Abstract
For Moloney murine leukemia virus (M-MuLV), sustained viral infections require expression from an integrated provirus. For many applications, non-integrating retroviral vectors have been utilized to avoid the unwanted effects of integration, however, the level of expression from unintegrated DNA is significantly less than that of integrated provirus. We find that unintegrated DNA expression can be increased in the presence of HDAC inhibitors, such as TSA, when applied in combination with integrase (IN) mutations. These mutants include an active site mutation as well as catalytically active INs bearing mutations of K376 in the MuLV C-terminal domain of IN. MuLV IN K376 is homologous to K266 in HIV-1 IN, a known substrate for acetylation. The MuLV IN protein is acetylated by p300 in vitro, however, the effect of HDAC inhibitors on gene expression from unintegrated DNA is not dependent on the acetylation state of MuLV IN K376.
Keywords: non-integrating vectors; Integrase; MuLV IN; HDAC inhibitors; pre-integration complex, PIC; intasome
Introduction
Integration of double-stranded viral DNA into the host chromatin, a hallmark of retroviral infection, is catalyzed by the virally encoded integrase (IN) protein. IN consists of three distinct domains as determined by limited proteolysis (Engelman and Craigie, 1992): the N-terminal Zn2+ binding domain (NTD), catalytic core domain (CCD), and the C-terminal domain (CTD). Various in vitro assays have demonstrated that the IN protein alone is sufficient to catalyze both the 3’ processing and strand transfer reactions (Craigie et al., 1990; Katz et al., 1990), however, in vivo, additional viral and cellular proteins associate with the DNA and IN protein to form the functional pre-integration complex (PIC) (Bowerman et al., 1989; Miller et al., 1997).
The PIC, a large nucleoprotein complex whose constituents vary depending on the retrovirus, minimally contains the viral DNA and the IN protein. Additional viral and cellular factors that have been identified as components of the PIC include the viral capsid (CA) and p12 proteins in MuLV (Bowerman et al., 1989; Prizan-Ravid et al., 2010) but not HIV (Bukrinsky et al., 1993), and the cellular host factor LEDGF, present in HIV (Cherepanov et al., 2003) but not MuLV (Llano et al., 2004). Common to PICs from both viruses is the cellular protein barrier-to-autointegration factor (BAF), an 89 amino acid cellular protein with non-specific DNA binding capabilities (Lee and Craigie, 1998; Lin and Engelman, 2003). Further analysis of PICs isolated from infected cells reveals that a protein structure is present at the viral DNA ends (Wei et al., 1997), conferring protection from DNase I cleavage up to ~20 bp from the termini (Wei et al., 1998). Evidence for an extended footprint, approximately 200-250 bp from the viral DNA ends, is uncovered when probed with Mu-mediated PCR (MM-PCR) (Chen et al., 1999; Wei et al., 1998). This complex will be referred to as the extended intasome, to distinguish it from the minimal intasome structure (Hare et al., 2010a) consisting of IN proteins and DNA only. To date, an extended intasome protein structure that could provide protection of up to ~200-250 bp of DNA has not been formed in vitro. While it is clear that the presence of IN is essential to formation of the extended intasome, the exact nature of its composition remains elusive (Chen et al., 1999; Wei et al., 1997, 1998).
The use of non-integrating retroviral vectors has been explored as an alternative avenue for gene delivery in gene therapy applications where expression is (generally) driven from internal promoters rather than the viral LTR (Bayer et al., 2008; Nightingale et al., 2006; Philippe et al., 2006; Rahim et al., 2009; Sloan and Wainberg, 2011; Yu et al., 2008). Although in theory, integration of a therapeutic target gene has many advantages over a non-integrating vector, the dangers of integration are widely recognized. Integration and alteration of the host DNA potentiates the risk of insertional mutagenesis and oncogene activation (Thomas et al., 2003). This risk materialized when several patients undergoing gene therapy developed leukemia as a direct result of integration near the LMO2 proto-oncogene (Hacein-Bey-Abina et al., 2003). Indeed, the use of integration-deficient vectors is a promising alternative, however, difficulty in obtaining adequate levels of sustained gene expression has been described in recent literature (Bayer et al., 2008; Yu et al., 2008).
Treating cells with histone deacetylase (HDAC) inhibitors has been reported to modulate the expression from the integrated provirus (Katz et al., 2007) as well as unintegrated HIV-1 genomes (Kantor et al., 2009). Acetylation of histone proteins is a well-known epigenetic marker for active transcription (Carrozza et al., 2003; Jenuwein and Allis, 2001) and response of viral promoters to the HDAC inhibitor trichostatin A (TSA) is promoter-dependent (Vanniasinkam et al., 2006). For MuLV, the chromatin state of the unintegrated LTR DNA is not defined, and MM-PCR suggests that the termini are protected within an extended footprint (Chen et al., 1999; Wei et al., 1998). The need to establish a transcriptionally active state in the LTR of the provirus in the normal life cycle has also been suggested (Bruce et al., 2008). It has previously been reported that the CTD of HIV IN is acetylated on four lysine residues by the histone acetyltransferases (HATs) GCN5 and p300 and is recognized by the host KAP1 protein (Allouch et al., 2011; Cereseto et al., 2005; Terreni et al., 2010; Topper et al., 2007), although the requirement for acetylation of HIV-1 IN has been debated (Topper et al., 2007). Additional DNA modeling of the HIV-1 IN CTD based on the structure of prototype foamy virus (PFV) CTDs, predicts that the lysine residues involved in acetylation are located on the interior of the intasome-DNA complex.
In an effort to further understand the effect HDAC inhibitors might have on expression from unintegrated DNA, we have analyzed several IN mutants of MuLV. We find that expression from the unintegrated viral promoter is enhanced by the presence of HDAC inhibitors in a temporally regulated fashion. Although MuLV IN can be acetylated in vitro by the HAT domain of p300, the enhancement is independent of MuLV IN K376. IN K376 is the only lysine present in the region analogous to HIV-1 IN where acetylation has been observed. Technologically, the ability to increase expression from unintegrated DNA would present a significant advantage when considering the use of non-integrating vectors for gene therapy applications.
