM FEN-1, FEN-1 cleavage was increased ∼15-fold for the two double-flap substrates (Figure 3B). A similar fold increase of FEN-1 cleavage in the presence of WRN was detected on the nick flap substrate when 500 pM human FEN-1 was present in the reaction mixture (Figure 3B). These results demonstrate that human FEN-1 preferentially cleaves double-flap substrates compared with nick flap substrates and that WRN effectively stimulates FEN-1 cleavage on the double-flap substrates as observed for the nick flap substrate. Importantly, the presence of WRN does not alter the cleavage specificity of human FEN-1.
Figure 3.
FEN-1 cleavage of the preferred double-flap substrate containing a 1-nt 3′ tail is stimulated by WRN. Reaction mixtures (20 μl) containing 10 fmol of the indicated double-flap DNA substrate, FEN-1 (31 pM), and WRN (4 nM) as indicated were incubated at 37°C for 15 min under standard conditions as described in MATERIALS AND METHODS. Products were resolved on 20% polyacrylamide urea-denaturing gels. (A) Phosphorimage of typical gel is shown. For each gel: lanes 1 and 5, no enzyme; lanes 2 and 6, FEN-1; lanes 3 and 7, FEN-1 + WRN; lanes 4 and 8, WRN. (B) % incision (mean value of at least three independent experiments with standard deviations [SD] indicated by error bars) for reactions containing FEN-1 (open symbol) or FEN-1 + WRN (filled symbol) on a conventional nick flap substrate (circle), 1-nt double-flap substrate with a complementary 3′ 1-nt tail (triangle) or 1-nt double-flap substrate with a noncomplementary 3′ 1-nt tail (square).
FEN-1 Cleavage of a 12-nt Equilibrating Flap Substrate Is Stimulated by WRN
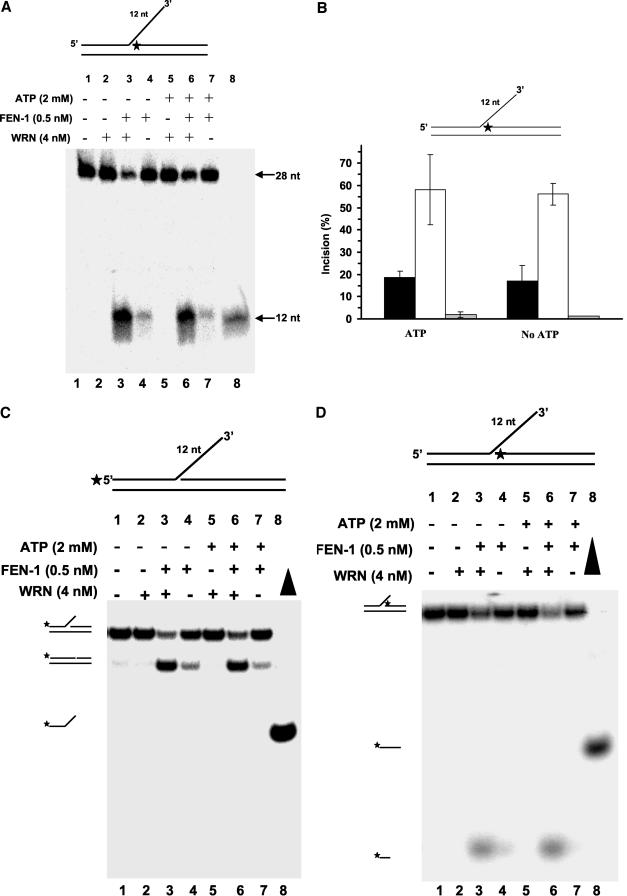
The maturation of Okazaki fragments during lagging strand synthesis requires that 5′ flaps arising from strand displacement DNA synthesis be processed. Because the 5′ flaps created are complementary to the template, the DNA intermediate can potentially equilibrate to form configurations of double-flap structures with varying 3′-tail and 5′-flap lengths. Yeast FEN-1 was shown to cleave the 1-nt 3′ flap conformer of displacement flap structures with complementary 3′ tails up to 12 nt in length precisely 1 nt into the downstream duplex region (Kao et al., 2002). We were interested in how WRN might affect the cleavage reaction catalyzed by human FEN-1 on this type of flap structure with overlapping complementary tails capable of branch migration. Using a DNA substrate initially constructed to have a 3′ flap of 12 nt complementary to the template, we observed that human FEN-1 cleavage resulted in a 12-nt product (Figure 4A, lane 4), identical to what was observed for yeast FEN-1. The 12-nt product is indicative of the ability of the substrate to equilibrate to a double-flap structure with a 3′ 1-nt tail that can be efficiently cleaved by FEN-1. WRN stimulated FEN-1 cleavage of the substrate as demonstrated by the approximately threefold increase in 12-nt product formed (Figure 4, A, lane 3, and B).
Figure 4.
WRN stimulates FEN-1 cleavage of a double-flap substrate with an equilibrating 3′ tail. Reaction mixtures (20 μl) containing 10 fmol of the indicated double-flap DNA substrate, FEN-1 (0.5 nM), and WRN (4 nM) in the absence (lanes 1-4) or presence (lanes 5-8) of ATP (2 mM) were incubated at 37°C for 15 min under standard conditions. Products were resolved on 20% polyacrylamide 7 M urea denaturing gel (A) or 12% polyacrylamide nondenaturing gel. (A) Phosphorimages of typical denaturing gels are shown. Lane 1, no enzyme; lanes 2 and 5, WRN; lanes 3 and 6, FEN-1 + WRN; lanes 4 and 7, FEN-1; lane 8, 12-nt marker. (B) % incision from A (mean value of at least three experiments) with SD indicated by error bars. Black bar, FEN-1; white bar, FEN-1 + WRN; gray bar, WRN. (C and D) Same as described for A with the indicated double-flap substrate, but the products were resolved on 12% polyacrylamide nondenaturing gels. Filled triangle, heat-denatured substrate control.
We next tested the effect of WRN on FEN-1 cleavage of the equilibrating flap substrate in the presence of 2 mM ATP. Under these reaction conditions, WRN unwinds a 3′ fixed flap DNA substrate with a short 19-base pair duplex region (Brosh et al., 2002b); however, WRN poorly unwinds a forked duplex substrate with a duplex region of 34 base pairs (Opresko et al., 2001). WRN (4 nM) did not unwind the three-stranded branch-migrating DNA structure (Figure 4, C and D) under conditions that the helicase is active on 3′ fixed flap or 5′ fixed flap 19-base pair duplex substrates (Brosh et al., 2002b). Neither the upstream 28-mer (Figure 4C) or the downstream 26-mer (Figure 4D) was unwound by WRN. These results suggest that the equilibrating double-flap structure is not suitable for WRN to unwind and/or the duplex region is sufficiently long that the enzyme fails to unwind because its mechanism is insufficiently processive. The latter reason may explain why the upstream oligonucleotide with a duplex region of 26-37 base pairs depending on the equilibrating state of the structure is not released by WRN. Alternatively, WRN may not efficiently recognize or bind the equilibrating flap structure, whereas it can unwind 3′ or 5′ fixed flap substrates (Brosh et al., 2002b). Despite its inability to unwind the equilibrating double-flap structure, WRN retained the ability to stimulate the site-specific FEN-1 cleavage reaction in the presence of ATP (Figure 4A, compare lanes 6 and 7) comparable to what was observed in the absence of ATP (lanes 3 and 4). From these results, it can be concluded that WRN stimulates FEN-1 cleavage of a double-flap structure that is the proposed cellular substrate of FEN-1. Furthermore this stimulation appears unrelated to the WRN helicase function.
