Abstract
Nitrogen catabolite repression (NCR) is a regulatory strategy found in microorganisms that restricts the utilization of complex and unfavored nitrogen sources in the presence of favored nitrogen sources. In fungi, this concept has been best studied in yeasts and filamentous ascomycetes, where the GATA transcription factors Gln3p and Gat1p (in yeasts) and Nit2/AreA (in ascomycetes) constitute the main positive regulators of NCR. The reason why functional Nit2 homologs of some phytopathogenic fungi are required for full virulence in their hosts has remained elusive. We have identified the Nit2 homolog in the basidiomycetous phytopathogen Ustilago maydis and show that it is a major, but not the exclusive, positive regulator of nitrogen utilization. By transcriptome analysis of sporidia grown on artificial media devoid of favored nitrogen sources, we show that only a subset of nitrogen-responsive genes are regulated by Nit2, including the Gal4-like transcription factor Ton1 (a target of Nit2). Ustilagic acid biosynthesis is not under the control of Nit2, while nitrogen starvation-induced filamentous growth is largely dependent on functional Nit2. nit2 deletion mutants show the delayed initiation of filamentous growth on maize leaves and exhibit strongly compromised virulence, demonstrating that Nit2 is required to efficiently initiate the pathogenicity program of U. maydis.
INTRODUCTION
Heterotrophic microorganisms are capable of utilizing a plethora of different carbon and nitrogen sources. Fungal saprophytes and bacteria strictly rely on the availability of (certain) organic carbon sources, while most have the ability to assimilate inorganic nitrogen in addition to utilizing organic nitrogen. The utilization of available nutrients by microorganisms is not unbiased, since carbon and nitrogen compounds that are easy to metabolize are preferred over more complex compounds that require a higher cost in terms of ATP and reducing equivalents (50).
Although the actual mechanisms or regulatory components of the utilization of complex nitrogen sources differ considerably in different classes of bacteria, the output of these signal transduction cascades is the same: the metabolization of complex nitrogen sources in the presence of a favored nitrogen source is repressed (reviewed in references 2 and 50). In eukaryotic organisms, this effect, called nitrogen catabolite repression (NCR) or nitrogen metabolite repression, has been best characterized in brewer's yeast (Saccharomyces cerevisiae) and in filamentous fungi of the phylum Ascomycota, where extensive work has been performed on Neurospora crassa and Aspergillus species (87).
In filamentous fungi, the availability of favored nitrogen sources (ammonium and Gln in Aspergillus nidulans; ammonium, Gln, and Glu in N. crassa) represses the utilization of other nitrogen sources, such as nitrate, peptides, or other free amino acids (57). The GATA transcription factors AreA and Nit2 are the central regulators of NCR and control the utilization of unfavored nitrogen sources in A. nidulans and N. crassa, respectively (reviewed in references 57 and 58). In the absence of favored nitrogen sources, Nit2 and AreA proteins activate the expression of nitrogen-metabolizing genes by binding to the respective promoters of these genes. Functional Nit2 proteins from ascomycetes harbor a highly conserved Zn finger domain in the C-terminal part of the protein as well as a Nit2/AreA-specific sequence motif (RMENLTWRMM) in the N-terminal part. The outermost C terminus is conserved as well (17, 87). The deletion of the nit2 or areA gene leads to the inability of filamentous fungi to utilize complex nitrogen sources (17, 47, 57, 66, 82). While Nit2/AreA proteins are required for the general derepression of nitrogen-metabolizing genes, additional pathway-specific activators are needed for the activation of most genes underlying NCR. For nitrate utilization, this activator is called Nit4 and NirA in N. crassa and A. nidulans, respectively, a Gal4-like transcription factor, which can only induce the expression of nitrate and nitrite reductase (NR) in the presence of Nit2/AreA (22). The regulation of AreA activity is best described in A. nidulans. Under N-sufficient conditions, AreA is evenly distributed between cytoplasm and nucleus and only strongly localizes to the nucleus under N starvation (80). Furthermore, AreA activity is regulated by mRNA stability (12, 62, 63), a positive autoregulation loop (49), and the negatively acting protein NmrA (3). No DNA-binding activity has been demonstrated for the N. crassa homolog Nmr1 (89), but Nmr1 interacts directly with Nit2 and thus inhibits the DNA binding of Nit2 (65, 88). The conserved outmost C terminus of Nit2 is required for the interaction with Nmr1 (65).