Results
Acetylation of MuLV IN
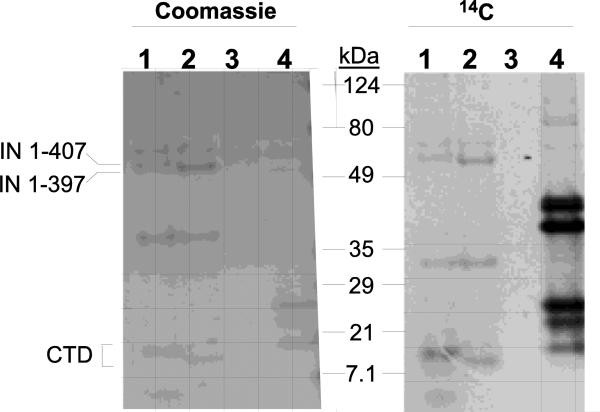
It has previously been reported that four lysine residues (K258, K264, K266, and K273) within the CTD of HIV-1 IN are a substrate for acetylation by host factors, and this acetylation has pleiotropic effects on viral replication (Allouch et al., 2011; Cereseto et al., 2005; Terreni et al., 2010). Here, the potential of the MuLV IN protein to serve as a substrate of the p300 catalytic domain was examined in vitro in the presence of 14C-labeled acetyl-CoA. Two bacterially expressed and purified MuLV IN constructs, IN 1-407 and 1-397 were tested and the modified proteins were then analyzed by SDS-PAGE and autoradiography. Shown in Fig. 1 (right lanes 1 and 2), both the full-length IN 1-407 and the C-terminal truncation, IN 1-397 corresponding to the predicted ~45kDa proteins were labeled by 14C-acetyl-CoA. The acetylation of histones served as a positive control for the reaction (Fig. 1, right, lane 4); as a negative control, no acetylation of BSA was detected (Fig 1, lanes 3). Interestingly, several minor protein break-down products visible by Coomassie staining (Fig. 1, left) were enriched for labeling with the 14C-acetylation reaction (Fig. 1, right). Of interest is the ~20 kDa fragments whose migration varied with the predicted truncation of the C-terminus suggesting that this fragment corresponds to the C-terminal third of the IN protein. These results indicate that at least one acetylation site maps within the MuLV IN CTD.
Figure 1. Acetylation of the MuLV IN protein.
Acetylation of MuLV IN with p300 catalytic domain was performed as described in Materials and Methods. Protein samples were subjected to SDS-PAGE and stained with Coomassie blue (left) or subjected to autoradiography (right). Lanes 1, MuLV IN 1-407; Lanes 2, MuLV IN 1-397; Lanes 3, BSA; Lanes 4, histone control. Position of migration of prestained molecular weight broad range markers (BioRad) are indicated.
Characterization of MuLV IN mutants
Sequence homology is low across the CTDs of various retroviral INs, however, structure-based sequence alignments between prototype foamy virus (PFV) (Hare et al., 2010b), HIV-1 (Chen et al., 2000; Hare et al., 2010b; Krishnan et al., 2010) and MuLV INs (P. Rossi personal communication) reveal that K370 in PFV and K266 in HIV-1 are homologous to K376 in MuLV IN (Fig 2A).
Figure 2. IN constructs and sequence alignments.
(A) Sequence alignment of MuLV, PFV, and HIV-1 CTDs. Secondary structural elements are shown above where E denotes residues in a beta conformation and H denotes residues in a helical conformation (β, beta strands; η, 310 helix). Asterisk denotes K376 in MuLV and the homologous lysine residues in PFV and HIV-1 reported. Amino acid numbering corresponds to MuLV IN. The MuLV CTD was structurally aligned (P. Rossi, personal communication) to PFV and HIV-1 CTDs with UCSF Chimera (Pettersen et al., 2004). (B) Diagram depicts the three domains of IN: Zn+ binding NTD, CCD, and CTD with residues of interest highlighted. Grey line in the NTD denotes the additional 50 amino acids in MLV IN not present in HIV IN. Grey line in CTD marks the non-essential extreme C-terminal 23 amino acids.
To determine whether K376 of MuLV IN affects viral replication, the residue was mutated to either alanine (K376A) or arginine (K376R). In addition, it has been suggested that the extreme C-terminus of HIV-1 IN can influence the phenotype of the lysine mutants (Topper et al., 2007). To further probe the role of the C-terminus of IN, we constructed several truncated IN proteins with and without the K376R/A mutations. The truncated IN, (ΔC), lacks 23 amino acids from the extreme C-terminus and has previously been shown to be viable in vivo (Roth, 1991). Similarly, for HIV-1 IN it has been shown that truncation of the extreme C-terminus allows for viral replication, however, the sequential deletion of amino acids negatively impacts IN activity (Dar et al., 2009; Mohammed et al., 2011). A catalytic site mutant (D184N) was also included as a control for IN defective in integration (Steinrigl et al., 2007). Fig 2B outlines the mutations within the CCD and CTD of MuLV IN utilized in these studies.
The HDAC inhibitor TSA affects viral propagation
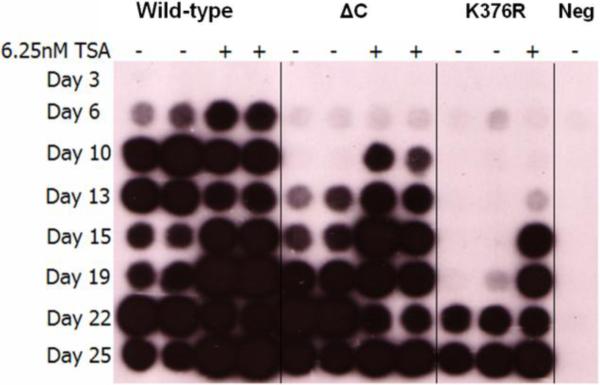
To determine what effect the HDAC inhibitor TSA would have on viral replication, wild-type (WT) ecotropic MuLV provirus, pNCA-C, along with the various mutant proviral constructs described above, were transiently transfected via DEAE-dextran into canine D17pJET cells (Goff et al., 1981). D17pJET cells stably express the ecotropic MuLV receptor (MCAT) and therefore allow for the spread of replication competent virus. To monitor viral fitness, the spread of WT and mutant virus was monitored over time by the presence of reverse transcriptase (RT) activity released into the supernatants. Figure 3 reveals that supernatant from cells transfected with the WT proviral DNA become RT positive at day 6. The presence of TSA during a WT MuLV infection did not increase the time course of viral passage, with initial detection of reverse transcriptase activity remaining at day 6. In contrast, the addition of TSA increases the replication kinetics of virus bearing the truncated (ΔC) IN by three days and accelerates the replication rate of the K376R mutant by a full week. DNA sequence analysis indicated the IN mutations were maintained in the TSA treated sample, ruling out the possibility of reversion. None of the additional IN mutant constructs, including K376RΔC, K376A, or D184N, became RT positive (data not shown). These results suggest that acetylation of IN K376 is not responsible for the enhanced viral kinetics, however, we cannot rule out the possibility that the HDAC inhibitors are acting on other Lys residues within IN. Alternatively, these results could be explained by the effects of HDAC inhibitors on cellular proteins.
Figure 3. RT assay.
Reverse transcriptase assay of viral supernatants from DEAE-dextran transfected cells at the indicated times post transfection with (+) and without (-) TSA. Results are representative of a minimum of three independent experiments.
