Abstract
Translation of goose parvovirus (GPV) 72 kDa Rep 1 is initiated from unspliced P9-generated mRNAs in ORF1 from the first in-frame AUG (537 AUG); however, this AUG is bypassed in spliced P9-generated RNA: translation of the 52 kDa Rep 2 protein from spliced RNA is initiated in ORF2 at the next AUG downstream (650 AUG). Usage of the 537 AUG was restored in spliced RNA when the GPV intron was replaced with a chimeric SV40 intron, or following specific mutations of the GPV intron which did not appear in the final spliced mRNA. Additionally, 650 AUG usage was gained in unspliced RNA when the GPV intron splice sites were debilitated. Splicing-dependent regulation of translation initiation was mediated in cis by GPV RNA surrounding the target AUGs. Thus, nuclear RNA processing of GPV P9-generated pre-mRNAs has a complex, but significant, effect on alternative translation initiation of the GPV Rep proteins.
INTRODUCTION
Goose parvovirus (GPV) is an autonomously replicating parvovirus that has been classified within the Dependovirus genus of the Parvovirinae (Cotmore and Tattersall, 2006a; Cotmore and Tattersall, 2006b; Zadori et al., 1995; Zadori et al., 2006). The genetic organization of GPV has recently been determined (Qiu et al., 2005), and interestingly, it shares features of both the Dependoviruses and members of the Parvovirus genus.
Detailed examination (Li, Qiu, and Pintel, 2009) has shown that both RNAs generated by the GPV left-most P9 promoter, which encode the GPV nonstructural proteins, are polyadenylated at high efficiency after reaching an AAUAAA signal at nucleotide (nt) 2434 in the center of the genome within the central intron. The larger mRNA remains unspliced while the smaller of the two is additionally spliced to remove a 384 nt intron (nts 814-1198) in the center of the nonstructural gene (Fig. 1). These mRNAs encode only two detectable nonstructural proteins in either 293T or goose CGBQ cells: ~72 kDa Rep1 and ~53 kDa Rep2, both of which terminate at the same site (nt 2418) 16 nts upstream of the internal polyadenylation site (pA)p. Rep1 is encoded from the large ORF 1 in the unspliced, larger rep-gene mRNA, utilizing the first available AUG (at nt 537) of that reading frame (Fig. 1A). Rep2 is encoded from the smaller, spliced mRNA; however its translation initiation bypasses the first AUG at nt 537, and instead utilizes a second AUG at nt 650. Translation from the second AUG at nt 650 proceeds in ORF 2 until the splice, after-which it resumes in ORF 1 which it shares with Rep1. Thus, these two proteins have different amino termini, and share a large portion of their carboxyl termini. The topography of these proteins can be seen in Fig. 1A.
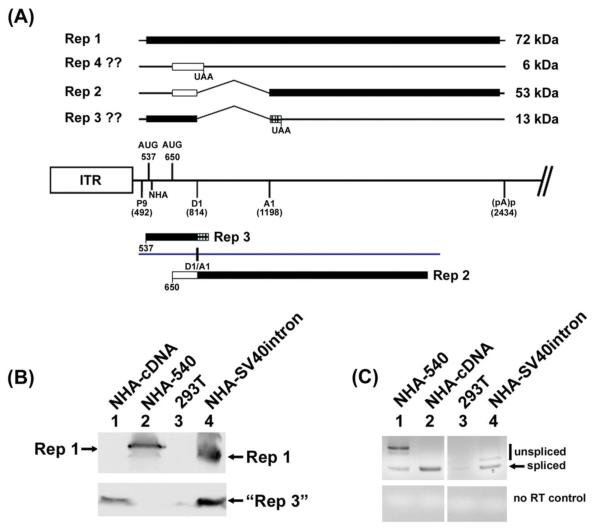
Figure 1. Substitution with a modified SV40 intron allowed usage of the 537 AUG in spliced RNA.