In yeasts, the utilization of complex nitrogen sources is mediated by two positively acting transcription factors: Gat1p, a closely related homolog of Nit2/AreA, and Gln3p, a GATA-type transcription factor that is not present in filamentous fungi (reviewed in references 15, 33, and 87). These two proteins act on a different subset of genes but also share commonly regulated genes.
The role of functional Nit2 homologs has been studied in plant pathogenic ascomycetes, where it also regulates nitrogen utilization and is required for full pathogenicity in most cases (9). Nit2 loss-of-function mutants of Colletotrichum lindemuthianum, Fusarium verticillioides, and Fusarium oxysporum showed reduced virulence on their respective hosts (17, 47, 66). The hemibiotrophic C. lindemuthianum clnr1 mutants were specifically affected in the transition from the biotrophic to the necrotrophic phase, while they showed no defects in the initial biotrophic phase itself (66). A depletion of Nit2 homologs in Magnaporthe grisea or in Cladosporium fulvum, on the other hand, had little or no effect on pathogenicity (23, 67). The reason why Nit2 homologs are required for full pathogenicity in some ascomycetous plant pathogens remains elusive. One common hypothesis is that plant pathogens are limited in nitrogen supply early in the infection process and that this limitation is an essential signal to start the infection cycle (74). This assumption is based on the observation that the expression of the C. fulvum effector Avr9 and the M. grisea pathogenicity factor Mpg1, as well as four other known M. grisea pathogenicity factors, are induced in axenic culture under N limitation (19, 77, 83). The expression of Avr9 is thereby under the control of the Nit2 homolog Nrf1 (67). However, only 1 out of 9 known C. fulvum effectors, Avr9, and only 5 out of 21 known M. grisea pathogenicity factors are induced by N limitation (19, 76, 77, 83). This implies on the one hand that cues other than N limitation play a role in starting the pathogenicity program and on the other hand that the reduced virulence observed in some Nit2 mutants is not solely an effect of reduced effector protein expression.
Although the regulation of nitrogen utilization has been well studied in ascomycetes and yeasts, only a few reports exist on these regulatory mechanisms in basidiomycete fungi. It is known that nitrate-metabolizing enzymes are transcriptionally repressed by ammonium in Hebeloma cylindrosporum and Ustilago maydis, but the mechanism behind this effect is unknown (4, 38, 39, 51). Gat1, the Nit2 homolog of the basidiomycete human pathogen Cryptococcus neoformans, regulates nitrogen utilization and glucuronoxylomannan secretion, but the deletion of Gat1 has no effect on virulence (48).
The basidiomycete Ustilago maydis is the causal agent of corn smut disease, and its dimorphic life style has been extensively characterized on a molecular level (see references 11 and 42 for excellent recent reviews on U. maydis). Haploid sporidia multiply by budding in a yeast-like manner and exhibit a saprophytic life style. After mating on hydrophobic surfaces, the growth of infectious filamentous dikaryotic hyphae is initiated. U. maydis then can infect its host, maize, via appressoria-like structures, and later it proliferates by intracellular and intercellular hyphae, inducing the formation of host-derived tumors. Since U. maydis shows a biotrophic lifestyle in planta, it does not kill its host but has to continuously acquire nutrients from living host tissue. We reported previously that the induction of tumors provides efficient means of rerouting organic nitrogen sources and soluble carbohydrates to the infection site at late infection stages (18, 36, 37). The nutritional situation in the early infection stages, however, remains unknown. Although there is no report of induced expression of potential effectors under N starvation in U. maydis, it has been shown that N limitation or low ammonium concentrations can induce the switch to the filamentous growth of haploid cells on artificial media (46, 73), an effect similar to the invasive growth of S. cerevisiae under nitrogen starvation (27). It was also shown that the N starvation-induced formation of conjugation tubes is dependent on compatible a mating loci in U. maydis, whereas the formation of true filaments is dependent on active a and b loci (5).