Mutation of IN K376 decreases integration
Many IN mutations can have pleiotropic effects (Engelman et al., 1995), including blocks at various stages of reverse transcription. To further investigate the defects in viral spread and uncover the mechanism by which TSA affects viral kinetics, we analyzed the levels of viral replicative intermediates as well as integrated provirus with qPCR. For qPCR analysis, VSV-G pseudotyped viral particles were used to infect TE671 cells in a single round-of-infection. To monitor replication intermediates we quantified minus strand strong stop products (MSSS), plus strand extension products (PSE), and 2-LTR circle junctions, a dead-end product known to increase in quantity in the presence of defective IN proteins (Colicelli and Goff, 1985, 1988; Donehower and Varmus, 1984; Roth et al., 1990). Integrated proviral DNA was quantified by Alu-PCR.
Virus producing cell lines were established in canine D17 cells containing the mutant viruses (NeoR) and packaging the reporter gene gfp (GIP). To confirm proper assembly of virus particles and verify that similar levels of mutant IN proteins were present, western blot analysis was performed on supernatant fractions from virus producing cells. Uniform levels of three viral proteins (RT, IN, and CA) were present among the viral constructs and sequence analysis of genomic DNA extracted from virus producing cells confirmed the presence of the predicted IN mutations (data not shown).
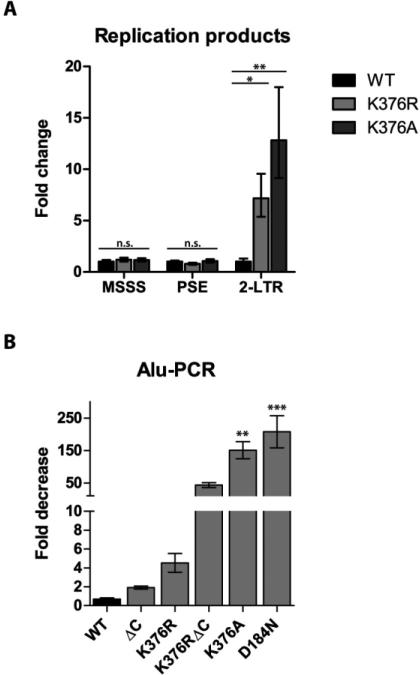
Figure 4A compares the relative levels of MSSS, PSE replication intermediates and 2-LTR circle junctions of virus bearing the IN K376R and IN K376A mutations to that of WT virus at 24 h post infection. Viral infections were normalized to the level of vRNA within particles, as determined by qPCR using the MSSS primers. Viral intermediates for MSSS and PSE were not statistically different from the WT infection indicating that a block in replication does not explain the delay in virus propagation observed in Fig. 3. In contrast, a significant increase in 2-LTR circular products accumulated for virus bearing the IN K376R and K376A mutants, ~7-fold and ~13-fold respectively. This data indicates that mutations in IN at K376 affect viral integration and correlates with the observed defects in virus propagation (Fig. 3).
Figure 4. qPCR of replication products and Alu-PCR.
(A) qPCR of replication products. Graph plots the fold-increase in the amount of PCR products relative to WT virus at 24 h post infection for minus strand strong stop (MSSS), plus strand extension (PSE) and 2-LTR circle products. (B) Alu-PCR. Graph plots the fold decrease in integrated provirus for IN mutants relative to WT IN at 18 days post infection. Results are mean with standard error, N=3. Statistical significance was determined by one-way ANOVA (***, P < 0.001; **, P < 0.01; *, P < 0.05; n.s., not significant).
To further confirm a defect in integration, Alu-PCR was performed to quantify integrated provirus. To eliminate the potential contribution of unintegrated viral DNA from the analysis, genomic DNA was obtained 18 days post infection. Figure 4B displays the fold decrease in integration, relative to WT infection, for the IN mutants described in Figure 2B. For the two IN mutants (IN ΔC and IN K376R), which were viable but displayed a delayed replication phenotype, we observed a 2-fold decrease in integration (IN ΔC) and a roughly 5-fold decrease in integration (IN K376R). Analysis of the integrants generated in TE671 cells by IN K376R indicated the global target site preference with respect to transcriptional start sites and CpG island did not differ significantly from that published in WT MuLV (Wu et al., 2003), eliminating chromosomal positional effects as the cause for decreased gene expression (data not shown).
A more dramatic decrease in viral integration was observed with IN mutants incapable of achieving a sustainable level of infection. The K376RΔC IN mutation imparted a roughly 50-fold decrease in integration, whereas the K376A CTD mutation and the catalytic mutant, D184N, showed a 150-200 fold decrease in the number of integrants. These results suggest that the ability of a virus to spread is dependent upon an integrated provirus. The effect of TSA on the kinetics of viral spread could be acting on either the integrated or unintegrated proviral DNA. To address this, we set out to define the expression of MuLV from unintegrated DNA and further characterize the mechanism by which TSA enhances viral spread.
HDAC inhibitors increase expression from unintegrated DNA
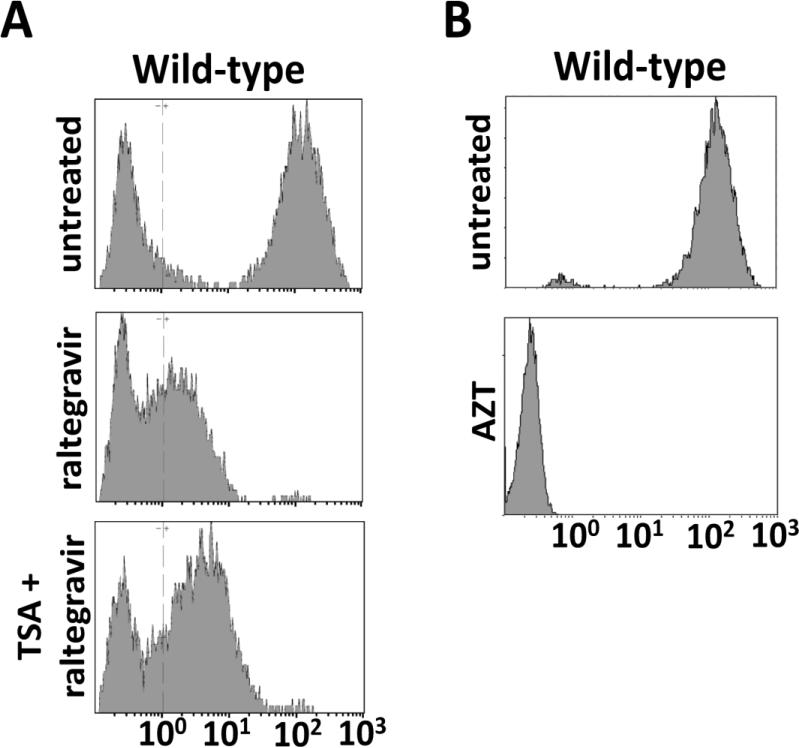
Gene expression from integration-defective retroviral vectors is significantly lower than the level of expression from integrated provirus (Bayer et al., 2008; Nightingale et al., 2006; Philippe et al., 2006; Rahim et al., 2009; Sloan and Wainberg, 2011; Yu et al., 2008). We hypothesized that gene expression can be driven from distinct DNA populations in infected cells: a low-level of gene expression driven from unintegrated DNA and a high level of gene expression, characteristic of retroviral vectors, driven from integrated provirus. To define these populations and determine if the unintegrated population is responsive to treatment with TSA, viral replication was performed using a single round-of-infection experiment in TE671 cells with WT IN virus packaging the gfp reporter gene. Infected cells were harvested at 72 hours post infection and analyzed by flow cytometry. The GFP expression profile of WT virus was examined in the presence and absence of the integrase inhibitor raltegravir (Fig. 5A). To compare the various panels of GFP expression in Fig. 5, we define four categories of cells based on the mean fluorescence intensity (MFI) of GFP: cells with no GFP expression (GFPnegative; MFI <100); cells expressing low levels of GFP (GFPlow 100≤ MFI ≤101); cells expressing medium levels of GFP (GFPmedium 101≤ MFI ≤102); and cells expressing high levels of GFP (GFPhigh; MFI >102). Virus bearing WT IN resulted in a uniform population that expressed GFPhigh (Fig 5, top). Raltegravir treatment results in a near complete loss of the GFPhigh population and the corresponding appearance of GFPlow and GFPmedium cells. In the presence of TSA, these cells show a positive shift in the MFI of GFP. Since the mechanism of action of raltegravir is to block the strand-transfer reaction, these results indicate that a population of unintegrated DNA exists that can allow low-levels of gene expression that is responsive to TSA.
