Abstract
Dictyostelium discoideum amoebas coordinate aggregation and morphogenesis by secreting cyclic adenosine monophosphate (cAMP) pulses that propagate as waves through fields of cells and multicellular structures. To retrace how this mechanism for self-organisation evolved, we studied the origin of the cAMP phosphodiesterase PdsA and its inhibitor PdiA, which are essential for cAMP wave propagation. D. discoideum and other species that use cAMP to aggregate reside in group 4 of the four major groups of Dictyostelia. We found that groups 1-3 express a non-specific, low affinity orthologue of PdsA, which gained cAMP selectivity and increased 200-fold in affinity in group 4. A low affinity group 3 PdsA only partially restored aggregation of a D. discoideum pdsA-null mutant, but was more effective at restoring fruiting body morphogenesis. Deletion of a group 2 PdsA gene resulted in disruption of fruiting body morphogenesis, but left aggregation unaffected. Together, these results show that groups 1-3 use a low affinity PdsA for morphogenesis that is neither suited nor required for aggregation. PdiA belongs to a family of matrix proteins that are present in all Dictyostelia and consist mainly of cysteine-rich repeats. However, in its current form with several extensively modified repeats, PdiA is only present in group 4. PdiA is essential for initiating spiral cAMP waves, which, by organising large territories, generate the large fruiting structures that characterise group 4. We conclude that efficient cAMP-mediated aggregation in group 4 evolved by recruitment and adaptation of a non-selective phosphodiesterase and a matrix component into a system for regulated cAMP degradation.
Keywords: Morphogenetic signalling, cAMP oscillations, Cyclic nucleotide phosphodiesterase, CTDC domain, Gene co-option, Evolution of development
INTRODUCTION
One of the more surprising outcomes of comparative genome analysis is that the phenotypic complexity of species is only marginally related to the number of protein-coding genes in their genome, as illustrated by the fact that both humans and single-celled protozoa, such as ciliates, have ∼25,000 protein-coding genes (Human Genome Sequencing Consortium, 2004; Eisen et al., 2006). This illustrates the fact that unicellular organisms are by no means simple. Their single cells can execute the multitude of functions, such as food uptake, metabolism, reproduction, directional movement, secretion, environmental sensing, prey hunting and predator evasion, for which higher organisms use many specialised cell types, organs and tissues. The genomes of complex multicellular organisms do contain much more non-coding DNA, with both a direct role in cis-regulation of gene expression, and an increasingly recognised enormous potential for post-transcriptional gene regulation (Zhou et al., 2010). This underpins the dependence of both the embryonic development and the adult physiology of multicellular organisms on accurately controlled expression of genes by their specialised cells at the right time and place. A hallmark of the evolution of multicellularity is the extensive use of communication between cells to regulate gene expression.
To understand how multicellular organisms evolved, it is therefore essential to retrace how complex intercellular communication emerged from more simple environmental sensing systems. We study the evolution of cell communication in the social amoebas, a highly tractable model system that combines a unicellular growth stage with an intricately orchestrated multicellular life style that is initiated by starvation. In the model organism D. discoideum, some starving cells emit pulses of cAMP, which elicit both chemotaxis and secretion of a cAMP pulse by surrounding cells. This causes a spiral or concentric cAMP wave to move outwards from the original source and the cells to move inwards, creating an aggregate. The aggregate tip continues to emit cAMP pulses, which cause the cell mass to project upwards to form a slug-shaped structure, which topples over and, after a period of migration, veers upwards once more to form a fruiting structure (Dormann and Weijer, 2001). During this process, the amoebas differentiate into spore cells and into four additional cell types that form a stalk and support structures, which carry the spore mass aloft (Williams, 2006).
The cAMP pulses are generated by a feedback network, in which secreted cAMP acts on G-protein-coupled cAMP receptors (cARs) to transiently activate cAMP synthesis by the adenylate cyclase (ACA) using a number of proteins as intermediates, such as PI3-kinase and the TORC complex (which also mediates cAMP induction of the chemotactic response) (Lee et al., 2005; Cai et al., 2010). Another essential requisite for dynamic cAMP signalling is the extracellular cAMP-phosphodiesterase PdsA, which hydrolyses cAMP between pulses. PdsA is extensively regulated at both the transcriptional and post-translational level. The PdsA gene is transcribed from three promoters during aggregation, growth and post-aggregative (late) development, respectively, with the late promoter being closest to the coding sequence (Faure et al., 1990). The PdsA protein can be both displayed on the exterior face of the plasma membrane, or cleaved off and released extracellularly. The released form facilitates cAMP wave propagation at low initial cell densities (Palsson, 2009). In the absence of extracellular cAMP, an inhibitor protein, PdiA, is released, which increases the affinity (Km) of the secreted form of PdsA from 5 μM to 5 mM. In the presence of cAMP, PdiA expression is inhibited, whereas PdsA expression is strongly enhanced (Yeh et al., 1978; Franke et al., 1987). pdsA-null mutants can no longer aggregate (Sucgang et al., 1997), whereas pdiA-null mutants can no longer produce spiral cAMP waves, and consequently have 50 times smaller aggregation territories than those of wild-type cells (Palsson et al., 1997).
D. discoideum is a member of group 4 of the Dictyostelid social amoebas (Schaap et al., 2006). Most, if not all, members in this group use cAMP to aggregate, but none of the species in groups 1-3 (Schaap et al., 2006) (A. Skiba and P.S., unpublished). Glorin, a modified dipeptide of glutamate and ornithine is used by Polysphondylium violaceum and a range of non-group 4 species (Shimomura et al., 1982; Asghar et al., 2012), whereas a folate derivative is used by D. minutum in group 3 (De Wit and Konijn, 1983). Remarkably, even species that do not use cAMP for aggregation, do use cAMP oscillations to coordinate slug and fruiting body morphogenesis (Schaap et al., 1984; Fukushima and Maeda, 1991). Consistent with their post-aggregative use of cAMP, non-group 4 species express cAR genes from a single promoter after aggregation (Alvarez-Curto et al., 2005). Group 4 species have an additional distal promoter that activates cAR expression just before aggregation (Louis et al., 1993; Alvarez-Curto et al., 2005). Proximal post-aggregative and more distal aggregative promoters are also hallmarks of the PdsA and ACA genes in D. discoideum (Faure et al., 1990; Galardi-Castilla et al., 2010), suggesting that the novel role of cAMP signalling during aggregation of group 4 species was achieved by distal insertion of new cis-regulatory regions in existing cAMP signalling genes. Such a scenario is in good agreement with a growing body of evidence showing that the evolution of new morphologies in animals and plants is largely dependent on elaboration of the cis-regulatory regions of genes, rather than their coding regions (Wray et al., 2003; Carroll, 2005).
To assess to what extent changes in gene regulation and protein function contributed to the evolution of cAMP signalling in the Dictyostelia, we studied conservation and change in both the regulation, function and biochemical properties of the PdsA genes throughout the Dictyostelium phylogeny. Our results show that elaboration of protein function also played a major role in enabling Dictyostelid amoebas to use cAMP oscillations to mediate efficient aggregation and formation of large fruiting structures.
MATERIALS AND METHODS
Cell culture
D. rosarium M45 (Dros), D. mucoroides S28 (Dmuc), P. violaceum P6 (Pvio), D. minutum 71-2 (Dmin), D. rhizopodium AusKY-4 (Drhi), P. pallidum PN500 (Ppal), Acytostelium anastomosans PP1 (Aana), D. aureo-stipes YA6 (Daus) and D. fasciculatum SH3 (Dfas) spores were grown in association with Escherichia coli 281 on 0.1% lactose/peptone (LP) agar plates (Raper, 1984). To obtain large quantities of cells, 1.5×108 amoebas were inoculated in 300 ml 10 mM sodium/phosphate buffer, pH 6.5 (PB), supplemented with a resuspended E. coli 281 pellet from a 300 ml stationary culture in LB. The suspension was shaken at 200 rpm until the amoeba had reached a density of ∼3×106 to ∼5×106 cells/ml. Ddis was grown in HL5 medium. For developmental time courses, growing cells were washed free from bacteria with PB, and incubated at 22°C on non-nutrient (NN) agar (1.5% agar in PB) at 5×106 cells/cm2. Activated charcoal was placed in the lids to improve synchronous development.