(A): The map shows features of the rep gene of GPV. The central line represents the genome and depicts the left end inverted terminal repeat (ITR), and the locations of the P9 promoter, the two AUGs used for initiation of Rep1 and Rep2, respectively; the location of the donor (D1) and acceptor (A1) of the rep-gene intron, the position of the HA tag at nt 540, and the proximal (internal) polyadenylation site (pA)p, taken from (Qiu et al., 2005). Above the line is depicted the potential protein products the rep-gene could encode from its known spliced and unspliced RNAs using the two initiating AUGs. Below the line is depicted the two protein products generated by the cDNA-like construct described in the text and in (Li, Qiu, and Pintel, 2009). The black bar indicates ORF1, the white bar indicates ORF2 and the stippled bar indicates ORF3. (B): The left lower panel is an immunoblot, using anti-HA antibody, of protein extracts from 293T cells either alone, or transfected with the constructs indicated as described in the text. All constructs contained an HA tag at nt 540 in ORF1. Rep1 of wild-type size (lane 2), and smaller due to the smaller SV40 intron (lane 4), are shown, as well as the “Rep3-like” protein (lanes 1 and 4) as described in the text. (C): The lower right panel shows an RT-PCR of RNA extracted from the same transfections probed at left, using primers spanning the splice junction. Note all products reflective of spliced RNAs are the same size, while the bands reflecting unspliced RNAs generated by the SV40 intron-containing construct (lane 4) are smaller than wild-type (lane 1).
The expression profile of the GPV nonstructural gene in both 293T and goose CGBQ cells exhibited some features surprising for a dependovirus, including the lack of a P19-like promoter and the presence of a spliced nonstructural gene mRNA (Qiu et al., 2005). In addition, it was notable that although the first in-frame AUG (the 537 AUG) was used to encode Rep1, it was by-passed in the spliced mRNA (a “Rep3-like protein” was not detected) (Li, Qiu, and Pintel, 2009; Qiu et al., 2005). Although readily used in spliced RNA, the second AUG (the 650 AUG) was not used in unspliced RNA. Further examination surprisingly showed that a cDNA construct expressing a “pre-spliced” RNA corresponding to the spliced smaller rep-gene mRNA utilized both AUGs (Li, Qiu, and Pintel, 2009). Additionally, when the GPV nonstructural-gene intron was replaced with that of the autonomous parvovirus minute virus of mice (MVM), both AUGs were efficiently used from the spliced mRNA (Li, Qiu, and Pintel, 2009). These results suggested that alternative translation initiation of the GPV rep-gene mRNAs was influenced by the splicing process itself, and further, upon the nature of the intervening intron.
In this report we have i) further characterized the nature of the intronic sequences that govern alternative translation initiation of spliced GPV mRNAs; ii) shown that the presence of splicing signals in the unspliced nonstructural gene RNA repress usage of the second AUG; and finally, iii) demonstrated that the splicing-dependent regulation of translation initiation was mediated in cis by GPV RNA surrounding the target AUGs. Over the last few years it has become increasingly clear that the nuclear splicing process can “mark” mRNAs with proteins that are retained during export and that can influence the localization and translation of mRNAs in the cytoplasm (Diem et al., 2007; Le Hir, Nott, and Moore, 2003; Le Hir and Seraphin, 2008; Ma et al., 2008; Michlewski, Sanford, and Caceres, 2008; Nott, Le Hir, and Moore, 2004; Swartz et al., 2007), although the detailed mechanism remains unclear (Dahan, Gingold, and Pilpel). Our results demonstrate that the nuclear processing of GPV RNAs influences the translation fate of these RNAs.