Figure 5. Effects of integrase reverse transcrtipase and HDAC inhibitors on transgene expression.
(A) WT viral infection in the presence or absence of the IN inhibitor raltegravir and TSA 3 dpi. (B) WT viral infection in the presence and absence of the RT inhibitor AZT 3dpi. GFP was measured by flow cytometry in cells infected with WT virus. Cell number is represented along the y-axis, and GFP intensity is measured on the x-axis.
To confirm that the GFP measured in the infected cells resulted from gene expression and not protein transfer of GFP in the viral particles, we treated target cells with the RT inhibitor AZT (Voelkel et al., 2010). In the presence of AZT we were unable to measure any GFP in the infected cells (Fig 5B). This indicates reverse transcription is required to generate the template for pol II dependent transcription and the GFP detected in infected cells is not a result of packaging GFP protein into the viral particles. These results from experiments with raltegravir and AZT are consistent with the conclusion that the GFP in the TSA-responsive GFPlow and GFPmedium cells originates from unintegrated viral DNA.
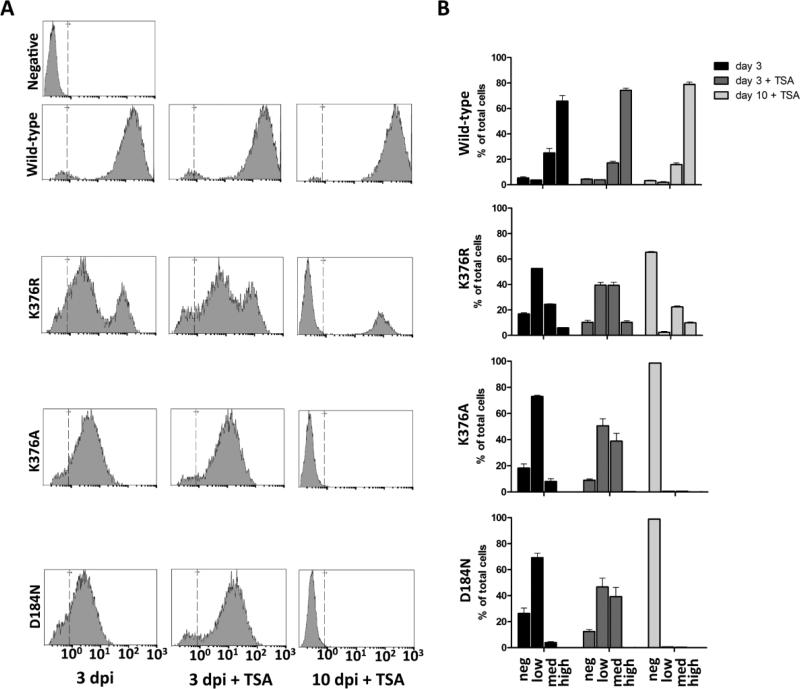
Using the single round-of-infection system, the effects of TSA-treatment on integrase-defective mutants was examined by monitoring GFP expression. Figure 6 compares WT with the integration defective D184N and K376A mutants along with IN K376R virus. Histogram plots in Figure 6A display the MFI of GFP at 3 days post infection (dpi) and 10 dpi and are quantified in Figure 6B. At 3 dpi, cells infected with WT virus fall exclusively into the GFPhigh population, whereas cells infected with IN K376R are present in two distinct populations (GFPlow/medium and GFPhigh). Cells infected with integration defective virus fall exclusively into the GFPlow and GFPmedium population. At 3 dpi when TSA is present in the culture medium, cells in the GFPlow and GFPmedium population show a shift in the MFI of GFP. These results are highlighted in Figure 6B by the increased percentage of cells in the GFPmedium category, having shifted from the GFPlow category when TSA is present. While the profile of WT virus remains largely unchanged, cells infected with IN K376R show an increase of ~15% in the GFPmedium category. Cells infected with the IN-defective mutants K376A and D184N show an even greater effect with the amount of GFPmedium cells increasing ~30-35%. The analysis of cells at 10 dpi, a time when the majority of unintegrated viral DNA is no longer present (Butler, Johnson, and Bushman, 2002) provides additional evidence supporting the hypothesis that cells populating the GFPlow and GFPmedium categories harbor unintegrated DNA. As would be expected for GFP expressed from integrated provirus, the GFP positive cells that persist until day 10 fall into the GFPhigh category. This category appears only when cells are infected with integration-competent virus. Additional HDAC inhibitors were also tested and yielded similar results (data not shown). The additional HDAC inhibitors included 4-phenylbutryate (PBA), valproic acid (VPA), and sodium butyrate. These results suggest that a general effect on HDACs is responsible for the observed phenotype.
Figure 6. GFP expression from unintegrated DNA.
(A) GFP expression was measured by flow cytometry in infected cells at 3 days post infection 3 (dpi) and 10 days post infection (10 dpi). The effects of adding TSA to the cell culture medium during infection were also monitored. Cell number is represented along the y-axis and GFP intensity is measured on the x-axis. (B) Quantitation of the results from panel A. Neg; MFI <100; Low: 100≤ MFI ≤101; Med: 101≤ MFI ≤102; and High; MFI >102.