Gene identification and analysis
Genomic DNA (gDNA) of species that represent all Dictyostelid groups was isolated as described earlier (Kawabe et al., 1999; Schaap et al., 2006). Degenerate oligonucleotide primers were designed to match amino acid sequences that are conserved between the PDE-II domains of Ddis PdsA and Saccharomyces cerevisiae PDE-II (Genbank id: EDV10448). Primers PDEF1 and PDER1 (supplementary material Table S1) yielded 633 nt products from Dmuc, Dros and Dmin gDNAs, which were validated by nested PCR with primer pair PDEF1 and PDERN1. The PCR products were sub-cloned in the pGEM-T Easy vector (Promega) and their sequence was determined from three independent clones. The Dmin PCR product was used to retrieve a full length Dmin PdsA coding sequence from a gDNA library (see below). Conserved regions between the Dmin- and Ddis PdsAs were used subsequently to design primers for nested PCR for amplification of PdsAs from more distant species. The first PCR was run using combinations of forward primers PdsA-51A and PdsA51B with reverse primers PdsA-3E and PdsA-3N, and the nested PCR used combinations of forward primers PdsA-51A and PdsA51B with reverse primer PdsA-32. The PCR program consisted of 35 cycles, with 30 seconds at 95°C, 50°C and 68°C each, and yielded products of about 300 bp from Pvio, Drhi, Aana, Daus and Dfas gDNAs. Full length D. purpureum (Dpur), Dfas, Ppal, Acytostelium subglobosum (Asub) and D. lacteum (Dlac) PdsA genes are recently available from ongoing genome sequencing projects (Heidel et al., 2011; Sucgang et al., 2011) (http://acytodb.biol.tsukuba.ac.jp/cgi-bin/index.cgi?.org=as) (P.S. and G. Gloeckner, unpublished results).
To determine the nucleotide sequence of the Ppal PdsA mRNAs, polyA+ RNA was isolated from Ppal cells developed for 12 hours. Full length cDNAs were subsequently synthesised by RNA-ligation mediated rapid amplification of 5′ and 3′ cDNA ends (RLM-RACE) and RT-PCR using the GeneRacer kit (Invitrogen) and primers PdsA-R54 to PdsA-R37 (supplementary material Table S1) according to the manufacturer’s instructions.
DNA constructs and transformation
Cloning and expression of Dmin PdsA
The Dmin PdsA PCR product was used to screen a custom-made Dmin λZAPII gDNA library (Alvarez-Curto et al., 2005), which yielded six positive plaques. Their pBluescript phagemids were isolated by in vitro excision and the inserts were sequenced. The sequences could be assembled into a 9 kb contig, which contained full length Dmin PdsA as well as one complete and two partial flanking genes (supplementary material Fig. S3).
To generate an expression construct for gene complementation, a 1.3 kb fragment consisting of the complete coding region of Dmin PdsA without its single intron, was amplified from λZAPII-derived plasmid P6 with primers Dmin-PdsA-Ex5′, which spans the intron that is positioned at 8 nt from the start codon, and Dmin-PdsA-Ex3′, which includes the stop codon. The primers contained a BamHI and a XhoI restriction site, respectively (supplementary material Table S1). After BamHI/XhoI digestion, the PCR product was cloned into the similarly digested vector pDV-CYFP (Meima et al., 2007), which deletes the YFP sequence from the vector and places Dmin PdsA downstream of the constitutively active Ddis actin15 promoter, yielding plasmid pA15::Dmin PdsA. This plasmid was transformed by electroporation into the Ddis PdsA-null mutant Uk7 (Sucgang et al., 1997).
Ppal PdsA gene disruption
To obtain a PdsA disruption vector for Ppal, a 1.18 kb 5′ PdsA fragment was amplified from Ppal gDNA with primers PdsA-53Xh and PdsA-33H, which contain XhoI and HindIII restriction sites, respectively, and a 1.41 kb 3′ UTR PdsA fragment was amplified with primers PdsA-52B and PdsA-32S, which contain BamHI and SacI restriction sites, respectively. Both PCR products were cloned into vector pLoxP-NeoIII, yielding pPpal PdsA-KO. To construct pLoxP-NeoIII, the XbaI site in pLoxP-NeoI (Kawabe et al., 2009) was destroyed by digestion with XbaI, fill-in with Klenow and self-ligation with T4-ligase. The A6Neo cassette with flanking LoxP sites was next excised with BamHI/HindIII and cloned into pBluescript SK–, creating pLoxP-NeoIII.
The Ppal PdsA-KO vector was digested with SacI and XhoI and co-electroporated into Ppal cells with 100 μM of primers PdsA-53Xh and PdsA-32S to enhance the efficiency of homologous recombination (Kuwayama et al., 2008). Transformants were selected on lawns of autoclaved bacteria, supported by agar containing 300 μg/ml G418 (Kawabe et al., 1999). Out of 60 clones, obtained from two separate experiments, 55 showed the same aberrant phenotype. Southern blots were performed on gDNAs from one normal and four aberrant clones, and from untransformed cells. All four aberrant clones showed disruption of PdsA, but two clones harboured an additional random vector integration. The normal clone did not harbour a PdsA disruption (supplementary material Fig. S4).
PdsA promoter-lacZ constructs and analysis
To construct gene fusions of the PdsA415 and PdsA802 promoters and lacZ, the 2.2 kb PdsA415 promoter was amplified from Ppal gDNA using primers PdsA-P51X and PdsA-P31B, whereas for the 3.6 kb PdsA802 promoter primers PdsA-P52X and PdsA-P32B were used. Both primer sets contain XbaI and BamHI restriction sites (supplementary material Table S1). After digestion with XbaI/BamHI, the 2.2 kb PCR product was ligated into the BamHI/XbaI digested pDdGal17 vector (Harwood and Drury, 1990), yielding vector P415-gal, and the 3.6 kb PCR product was ligated into the BamHI/XbaI digested pDdGal16 vector (Harwood and Drury, 1990), yielding vector P802-Gal. Ppal cells were transformed with either of the vectors. β-Galactosidase activity was visualised with X-gal in developing Ppal structures as described previously (Dingermann et al., 1989; Kawabe et al., 2009).
PDE assays
To measure PDE-II activity, intact cells were serially diluted between 107 and 3×105 cells/ml in PB to yield activities that hydrolysed <60% of the substrate. Alternatively, cells were lysed through nuclepore filters (pore size 3 μm) at 108 cells/ml. Lysate (1 ml) was centrifuged for 10 minutes at 14,000 g to separate the cytosol and particulate fraction, with the latter being resuspended in 1 ml PB, and both diluted to the equivalent of 107 cells/ml. Cells or cell fractions were incubated for 30 minutes at 22°C with 10 nM or 100 nM [2,8-3H]cAMP (3HcAMP, Amersham, UK) in a total volume of 20 μl in the presence of 0.2 mM of the PDE-I inhibitor 3-isobutyl-1-methylxanthine (Sigma). Unlabelled cAMP or cGMP was added as indicated in the figure legends. Reactions were terminated by boiling and [2,8-3H] 5′AMP was hydrolysed further with 10 μg of Naja messambica snake venom (SA Venom Suppliers, Louis Trichardt, SA, USA) to [2,8-3H]adenosine, which was separated from 3HcAMP by adsorption of the latter to Dowex anion exchange resin.
RESULTS
Identification of PdsA genes in group representative taxa
Most Dictyostelia are members of four major taxon groups that combine to form two branches, containing groups 1 and 2 and groups 3 and 4, respectively (Schaap et al., 2006) (A. Skiba and P.S., unpublished). Only group 4 taxa use cAMP to aggregate (Schaap et al., 2006) (A. Skiba and P.S., unpublished) and require PdsA to generate steep chemotactic gradients (Darmon et al., 1978). To investigate whether PdsA is conserved in the Dictyostelids, we used a PCR approach with degenerate oligonucleotide primers to amplify homologues of the D. discoideum (Ddis) PdsA gene from group-representative taxa. Figure 1A shows a schematic representation of the SSU rRNA phylogeny of the Dictyostelia and illustrates the position of the test species. The first set of primers was designed to match regions conserved between the PDE-II domain of Ddis PdsA and non-dictyostelid PDE-II type enzymes. This set only yielded products from two other group 4 taxa, D. mucoroides (Dmuc) and D. rosarium (Dros), and from a group 3 taxon D. minutum (Dmin). The Dmin PdsA product was subsequently used to screen a genomic library. This yielded a full-length PdsA gene (see below), the sequence of which was used to design more specific primers for dictyostelid PdsAs.