RESULTS AND DISCUSSION
Substitution with a modified SV40 intron allowed usage of the 537 AUG in spliced RNA
We have previously shown that when intronic sequences were precisely removed from the GPV rep gene such that it expressed a “pre-spliced” cDNA-like RNA, or when the GPV intron was precisely substituted with the small intron from MVM, the first in-frame AUG (the 537 AUG) was no longer bypassed in spliced P9-generated RNA following transfection of these constructs in either 293 or goose CGBQ cells (Li, Qiu, and Pintel, 2009). That is, a “Rep3-like” protein was efficiently produced (see map in Fig. 1A). This suggested that the splicing process itself may have influenced the choice of initiating AUGs in this RNA, and importantly, that this regulation was dependent upon the nature of the intron being excised from the pre-mRNA. To generalize this observation, and rule out that usage of the 537 AUG was linked specifically to insertion of the MVM small intron, we asked whether a similar result would be obtained by substituting another intron. Therefore, we precisely replaced the GPV intron with a commonly used 132 nt modified SV40 early region intron from the plasmid pCI (Promega). The intron was further modified to maintain ORF 1 in unspliced RNA. The Rep1 translated from unspliced RNA generated by this construct would be 84 amino acids smaller than wild-type GPV Rep1. Expression was monitored following transfection of 293T cells using a construct tagged at the N-terminus as depicted, and the SV40 intron was very efficiently spliced in this context (NHA-SV40intron, Fig.1, lane 4, bottom right panel). [We have previously shown that the HA tag itself at this position has no effect translation of these RNAs (Li, Qiu, and Pintel, 2009)]. As previously seen with the MVM chimera (Li, Qiu, and Pintel, 2009) - and in contrast to the wild-type situation - the 537 AUG was efficiently used following splicing of the SV40 intron. This was demonstrated by the abundant presence of the “Rep3-like” protein (Fig. 1B, lane 4). Rep1, smaller due to the smaller intron retained in unspliced RNA, was also detected. As controls, expression of the singly N-terminal-HA-tagged cDNA clone (NHA-cDNA) revealed the expected utilization of the 537 AUG from the “pre-spliced RNA” indicated by the presence of the “Rep3-like” protein (Fig. 1B, lane 1; Fig. 1C, lane 2). Furthermore, usage of the 537 AUG following transfection of a singly N-terminal HA-RepCap construct (NHA-540) was restricted to the unspliced RNA, as indicated by the presence of Rep1 but the absence of the “Rep3-like” protein (Fig. 1B, lane 2; Fig. 1C, lane 1). Our results suggested that how the GPV intron is processed exerts a specific inhibition of usage of the 537 AUG, and that either in its absence, or when the pre-mRNAs are spliced using other introns, both AUGs are efficiently used to initiate translation.
Sequences within the 3′ region of the GPV intron influenced translation initiation site choice of the spliced mRNA product
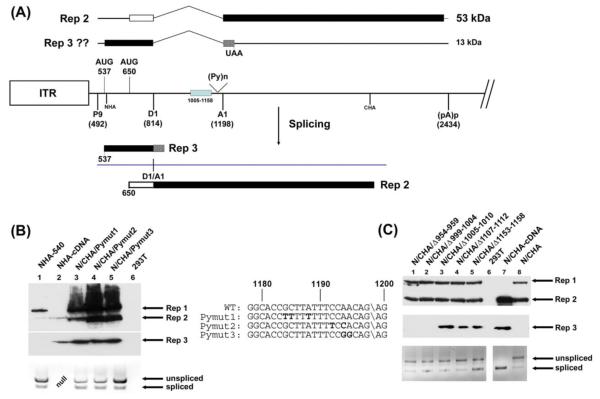
It is known that although the basic splicing machinery is used for all major class introns, different introns can program the assembly of different accessory splicing factors on pre-mRNAs (Michlewski, Sanford, and Caceres, 2008). A subset of these can remain with the processed RNA during transit to the cytoplasm (Le Hir and Seraphin, 2008; Swartz et al., 2007). Since the nature of the excised intron seems to influence its translational fate, it seemed reasonable to investigate whether there were cis-acting sequences which might mediate this process by programming the assembly and deposition of such factors. To begin to determine the cis-acting signals within the GPV intron that might explain the by-pass of the 537 AUG in spliced P9-generated RNA, we initiated a mutational analysis of known splicing signals within the intron. [Although these mutations also affected the large Rep protein expressed from unspliced P9-generated RNA, we have previously shown that this protein itself plays no detectable role in the processing or translation