Here, we report the identification of a functional Nit2 homolog in U. maydis which controls nitrogen utilization, and we show that it is required for the efficient initiation of filamentous growth and for full virulence on maize leaves.
MATERIALS AND METHODS
Strains and culture conditions.
Fungal strains used in this study are given in Table 1. U. maydis sporidia were propagated at 28°C on potato dextrose plates or in liquid YEPSlight medium (81) if not specified otherwise. For the determination of nitrogen utilization, minimal medium (MM) plates (35) were supplemented with 2% (wt/vol) glucose and 0.3% (wt/vol) KNO3 (NMM) or (NH4)2SO4 (AMM) or 10 mM the respective nitrogen source. Spermine and spermidine were provided at 2 mM, and guanine at 5 mM; for nitrogen starvation medium (−N), no nitrogen source was added. For liquid medium, agar was omitted. Resistance to chlorate was tested on MM plates supplemented with 10 mM proline and 250 mM KClO3. For transcript and enzyme activity analyses, sporidia were grown in liquid ammonium MM to an optical density at 600 nm (OD600) of ∼1.0, harvested by centrifugation, and washed 2 times in H2O. Aliquots were transferred to liquid AMM, NMM, or −N medium. Cells were harvested by centrifugation at different time points after transfer to fresh medium as indicated in the text and immediately snap-frozen in liquid nitrogen.
Table 1.
Ustilago maydis strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| SG200 | a1mfa2bW2bE1 | 45 |
| SG200-Δnit2 | a1mfa2bW2bE1 Δum10417 | This study |
| SG200-ΔN-nit2 | a1mfa2bW2bE1 ΔN terminus-um10417 | This study |
| SG200-Δnit2-Nit2 | a1mfa2bW2bE1 Δum10417 ipr[Pum10417-um10417] | This study |
| SG200-Δton1 | a1mfa2 bW2bE1 Δum10005 | This study |
| AB31 | Pcrg::bw2, Pcrg::bE1 | 10 |
| AB31-Δnit2 | Pcrg::bw2, Pcrg::bE1 Δum10417 | This study |
P, promoter; ipr, allele of ip conferring resistance to carboxin.
Infection assay and assessment of pathogenicity.
Plant infection was conducted as previously described (26). In brief, a suspension of fungal sporidia (OD600 of 1) or the same volume of water for mock controls was injected by syringe into the stems of 7- to 10-day-old maize seedlings (cv. Early Golden Bantam), which resulted in local infections of leaves 3 to 5. Infection symptoms were documented 8, 10, 14, and 20 days postinfection (dpi).
The pathogenicity of the indicated U. maydis strains was assessed by generating a disease index at the indicated time points by classifying symptom severity according to predefined categories as described in reference 21.
Construction of U. maydis transgenics.