To further define the temporal effect of TSA on unintegrated DNA, single round-of-infection studies were performed with IN K376R virus packaging the gfp reporter gene. In the virus replication assay shown in Figure 3, the HDAC inhibitor TSA was present throughout the course of infection. To determine whether TSA was required in a time-dependent manner, target cells were treated with TSA either 12 h prior to infection, during the infection, or 48 h post infection. Infected cell cultures were harvested at 72 h post infection and analyzed by flow cytometry (Fig. 7). In this experiment, IN K376R the GFPlow and GFPmedium population of cells predominate with a smaller population comprised of GFPhigh cells. The GFPlow and GFPmedium cells corresponding with the proposed unintegrated DNA pool are responsive to TSA, showing a positive shift in MFI. Cells treated with TSA 12 h prior to infection display similar expression profiles to those treated at the time of infection. In contrast, TSA had no discernible effect on GFP expression in cells treated 48 h post infection. Time course analyses have indicated viral integration occurs within the first 12-24 hours after infection (Roe et al., 1997). These studies support the results that the HDAC inhibitors are not acting through the integrated provirus, but rather, are required at the time when the unintegrated provirus is established in the newly infected cells.
Figure 7. Temporal requirement for TSA effect on transgene expression.
(A) Comparison of cells infected with WT and IN K376R mutant virus and treated with the HDAC inhibitor TSA at various time points. Untreated, no TSA added; Pre-treatment, TSA added 12 h prior to infection; Time of, TSA added with viral infection; Post-treatment, TSA added 48 h post infection. GFP was measured by flow cytometry at 3 dpi. Cell number is represented along the y-axis, and GFP intensity is measured on the x-axis.
Discussion
Post-translational modification of proteins is a well-established cellular mechanism to modulate function in response to various spatial, temporal, and environmental cues. Acetylation of lysine residues within proteins is one such modification, most notably recognized as an epigenetic modifier of histone proteins. The presence of acetylated histones is generally associated with active transcription (Carrozza et al., 2003). Previous studies aimed at understanding transcriptional repression of integrated provirus has shown that HDAC inhibitors are capable of re-activating expression from a subpopulation of integrants, presumably through modification of the histone proteins (Katz et al., 2007). In a WT MuLV infection, transcription is driven from the integrated proviral DNA and sustained viral infection is not observed in the absence of IN protein. A recent interest in utilizing non-integrating retroviral vectors for gene expression, however, has raised the question as to whether protein acetylation plays a role in transient expression from unintegrated viral DNA.
In addition to the role of histone acetylation in retroviral gene expression, it has been demonstrated that the HIV IN CTD itself is targeted for acetylation (Cereseto et al., 2005; Terreni et al., 2010), although this acetylation appears to be dispensable for viral replication (Topper et al., 2007). Data presented in this study indicates the MuLV IN is also a substrate for in vitro acetylation by the p300 protein. To test whether mutation of the CTD from MuLV IN would have an effect on transgene expression, GFP expression was monitored in infected cells. When tested, there was in fact a clear decrease in gene expression following viral infection with IN K376 mutant viruses. This effect, however, was modulated by HDAC inhibitors and therefore, our results indicate that acetylation of K376 in MuLV IN is not responsible for this phenotype. We cannot eliminate the possibility that acetylation of alternative lysines beyond K376 of MuLV IN could be regulated.
Utilizing the IN inhibitor raltegravir, as well as a catalytically inactive IN mutant (D184N), we determined that the HDAC inhibitors were acting on unintegrated viral DNA. From this, we infer that upon addition of HDAC inhibitors, changes in chromatin structure of the unintegrated DNA alter the accessibility of viral DNA to host transcription factors. Currently there is no direct evidence that DNA within the MuLV PICs is chromatinized. Alternatively, the effects of HDAC inhibitors could affect cellular proteins required for the infectious process. Various cellular proteins are targeted for acetylation and it has previously been shown that HDAC inhibitors can affect intracellular trafficking (Barua and Rege, 2010).
These studies are directly relevant to the field of non-integrating viral vectors (Banasik and Jr, 2010). Arguably, one of the greatest benefits to using a retroviral-based vector is the high-level of gene expression associated with an integrated virus. However, the shortfalls are also established, notably the eventual transcriptional repression that occurs over time and integration of the DNA itself can lead to gene disruption and/or oncogene activation. Various studies have recently explored the potential use of non-integrating retroviral vectors, where the IN protein contains point mutations that render it inactive. For non-integrating retroviral vectors, a bottleneck that remains is the low level of expression obtained from these vectors in comparison with the integrated provirus. The results from these studies identify a means to transiently improve the level of expression, using a combination of specific MuLV IN mutations and HDAC inhibitors. Notably, transgene expression was detected from the majority of the infected cells, indicating expression from an abundant proviral DNA species. The addition of TSA at two days post-infection, a time when circular products are abundant, has no effect on GFPlow and GFPmedium expression. We interpret this time dependence of TSA addition to suggest that it is required early during viral replication to prevent the formation of a repressed state within the viral LTR. In the current study, the effect was only measured on expression from the viral LTR promoter. Whether the observed activation/repression would extend to internal promoters within self-inactivating (SIN) vectors is of interest and is currently being examined.
Conclusion
Productive retroviral infection is driven by expression from the integrated virus. In these studies, we demonstrate increased transgene expression from the unintegrated proviral DNA in the presence of HDAC inhibitors. Furthermore, treating cells harboring unintegrated viral DNA with HDAC inhibitors is capable of altering the kinetics of viral replication, however, it is not sufficient to achieve a sustained infection of a non-integrating vector through multiple rounds of infection in a dividing cell. Utilization of the described IN mutants in combination with HDAC inhibitors can increase the expression of transgenes from non-integrating vectors and has direct applications for retroviral vectors in molecular biology or gene therapy applications, for example in iPS formation, where transient expression of reprogramming factors is required.
Materials and Methods
Generation of plasmids
Construction and analysis of pNCA-C, a viable, replication-competent M-MuLV proviral construct has been previously described (Felkner and Roth, 1992). The pNCA-C-XN-SU8 M-MuLV proviral construct (ΔC) was derived from pNCA-C (Roth, 1991; Seamon et al., 2000). This contains a Not I linker within the Xba I site at the 3’ terminus of the M-MuLV pol gene, yielding a 23-amino-acid C-terminal truncation of the MuLV IN protein plus a suppressor tRNA in the 3’ long terminal repeat (LTR) (SU8) (Lobel and Goff, 1984).