of GPV RNA (Li, Qiu, and Pintel, 2009; Qiu et al., 2005).] We found that certain nucleotide changes within the GPV 3′ splice site, which allow levels of intron excision either similar to or greater than wild-type, resulted in the use of both the 537 AUG and the 650 AUG in the final spliced product, as seen by the production of the “Rep3-like” protein in these experiments. These mutations included changes which improved the polypyrimidine tract (Pymut 1 and Pymut2, Fig. 2B, lanes 3-4; RNA splicing levels modestly greater than wild-type shown at bottom; mutations shown in central panel), or were within the extended 3′ cleavage site (Pymut3, Fig.2B, lane 3; RNA splicing levels similar to wild-type shown at bottom; mutations shown in central panel). Furthermore, splicing proficient small deletions between nt 1005-1158 (Fig. 2C, lanes 3-5; RNA splicing levels shown at bottom), but not adjacent mutations further upstream between nts 954-1004 (Fig. 2C, lanes 1-2; RNA splicing levels shown at bottom), had a similar effect. It is important to note that these mutations, while presumably having an effect on the nature of the assembled spliceosome, do not appear in the final spliced mRNA product whose translation is affected, implicating an antecedent event. Mutations around the intron donor had no detectable effects on translation of spliced P9-generated RNAs (data not shown). Thus it is likely that in addition to basic spliceosome formation, the 3′ region of the GPV intron specifically programmed the assembly of splicing-associated factors that affect the translation initiation choice of spliced mRNA in the cytoplasm. Further mapping using chimeras between the GPV and MVM or SV40 introns was attempted, but these constructs generated multiple artifactual splicing products confounding this approach.
Figure 2. Sequences within the 3′ region of the GPV intron influenced translation initiation site choice of the spliced mRNA product.
(A): The map shows potential RNAs and proteins generated by spliced P9-generated RNAs. The central line represents the genome as described in the legend to Fig. 1, with an additional box to indicate the region within which deletions allow 537 AUG usage as described in the text, indication of the location of the intron polypyrimidine tract: (Py)n, and the locations of the N-terminal and C-terminal HA tags at nts 540 and 2241, respectively. Above and below the genome line are depicted the potential protein products of the spliced RNA using the two initiating AUGs. (B): The left lower panel is an immunoblot, using anti-HA antibody, of protein extracts from 293T cells either alone, or transfected with the 3′ splice site mutation constructs indicated and as described in the text. The mutant region sequences are shown to the right with the mutations in bold. All of the mutant constructs contained both the N- and C-terminal HA tag in ORF 1 at nt 540 and nt 2241, respectively. The wild-type RepCap clone (NHA-540) as well as the cDNA-like clone (NHA-cDNA), both singly tagged in ORF1 at the amino terminus and described in the text, were included as controls. Rep1, Rep 2 and the “Rep3-like” proteins are indicated. Below the immunoblot is an RT-PCR of RNAs extracted from the same transfection indicating splicing levels using primers spanning the splice junction. In the lane marked “NULL”, no sample was run. (C): The right lower panel is an immunoblot, using anti-HA antibody, of protein extracts from 293T cells either alone, or transfected with the deletion mutation constructs indicated. Deletion limits are shown in their respective titles and are described in the text. All of these constructs contained both a N- and C-terminal HA tag in ORF 1 at nt 540 and nt 2241, respectively. The wild-type RepCap clone, as well as the cDNA-like clone, both also doubly tagged (N/CHA, and N/CHA-cDNA, respectively) were included as controls. Rep1, Rep 2 and the “Rep3-like” proteins are indicated. Below the immunoblot is an RT-PCR of RNAs extracted from the same transfection indicating splicing levels using primers spanning the splice junction.