Processing of a HJ Structure by WRN and FEN-1
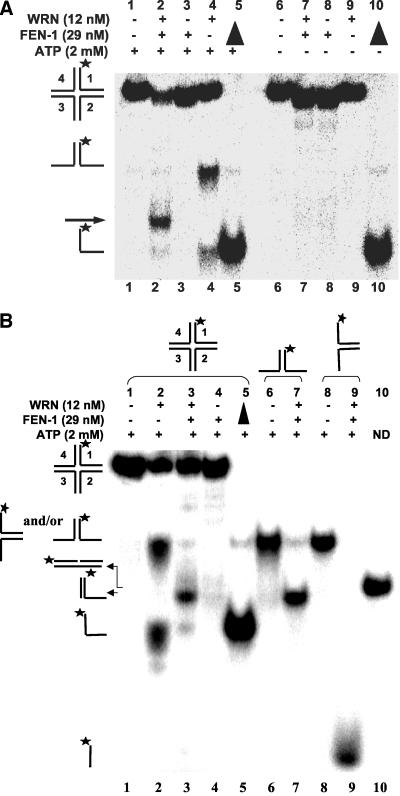
The colocalization and FRET data showing that WRN and FEN-1 interact in a complex in vivo at arrested replication fork foci suggested that the two proteins function together to process a specific DNA structure that forms at the blocked fork. DNA damage can induce replication fork regression, which results in a chicken-foot HJ intermediate. It has been proposed that the reversed replication fork intermediate may be subsequently stabilized by recombination proteins and processed by the RecQ-RecJ helicase-nuclease in E. coli (Courcelle and Hanawalt, 1999; Courcelle et al., 2003). This permits stabilization of the replication fork intermediate before lesion removal and replication restart. To determine whether WRN and FEN-1 can process the HJ intermediate, we performed a series of biochemical experiments using purified recombinant wild-type and mutant proteins (WRN and FEN-1) and various oligonucleotide-based HJs.
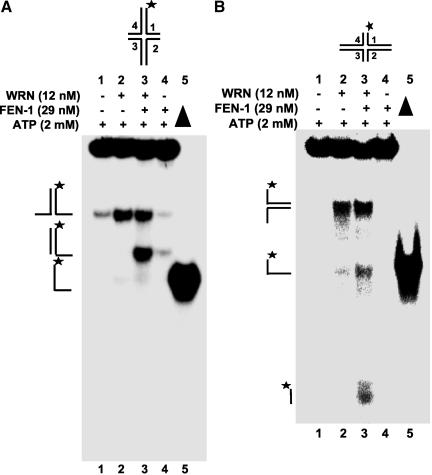
In the presence of ATP (2 mM), WRN helicase activity unwound HJ(X12-1) yielding a forked duplex and single-stranded DNA (Figure 5A, lane 4). When both WRN and FEN-1 are present in the reaction, a unique band was detected (Figure 5A, lane 2) that migrated between the forked duplex and single-stranded DNA species. HJ incision by FEN-1 alone was hardly detectable (≤1%; lane 3), indicating that FEN-1 cleavage is dependent on the presence of WRN. A 60-fold rate increase in HJ incision was observed when WRN was present in the FEN-1 cleavage reaction (our unpublished results). Because WRN-catalyzed unwinding of HJ and other DNA structures requires ATP (Constantinou et al., 2000), we sought to determine the outcome if ATP were omitted from the reactions containing WRN and/or FEN-1. WRN did not unwind HJ in the absence of ATP (Figure 5A, lane 9). The uniquely migrating DNA species was minimally detected from reactions containing WRN and FEN-1 but lacking ATP (lane 7), suggesting that ATP-dependent WRN helicase activity is necessary for processing of the HJ structure by WRN and FEN-1. Similar results were obtained for HJ(X12-3) (unpublished data).
Figure 5.
WRN stimulates FEN-1 cleavage of a Holliday junction. (A) FEN-1 cleavage of a HJ is dependent on WRN and ATP. (A) Reactions (20 μl) containing 2.5 fmol of HJ(X12-1), FEN-1 (29 nM), and/or WRN (12 nM) were incubated at 37°C under standard reaction conditions as described in MATERIALS AND METHODS in the presence or absence of 2 mM ATP as specified. Phosphorimage of a typical native gel is shown. (▴), heat denatured substrate control. The position of HJ, forked duplex, and single-stranded DNA are indicated. An arrowhead indicates the cleavage product. (B) Reactions containing 2.5 fmol of HJ(X12-1), X12-1: X12-4 forked duplex, or X12-1: X12-2 forked duplex and the indicated proteins were performed as described above. The nicked duplex (ND; lane 10) was constructed in vitro as described in MATERIALS AND METHODS.
FEN-1 endonucleolytic cleavage is dependent on the ability of the enzyme to track down unannealed 5′ ssDNA to the fork junction (Murante et al., 1995). The ability of WRN to catalyze branch fork migration and the requirement for ATP in the WRN-FEN-1 cleavage reactions of the HJ substrate suggested that WRN unwinds the HJ structure thereby providing a free 5′ ssDNA loading site for FEN-1 to track to the cleavage sites. In this proposed mechanism, the product of the WRN-FEN-1 reaction would be a linear partial duplex with a 3′ ssDNA flanking tail. Alternatively, FEN-1 might introduce incisions in juxtaposed positions across the HJ during WRN branch fork migration similar to a HJ resolvase and the resultant product would be a nicked duplex. To address these possibilities, we compared the products of a FEN-1 cleavage reaction conducted in the presence of WRN on a forked duplex (Figure 5B, lane 7) with the products of a HJ(X12-1) cleavage reaction (Figure 5B, lane 3). The comigration of the major HJ cleavage product and forked duplex (5′-32P-X12-1: X12-4) cleavage product and demonstration that a nicked duplex (ND) of the same sequence constructed in vitro migrated slightly slower than either the HJ or forked duplex cleavage products (Figure 5B, compare lanes 3 and 7 to lane 10) indicated that the cleavage product of reactions containing WRN, FEN-1, and HJ(X12-1) was a 3′ tailed linear duplex rather than a nicked duplex. A very minor fast migrating product was detected in some WRN-FEN-1 HJ cleavage reactions (lane 3) that comigrated with the 5′-32PX12-1: X12-2 forked duplex cleavage product (lane 9). Despite the strong ability of FEN-1 to cleave the 5′-32P-X12-1: X12-2 forked duplex in the presence of WRN (compare lanes 8 and 9), this cleavage product was hardly detected in reactions containing WRN, FEN-1, and HJ(X12-1). The relative abundance of the linear partial duplex product compared with the incised oligonucletide 1 suggests that the synthetic four-way HJ has a biased isomeric form influenced by localized sequence effects at the cross-over point, as previously recognized (Murchie et al., 1991).