To generate pNCA-C-Neo and pNCA-C-XN-Neo (ΔC), vectors pNCA-C and pNCAC-XN-SU8 were used as templates. The A nucleotide in the ATG start codon of Env was mutated to a C, via overlapping PCR, maintaining the coding sequence in IN. The coding region of the neo gene obtained from the vector pRVL (Bupp and Roth, 2002) was then cloned in such that the start codon overlapped with the stop codon of the IN gene. Specifically, the last base in IN along with the stop codon, CTAA, was mutated to ATGA, rendering the IN protein product unaltered. The neo gene is therefore expressed as a spliced message in place of env. Nucleotide residues ATAAAATAAA, identical to that immediately following the Env stop codon, along with a NsiI restriction site were added adjacent to the neomycin stop codon via PCR. The resulting overlapping PCR product was then digested with HindIII and NsiI and cloned into the pNCA-C vector. PCR reactions were carried out with KOD polymerase (Novagen).
The pNCA-C K376R/A and D184N mutations were generated by overlapping PCR with pNCA-C, pNCA-C-XN-SU8, pNCA-C-Neo, and/or pNCA-C-XN-Neo serving as the template. The PCR reactions were carried out with KOD polymerase (Novagen) and full-length insert products were gel isolated. The K376 mutant PCR products were digested, and cloned into pNCA-C, pNCA-C-XN-SU8, pNCA-C Neo, and pNCA-C-XNNeo accordingly at the HindIII and NsiI sites. The D184N mutant PCR product was digested with HindIII and SfiI and cloned into pNCA-C and pNCA-C-Neo.
IN purification
pGV358 (gift from Kushol Gupta, Univ. of Pennsylvania) is a pETDuet based vector that has an Mxe intein in frame at the C terminus followed by the non-cleavable hexahistidine tagged chitin binding domain. M-MuLV integrase from positions 1-407 and 1-397 were PCR amplified from the pNCA-C (Felkner and Roth 1992) template in which a single point mutation was introduced to abolish the NdeI site in the integrase coding region. The PCR fragment was cloned between the NdeI and XhoI sites of pGV358. The following primers were used: 1-407 IN and 1-397 IN forward primer (NdeI site underlined) – 5’ GGAATTCCATATGATAGAAAATTCATCACCCTACACCTCAG 3’. 1-407 IN reverse primer (XhoI site underlined) – 5’ CCGGCTCGAGGGCCTCGCGGGTTAACC 3’. 1-397 IN reverse primer (XhoI site underlined) – 5’ CCGGCTCGAGGTTTTGAGAGCGTTGAACGCGCC 3’. Plasmids were introduced into BL21(DE3) E. coli and protein expression was induced for a 1 liter culture at an O.D.600 of 0.7 for 24 hours at 17°C. Cells were harvested and lysed in ice cold buffer containing 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 10 mM CHAPS, complete EDTA protease inhibitor tablets (Roche). Fusion proteins were purified using Ni NTA (Qiagen) and chitin beads (New England Biolabs) as per the manufacturer's recommendations. IN 1-407 and IN 1-397 were released from the fusion protein by incubating for 48 hours at 4°C with 20 mM HEPES pH 8.0, 1M NaCl, 10% glycerol, 0.1mM EDTA, 50 mM DTT (Fluka), 0.01% Igepal CA -630 (Sigma) and 10 μM zinc sulfate. Cleaved protein was then further purified using SP-Sepharose chromatography (GE Healthcare). The purified fraction obtained against a salt gradient of 250 mM – 2M NaCl was then concentrated using Ultracel 30K concentrator (Millipore) in the presence of 0.01% Igepal CA–630 to ~0.5–1 mg/ml and used for the acetylation assays. The final buffer composition of the protein is 20 mM HEPES pH 7.0, 250-750 mM NaCl, 10% glycerol, 1mM DTT, 0.1mM EDTA, 0.01% Igepal CA–630, 10μM ZnSO4.
HAT assay
4 μg recombinant truncated IN (1-397) or full length IN (1-407) protein was incubated for 30 min at 30°C with 3.8 μg p300 (BML-SE451, Enzo Life Sciences) in 25 μl Tris-HCl (50 mM, pH 8.0) containing 10% glycerol, 0.1mM EDTA, 0.08 uCi of [14 C] acetyl-CoA (5 mCi/mmole, MP/ICN), and 0.16 μg Acetyl-CoA (20-192A, Millipore). 4 μg Core Histone (13-107B, Millipore) and BSA (Sigma) were used as positive and negative controls. After protein acetylation, reaction mixtures were heated at 95°C for 5 min with 1X LDS sample buffer (Invitrogen) and subjected to electrophoresis on a SDS 4-20% gradient polyacrylamide gel (Pierce). The gel was dried on a gel dryer (Model 583, BioRad) and exposed to Kodak XAR film.
Cell Culture
The generation and maintenance of canine D17pJET cells stably expressing the ecotropic M-MuLV receptor (MCAT) (Albritton et al., 1989) has been previously described (O'Reilly and Roth, 2000). Proviral DNA was transiently introduced into D17pJET cells to monitor the time course of virus replication and to generate cell lines producing modified IN containing virus. D17pJET cells were split at 1 × 105 cells per 60 mm dish and cultured in DMEM (Gibco) with 10% serum. The following day proviral DNA was transiently introduced in duplicate with DEAE-dextran using 250 ng of DNA (McCutchan and Pagano, 1968). The cells were then incubated at 37°C in 5 ml of fresh medium and maintained with and without 6.25 nM trichostatin A (HDAC inhibitor). Upon confluence, supernatant was collected and cells were passed to 10 cm plates. Supernatant was collected at confluence over the course of several weeks and assayed for RT activity (Goff et al., 1981). Virus producing cell lines were maintained in culture for replication kinetics experiments.
To generate virus producing D17 canine cell lines, 293T CeB cells (Cosset et al., 1995) were transfected with 3 μg of WT or IN mutant pNCA-C-Neo plasmids in combination with 3 μg of pHIT-G plasmid DNA (VSV-G) using the Mammalian Transfection Kit [stable CaPO4] (Stratagene) as described by the manufacturer. The following morning the medium was replaced with 5 ml of fresh medium supplemented with 10 mM Na-butyrate for 6 h to enhance expression from plasmid DNA (Soneoka et al., 1995). The medium was again changed and virus was collected after approximately 36 h. Viral supernatant was used to infect D17 cells for 72 h thereafter cells were treated with G418 (400 μg/ml). Upon selection of G418 resistant producer cell lines, the proviral reporter construct, pGIP (Chen et al., 2001), was then introduced into these D17-Neo cell lines in a similar fashion as described above. D17-Neo virus producing cell lines were selected with 2.5 μg/ml of puromycin yielding the D17-Neo-GIP cell lines for subsequent viral infection assays.