Competent splice sites prevented 650 AUG usage in unspliced RNA
The 650 AUG, as the second in-frame AUG, was not used in unspliced P9-generated RNA (a “Rep 4-like” protein was not produced, see Fig. 1A); however, because it is used in preference to the first AUG (the 537 AUG) in spliced P9-generated RNA, we suspected that by-pass of the 650 AUG in unspliced RNA may have been regulated in this context also. Surprisingly, using a doubly-tagged construct as described below (see Fig. 3C), we found that 650 AUG usage in unspliced RNAs was influenced by the presence of competent splicing sites.
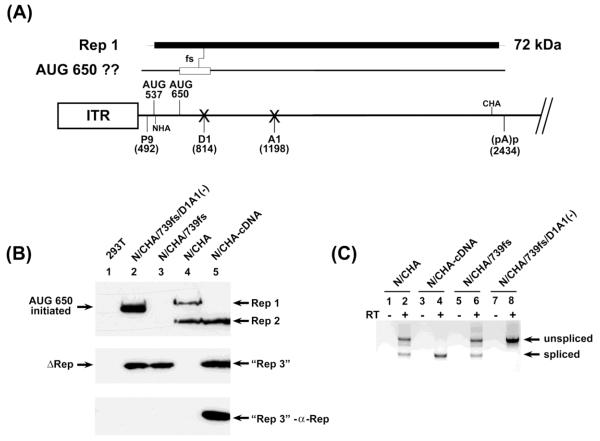
Figure 3. Competent splice sites prevented 650 AUG usage in unspliced RNA.
(A): The map shows potential products from unspliced RNAs and the frame shift mutant described. The central line depicts the genome as described in Fig. 1, with “Xs” at the donor and acceptor site to indicate their mutation, and positions of both the N-terminal and C-terminal HA tags, at nts 540 and 2241, respectively. Above the line is depicted the coding region for Rep1 and the site of the frame shift described, and the potential protein generated by this mutant if the 650 AUG is used. (B): The left panel is an immunoblot of protein extracts from 293T cells either alone, or transfected with the constructs indicated and as described in the text. The upper two panels are reacted with anti-HA antibody. All of these constructs contained both the N- and C-terminal HA tag in ORF 1 at nt 540 and nt 2241, respectively. The wild-type RepCap clone as well as the cDNA-like clone, both also doubly tagged (N/CHA and N/CHA-cDNA, respectively), were included as controls. Rep1, Rep 2 and the “Rep3-like” proteins, reacting with anti-HA antibody, are indicated. The bottom-most panel shows that the “Rep3-like” protein (lane 4), but not the similarly migrating Δ-Rep proteins (lane 2 and 3), is authentic Rep3, as it also reacts with an anti-Rep antibody raised to the unique portion of Rep3 shown in Figure 1 and as described in the text and Materials and Methods. (C): The panel to the lower right is an RT-PCR of RNAs extracted from the same transfection indicating splicing levels using primers spanning the splice junction.