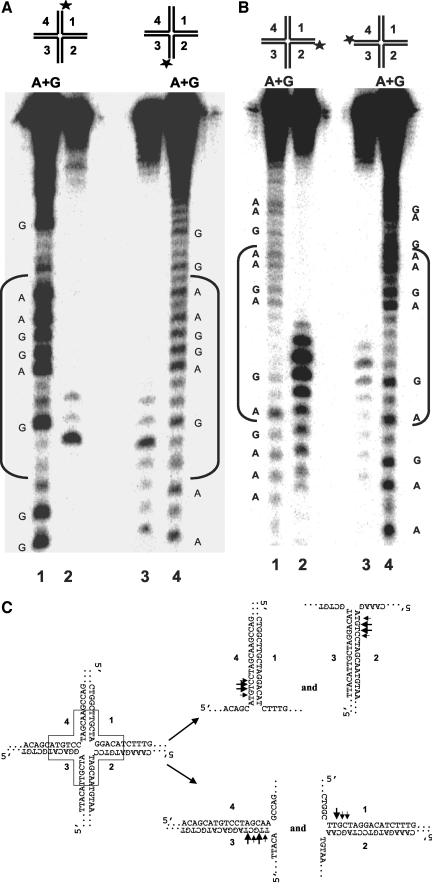
To map the cleavage sites, we examined on denaturing gels the products of reactions containing WRN, FEN-1, and the HJ substrate with a 5′32P label on strands 1, 2, 3, or 4. A single major cut site TT^GC and two neighboring minor cut sites were observed for strand 1 (lane 2 in Figure 6A and 6C). FEN-1 incision of HJ(X12-3) with a 5′32P label on strand 3 was also dependent on WRN (our unpublished results). A single major cleavage site TT^GC was detected along with a number of minor cut sites flanking both sides of the major cleavage site (lane 3 in Figure 6A and 6C). Two major cleavage sites, C^TTC and GT^TC, within the homologous core were detected in strands 2 and 4 (Figure 6B, lanes 2 and 3, and 6C). Several minor incision sites in strands 2 and 4 were also detected both inside and outside the region of homology. These incision mapping data, summarized in Figure 6C, indicate that WRN and FEN-1 process the HJ substrate in a structure-dependent manner that is dependent on ATP.
Figure 6.
Mapping of cleavage sites on each arm of the Holliday junction. HJ structures labeled in strand 1 (X12-1), strand 2 (X12-2), strand 3 (X12-3), or strand 4 (X12-4) were incubated with WRN and FEN-1 under standard conditions. Products were resolved on 15% polyacrylamide, 7 M urea denaturing gels. (A) Lane 1, A+G sequence ladder produced from X12-1; lane 2, cleavage products from HJ(X12-1); lane 3, cleavage products from HJ(X12-3); lane 4, A+G sequence ladder from X12-3. (B) Lane 1, A+G sequence ladder produced from X12-2; lane 2, cleavage products from HJ(X12-2); lane 3, cleavage products from HJ(X12-4); lane 4, A+G sequence ladder from X12-4. (C) Schematic representation of major and minor FEN-1 cleavage sites (shown by large and small arrows, respectively) within the homologous core on forked duplex products of WRN HJ branch migration.
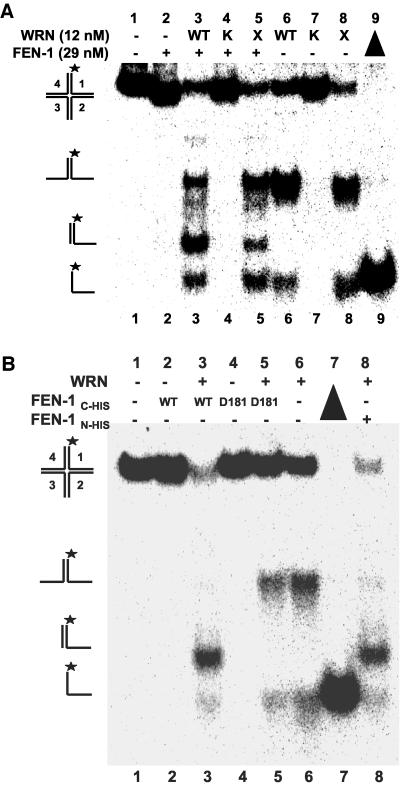
WRN Helicase Activity and FEN-1 Nuclease Activity Are Responsible for HJ Cleavage
To assess if the linear partial duplex product of the FEN-1 cleavage reaction is dependent on the branch migration activity of WRN, we tested the effect of a recombinant WRN protein with a site-directed mutation in the active site of its ATPase domain on FEN-1 cleavage. The WRN-K577M mutant, devoid of ATPase or helicase activity (Gray et al., 1997; Brosh et al., 1999), failed to unwind the HJ (Figure 7A, lane 7) and also failed to stimulate FEN-1 cleavage (lane 4). Thus WRN ATP hydrolysis/DNA unwinding was required for the FEN-1 cleavage reaction. The WRN exonuclease defective mutant WRN-E84A (Huang et al., 1998) retained the ability to unwind the HJ (Figure 7A, lane 8) and stimulate FEN-1 incision (lane 5), indicating that the exonuclease activity of WRN is not required for FEN-1 cleavage of the unwound HJ products.
Figure 7.
WRN helicase activity and FEN-1 nuclease activity are responsible for HJ cleavage. (A) WRN ATPase/helicase but not exonuclease activity is required for stimulation of FEN-1 HJ resolution. Reactions containing 2.5 fmol of HJ(X12-1), wild-type or mutant WRN (12 nM) and FEN-1 (29 nM) were conducted under standard conditions. Phosphorimage of a typical native gel is shown. WT, wild-type WRN; K, WRN-K577M; X, WRN-E84A. (B) FEN-1 nuclease activity is responsible for HJ resolution. Reactions were conducted under standard conditions using either wild-type FEN-1N-HIS, wild-type FEN-1C-HIS, or nuclease defective FEN-1C-HIS-D181A proteins as indicated. Phosphorimage of a typical native gel is shown. (▴), heat-denatured substrate control.
It was conceivable that the purified recombinant N-terminal histidine-tagged FEN-1 might contain a minor contaminant responsible for the observed HJ cleavage activity. To address this concern, a C-terminal histidine-tagged FEN-1 with either wild-type sequence or bearing a site-directed mutation D181A in the active site of the nuclease domain was tested for cleavage of the products from WRN HJ unwinding (Figure 7B). In control studies, wild-type FEN-1C-HIS effectively cleaved a conventional 1- or 26-nt 5′ flap substrate to yield the expected products, whereas the FEN-1 with the D181A mutation (FEN-1C-HIS-D181A; Shen et al., 1996) was confirmed to be nuclease defective on either 5′ flap substrate (unpublished data). FEN-1C-HIS cleaved in the presence of WRN (Figure 7B, lane 3), yielding the linear partial duplex that comigrated with the product obtained in reactions containing FEN-1N-HIS and WRN (lane 8). No FEN-1 cleavage activity was detected in reactions conducted in the absence of WRN (lane 2), consistent with results using N-terminal tagged FEN-1. The D181A FEN-1 mutant failed to cleave in the presence or absence of WRN (lanes 4 and 5); rather, WRN helicase activity prevailed (lane 5), similar to that observed in reactions containing only WRN (lane 6). These results demonstrate that the intrinsic nuclease activity of FEN-1 is responsible for DNA cleavage in a reaction dependent on WRN ATPase/helicase activity.