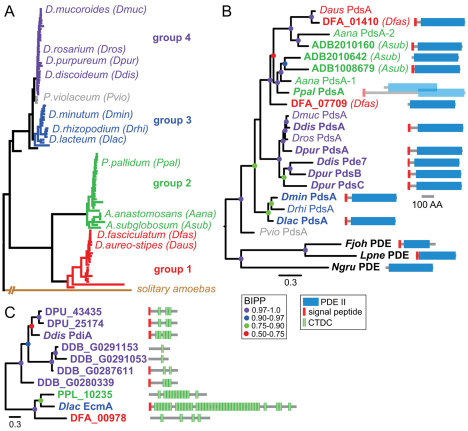
Fig. 1.
Identification of PdsA and PdiA genes across the Dictyostelid phylogeny. (A) Schematic representation of the Dictyostelid phylogeny as inferred from SSU rRNA sequences and 18 orthologous genes obtained from fully sequenced genomes (Schaap et al., 2006) (A. Skiba and P.S., unpublished). The positions of species that were used in this work are shown. (B) Partial (regular text) or complete (bold text) PdsA coding sequences were obtained by PCR, cloning from a genomic library or BLAST query of genome sequencing projects. The multiple Asub (ADB), Dfas (DFA) and Dpur (DPU) sequences are named by their locus tags. The PDE-II domains of the derived protein sequences were aligned with the PDE-II domains of three related non-dictyostelid PDE-II proteins using CLUSTALW (Chenna et al., 2003). The alignment was used to assess phylogenetic relationships between the sequences by Bayesian inference (Ronquist and Huelsenbeck, 2003) using a mixed amino acid model with rate variation across sites estimated by a gamma distribution with a proportion of invariable sites. Posterior probabilities for the position of the tree nodes are indicated by coloured dots. The complete coding sequences are annotated with the domain architecture of the proteins. Ppal PdsA is transcribed from multiple promoters and generates two different proteins by alternative splicing (see Fig. 4). (C) The closest Ddis PdiA homologues in group-representative taxa were obtained by BLAST query of genome sequencing projects. An alignment of the full length protein sequences was used to construct a phylogenetic tree by Bayesian inference, which was decorated with the domain architecture of the proteins.
The new primers yielded a single sequence from another group 3 taxon D. rhizopodium (Drhi), the group-intermediate taxon P. violaceum (Pvio), the group 1 taxa D. aureo-stipes (Daus) and D. fasciculatum (Dfas), and two sequences from the group 2 taxon A. anastamosans (Aana). Complete coding sequences for D. purpureum (Dpur), D. lacteum (Dlac), A. subglobosum (Asub), P. pallidum (Ppal) and D. fasciculatum (Dfas) PdsAs and for the Ddis PdsA homologue, Pde7 (Bader et al., 2007), were recently obtained from ongoing (Asub, Dlac) and completed genome sequencing projects (Eichinger et al., 2005; Heidel et al., 2011; Sucgang et al., 2011). All derived partial and complete PdsA amino acid sequences were aligned with the closest PdsA homologues in non-dictyostelid organisms, which are three PDE-II sequences from Naegleria gruberi, Legionella pneumophila and Flavobacterium johnsoniae (supplementary material Fig. S1).
The sequence alignment was used to determine phylogenetic relationships between the PdsA sequences (Fig. 1B). The analysis shows that the group 4 PdsAs form two sister clades, whereas the group 3 PdsAs cluster together. The single Ppal PdsA clusters with one of the Aana and two Asub PdsAs, whereas the other Asub- and Aana PdsAs cluster with the group 1 PdsAs from Dfas and Daus. The second Dfas PdsA is more related to the cluster containing Ppal PdsA. These data suggest that two PdsA genes were present in the last common ancestor (LCA) to groups 1 and 2; one gene was lost in Ppal, whereas the other was duplicated in Asub. The LCA to groups 3 and 4 contained a single PdsA, which was duplicated twice in group 4.
Ddis PdsA is inhibited by a secreted inhibitor protein, PdiA, which, apart from a hydrophobic leader peptide, also contains five 24-amino acid (aa) cysteine-rich repeats that are found in large numbers in extracellular matrix proteins, such as EcmA and EcmB (Williams et al., 1987; Wu and Franke, 1990). These repeats were recently classified as Dicty-CTDC domains (PFAM accession number PF00526). BLAST queries with PdiA identified two and four PdiA-like genes in the genomes of the group 4 taxa Ddis and Dpur, respectively (Fig. 1C). However, in group 1, 2 and 3 genomes only large EcmA-like proteins were hit. The two Dpur proteins are the closest homologues of Ddis PdiA, but seem to have fewer CTDC domains. However, the aligned Ddis and Dpur sequences show only small variations over the regions that encompass the missing repeats (supplementary material Fig. S2) and high (62%) sequence identity over the entire length of the three proteins. In short, it appears that Dpur probably has one or two functional PdiAs, but that PdiA is not present in group 1, 2 or 3 taxa.
Functional analysis of a group 3 PdsA
The partial PdsA PCR product from the group 3 taxon Dmin was used to screen a Dmin λZAP genomic library. Six positive clones were identified, which could be assembled into a 9.02 kb contig (supplementary material Fig. S3). This contig contains Dmin PdsA and three flanking genes, which are most similar to the Ddis genes rbm8A, DDB_G0286003 and AbcF1, which occupy the same positions relative to Ddis PdsA on chromosome 4, indicating that Dmin PdsA is a true orthologue of Ddis PdsA.
To investigate whether the PdsAs from taxa that do not use cAMP as attractant are functional cAMP phosphodiesterases, we expressed the Dmin PdsA gene from the constitutive A15 promoter in the Ddis pdsA-null mutant Uk7. This mutant is defective in aggregation and fruiting body formation, and is fully restored by expression of Ddis PdsA (Sucgang et al., 1997). At a high cell density, the Uk7 cells formed a few small aggregates and misshapen fruiting bodies (Fig. 2A). Aggregation of Uk7/A15::Dmin PdsA cells, though still incomplete, was improved and more normal fruiting bodies were formed. At three- to tenfold lower cell densities, A15::Dmin PdsA became progressively less efficient in restoring Uk7 aggregation (data not shown).
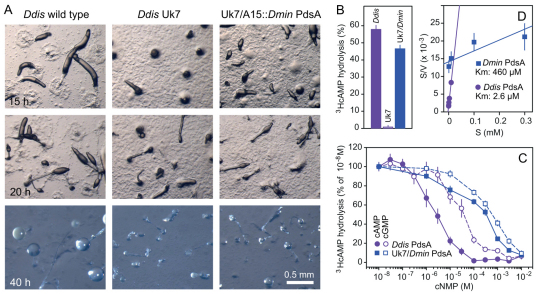
Fig. 2.
Complementation of a D. discoideum pdsA– mutant by D. minutum PdsA. (A) Restoration of development. The Ddis pdsA– mutant Uk7 was transformed with a gene fusion of the constitutive Ddis A15 promoter and a Dmin PdsA cDNA. The Uk7 parent strain DH1 (wild type), Uk7 and Uk7/A15::DminPdsA were incubated at 1.5×106 cells/cm2 on NN agar and photographed every 5 hours until fruiting formation no longer progressed. Scale bar: 0.5 mm. (B) PdsA activity. Wild-type, Uk7 and Uk7/A15::DminPdsA cells were starved for 6 hours and subsequently assayed for hydrolysis of 100 nM 3HcAMP at 8×105 cells/ml. (C) Competition curves. Starved DH1 and Uk7/A15::DminPdsA cells were incubated for 30 minutes with 10 nM 3HcAMP and increasing concentrations of cAMP or cGMP and assayed for 3HcAMP hydrolysis. Results are plotted as percentage of hydrolysis obtained at 10 nM 3HcAMP. (D) Hanes plot. Data obtained from the competition experiments were converted into absolute conversion rates (V; moles 5′AMP/minute107 cells). The ratio of S (cAMP concentration) over V was plotted against S. After linear regression analysis this yields a curve with an X-intercept equal to –Km. All experiments represent mean and s.e.m. of at least two experiments performed in triplicate.