Infections
Single round of infection experiments into human TE671 cells for FACS analysis and Alu-PCR were performed by transfecting D17-Neo-GIP producer cell lines with 3 μg of pHIT-G plasmid DNA (VSV-G). Approximately 2 × 105 cells were plated on 10 cm dishes the evening prior to transfection. The VSV-G pseudotyped virus was collected in 10 ml and equivalent virus production was confirmed by anti-CA western blot analysis using a 1/1000 dilution of goat anti-MuLV CA (75S-287) from the NCI-BCB Repository (distributed by Microbiological Associates, Inc.). Similarly, to detect RT and IN from viral particles, 10 ml of viral supernatant was collected from confluent producer cell lines and centrifuged and prepared as above, then run on a 10% SDS-PAG. A 1/1000 dilution of a rabbit polyclonal antibody recognizing both RT and IN was used to detect protein from the mutant IN virus producing cell lines (Tanese et al., 1986). Approximately 1 × 105 TE671 cells were plated per 60 mm dish the evening prior to infection. The following day virus was added to cells at 37°C for 72 h. Where appropriate, HDAC inhibitors were added to the TE671 target cells either the evening prior to infection (pre-treatment), at the time of infection, or 48 h after the virus was added (post-treatment). HDAC concentrations used during infection were as follows: trichostatin A (TSA), 20 nM; sodium 4-phenylbutryate (PBA), 5 mM; valproic acid (VPA), 2 mM; sodium butyrate, 2 mM. Where appropriate, the IN inhibitor raltegravir was added to a final concentration of 100 nM at the time of infection. Raltegravir was obtained through the NIH AIDS Research and Reference Reagent Program (www.aidsreagent.org). Where indicated, AZT (Sigma A2169) was added at 50 μM (Voelkel et al., 2010) to target cells 24h prior to infection and remained in the medium for the duration of the experiment.
qPCR
Single round of infection experiments into human TE671 cells were performed by transfecting D17-Neo-GIP WT IN, IN-K376R IN-K376A, and IN D184N producer cell lines with 2 μg of pHIT-G plasmid DNA (VSVG) using the Fugene 6 (Roche) as described by the manufacturer. Approximately 5 × 105 cells were plated on 10 cm dishes the evening prior to transfection. The VSV-G pseudotyped virus was collected at 48 h post transfection and stored at -80 degrees. Viral particles were quantified and normalized by qPCR using primers/probe set for quantification of minus strand strong stop (MSSS) intermediates (see below). pNCA-C-Neo plasmid was used to establish the standard curve. Viral RNA was purified with the QIAamp Viral RNA mini kit (Qiagen) and cDNA synthesis was performed with using Superscript III Reverse Transcriptase primed with Random Hexamers (Invitrogen).
The evening prior to infection, TE671 cells were seeded on 60 mm dishes at approximately 2 × 105 cells per dish. The following day, cells were infected in triplicate with 2 mL viral supernatants (6.7 × 109 copies/mL) per dish in the presence of 8 μg/mL polybrene (Sigma, H9268) at 37°C. Infected cells were harvested at 24 h post-infection, and genomic DNA was purified with the DNeasy Blood & Tissue Kit (Qiagen). and applied for qPCR.
qPCR reactions were prepared using the TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems) and analyzed on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Each reaction was performed in triplicate in a total volume of 25 μl using 300 ng of genomic DNA as template. The reaction conditions were as follows: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, followed by 95°C for 15 sec and 60°C for 1 min for 40 cycles. Primers were used at a concentration of 900 nM and probes were used at 250 nM. The primers and probe used for quantification were as follows: Primer A, (5’- GCCAGTCCTCCGATTGACTG -3’); Primer B, (5’- TGACGGGTAGTCAATCACTCAGAG -3’); Primer C, (5’- AATCGGACAGACACAGATAAGTTGC -3’); Probe E, (5’- /56-FAM/CATCCGACTTGTGGTCTCGCTGTTCC/36-TAMSp/ -3’). Primers for quantification of minus strand strong stop (MSSS) intermediates were Primer A and Primer B. Primers A and C were used for quantification of plus strand extension (PSE). The probe used for -SSS and +SE was Probe E. Primers used for quantification of 2-LTR circles were 2-LTR forward: 5'-CTT GTG GTC TCG CTG TTC CTT-3' and 2-LTR Reverse: 5'-TGC AAA ATG GCG TTA CTT AAG C-3' and 2-LTR probe: 5'-/56-FAM/TGA AAG ACC CCA CCT GTA GGT TTG GCA AG/36-TAMSp/-3'. Reactions were normalized to GAPDH using primers: 5'- TCGACAGTCAGCCGCATCT -3' and 5'- CTAGCCTCCCGGGTTTCTCT -3' and probe: 5'- /56-FAM/TCGCCAGGTGAAGACGGGCG/3IABIk_FQ/ -3'.
For Alu-PCR reactions in human TE671 cells to quantify integrated provirus, single round of infection experiments were performed using VSV-G pseudotyped virus as described above. Genomic DNA was harvested from cells 18 days post infection (Bruce et al., 2008) and 4 ul (~400 ng) of genomic DNA was used as template for an initial amplification under the following reaction conditions: 1 cycle at 95°C for 5 min, followed by 12 cycles at 95°C for 15 sec, 66.8°C for 30 sec, and 68°C for 1 min 30 sec. Primers were used at a concentration of 300 nM each. LTR forward bp282 primer (5’- CAGCCCTCAGCAGTTTCTAGAGA -3’). Alu primer (5’- AGCTACTCGGGAGGCTGAGG -3’). PCR reactions were carried out with KOD polymerase (Novagen) and the resulting PCR product was used for a subsequent nested qPCR reaction. qPCR reactions to quantitate proviral DNA followed the qPCR conditions outlined above with the following primers/probes: LTR taqman forward primer: (5’- TCAGTTCGCTTCTCGCTTCTGT -3’). LTR taqman reverse primer: (5’- GGGTTGTGGGCTCTTTTATTGA -3’). LTR taqman probe: (5’-/56-FAM/CGCGCGCTTCTGCTCCCC/36-TAMSp/ -3). Primers specific for human β-actin were used for an internal control to normalize for input DNA. β-actin forward: (5’- TCACCCACACTGTGCCCATCTACGA -3’). β-actin reverse: (5’- CAGCGGAACCGCTCATTGCCAATGG -3’). β-actin probe: (5’- /56-FAM/ATGCCCTCCCCCATGCCATCCTGCGT/TAMSp/ -3’).
Flow Cytometry
Infected cells were trypsinized, centrifuged, and resuspended in 1 ml of PBS for analysis of GFP expression by flow cytometry. All flow cytometry experiments were carried out on a Coulter Cytomics FC500 Flow Cytometer at the Analytical Cytometry Image Analysis Core Facility at the Environmental and Occupational Health Sciences Institute, 170 Frelinghuysen Road, Piscataway, NJ 08854.
Acknowledgements
This work is supported by National Institutes of Health grants RO1 GM070837 and RO1 GM088808, awarded to M.J.R. W.M.S. was supported by NIH training grants T32 GM08360 and T32 A1007403. We thank Anindita Sarangi for technical assistance.