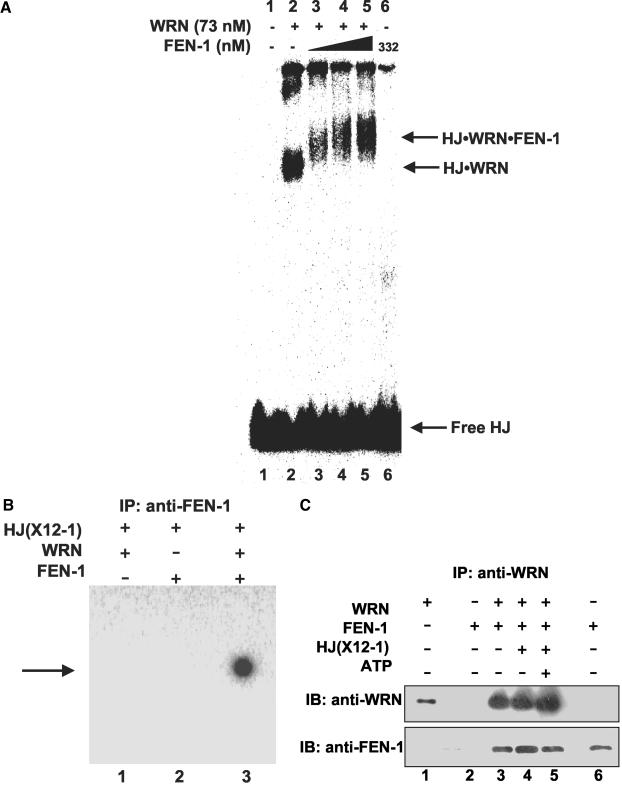
WRN Recruits FEN-1 to the HJ
Although FEN-1 has been implicated in 5′ flap processing that occurs during maturation of lagging strands in DNA replication, FEN-1 cleavage activity is inhibited by double-stranded DNA flaps because the enzyme cannot bind to a free 5′ ssDNA end and track to the “elbow” of the flap (Murante et al., 1995). FRET analysis suggested that WRN and FEN-1 associate at a DNA structural intermediate found at arrested replication foci. Replication fork regression that can occur at a blocked replication fork gives rise to a HJ structure. The ability of WRN to bind synthetic HJ structures (Constantinou et al., 2000) and physically interact with FEN-1 (Brosh et al., 2001) suggested that WRN recruits FEN-1 to the HJ. To test this, gel-shift assays were performed with radiolabeled HJ, WRN, and/or FEN-1. WRN-HJ complexes were detected by the appearance of a retarded species (Figure 8A, lane 2), whereas FEN-1 alone did not significantly bind HJ (lane 6). When both WRN and FEN-1 were incubated with HJ, we observed a FEN-1 super-shifted species compared with the WRN-HJ complex (lanes 3-5). To confirm that FEN-1 was present in the super-shifted complex, the binding reaction mixtures were immunoprecipitated with anti-FEN-1 antibody-coated beads. Radiolabeled HJ was immunoprecipitated for those reactions containing WRN and FEN-1 (Figure 8B, lane 3), whereas HJ failed to be immunoprecipitated in binding reactions containing only WRN or FEN-1 (Figure 8B, lanes 1 and 2). HJ also failed to precipitate in the absence of WRN and FEN-1 or when anti-FEN-1 antibody was omitted in the pull-down experiment (unpublished data). These results verify that FEN-1 was present in the protein-HJ super-shifted complex and suggest that WRN binding to the HJ is required in order for FEN-1 to bind HJ.
Figure 8.
WRN recruits FEN-1 to the Holliday junction. (A) WRN-HJ complex is super-shifted by FEN-1. Binding mixtures (20 μl) containing 25 fmol of HJ(X12-1), 1 mM ATPγS, WRN (73 nM), and FEN-1 (116, 174, and 332 nM, lanes 3, 4 and 5, respectively) were incubated at 24°C for 20 min followed by 0.25% glutaraldehyde cross-linking. Protein-DNA complexes were analyzed on nondenaturing 5% polyacrylamide gels. A phosphorimage of a typical gel is shown. (B) Identification of FEN-1 in the super-shifted protein-HJ complex. HJ(X12-1) was incubated with WRN and/or FEN-1 under standard binding conditions. Protein-HJ complexes were UV-cross-linked, incubated with anti-FEN-1 antibody, precipitated with protein-G agarose beads, and electrophoresed on 8% polyacrylamide SDS gels. A phosphorimage of a gel from a typical experiment is shown. (C) Purified WRN and FEN-1 interact directly under HJ resolution reaction conditions. Purified WRN (12 nM; lanes 3, 4, 5) and FEN-1 (29 nM; lanes 2, 3, 4, 5) were incubated in HJ resolution buffer in the absence or presence of HJ(X12-1) (37.5 fmol) and/or ATP (2 mM) as indicated. Binding mixtures were subsequently incubated with anti-WRN antibody and adsorbed to protein-G agarose beads. Beads were extensively washed and bound proteins were eluted and resolved on 8-16% SDS-PAGE. Proteins were transferred to PVDF membranes and probed with either mouse anti-WRN or rabbit anti-FEN-1 antibodies as noted. In both panels, markers for recombinant WRN (50 ng; lane 1) and recombinant FEN-1 (50 ng; lane 6) are shown.
A physical interaction between WRN and FEN-1 was previously demonstrated by their coimmunoprecipitation from HeLa cell lysate and affinity pull-down experiments using GST-WRN recombinant protein fragments (Brosh et al., 2001). Because WRN was able to recruit FEN-1 to the HJ, we wanted to ascertain that WRN and FEN-1 physically interacted with each other in solution under the HJ resolution reaction conditions. To test for the direct interaction, purified WRN and FEN-1 were incubated together, precipitated with anti-WRN antibody and subsequently examined for the presence of FEN-1 and WRN by Western blot analysis. The anti-WRN antibody coimmunoprecipitated WRN and FEN-1 in the HJ resolution reaction buffer (Figure 8C, lane 3). Both WRN and FEN-1 were also coimmunoprecipitated in HJ reaction buffer supplemented with the HJ substrate in the absence (lane 4) or presence (lane 5) of ATP. FEN-1 failed to be precipitated when WRN was omitted from the binding incubation (lane 2) or using a goat normal antiserum (unpublished data). These results indicate that WRN and FEN-1 directly interact with each other under the cleavage reaction and gel-shift conditions supporting the conclusion that WRN recruits FEN-1 to the HJ by a direct protein interaction.