To assess whether Dmin PdsA is fully functional, we measured the activity of the expressed enzyme. Figure 2B shows that intact Uk7/A15::DminPdsA cells hydrolyse 100 nM 3HcAMP almost as rapidly as wild-type Ddis cells, whereas Uk7 cells show only negligible activity. To determine whether Dmin PdsA has a similar affinity and selectivity for cAMP as Ddis PdsA, we measured hydrolysis of 10 nM 3HcAMP in the presence of increasing concentrations of unlabelled cAMP or cGMP. Figure 2C shows that about 200-fold higher cAMP concentrations are required to compete for 3HcAMP hydrolysis by Dmin PdsA than for Ddis PdsA, and Dmin PdsA has also lost most of its selectivity for cAMP over cGMP. Km values for Ddis and Dmin PdsA are 2 μM and 460 μM, respectively, as derived from a Hanes plot (Fig. 2D) (Biswanger, 2002). The poor ability of Dmin PdsA to restore Uk7 aggregation is very likely to be a result of its low affinity for cAMP.
Developmental regulation of PdsA expression
Ddis PdsA is transcribed from three different promoters during growth, aggregation and post-aggregative development that yield mRNAs of 1.9, 2.4 and 2.2 kb, respectively (Faure, et al., 1990). For at least one synchronously developing species in each of the taxon groups, we investigated whether its PdsA gene was under similar complex regulation (Fig. 3). The Ddis 2.4 and 2.2 kb mRNA bands were not clearly resolved, but at least two differentially regulated PdsA mRNA species are present in another group 4 taxon Dros. Group 3 Dmin PdsA mRNA appears shortly after aggregation. Ppal in group 2 transcribes a >2.2 kb mRNA throughout development and a <1.9 kb mRNA after aggregation. In group 1, Daus transcribes two mRNA species during growth and early development, with one transcript re-appearing during aggregation, whereas Dfas transcribes two or three mRNAs during growth and retains the largest during post-aggregative development. It should be noted that northern blots are indicative, but not reliably diagnostic for mRNA heterogeneity; mRNAs of similar size might not be resolved, and band displacement artefacts can occur when mRNAs have the same size as the highly abundant ribosomal RNAs. This seems to be the case for the two identically regulated Dmin mRNA bands. Nevertheless, there appears to be considerable heterogeneity in the complexity of PdsA transcription that does not show any obvious group-specific trend.
Fig. 3.
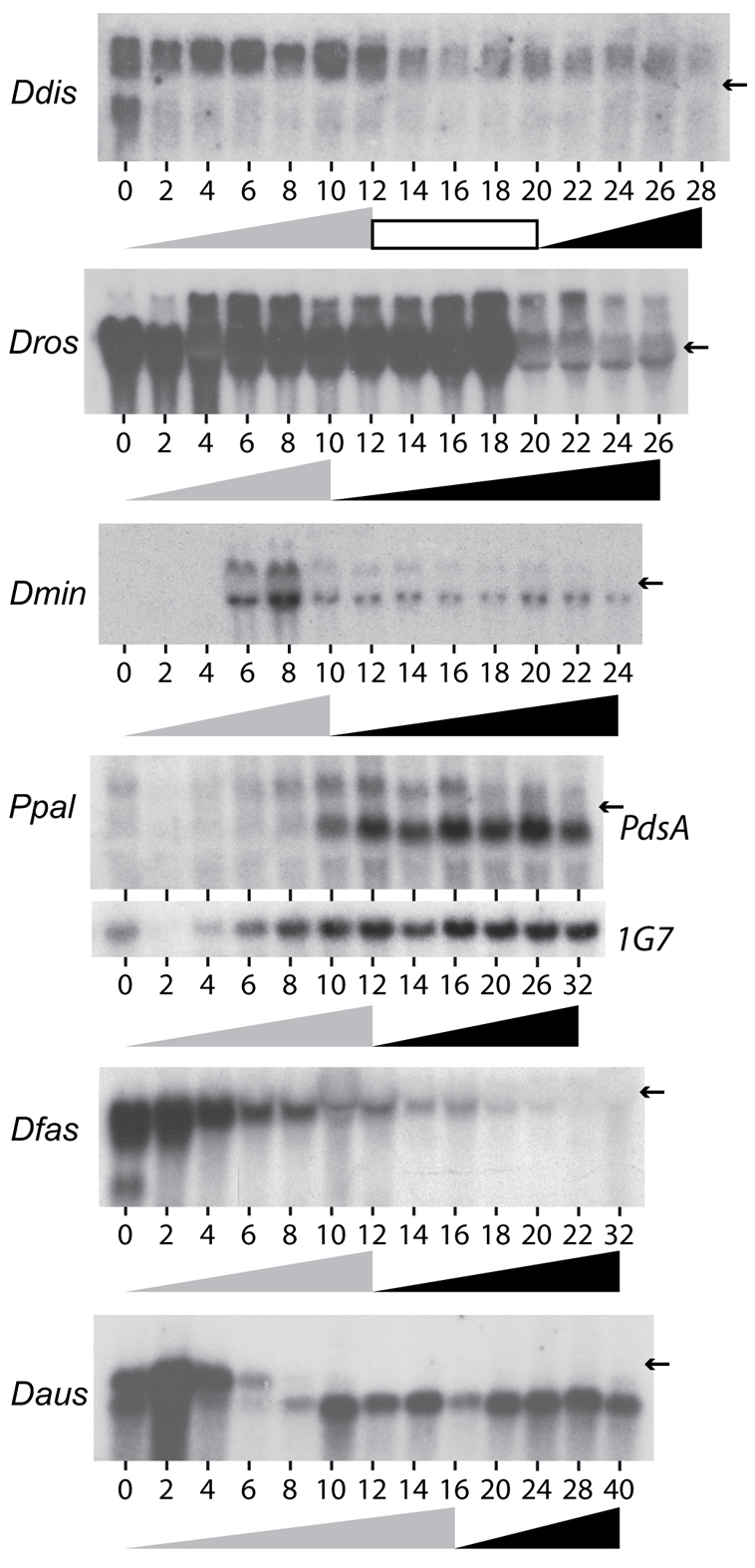
Developmental regulation of PdsA transcription in group-representative taxa. Ddis, Dros, Dmin, Ppal, Dfas and Daus cells were freed from bacteria and incubated at 22°C on NN agar until fruiting bodies had formed. Total RNA was extracted at 2-hour intervals and northern transfers were probed at high stringency with [32P]dATP-labelled PCR products amplified from the PdsA genes in each of the species. The same blots were re-probed at medium stringency with the constitutively expressed Ddis IG7 gene. All transfers showed equal sample loading (data not shown), except Ppal, for which the first four slots received less total RNA. The progression of aggregation and fruiting body formation are outlined underneath the time courses by grey and black triangles, respectively, and an intervening migrating slug stage is indicated by a white box. Arrows indicate the position of the 1.9 kb 17S rRNA band.
P. pallidum PdsA gene structure and expression pattern of isoforms
Ppal is thus far the only non-group 4 species that is amenable to genetic manipulation and we therefore focussed research into the role and regulation of PdsA on this species. To define the promoters and determine the sequences of the two PdsA mRNAs that are transcribed in Ppal (Fig. 3), we performed RT-PCR and oligo-capping RACE on mRNA isolated at 12 hours of development. Four different PdsA mRNA sequences were obtained, encoding PdsA proteins with different N-termini (Fig. 4, supplementary material Fig. S5). The shortest 1.35 kb mRNA encodes a 415 aa protein (PdsA415) that is somewhat smaller than Ddis PdsA (452 aa). Ppal PdsA415 has no obvious N-terminal signal peptide (supplementary material Fig. S1), which is essential for cell surface and extracellular localisation of Ddis PdsA (Franke et al., 1991). Of the others, the 2.55 and 2.81 kb mRNAs encode the same 802 aa protein (PdsA802), owing to extension of the N-terminal region, which now also includes a signal peptide. The position of the first intron downstream of the start codon is the same in the longer mRNAs, but the upstream intron boundaries vary. The longest 2.82 kb mRNA contains an ATG in the first exon, but this open reading frame terminates immediately at the second exon, suggesting that this mRNA is not functional.
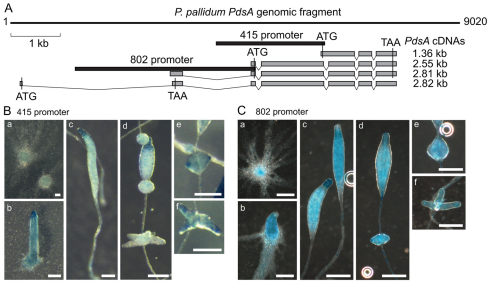
Fig. 4.