As expected from previous results, GPV RepCap HA-tagged in both the C-terminal and N-terminal region of the rep gene (N/CHA) generated both Rep1 and Rep2 (Fig. 3B, lane 4), and a cDNA construct doubly-HA-tagged in both the N- and C-terminal regions (N/CHA-cDNA) generated Rep2 from the 650 AUG as well as a “Rep3-like” protein from the 537 AUG (Fig. 3B, lane 5). The “Rep3-like” protein was confirmed as such by a Rep3-specific antibody directed to the unique C-terminal region of Rep3 (Fig. 3C, lowest panel, lane 5; also see diagram in Fig. 1A). In order to more conveniently test whether the 650 AUG could be used in unspliced RNA we introduced a frame shift in the doubly-HA-tagged RepCap construct fusing the 650 AUG-initiated ORF2 into the Rep1 ORF in unspliced RNA by insertion of a single nucleotide at nt 739 (upstream of the intron – N/CHA/739fs, Fig. 3A). As expected, use of the 650 AUG in this construct could not be detected (Fig. 3B, lane 3). Also as expected, Rep1 was terminated by the frame-shift, and although this generated a protein of similar size to the “Rep3-like protein” it was not Rep3, as it was not detected by the Rep3-specific antibody (Fig. 3B, lowest panel, lane 3). Surprisingly, however, when the donor and acceptor were mutated in this construct [N/CHA/739fs/D1A1(-)], an abundant protein generated from the 650 AUG was produced (Fig. 3B, lane 2). Rep1, prematurely terminated as expected due to the insertion-mediated frame shift (and which did not interact with the anti-Rep3 antibody, Fig. 3B, lowest left panel, lane 2) was also generated. The RNAs generated by these constructs were spliced (or not) as expected (Fig. 3C, right panel). These results indicated that either the presence of viable splice sites in the P9-generated pre-mRNA played a role in suppressing usage of the potent 650 AUG from unspliced RNA, perhaps by preventing engagement of the spliceosome or associated splicing factors, or that the splicing process itself enhanced 650 AUG usage.
Sequences surrounding the 537 AUG mediated its by-pass in spliced RNA
The bypassing of the 537 AUG in spliced RNAs was not mediated in cis by the nature of the immediate sequence comprising those initiation signals: neither improving the 537 AUG to Kozak consensus (Kozak, 1986), nor knocking out the downstream 650 AUG, nor making both positions the same – each either the 9nts surrounding the 537 AUG or the 650 AUG, nor switching the two initiation sequences, increased the usage of the 537 AUG in spliced RNA (data not shown). However, larger regions of GPV RNA upstream of the 537 and 650 AUGs were found to play a role in bypassing the 537 AUG in spliced RNA. In RepCap constructs that were both N-terminally (at nt 540) and C-terminally (at nt 2241) tagged (see Fig. 4A), replacing the full region upstream of the 537 AUG [N/CHA-AAV5(493-527), Fig. 4B, lane 1], the region between the two AUGs [N/CHA-AAV5(543-645), Fig. 4B, lane 2], or the region downstream of the 650 AUG up to the splice donor (data not shown), with heterologous sequence from the AAV5 capsid gene known to have no splice sites, had little effect on relative protein levels. However, when regions upstream of the 537 AUG and the 650 AUG were replaced together [N/CHA-AAV5(493-641), Fig. 4B, lane 5], the 537 AUG was efficiently expressed from spliced RNA. These results suggest that the inhibition of the use of the 537 AUG dictated by the splicing process was at least in part mediated by partner cis-acting GPV signals in the translated RNA.
Figure 4. Sequences surrounding the 537 AUG mediated its by-pass in spliced RNA.
(A): The map shows a schematic of the GPV genome between the RNA initiation site at nt 492 and the splice donor at nt 814. The 537 AUG and the 650 AUG are shown, as well as the regions of the GPV genome substituted with heterologous AAV5 capsid regions sequence (stippled) in the various constructs analyzed. As described in the text, all inserts were constructed to maintain the ORF1 reading frame throughout the insertion, and to maintain the HA tag at nt 540, which is also indicated. In addition, they contained a C-terminal ORF1 HA tag at nt 2241, which does not appear on this map. (B): Below is shown an immunoblot of protein extracts from 293T transfected with the constructs indicated and as diagrammed above and further described in the text. The doubly-tagged wild-type RepCap clone (N/CHA), as well as the cDNA-like clone singly N-terminally HA-tagged at nt 540 (NHA-cDNA), were included as controls. Rep1, Rep2, and Rep3, indicating usage of the 537 AUG in spliced RNA, are shown.
Taken together, the results presented in this manuscript extend further the observation that nuclear RNA processing of GPV P9-generated pre-mRNAs has a complex, and significant, effect on alternative translation initiation of the GPV Rep proteins.