The HJ-specific Binding Protein RuvA Blocks WRN Branch Fork Migration and FEN-1 Cleavage of the Branch Fork Migration Products
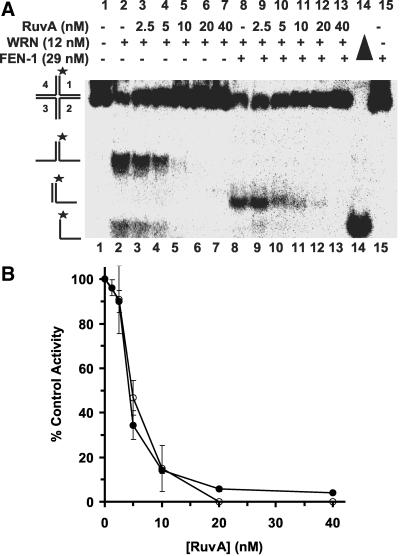
E. coli RuvA has a high affinity for the HJ (Parsons et al., 1992) and blocks WRN branch migration activity on α-structures (Constantinou et al., 2000). To establish that WRN helicase activity is initiated at the junction, we examined the effect of RuvA on WRN-catalyzed DNA unwinding. With increasing concentrations of RuvA, we observed dose-dependent inhibition of WRN branch migration activity (Figure 9, A and B). At a RuvA concentration of 10 nM (Figure 9A, lane 5), we detected 85% inhibition of HJ unwinding by WRN (Figure 9B). In contrast, RuvA did not inhibit WRN helicase activity on forked duplex substrates (X12-1: X12-2 or X12-1: X12-4; unpublished data). At 10 nM RuvA, FEN-1 cleavage was inhibited by 85% (Figure 9A, lane 10, and B). RuvA inhibited WRN-dependent cleavage by FEN-1 in a dose-dependent manner closely resembling the profile for inhibition of WRN branch migration (Figure 9, A and B). These results suggest that WRN initiates unwinding at the junction cross-over and that RuvA prevents FEN-1 cleavage by blocking WRN-catalyzed branch migration of the HJ.
Figure 9.
RuvA inhibits WRN-stimulated FEN-1 cleavage of Holliday junction by blocking WRN branch-migrating activity. Reactions (20 μl) containing 2.5 fmol of HJ(X12-1) and WRN (12 nM) in the presence or absence of FEN-1 (29 nM) and increasing amounts of RuvA (0-40 nM) were incubated at 37°C under standard conditions. (A) Phosphorimage of a typical native gel is shown. Filled triangle, heat denatured substrate control. (B) Helicase (○) and cleavage (•) activities are expressed as percent of control reactions in which RuvA was omitted.
Orientation of WRN Branch Fork Migration Is Influenced by the Length of Duplex Arms in the HJ
WRN has been previously shown to catalyze branch fork migration of recombination intermediate α structures as well as bind and unwind oligonucleotide-based HJ structures. To better understand the role of HJ DNA structural elements that influence the ability of WRN to catalyze branch fork migration, we tested two synthetic HJ with two oppositely positioned long arms (45 base pairs) and two oppositely positioned short arms (21 base pairs) for unwinding by WRN and subsequent cleavage by FEN-1. For the first HJ in which the long duplex arms are vertical (HJ(X1)), WRN unwound the horizontal arms of the HJ structure to yield the forked duplex product (Figure 10A, lane 2), which could be cleaved by FEN-1 to generate the linear partial duplex product (Figure 10A, lane 3). For the second HJ substrate with long horizontal duplex arms and short vertical arms (HJ(X2)), WRN unwound the HJ to give forked duplex product (Figure 10B, lane 2), which was then cleaved by FEN-1 to produce a short incised oligonucleotide (Figure 10B, lane 3). Denaturing gel analysis of the products from FEN-1 cleavage of HJ(X1) or HJ(X2) conducted in the presence of WRN and ATP revealed that the product was a 21-mer, the predicted size of a FEN-1 cleavage reaction on a forked duplex whose short vertical arms had been unwound (our unpublished results). These results show that the length of duplex arms in the HJ confers a bias for the orientation that WRN uses to unwind the HJ. A similar observation was made for the E. coli branch fork motor RuvAB (van Gool et al., 1999).
Figure 10.
The length of duplex arms in the HJ influences orientation of WRN branch fork migration. Reactions (20 μl) containing 2.5 fmol of HJ(X1) (A) or HJ(X2) (B), FEN-1 (29 nM), and/or WRN (12 nM) were incubated at 37°C under standard reaction conditions in the presence of 2 mM ATP. Phosphorimage of a typical native gel is shown for each substrate reaction. Filled triangle, heat denatured substrate control. The position of HJ, forked duplex, single-stranded DNA and FEN-1 cleavage product is indicated.
WRN Unwinds a Chicken Foot Structure Enabling FEN-1 to Cleave the Unwound Structure
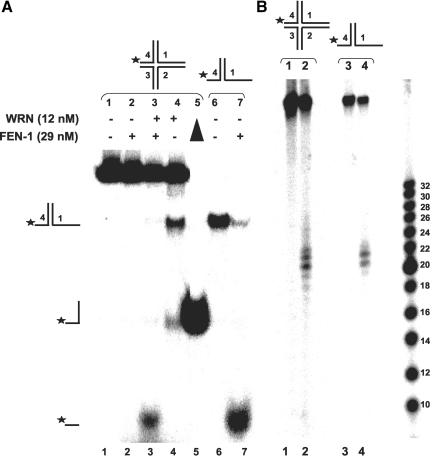
The effect of duplex arm length in the HJ structure on orientation of WRN helicase activity suggested that WRN and FEN-1 might process a regressed chicken foot HJ intermediate in a specific manner. To address this possibility in vitro, we examined the effects of WRN and/or FEN-1 on a four-stranded structure with three duplex arms of sufficient length to confer an effect on orientation of HJ unwinding by WRN, i.e., 45 base pairs, as demonstrated in the previous section, and a single short duplex arm of 21 base pairs. WRN was found to unwind the chicken foot HJ, yielding primarily the two-stranded forked duplex and to a lesser extent the single-stranded 46-mer (Figure 11A, lane 4). As expected, FEN-1 alone did not cleave the four-stranded structure, but in the presence of WRN and ATP cleaved the short arm to give a fast migrating radiolabeled species (Figure 11A, lane 3) that comigrated with the FEN-1 cleavage product from reactions containing the X1-1 + 25:X1-4 forked duplex (Figure 11A, lane 7). The FEN-1 cleavage products from the synthetic HJ structure were verified by denaturing gel analysis (Figure 11B, lane 2) to comigrate with the FEN-1 cleavage products from reactions containing the forked duplex (X1-1 + 25:X1-4; Figure 11B, lane 4). The WRN-FEN-1 incision reaction on the chicken foot HJ structure was dependent on the presence of ATP (our unpublished results), as observed previously with the symmetrical HJ structures, indicating that FEN-1 cleavage was dependent on WRN helicase activity. These results indicate that WRN can unwind the chicken foot structure enabling FEN-1 to cleave the unwound 5′ ssDNA arm endonucleolytically.
Figure 11.
WRN unwinds a chicken foot DNA structure enabling FEN-1 to cleave the unwound product. Reactions (20 μl) containing 2.5 fmol of HJ (chicken foot) (A) or X1-1 + 25: X1-4 forked duplex (B), FEN-1 (29 nM), and/or WRN (12 nM) were incubated at 37°C under standard reaction conditions in the presence of 2 mM ATP. Phosphorimage of a typical native gel is shown for each substrate reaction. Filled triangle, heat denatured substrate control. The position of HJ (chicken foot), forked duplex, single-stranded DNA, and FEN-1 cleavage product is indicated.