Structure and expression patterns of the Ppal PdsA gene. (A) Gene structure. Four different PdsA mRNA sequences were obtained by RT-PCR and oligo-capping RACE (Maruyama and Sugano, 1994) performed on Ppal mRNA isolated at 12 hours of development. The exons that make up the four transcripts are outlined in grey boxes and the positions of the putative start and stop codons are indicated. The 1.36 kb mRNA encodes a protein of 415 aa, whereas the 2.55 and 2.81 kb mRNAs yield a 802 aa protein. The largest mRNA has its start and stop codons in the first and second exon, respectively. The putative promoter sequences for the 415 and 802 aa proteins are outlined by black boxes. (B,C) Spatial regulation of promoter activity. The 415 and 802 promoter regions as outlined in panel A were fused to lacZ, yielding P415-gal and P802-gal, respectively, and expressed in Ppal. The P415-gal (B) and P802-gal (C) cells were developed on NN agar and intact developing structures were fixed and stained with X-gal. a, aggregate; b, tip formation; c, primary sorogen formation; d, segregation of whorls; e,f, formation of secondary sorogens. Scale bars: 0.1 mm.
We next examined the spatial expression patterns of the PdsA415 and PdsA802 isoforms. For the PdsA415 isoform, we amplified 2.18 kb of 5′UTR, upstream from its putative start codon (Fig. 4A, 415 promoter) and for the PdsA802 isoform we amplified 3.62 kb of 5′UTR upstream of the putative start codon in the 2.55 and 2.81 kb mRNAs (Fig. 4A, 802 promoter). These sequences were fused to the lacZ reporter gene, yielding P415-gal and P802-gal, and transformed into Ppal cells. Intact developing structures were stained with X-Gal to visualise the lacZ expression pattern (Fig. 4B,C). P415-gal was first expressed weakly in streaming aggregates (Fig. 4Ba). In the newly formed sorogen, expression was strongest at the tip region (Fig. 4Cb), disappearing almost completely from cells below the tip (Fig. 4Bc,d). The secondary sorogens that are formed from cell masses at the rear of the main sorogen usually showed enriched 415-gal expression at their tips (Fig. 4Be,f). P802-gal was already expressed in aggregation streams (Fig. 4Ca,b) and remained expressed in the whole structure throughout fruiting body formation, with somewhat higher expression in the tip region (Fig. 4Dc-f). It should be noted that P802-gal could contain the promoters for both the 2.55 and 2.81 kb mRNAs.
Developmental role of Ppal PdsA
To understand the role of PdsA in species that do not use cAMP for aggregation, we disrupted the single PdsA gene of Ppal by homologous recombination (supplementary material Fig. S4). The pdsA– cells showed normal aggregation with inflowing streams of amoebas (compare Fig. 5A and 5B). However, thereafter, the formation of slugs and fruiting bodies was slow and aberrant. Only small club-like structures were formed (Fig. 5C), without an obvious stalk or spore head, or the whorls of side branches that are characteristic for this species (Fig. 5D). The aberrant pdsA– fruiting structures were stained with Calcofluor, which stains the cellulose-rich wall of mature spores and stalk cells, to assess whether terminal cell differentiation had occurred. Wild-type fruiting bodies form stalks that are one cell thick, and elliptical spores (Fig. 5E). In pdsA– fruiting bodies, the stalk cells are more disorganised, and the sori or the cell masses that are clinging to the stalk contain encapsulated round cells (Fig. 5F). Ppal amoebas can differentiate directly into spherical cysts without aggregating. However, electron-microscopic examination of the pdsA–spores revealed that they display the condensed cytoplasm and thick three-layered cell wall that characterises wild-type spores (compare Fig. 5G and 5I). Cysts are less dense and are surrounded by a thinner two-layered cell wall (Fig. 5H) (Hohl et al., 1970; Kawabe et al., 2009).
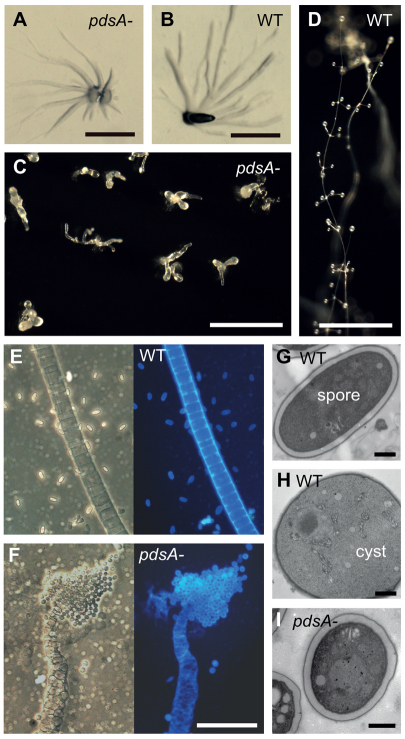
Fig. 5.
Phenotype of the P. pallidum pdsA– mutant. (A-D) Multicellular structures. Wild-type (WT) and pdsA– cells were harvested from bacterial growth plates, plated on water agar or charcoal agar, and incubated at 22°C until aggregates (A,B) had formed at 12 hours, or mature fruiting bodies (C,D) at 38 hours. Scale bars: 0.5 mm. (E,F) Terminal cell differentiation. Fruiting bodies of wild-type (E) and pdsA– (F) Ppal cells were transferred to a droplet of 0.001% Calcofluor on a slide glass and photographed under phase contrast (left) and UV illumination (right). Scale bar: 50 μm. (G-I) Fruiting structures were prepared for transmission electron microscopy as described previously (Kawabe et al., 2009) and sectioned. The images show an elliptical wild-type spore (G) and spherical wild-type cyst (H), and a spherical pdsA– spore (I). Scale bars: 1 μm.
We examined next whether the phenotype of the pdsA– mutant is cell autonomous by mixing pdsA– and wild-type cells at different ratios and allowing the mixtures to form fruiting bodies (Fig. 6). Chimeric fruiting bodies with 50% wild-type cells showed wild-type morphology (compare Fig. 6A and 6B). With only 10% wild-type cells, the chimeric fruiting bodies showed normal spore heads and main stalks. However, the number of side branches on fruiting bodies was reduced (Fig. 6C). Calcofluor staining revealed that elliptical spore morphology and regular stalk formation were fully restored by mixing pdsA– with 10% wild-type cells (Fig. 6D). We also measured the ratio of wild-type to pdsA– spores in the chimeric fruiting bodies. Figure 6E shows that chimeric fruiting bodies formed from mixtures with 10% or 50% wild-type cells contained 9% or 47% wild-type spores, respectively. This means that the tendency of wild-type and pdsA– cells to differentiate into either spore or stalk cells in chimeric structures is almost the same. Combined, these data show that PdsA is required for post-aggregative morphogenesis and spore elongation in Ppal in a non-cell autonomous manner.
Fig. 6.