MATERIALS and METHODS
293T and CGBQ cells were propagated as previously described(Li, Qiu, and Pintel, 2009). Transfections of cloned DNA was performed using Lipofectamine (Invitrogen) as recommended by the supplier.
Most constructs have been previously described (Li, Qiu, and Pintel, 2009). Additional constructs were prepared by standard overlapping PCR techniques using the previously described GPV infectious plasmid clone as a parent (Qiu et al., 2005), and were fully sequenced to ensure that only the predicted mutations were present. For the donor knock-out the 5′ intron G residue was deleted changing wild-type GCA/GTGAGT to GCATGAGT, and for the acceptor knock-out the wild-type ACAG\AGCG was changed to ACCGCGCG. For the SV40 intron insertion, the modified SV40 intron from the plasmid pCI was precisely cloned into the GPV genome such that the spliced P9-generated product was wild-type. In addition two nucleotide changes were introduced to maintain ORF1 in unspliced RNA. The AAV5 insertion mutations contained regions of the AAV5 capsid coding sequence as described, and were constructed so that ORF1 was maintained throughout the insertion, and that the HA tag at nt 540 was retained.
RNA was extracted, and RT PCR was performed as previously described (Li, Qiu, and Pintel, 2009). Westerns were performed as previously described, using either anti-HA antibody (Sigma, St. Louis, MO), anti-FLAG antibody (Sigma, St. Louis, MO), or anti-Rep3 antibody raised in rabbits to a peptide contained within the unique Rep3 region in ORF3 downstream of the splice acceptor (see Fig. 1). This antibody was made commercially by New England Peptide (Gardner, MA).
HIGHLIGHTS.
Alternative translation initiation of goose parvovirus mRNA is splicing dependent
Substitution or modification of the GPV intron affected translation initiation choice
Use of an internal AUG in unspliced RNA was gained following splice site debilitation
ACKNOWLEDGEMENTS
We thank Lisa Burger for excellent technical assistance, Dave Farris for help with figure preparation, and Jianming Qiu for discussion. This work was supported by PHS grants AI 46458 and AI 56310 from NIH to DJP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cotmore SF, Tattersall P. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses. In: Kerr J, et al., editors. “Parvoviruses”. Hodder Arnold; London, UK: 2006a. pp. 171–188. [Google Scholar]
- Cotmore SF, Tattersall P. Structure and organization of the viral genome. In: Kerr J, et al., editors. “Parvoviruses”. Hodder Arnold; London, UK: 2006b. pp. 73–94. [Google Scholar]
- Dahan O, Gingold H, Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 27(8):316–22. doi: 10.1016/j.tig.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14(12):1173–9. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Kozak M. “ In Advances in Virus Research”. Academic Press, Inc.; 1986. Regulation of protein synthesis in virus-infected animal cells; pp. 229–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28(4):215–20. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133(2):213–6. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Li L, Qiu J, Pintel DJ. The choice of translation initiation site of the rep proteins from goose parvovirus P9-generated mRNA is governed by splicing and the nature of the excised intron. J Virol. 2009;83(19):10264–8. doi: 10.1128/JVI.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133(2):303–13. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30(2):179–89. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18(2):210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Yoto Y, Zadori Z, Pintel D. The expression strategy of goose parvovirus exhibits features of both the Dependovirus and Parvovirus genera. J Virol. 2005;79(17):11035–44. doi: 10.1128/JVI.79.17.11035-11044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282(27):19844–53. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- Zadori Z, Stefancsik R, Rauch T, Kisary J. Analysis of the complete nucleotide sequences of goose and muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2. Virology. 1995;212(2):562–73. doi: 10.1006/viro.1995.1514. [DOI] [PubMed] [Google Scholar]
- Zadori Z, Szelei J, Kiss I, Tijssen P. In: “Waterfowl parvoviruses.” Parvoviruses. Kerr J, et al., editors. Hodder Arnold; London, UK: 2006. [Google Scholar]