DISCUSSION
The critical importance of RecQ helicases in maintaining genomic integrity is thought to derive from their ability to suppress recombination (Wu and Hickson, 2002; Cobb et al., 2002); however, the precise mechanisms that various RecQ helicases employ to serve as “antirecombinases” are not well understood. Mutations in the WRN and BLM helicases are associated with a number of replication defects that include impaired fork progression, accumulation of abnormal replication intermediates, and aberrant homologous recombination. It has been proposed that at least one function of the WRN or BLM DNA helicases is to prevent aberrant deleterious recombinogenic pathways when replication is perturbed by DNA damage, alternate DNA structure, or impaired DNA synthesis. The processing of aberrant DNA structures by RecQ helicases is likely to counter their potential toxicity incurred by recombinogenic pathways. Evidence that replication forks of presumably damaged chromosomes stall normally in vivo suggests that cells are likely to have mechanisms to deal with the aberrant DNA structures. The ability of helicases to unwind duplex and alternate DNA structures may increase access to repair and replication proteins important for the resolution of abnormal DNA structure.
On the basis of our recent work demonstrating a physical and functional interaction between WRN and the replication-associated structure-specific endonuclease FEN-1, we hypothesized that WRN and FEN-1 associate with each other to process certain DNA structures that arise during replication. The copurification of WRN with a DNA replication complex (Lebel et al., 1999) and interaction of WRN with proteins involved in DNA replication (Brosh and Bohr, 2002) is consistent with the notion that WRN plays a direct role in some aspect of DNA replication. To test for an interaction in vivo between WRN and FEN-1, we examined the localization of the two proteins by FRET analysis and determined that the WRN:FEN-1 association is strongly enhanced when replication forks are blocked by DNA damage. In these spots WRN and FEN-1 directly interact or are very close to each other (<100 Å) in the same complex. These results suggest that WRN and FEN-1 work together for a specialized function in replicating foci.
Genetic and biochemical evidence strongly suggests that the physiological substrate of FEN-1 is the double-flap structure that arises as a consequence of strand displacement during DNA synthesis (Xie et al., 2001; Kao et al., 2002). The results presented in this study demonstrate that WRN stimulates FEN-1 cleavage on the double-flap substrate; furthermore, the cleavage specificity of FEN-1 is retained in the presence of WRN. These results would suggest that WRN participates with FEN-1 in replication (Figure 12A) or repair. Interestingly, very recent evidence suggests that the BLM helicase functions in replication and repair pathways that involve yeast FEN-1 and yeast Dna2 (Imamura and Campbell, 2003).
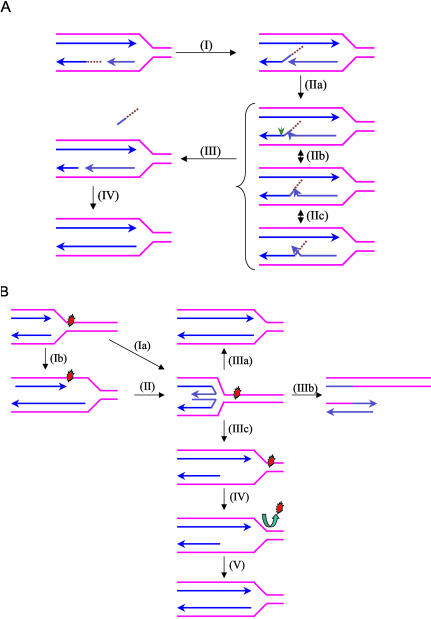
Figure 12.
Proposed models for the biological roles of the WRN helicase-FEN-1 nuclease interaction. (A) Proposed role of WRN to stimulate FEN-1 cleavage of double-flap structures during DNA replication. During Okazaki fragment processing, the 5′ flap intermediate without or with initiator RNA (as shown) that is created by strand displacement (I) can branch migrate to form numerous interconverting structures (IIa, IIb, IIc) based on sequence complementarity. In a reaction stimulated by WRN, FEN-1 specifically recognizes and cleaves (green arrow) the double-flap structure containing a 1-nt 3′ tail at the position between the first two base pairs of the downstream duplex (III). The resulting nick product is subsequently sealed by DNA ligase to restore the integrity of the replicated lagging strand (IV). (B) Proposed roles of WRN and possibly other RecQ helicases at stalled replication forks to restart replication. Progression of a replication fork is blocked by a DNA lesion. A stalled replication fork can regress to a chicken foot HJ structure by annealing of the leading and lagging strands (Ia). Leading and lagging strand synthesis can become uncoupled when only leading strand polymerization is stalled by a DNA lesion (Ib). The chicken foot structure is generated by annealing of the leading and laggin strand and resumption of leading strand synthesis using the nascent lagging strand as template (II). The chicken foot can be acted upon by different pathways leading to either repair of the lesion or lesion bypass. A potential role of WRN or a related RecQ helicase in the rescue of a stalled fork is to catalyze reverse branch migration past the lesion to reset the replication fork and the lesion can be subsequently corrected by DNA repair (IIIa). In a second scenario, resolution of the chicken foot by a HJ resolvase leads to fork cleavage (IIIb). Repair of the broken fork is then achieved by homologous recombination and enables replication to restart. Alternatively, replication can be resumed by a nonrecombinogenic mechanism involving the concerted action of a RecQ helicase and a 5′ flap endonuclease/exonuclease (IIIC-V). WRN unwinds the duplex arm of the chicken foot structure and stimulates FEN-1 cleavage of the newly synthesized lagging strand to create a region of single-stranded DNA (IIIc). The newly formed three-stranded DNA structure can be stabilized by a Rad51-like protein while the lesion is repaired (IV). This leads to the accurate restart of replication (V) by a process that does not involve recombination.
In addition to double-flap structures at replicating foci, stalled or reversed replicating forks are likely to contain a variety of other DNA structures (Whitby et al., 2003). Initially, a blocked replication fork can potentially reverse to expose a 3′ leading nascent ssDNA or 5′ nascent lagging ssDNA depending on which template strand is blocked. The latter structure with a 5′ ssDNA tail can be endonucleolytically cleaved by human FEN-1 (our unpublished results). Fork reversal can also result in the reannealing of template strands and give rise to a chicken foot intermediate resembling an HJ structure (Figure 12B). Aberrant DNA structures at stalled or reversed replication forks have been observed in vivo (Sogo et al., 2002) and may be processed in nonrecombinogenic or recombinogenic modes. Accurate recovery of replication in E. coli was shown to require several proteins in the recF pathway of recombination, including the RecQ helicase and RecJ 5′ to 3′ exonuclease (Courcelle and Hanawalt, 1999; Courcelle et al., 2003). By pulse-chase analysis, RecQ and RecJ were determined to be responsible for the degradation of the nascent lagging strand of the blocked replication forks before their resumption (Courcelle and Hanawalt, 1999). It was proposed that RecQ and RecJ act together at blocked replication forks to create a ssDNA gap large enough for RecA to bind and stabilize the fork. More recently, it was demonstrated by two-dimensional gel electrophoresis that RecQ and RecJ cooperate to process a regressed replication fork X-shaped intermediate that is induced by DNA damage, which can then be maintained by RecA RecFOR (Courcelle et al., 2003). A similar mechanism for fork reversal and accumulation of ssDNA at stalled replication forks has been proposed in eukaryotic cells with a checkpoint defect in Rad53 kinase (Sogo et al., 2002). Thus growing evidence suggests that the accumulation of HJ structures through fork reversal produces abnormal replication intermediates that become processed by recombination pathways that can lead to genomic instability in certain mutant backgrounds. An intact replication checkpoint response insures that replication and recombination are coordinated with each other.