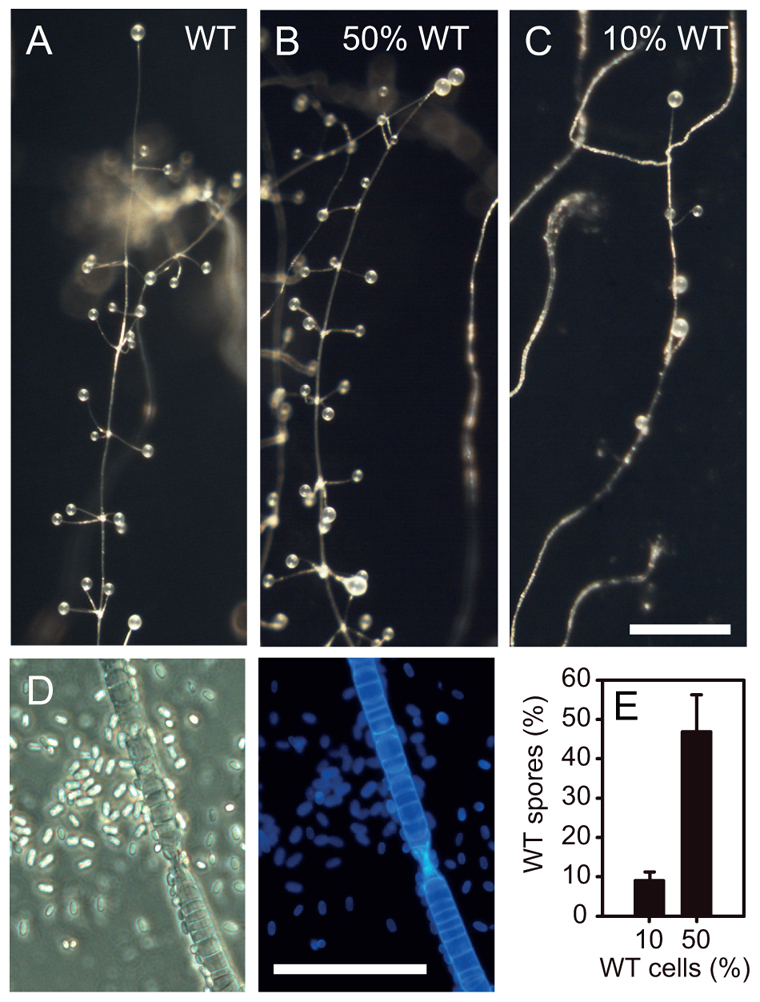
Synergistic development of pdsA– and wild-type cells. (A-D) Pdsa– cells were mixed with wild-type cells at ratios of 0:1 (A), 1:1 (B) and 9:1 (C,D) and plated on charcoal agar to develop into fruiting bodies. To observe spore and stalk cell morphology (D) structures were stained with Calcofluor. Even with only 10% wild-type cells, which partially rescued fruiting body formation, normal stalk and elliptical spore cell differentiation were fully restored. Scale bars: in A-C, 0.5 mm; in D, 50 μm. (E) Spores were harvested from chimeric fruiting bodies and clonally plated with K. aerogenes on agar. The number of clones showing wild-type or pdsA– fruiting body morphology were counted and recalculated to express the percentage of wild-type to total spores. Mean and s.d. of three experiments are shown.
Activity and cellular localisation of the P. pallidum PdsA
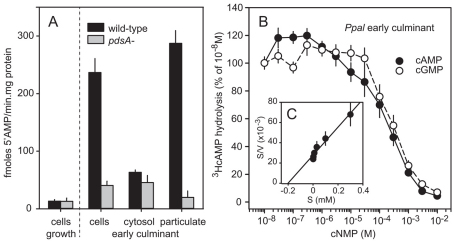
The non-cell autonomous phenotype of the Ppal pdsA-null mutant suggests that PdsA is only essential for extracellular cAMP signalling, despite the fact that the PdsA415 isoform does not have a signal peptide. To confirm that the Ppal pdsA-null mutant is defective in extracellular cAMP hydrolysis, we measured 3HcAMP hydrolysis in intact and fractionated cells from early culminants. Figure 7A shows that wild-type culminant cells hydrolyse 3HcAMP about six times faster than pdsA– culminant cells. In fractionated cells, all PdsA specific 3HcAMP hydrolysis is localised in the particulate fraction, indicating that all measured PdsA activity is membrane associated. Growth stage cells show no PdsA specific 3HcAMP hydrolysis. We also determined the Km and substrate selectivity of Ppal PdsA (Fig. 7B,C). Similar to Dmin PdsA, the affinity of Ppal PdsA for cAMP is very low (Km: 200 μM) and the enzyme is not selective for cAMP over cGMP.
Fig. 7.
Localisation and substrate specificity of Ppal PdsA. (A) Stage specificity and localisation. Ppal wild-type or pdsA– cells were harvested at 0 hours (growth stage) or 16 hours of development (early culminants) and assayed for 3HcAMP hydrolysis as intact cells, or cells were lysed and assayed as separated soluble and particulate fractions. 5′AMP production was standardised on the protein content of the cell suspension or cell lysate. (B) Selectivity and affinity. Intact early culminant cells were incubated for 30 minutes with 10 nM 3HcAMP and increasing concentrations of cAMP or cGMP and assayed for 3HcAMP hydrolysis. Results are plotted as percentage of hydrolysis obtained at 10 nM 3HcAMP. (C) Hanes plot. Data from B were recalculated into absolute conversion rates (V) and plotted as a Hanes plot to estimate the Km of Ppal PdsA. All experiments represent mean and s.e.m. of at least two experiments performed in triplicate.
DISCUSSION
PdsA gene regulation only partially reflects its utilisation
PdsA is an essential component of the network that generates cAMP oscillations and enables aggregation and morphogenesis of the model organism D. discoideum (Wu et al., 1995; Sucgang et al., 1997). Its inhibitor protein PdiA is instrumental for long range propagation of spiral cAMP waves in a field of starving cells (Palsson and Cox, 1996; Palsson et al., 1997), but is not expressed during post-aggregative development (Franke et al., 1991). PdsA genes could be identified in all selected test species across the four Dictyostelid taxon groups, indicating that PdsA is likely to be conserved in all Dictyostelia. By contrast, PdiA was only detected in group 4 taxa (Fig. 1).
Ddis PdsA is transcribed as three alternatively spliced mRNA species from three different promoters, which direct expression during growth, aggregation and in the anterior prestalk region of multicellular structures, respectively (Faure et al., 1990; Hall et al., 1993). Group 1 and 2 taxa also transcribed two to three mRNA species, of which at least one was already expressed during growth and aggregation. Only the group 3 taxon Dmin did not express PdsA during growth and early aggregation (Fig. 3). This evolutionary progression in expression patterns differs strongly from that of the cAR genes, which are also essential for oscillatory cAMP signalling. cARs are expressed after aggregation in group 1 and 2 taxa, during both aggregation and late development in group 4 taxa and during growth and late development in Dmin (Alvarez-Curto et al., 2005). At least for groups 1 and 2, the cAR expression pattern is consistent with its exclusive role in post-aggregative development (Kawabe et al., 2009). However, both the group 3 cAR and group 1 and 2 PdsAs, are expressed at stages at which they do not appear to be required.
PdsA is essential for post-aggregative development in non-group 4 species
Disruption of the PdsA gene in the group 2 taxon Ppal did not affect aggregation, but resulted in formation of highly abnormal fruiting bodies with disorganised stalk cells and round spores. This phenotype resembles that of a Ppal car-null mutant, but there are also differences. The car– spores turned out to be cysts (Kawabe et al., 2009), whereas the pdsA– spores showed otherwise normal spore morphology (Fig. 5I). Additionally, in contrast to the car– phenotype, the pdsA– phenotype is non-cell autonomous, as both fruiting body formation and elliptical spore morphology can be restored by mixing pdsA– cells with wild-type cells (Fig. 6).
The pdsA– and car– phenotypes substantiate conclusions drawn from pharmacological experiments that all non-group 4 taxa use oscillatory cAMP signalling for organisation of post-aggregative morphogenesis, but not for aggregation (Alvarez-Curto et al., 2005; Kawabe et al., 2009) (A. Skiba and P.S., unpublished). The non-cell autonomous nature of the pdsA– phenotype indicates that PdsA is involved in mediating cAMP signalling between cells, but not in transduction of the cAMP signal, as is the case for cARs. It is unclear what causes the round spore morphology. The car– mutants formed cysts in their fruiting bodies, because they lack cAR-mediated cAMP induction of pre-spore differentiation, but have retained PKA-mediated induction of encystation (Kawabe et al., 2009). The pdsA– structures are likely to contain excessively high extracellular cAMP levels, which possibly accelerate spore wall assembly relative to spore elongation.
PdsA is an opportunistic cAMP phosphodiesterase
In contrast to the Dmin cAR, which can fully complement the aggregation deficient phenotype of a Ddis car-null mutant (Kawabe et al., 2009), Dmin PdsA can only partially rescue aggregation of a Ddis pdsA– mutant (Fig. 2). This is most likely due to its 200-fold reduced affinity for cAMP, which it shares with the Ppal PdsA from group 2 (Fig. 7B). Both the Dmin and Ppal PdsAs show no selectivity for cAMP over cGMP. PdsA is a PDE-II type enzyme, which have a catalytic domain that is similar to that of the metallo-beta lactamases, but not to the metazoan PDE-I domain (Conti and Beavo, 2007). In fungi, PDE-II enzymes generally have a lower affinity for cAMP than do PDE-I enzymes (Nikawa et al., 1987; Zhang et al., 2011). PDE-II type genes are present in most major clades of bacteria, but only two genes have been analysed functionally. Vibrio fischeri, a symbiotic marine bacterium, exposes a PDE-II type enzyme, CpdP, outward into the periplasm. This enzyme hydrolyses cyclic nucleotides indiscriminately at a very high rate, supposedly to enable V. fischeri to utilise cyclic nucleotides, secreted by its hosts, as a carbon source (Callahan et al., 1995). Myxococcus xanthus PdeE is the other characterised PDE-II. Apart from cAMP and cGMP, it also hydrolyses 5′nucleotides (Kimura et al., 2011). Evidently, the roles of prokaryote PDE-II enzymes are not confined, or perhaps not even related, to cAMP signal transduction.