Abbreviations
- IN
Integrase
- M-MuLV
Moloney murine leukemia virus
- PIC
pre-integration complex
- TSA
Trichostatin A
- HDAC
histone deacetylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albritton LM, Tweng L, Scadden D, Cunningham JM. A putative murine retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Allouch A, Primio CD, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host & Microbe. 2011;9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Banasik M, Jr, P.M. Integrase-defective lentiviral vectors: progress and applications. Gene Therapy. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- Barua S, Rege K. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 2010;31:5894–5902. doi: 10.1016/j.biomaterials.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Bayer M, Kantor B, Cockrell A, Ma H, Zeithaml B, Li X, McCown T, Kafri T. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Bruce JW, Ahlquist P, Young JA. The host cell sulfonation pathway contributes to retroviral infection at a step coincident with provirus establishment. PLoS Pathog. 2008;4:e1000207. doi: 10.1371/journal.ppat.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci U S A. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp K, Roth MJ. Altering retroviral tropism using a random-display envelope library. Mol. Ther. 2002;5:329–335. doi: 10.1006/mthe.2002.0546. [DOI] [PubMed] [Google Scholar]
- Butler SL, Johnson EP, Bushman FD. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J Virol. 2002;76(8):3739–47. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. The EMBO Journal. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Rivera A, Ron N, Dougherty JP, Ron Y. A gene therapy approach for treating T-cell-mediated autoimmune diseases. Blood. 2001;97:886–894. doi: 10.1182/blood.v97.4.886. [DOI] [PubMed] [Google Scholar]
- Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- Chen J,C-H, Krucinski J, Miercke L,JW, Finer-Moore J,S, Tang A,H, Leavitt A,D, Stroud R,M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc. Natl. Acad. Sci. USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Colicelli J, Goff SP. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Colicelli J, Goff SP. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Cosset F-L, Takeuchi Y, Battini J-L, Weiss R,A, Collins KLM. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Dar M, Monel B, Krishnan L, Shun M, Nunzio FD, Helland D, Engelman A. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology. 2009;6:94. doi: 10.1186/1742-4690-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc. Natl. Acad. Sci. USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkner RH, Roth MJ. Mutational analysis of N-linked glycosylation sites of the SU protein of Moloney murine leukemia virus. J. Virol. 1992;66:4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants; use of a rapid assay for release of virion reverse transcriptase. J. Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010a;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A. 2010b;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kantor B, Ma H, Webster-Cyriaque J, Monahan PE, Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc. Natl. Acad. Sci. USA. 2009;106:18786–18791. doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger JG, Skalka AM. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol. 2007;81:2592–2604. doi: 10.1128/JVI.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proceedings of the National Academy of Sciences. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci U S A. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel LI, Goff SP. Construction of mutants of Moloney murine leukemia virus by suppressor-linker insertion mutagenesis: positions of viable insertion mutations. Proc. Natl. Acad. Sci. USA. 1984;81:4149–4153. doi: 10.1073/pnas.81.13.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan JH, Pagano JS. Enhancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J. Natl. Cancer Inst. 1968;41:351–357. [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed KD, Topper MB, Muesing MA. Sequential deletion of the Integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J. Virol. 2011;85:4654–4666. doi: 10.1128/JVI.02374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, Bahner I, Kohn DB. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- O'Reilly L, Roth MJ. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 2000;74:899–913. doi: 10.1128/jvi.74.2.899-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, Jacques M, Serguera C. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prizan-Ravid A, Elis E, Laham-Karam N, Selig S, Ehrlich M, Bacharach E. The Gag cleavage product, p12, is a functional constituent of the murine leukemia virus pre-integration complex. PLoS Pathogens. 2010;6:e1001183. doi: 10.1371/journal.ppat.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Howe SJ, Buckley SM, Acosta-Saltos AD, Elston KE, Ward NJ, Philpott NJ, Cooper JD, Anderson PN, Waddington SN, Thrasher AJ, Raivich G. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009;16:509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- Roe T, Chow SA, Brown PO. 3'-end processing and kinetics of 5'-end joining during retroviral integration in vivo. J. Virol. 1997;71:1334–1340. doi: 10.1128/jvi.71.2.1334-1340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MJ. Mutational analysis of the carboxy terminus of the Moloney murine leukemia virus integration protein. J. Virol. 1991;65:2141–2145. doi: 10.1128/jvi.65.4.2141-2145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MJ, Schwartzberg P, Tanese N, Goff SP. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J. Virol. 1990;64:4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon JA, Adams M, Sengupta S, Roth MJ. Differential effects of C-terminal molecular tagged integrase on repication competent Moloney-murine leukemia virus. Virology. 2000;274:412–419. doi: 10.1006/viro.2000.0481. [DOI] [PubMed] [Google Scholar]
- Sloan RD, Wainberg MA. The role of unintegrated DNA in HIV infection. Retrovirology. 2011;8:52. doi: 10.1186/1742-4690-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman MS, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:629–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrigl A, Nosek D, Ertl R, Günzburg WH, Salmons B, Klein D. Mutations in the catalytic core or the C-terminus of murine leukemia virus (MLV) integrase disrupt virion infectivity and exert diverse effects on reverse transcription. Virology. 2007;362:50–59. doi: 10.1016/j.virol.2006.11.037. [DOI] [PubMed] [Google Scholar]
- Tanese N, Roth MJ, Goff SP. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol. 1986;59:328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terreni M, Valentini P, Liverani V, Gutierrez MI, Primio CD, Fenza AD, Tozzini V, Allouch A, Albanese A, Giacca M, Cereseto A. GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology. 2010;7:18. doi: 10.1186/1742-4690-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Topper M, Luo Y, Zhadina M, Mohammed K, Smith L, Muesing MA. Posttranslational acetylation of the human immunodeficiency virus type 1 Integrase carboxyl-terminal domain is dispensable for viral replication. J. Virol. 2007;81:3012–3017. doi: 10.1128/JVI.02257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanniasinkam T, Ertl H, Tang Q. Trichostatin-A enhances adaptive immune responses to DNA vaccination. J. Clinical Virol. 2006;36:292–297. doi: 10.1016/j.jcv.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Voelkel C, Galla M, Maetzig T, Warlich E, Kuehle J, Zychlinski D, Bode J, Cantz T, Schambach A, Baum C. Protein transduction from retroviral Gag precursors. Proc Natl Acad Sci U S A. 2010;107:7805–7810. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SQ, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci U S A. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yu SS, Dan K, Chono H, Chatani E, Mineno J, Kato I. Transient gene expression mediated by integrase-defective retroviral vectors. Biochem Biophys Res Commun. 2008;368:942–947. doi: 10.1016/j.bbrc.2008.02.012. [DOI] [PubMed] [Google Scholar]