Analogous to the coordinate activity of bacterial RecQ and RecJ in replication restart, we hypothesized that WRN and FEN-1 might together process stalled forks that have regressed (Figure 12B). This led us to investigate the action of WRN and FEN-1 on the four-stranded DNA structural intermediate of the regressed replication fork. Biochemical characterization of the products from reactions containing model synthetic HJ molecules and purified recombinant proteins enabled us to determine how the four-stranded DNA structure is metabolized by WRN and FEN-1 (Figure 12B). We conclude from our biochemical studies that WRN helicase activity initiating from the HJ core can provide a suitable DNA molecule with a free 5′ ssDNA end on which FEN-1 can load to ultimately catalyze structure-specific cleavage of the unwound 5′ ssDNA arm. Orientation of WRN branch fork migration is influenced by the length of the duplex arms in the HJ structure such that short (21 base pairs) duplex arms residing across from each other are preferentially unwound by WRN helicase compared with the longer 45-base pair duplex arms. The unwound 5′ ssDNA tracts of the short arms can be subsequently cleaved by FEN-1. Thus the catalytic cleavage of the regressed fork intermediate by FEN-1 requires WRN branch fork migration to provide the suitable substrate for FEN-1 loading. The physical interaction of WRN with FEN-1 is likely to facilitate FEN-1 endonucleolytic cleavage.
WRN-/- cells display a reduced rate of repair, elevated apoptotic cell death, and increased DNA strand breaks after replication arrest by nucleotide deletion or DNA damage induced in S-phase (Pichierri et al., 2001). The observation that expression of a HJ resolvase rescues both the recombination defect and cell survival after DNA damage in WRN-/- cells (Saintigny et al., 2002) suggests that four-way junctions are a target in vivo of the WRN helicase. This is further supported by the observation that suppression of RAD51-dependent recombination significantly improved survival of WRN-/- cells after DNA damage (Saintigny et al., 2002). Collectively these observations suggest that the inappropriate processing of stalled replication fork intermediates directly contributes to the aberrant homologous recombination characteristic of WRN-/- cells. Elevated sensitivity to replication inhibitors (Pichierri et al., 2001), hypersensitivity to camptothecin (Poot et al., 1999; Pichierri et al., 2000), and the prolonged S phase (Poot et al., 1992) in WRN-/- cell lines point to an underlying deficiency when replication fork progression is abnormal. Furthermore, WRN protein foci formation by DNA damaging agents is dependent on replication and shows colocalization with RAD51 (Sakamoto et al., 2001). Altogether the cellular and biochemical data support a model in which the genomic instability of WS stems from the uncoordinated processing of replication intermediates that are channeled to recombination pathways that are ultimately deleterious. The coordinate action of WRN and FEN-1 is likely to be important for the maintenance of genomic integrity absent in WS.
Acknowledgments
We thank our colleagues in the Laboratory of Molecular Gerontology (National Institute on Aging, National Institutes of Health) and the Bambara Lab (Department of Biochemistry and Biophysics, University of Rochester Medical Center) for helpful discussion. We also thank Dr. E. Warbrick for the plasmid construct ECFP-FEN-1, Dr. D. Wilson III for FEN-1C-HIS and FEN-1C-HISD181A recombinant proteins, and Dr. M. Cox for RuvA. This work was supported in part by NIH Grant GM24441.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-08-0567. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0567.
References
- Bambara, R.A., Murante, R.S., and Henricksen, L.A. (1997). Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272, 4647-4650. [DOI] [PubMed] [Google Scholar]
- Baynton, K., Otterlei, M., Bjoras, M., von Kobbe, C., Bohr, V.A., and Seeberg, E. (2003). WRN interacts physically and functionally with the recombination mediator protein RAD52. J. Biol. Chem. (Epub). [DOI] [PubMed]
- Brosh, R.M. Jr., and Bohr, V.A. (2002). Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol. 37, 491-506. [DOI] [PubMed] [Google Scholar]
- Brosh, R.M. Jr., Driscoll, H.C., Dianov, G.L., and Sommers, J.A. (2002a). Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry 41, 12204-12216. [DOI] [PubMed] [Google Scholar]
- Brosh, R.M., Jr., Orren, D.K., Nehlin, J.O., Ravn, P.H., Kenny, M.K., Machwe, A., and Bohr, V.A. (1999). Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 274, 18341-18350. [DOI] [PubMed] [Google Scholar]
- Brosh, R.M., Jr., Waheed, J., and Sommers, J.A. (2002b). Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 277, 23236-23245. [DOI] [PubMed] [Google Scholar]
- Brosh, R.M., Jr., von Kobbe, C., Sommers, J.A., Karmakar, P., Opresko, P.L., Piotrowski, J., Dianova, I., Dianov, G.L., and Bohr, V.A. (2001). Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 20, 5791-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, J., Bjergbaek, L., and Gasser, S. (2002). RecQ helicases: at the heart of genetic stability. FEBS Lett. 529, 43-48. [DOI] [PubMed] [Google Scholar]
- Constantinou, A., Tarsounas, M., Karow, J.K., Brosh, R. M., Jr., Bohr, V.A., Hickson, I.D., and West, S.C. (2000). Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle, J., Donaldson, J.R., Chow, K.H., and Courcelle, C.T. (2003). DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299, 1064-1067. [DOI] [PubMed] [Google Scholar]
- Courcelle, J., and Hanawalt, P.C. (1999). RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262, 543-551. [DOI] [PubMed] [Google Scholar]
- Ellis, N.A., Groden, J., Ye, T.Z., Straughen, J., Lennon, D.J., Ciocci, S., Proytcheva, M., and German, J. (1995). The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83, 655-666. [DOI] [PubMed] [Google Scholar]
- Freudenreich, C.H., Kantrow, S.M., and Zakian, V.A. (1998). Expansion and length-dependent fragility of CTG repeats in yeast. Science 279, 853-856. [DOI] [PubMed] [Google Scholar]
- Gray, M.D., Shen, J.C., Kamath-Loeb, A.S., Blank, A., Sopher, B.L., Martin, G.M., Oshima, J., and Loeb, L.A. (1997). The Werner syndrome protein is a DNA helicase. Nat. Genet. 17, 100-103. [DOI] [PubMed] [Google Scholar]
- Gray, M.D., Wang, L., Youssoufian, H., Martin, G.M., and Oshima, J. (1998). Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res. 242, 487-494. [DOI] [PubMed] [Google Scholar]
- Greene, A.L., Snipe, J.R., Gordenin, D.A., and Resnick, M.A. (1999). Functional analysis of human FEN1 in Saccharomyces cerevisiae and its role in genome stability. Hum. Mol. Genet. 8, 2263-2273. [DOI] [PubMed] [Google Scholar]
- Huang, S., Li, B., Gray, M.D., Oshima, J., Mian, I.S., and Campisi, J. (1998). The premature ageing syndrome protein, WRN, is a 3′->5′ exonuclease. Nat. Genet. 20, 114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, O., and Campbell, J.L. (2003). The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl. Acad. Sci. USA 100, 8193-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.E., Kovvali, G.K., Prakash, L., and Prakash, S. (1995). Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269, 238-240. [DOI] [PubMed] [Google Scholar]
- Kao, H.I., Henricksen, L.A., Liu, Y., and Bambara, R.A. (2002). Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277, 14379-14389. [DOI] [PubMed] [Google Scholar]
- Kim, K., Biade, S., and Matsumoto, Y. (1998). Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273, 8842-8848. [DOI] [PubMed] [Google Scholar]