Ppal and Dmin PdsA show Kms for cAMP of 0.2 and 0.46 mM respectively, indicating a 100-200 fold lower affinity for their substrate as Ddis PdsA (Fig. 2D, Fig. 7B). Combined with their lack of selectivity for cAMP, this strongly suggests that PdsA originally had other substrates, or is a broad-spectrum hydrolase. One can only guess what its actual substrate is or might have been. Because PdsA is exposed on the cell surface, roles in environmental signalling or detoxification seem likely. Such roles might explain why so many species express PdsA during growth (Fig. 3), without there being an obvious requirement for secreted cAMP at this stage. The low affinity of the non-group 4 PdsAs for cAMP might not be problematic for its role in post-aggregative development, where cAMP released between the tightly packed cells can easily reach the supramicromolar concentrations that are, for example, required for pre-spore gene induction (Schaap and Van Driel, 1985; Oyama and Blumberg, 1986). However, in a field of dispersed starving cells that respond chemotactically to as little as 0.1 nM cAMP (Van Haastert and Konijn, 1982), an enzyme with a Km of 0.4 mM will not be efficacious to generate steep chemotactic gradients. The 200-fold increase in affinity for cAMP of the group 4 PdsAs is therefore likely to have been an essential adaptation to enable cAMP-mediated aggregation in this group.
Improvement of cAMP signalling dynamics by recruitment of a matrix protein
The evolutionary history of the PdsA inhibitor PdiA is equally intriguing. PdiA consists of eight 24 aa cysteine-rich repeats, of which five are recognised as Dicty-CTDC domains and the other three are degenerate CTDC domains (supplementary material Fig. S2). Apart from a few prokaryote proteins, CTDC domains are unique for Dictyostelia. They are typically present in long arrays in extracellular matrix proteins, such as ecmA and ecmB (McRobbie et al., 1988), cysteine oxidation of which will form intra- and interprotein crosslinks. The Ddis genome contains 20 CTDC domain proteins; most include a signal peptide and ten include a PA14 domain with a proposed role in carbohydrate binding (http://dictybase.org/). The Ppal and Dfas genomes contain at least 25 and 19 CTDC proteins, respectively (H. Lawal and P.S., unpublished results). Three of the Ddis proteins, PsiA, and PsiF(DicA) and cyrA, act as external signals controlling gene expression and cell motility (Kolbinger et al., 2005; Yamada et al., 2010; Suarez et al., 2011). Evidently, CTDC domain-containing genes were strongly amplified in early Dictyostelid evolution, and from this pool of matrix components, genes with novel signalling roles emerged. This scenario bears similarity to the evolution of the hedgehog proteins, important signal molecules in animal development, which originated from a cleavable substrate adhesion protein in the unicellular choanoflagellate ancestor (Snell et al., 2006).
PdiA probably emerged late in Dictyostelid evolution, as a putative duplicated orthologue could only be recognised in Dpur, another group 4 species (Fig. 1C). PdiA was theoretically predicted and experimentally confirmed to generate the conditions that allow spiral cAMP waves, which cover large territories, to form in preference to concentric waves, which organise fewer cells into aggregates (Palsson and Cox, 1996; Palsson et al., 1997). Group 4 species stand out by forming relatively large aggregates and fruiting structures (Schaap et al., 2006). It is not unlikely that they owe this feature to recruitment of PdiA from the pool of matrix proteins.
Opportunistic exploitation of existing genes limits gene number expansion
The evolution of the PdsA-PdiA system illustrates in a fascinating manner how a sophisticated intercellular communication system, capable of organising the movement of up to a million cells, emerged by modification of existing genes with fairly mundane or redundant functions. These modifications involved both elaboration of promoter regions as well as mutations in the coding sequence that allowed a non-selective low affinity phosphodiesterase to selectively and efficiently hydrolyse cAMP, and a matrix protein to bind to this enzyme with nanomolar affinity and regulate its activity. The extreme versatility by which evolutionary processes can multiply the functionality of existing genes probably explains why dictyostelids and humans, with their complex multicellular lifestyles, do not require a proportionally greater gene pool than their unicellular ancestors.
Supplementary Material
Acknowledgments
We thank Dr Daniel Rozen, who amplified the Dmuc and Dros PdsA genes in an early phase of the project, as well as the Asub genome sequencing project for providing gene sequences. We thank John James and Dr Alan Prescott for expert electron microscopy.
Footnotes
Funding
This research was funded by Biotechnology and Biological Sciences Research Council (BBSRC) project grants [BB/D013453/1, BB/E016308/1 and BB/G020426/1] and a Wellcome Trust Project grant [090276]. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.077099/-/DC1
References
- Alvarez-Curto E., Rozen D. E., Ritchie A. V., Fouquet C., Baldauf S. L., Schaap P. (2005). Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc. Natl. Acad. Sci. USA 102, 6385–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar A., Groth M., Siol O., Gaube F., Enzensperger C., Glockner G., Winckler T. (2012). Developmental gene regulation by an ancient intercellular communication system in social amoebae. Protist 163, 25–37 [DOI] [PubMed] [Google Scholar]
- Bader S., Kortholt A., Van Haastert P. J. M. (2007). Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP. Biochem. J. 402, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswanger H. (2002.). Enzyme kinetics. Principles and methods. Weinheim, Germany: Wiley-VCH; [Google Scholar]
- Cai H., Das S., Kamimura Y., Long Y., Parent C. A., Devreotes P. N. (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol. 190, 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan S. M., Cornell N. W., Dunlap P. V. (1995). Purification and properties of periplasmic 3′:5′-cyclic nucleotide phosphodiesterase. J. Biol. Chem. 270, 17627–17632 [DOI] [PubMed] [Google Scholar]
- Carfi A., Pares S., Duee E., Galleni M., Duez C., Frere J. M., Dideberg O. (1995). The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14, 4914–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B. (2005). Evolution at two levels: On genes and form. PLoS Biol. 3, e245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M., Beavo J. (2007). Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Ann. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- Darmon M., Barra J., Brachet P. (1978). The role of phosphodiesterase in aggregation of Dictyostelium discodeum. J. Cell Sci. 31, 233–243 [DOI] [PubMed] [Google Scholar]
- De Wit R. J. W., Konijn T. M. (1983). Identification of the acrasin of Dictyostelium minutum as a derivative of folic acid. Cell Differ. 12, 205–210 [DOI] [PubMed] [Google Scholar]
- Dingermann T., Reindl N., Werner H., Hildebrandt M., Nellen W., Harwood A., Williams J., Nerke K. (1989). Optimization and in situ detection of Escherichia coli beta-galactosidase gene expression in Dictyostelium discoideum. Gene 85, 353–362 [DOI] [PubMed] [Google Scholar]
- Dormann D., Weijer C. J. (2001). Propagating chemoattractant waves coordinate periodic cell movement in Dictyostelium slugs. Development 128, 4535–4543 [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J. A., Glockner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure M., Franke J., Hall A. L., Podgorski G. J., Kessin R. H. (1990). The cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum contains three promoters specific for growth, aggregation, and late development. Mol. Cell. Biol. 10, 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J., Podgorski G. J., Kessin R. H. (1987). The expression of two transcripts of the phosphodiesterase gene during the development of Dictyostelium discoideum. Dev. Biol. 124, 504–511 [DOI] [PubMed] [Google Scholar]
- Franke J., Faure M., Wu L., Hall A. L., Podgorski G. J., Kessin R. H. (1991). Cyclic nucleotide phosphodiesterase of Dictyostelium discoideum and its glycoprotein inhibitor: Structure and expression of their genes. Dev. Gen. 12, 104–112 [DOI] [PubMed] [Google Scholar]
- Fukushima S., Maeda Y. (1991). Whorl formation in Polysphondylium violaceum: Relevance to organization by cyclic AMP. Dev. Growth Differ. 33, 525–533 [DOI] [PubMed] [Google Scholar]