- Kitao, S., Ohsugi, I., Ichikawa, K., Goto, M., Furuichi, Y., and Shimamoto, A. (1998). Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54, 443-452. [DOI] [PubMed] [Google Scholar]
- Kitao, S., Shimamoto, A., Goto, M., Miller, R.W., Smithson, W.A., Lindor, N.M., and Furuichi, Y. (1999). Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22, 82-84. [DOI] [PubMed] [Google Scholar]
- Klungland, A., and Lindahl, T. (1997). Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16, 3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska, R.J., Stefanovic, L., Tran, H.T., Resnick, M.A., Gordenin, D.A., and Petes, T.D. (1998). Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell Biol. 18, 2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel, M., Spillare, E.A., Harris, C.C., and Leder, P. (1999). The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 274, 37795-37799. [DOI] [PubMed] [Google Scholar]
- Lieber, M.R. (1997). The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19, 233-240. [DOI] [PubMed] [Google Scholar]
- Martin, G.M. (1978). Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig. Artic. Ser. 14, 5-39. [PubMed] [Google Scholar]
- Matyus, L. (1992). Fluorescence resonance energy transfer measurements on cell surfaces. A spectroscopic tool for determining protein interactions. J. Photochem. Photobiol. B 12, 323-337. [DOI] [PubMed] [Google Scholar]
- Maxam, A.M., and Gilbert, W. (1980). Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65, 499-560. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., Lloyd, R.G., and Marians, K.J. (2001). Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 98, 8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh, P., Karow, J.K., Brosh, R.M., Jr., Bohr, V.A., and Hickson, I.D. (2001). The Bloom's and Werner's syndrome proteins are DNA structurespecific helicases. Nucleic Acids Res. 29, 2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murante, R.S., Rust, L., and Bambara, R.A. (1995). Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 270, 30377-30383. [DOI] [PubMed] [Google Scholar]
- Murchie, A.I., Portugal, J., and Lilley, D.M. (1991). Cleavage of a four-way DNA junction by a restriction enzyme spanning the point of strand exchange. EMBO J. 10, 713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negritto, M.C., Qiu, J., Ratay, D.O., Shen, B., and Bailis, A.M. (2001). Novel function of Rad27 (FEN-1) in restricting short-sequence recombination. Mol. Cell Biol. 21, 2349-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, J.P., Shen, B., Park, M.S., and Sklar, L.A. (1996). Kinetic analysis of human flap endonuclease-1 by flow cytometry. Biochemistry 35, 11668-11676. [DOI] [PubMed] [Google Scholar]
- Opresko, P.L., Laine, J.P., Brosh, R.M., Jr., Seidman, M.M., and Bohr, V.A. (2001). Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein. J. Biol. Chem. 277, 41110-41119. [DOI] [PubMed] [Google Scholar]
- Orren, D.K., Brosh, R.M., Jr., Nehlin, J.O., Machwe, A., Gray, M.D., and Bohr, V.A. (1999). Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 27, 3557-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei, M. et al. (1999). Post-replicative base excision repair in replication foci. EMBO J. 18, 3834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau, J., and Wellinger, R.J. (1999). Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell Biol. 19, 4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, C.A., Tsaneva, I., Lloyd, R.G., and West, S.C. (1992). Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl. Acad. Sci. USA 89, 5452-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri, P., Franchitto, A., Mosesso, P., and Palitti, F. (2000). Werner's syndrome cell lines are hypersensitive to camptothecin-induced chromosomal damage. Mutat. Res. 456, 45-57. [DOI] [PubMed] [Google Scholar]
- Pichierri, P., Franchitto, A., Mosesso, P., and Palitti, F. (2001). Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12, 2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot, M., Gollahon, K.A., and Rabinovitch, P.S. (1999). Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 104, 10-14. [DOI] [PubMed] [Google Scholar]
- Poot, M., Hoehn, H., Runger, T.M., and Martin, G.M. (1992). Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cells. Exp. Cell. Res. 202, 267-273. [DOI] [PubMed] [Google Scholar]
- Postow, L., Ullsperger, C., Keller, R.W., Bustamante, C., Vologodskii, A.V., and Cozzarelli, N.R. (2001). Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276, 2790-2796. [DOI] [PubMed] [Google Scholar]
- Prince, P.R., Emond, M.J., and Monnat, R.J., Jr. (2001). Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 15, 933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., Li, X., Frank, G., and Shen, B. (2001). Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 276, 4901-4908. [DOI] [PubMed] [Google Scholar]
- Reagan, M.S., Pittenger, C., Siede, W., and Friedberg, E.C. (1995). Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177, 364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny, Y., Makienko, K., Swanson, C., Emond, M.J., and Monnat, R.J., Jr. (2002). Homologous recombination resolution defect in Werner syndrome. Mol. Cell Biol. 22, 6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, S., Nishikawa, K., Heo, S.J., Goto, M., Furuichi, Y., and Shimamoto, A. (2001). Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells 6, 421-430. [DOI] [PubMed] [Google Scholar]
- Schweitzer, J.K., and Livingston, D.M. (1998). Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7, 69-74. [DOI] [PubMed] [Google Scholar]
- Shen, B., Nolan, J.P., Sklar, L.A., and Park, M.S. (1996). Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J. Biol. Chem. 271, 9173-9176. [DOI] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599-602. [DOI] [PubMed] [Google Scholar]
- Sommers, C.H., Miller, E.J., Dujon, B., Prakash, S., and Prakash, L. (1995). Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270, 4193-4196. [DOI] [PubMed] [Google Scholar]
- Tishkoff, D.X., Filosi, N., Gaida, G.M., and Kolodner, R.D. (1997). A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88, 253-263. [DOI] [PubMed] [Google Scholar]
- Vallen, E.A., and Cross, F.R. (1995). Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell Biol. 15, 4291-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool, A.J., Hajibagheri, N.M., Stasiak, A., and West, S.C. (1999). Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev. 13, 1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M.C., Osman, F., and Dixon, J. (2003). Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 278, 6928-6935. [DOI] [PubMed] [Google Scholar]
- Wu, L., and Hickson, I.D. (2002). RecQ helicases and cellular responses to DNA damage. Mutat. Res. 509, 35-47. [DOI] [PubMed] [Google Scholar]
- Wu, X., Wilson, T.E., and Lieber, M.R. (1999). A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96, 1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z., and Liu, Y. (2001). Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81, 2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., Liu, Y., Argueso, J.L., Henricksen, L.A., Kao, H.I., Bambara, R.A., and Alani, E. (2001). Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability, and flap cleavage. Mol. Cell Biol. 21, 4889-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.E. et al. (1996). Positional cloning of the Werner's syndrome gene. Science 272, 258-262. [DOI] [PubMed] [Google Scholar]