- Galardi-Castilla M., Garciandía A., Suarez T., Sastre L. (2010). The Dictyostelium discoideum acaA gene is transcribed from alternative promoters during aggregation and multicellular development. PLoS ONE 5, e13286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. L., Franke J., Faure M., Kessin R. H. (1993). The role of the cyclic nucleotide phosphodiesterase of Dictyostelium discoideum during growth, aggregation, and morphogenesis: Overexpression and localization studies with the separate promoters of the pde. Dev. Biol. 157, 73–84 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98 [Google Scholar]
- Harwood A. J., Drury L. (1990). New vectors for expression of the E. coli lacZ gene in Dictyostelium. Nucleic Acids Res. 18, 4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel A., Lawal H., Felder M., Schilde C., Helps N., Tunggal B., Rivero F., John U., Schleicher M., Eichinger L., et al. (2011). Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 21, 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl H. R., Miura-Santo L. Y., Cotter D. A. (1970). Ultrastuctural changes during formation and germination of microcysts in Polysphondylium pallidum, a cellular slime mould. J. Cell Sci. 7, 285–306 [DOI] [PubMed] [Google Scholar]
- Human Genome Sequencing Consortium (2004). Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 [DOI] [PubMed] [Google Scholar]
- Kawabe Y., Enomoto T., Morio T., Urushihara H., Tanaka Y. (1999). LbrA, a protein predicted to have a role in vesicle trafficking, is necessary for normal morphogenesis in Polysphondylium pallidum. Gene 239, 75–79 [DOI] [PubMed] [Google Scholar]
- Kawabe Y., Morio T., James J. L., Prescott A. R., Tanaka Y., Schaap P. (2009). Activated cAMP receptors switch encystation into sporulation. Proc. Natl. Acad. Sci. USA 106, 7089–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Yoshimi M., Takata G. (2011). Enzymatic and mutational analyses of a class II 3′,5′-cyclic nucleotide phosphodiesterase, PdeE, from Myxococcus xanthus. J. Bacteriol. 193, 2053–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbinger A., Gao T., Brock D., Ammann R., Kisters A., Kellermann J., Hatton D., Gomer R. H., Wetterauer B. (2005). A cysteine-rich extracellular protein containing a PA14 domain mediates quorum sensing in Dictyostelium discoideum. Eukaryotic Cell 4, 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama H., Yanagida T., Ueda M. (2008). DNA oligonucleotide-assisted genetic manipulation increases transformation and homologous recombination efficiencies: evidence from gene targeting of Dictyostelium discoideum. J. Biotechnol. 133, 418–423 [DOI] [PubMed] [Google Scholar]
- Lee S., Comer F. I., Sasaki A., McLeod I. X., Duong Y., Okumura K., Yates J. R., Parent C. A., Firtel R. A. (2005). TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16, 4572–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J. M., Saxe C. L., III, Kimmel A. R. (1993). Two transmembrane signaling mechanisms control expression of the cAMP receptor gene cAR1 during Dictyostelium development. Proc. Natl. Acad. Sci. USA 90, 5969–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sugano S. (1994). Oligo-capping:a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138, 171–174 [DOI] [PubMed] [Google Scholar]
- McRobbie S. J., Jermyn K. A., Duffy K., Blight K., Williams J. G. (1988). Two DIF-inducible, prestalk-specific mRNAs of Dictyostelium encode extracellular matrix protein of the slug. Development 104, 275–284 [DOI] [PubMed] [Google Scholar]
- Meima M. E., Weening K. E., Schaap P. (2007). Vectors for expression of proteins with single or combinatorial fluorescent protein and tandem affinity purification tags in Dictyostelium. Protein Expr. Purif. 53, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Sass P., Wigler M. (1987). Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 3629–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M., Blumberg D. D. (1986). Interaction of cAMP with the cell-surface receptor induces cell-type specific mRNA accumulation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 83, 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson E. (2009). A cAMP signaling model explains the benefit of maintaining two forms of phosphodiesterase in Dictyostelium. Biophys J. 97, 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson E., Cox E. C. (1996). Origin and evolution of circular waves and spirals in Dictyostelium discoideum territories. Proc. Natl. Acad. Sci. USA 93, 1151–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson E., Lee K. J., Goldstein R. E., Franke J., Kessin R. H., Cox E. C. (1997). Selection for spiral waves in the social amoebae Dictyostelium. Proc. Natl. Acad. Sci. USA 94, 13719–13723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper K. B. (1984). The Dictyostelids. Princeton, New Jersey: Princeton University Press; [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Schaap P., Van Driel R. (1985). Induction of post-aggregative differentiation in Dictyostelium discoideum by cAMP. Evidence of involvement of the cell surface cAMP receptor. Exp. Cell Res. 159, 388–398 [DOI] [PubMed] [Google Scholar]
- Schaap P., Konijn T. M., Van Haastert P. J. M. (1984). cAMP pulses coordinate morphogenetic movement during fruiting body formation of Dictyostelium minutum. Proc. Natl. Acad. Sci. USA 81, 2122–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P., Winckler T., Nelson M., Alvarez-Curto E., Elgie B., Hagiwara H., Cavender J., Milano-Curto A., Rozen D. E., Dingermann T., et al. (2006). Molecular phylogeny and evolution of morphology in the social amoebas. Science 314, 661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O., Suthers H. L. B., Bonner J. T. (1982). Chemical identity of the acrasin of the cellular slime mold Polysphondylium violaceum. Proc. Natl. Acad. Sci. USA 79, 7376–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell E. A., Brooke N. M., Taylor W. R., Casane D., Philippe H., Holland P. W. H. (2006). An unusual choanoflagellate protein released by Hedgehog autocatalytic processing. Proc. R. Soc. Biol. Sci. 273, 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez A., Huber R. J., Myre M. A., O’Day D. H. (2011). An extracellular matrix, calmodulin-binding protein from Dictyostelium with EGF-like repeats that enhance cell motility. Cell Signal. 23, 1197–1206 [DOI] [PubMed] [Google Scholar]
- Sucgang R., Weijer C. J., Siegert F., Franke J., Kessin R. H. (1997). Null mutations of the Dictyostelium cyclic nucleotide phosphodiesterase gene block chemotactic cell movement in developing aggregates. Dev. Biol. 192, 181–192 [DOI] [PubMed] [Google Scholar]
- Sucgang R., Kuo A., Tian X., Salerno W., Parikh A., Feasley C. L., Dalin E., Tu H., Huang E., Barry K., et al. (2011). Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 12, R20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J. M., Konijn T. M. (1982). Signal transduction in the cellular slime molds. Mol. Cell. Endocrinol. 26, 1–17 [DOI] [PubMed] [Google Scholar]
- Williams J. G. (2006). Transcriptional regulation of Dictyostelium pattern formation. EMBO Rep. 7, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Ceccarelli A., McRobbie S., Mahbubani H., Kay R. R., Early A., Berks M., Jermyn K. A. (1987). Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell 49, 185–192 [DOI] [PubMed] [Google Scholar]
- Wray G. A., Hahn M. W., Abouheif E., Balhoff J. P., Pizer M., Rockman M. V., Romano L. A. (2003). The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 [DOI] [PubMed] [Google Scholar]
- Wu L., Franke J. (1990). A developmentally regulated and cAMP-repressible gene of Dictyostelium discoideum: cloning and expression of the gene encoding cyclic nucleotide phosphodiesterase inhibitor. Gene 91, 51–56 [DOI] [PubMed] [Google Scholar]
- Wu L., Franke J., Blanton R. L., Podgorski G. J., Kessin R. H. (1995). The phosphodiesterase secreted by prestalk cells is necessary for Dictyostelum discoideum. Dev. Biol. 167, 1–8 [DOI] [PubMed] [Google Scholar]
- Yamada Y., Minamisawa H., Fukuzawa M., Kawata T., Oohata A. A. (2010). Prespore cell inducing factor, psi factor, controls both prestalk and prespore gene expression in Dictyostelium development. Dev. Growth Differ. 52, 377–383 [DOI] [PubMed] [Google Scholar]
- Yeh R. P., Chan F. K., Coukell M. B. (1978). Independent regulation of the extracellular cyclic AMP phosphodiesterase-inhibitor system and membrane differentiation by exogenous cyclic AMP in Dictyostelium discoideum. Dev. Biol. 66, 361–374 [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu K., Zhang X., Tang W., Wang J., Guo M., Zhao Q., Zheng X., Wang P., Zhang Z. (2011). Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS ONE 6, e17241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Hu H., Lai M. (2010). Non-coding RNAs and their epigenetic regulatory mechanisms. Biol. Cell 102, 645–655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.