U. maydis transgenics were generated in the solopathogenic strain SG200 (45), with the exception of AB31-Δnit2, which was in the AB31 background (10), using a PCR-based technique (44). A 1-kb flanking region upstream (LB) and downstream (RB) of the nit2 gene was amplified using primers 5′-TTTCAGCCTTGTCTTCTGTGC-3′ and 5′-CACGGCCTGAGTGGCCGAAAGAATCGCAAGACGGAG-3′ (LB for both constructs), 5′-GTGGGCCATCTAGGCCTTCGGGGTCCGCATCTTC-3′ and 5′-GCAACAAAGCCGTAATATCACC-3′ (RB ΔN-nit2), or 5′-GTGGGCCATCTAGGCCGGTTTTGTTTAGTGTACCGTCTTTCT-3′ and 5′-TCCATCCATGCCAGAATCTCG-3′ (RB Δnit2). PCR products were digested with SfiI and ligated to the hygromycin resistance cassette of pBS-hhn (44). Primers 5′-TTCGTGAATGGAAGACGTGAGG-3′ and 5′-CACGGCCTGAGTGGCCGAAGACGGTGGCAAGATGGAC-3′ (LB) and 5′-GTGGGCCATCTAGGCCATCGCTTTCGCTGTTGGTCTCG-3′ and 5′-TGCCTGCTGGTGCGGATTGTC-3′ (RB) were used to generate the Δton1 construct. For the nit2 complementation assay, a genomic fragment, including the Nit2 gene and 1 kb upstream of the start codon, was amplified by PCR using primers 5′-CACAAGCTTCGCTCGGTCCTCCTACTCGT-3′ and 5′-CACGCGGCCGCACAAAACCTCACTTGCGCTGTC-3′ and cloned into the HindIII and NotI sites of p123 (75), a vector that integrates into the ip locus of U. maydis and confers resistance to carboxin. The transformation of U. maydis was performed as previously described (72), and the selection of transformants occurred on hygromycin or carboxin.
Sequence analysis.
The N. crassa Nit2 protein sequence (P19212) was used in a BLASTP search (1) to identify Um10417 as a sequence homolog. Domains were identified using the Conserved Domain Database (56). Protein alignments were performed using the MUSCLE algorithm (20), and distance trees were visualized with Geneious 5.0.4 software (Biomatters Ltd., New Zealand). The following strains (with accession numbers) were used: A. nidulans XP_681936, Gibberella fujikuroi P78688, F. oxysporum f. sp. lycopersici ABD60578, C. lindemuthianum AAN65464, Magnaporthe oryzae XP_366679, Schizophyllum commune BAA96108, C. neoformans XP_566547, and N. crassa P19212.2.
Transcript analyses.
Total RNA from sporidia grown on the indicated media for the given times after transfer from AMM was extracted using the RNase-all method (14), and RNA quality was assessed after separation on formaldehyde gels or using an Agilent 2100 Bioanalyzer. For RNA blot analysis, standard molecular techniques were used. Ten μg total RNA was loaded, separated, and probed with 32P-labeled, gene-specific probes. nar1 and nrt probes were amplified using primers 5′-TTCCTCGATCCCAAGAAATG-3′ and 5′-CTGGATGGGGTCTAAAGCAG-3′ (nar1) and 5′-CGCACTGCCGACAACGAAA-3′ and 5′-GCAGGGATACCGTAGTTGGCA-3′ (nrt). Tomato 18S rRNA was used as a loading control.
Whole-genome transcript analyses with 400 ng total RNA extracted from sporidia cultivated in the indicated media for the given times after transfer from AMM were performed using custom U. maydis 8×15K microarrays (Agilent, Santa Clara, CA). Annotated genes were downloaded from the MUMDB homepage (http://mips.helmholtz-muenchen.de/genre/proj/ustilago/), and specific probes were designed using eArray software (Agilent). Sample labeling, chip hybridization and scanning, and feature extraction were performed with the one-color spike mix (Agilent) according to the manufacturer's protocol. Data were analyzed using GeneSpring XI software with standard settings. Statistically significantly deregulated genes (>2-fold change in from values for the control and P < 0.05 by t test, with unequal variance after Benjamini-Hochberg correction) were identified with the volcano plot option of GeneSpring XI for the comparisons of conditions and genotypes as indicated.
The expression pattern of selected genes was verified by quantitative reverse transcription-PCR (qRT-PCR) using the Brilliant II SYBR green quantitative PCR (qPCR) master mix and Mx3000P qPCR system (Stratagene). Gene expression from three biological replicates per condition tested was normalized against U. maydis gapDH. The following primers were used: gapDH, 5′-CTCAGGTCAACATCGGTATCAACG-3′ and 5′-CCGTGGGTGGAGTCGTACTTG-3′; um01756.2, 5′-CGGTTGCCCTGCTAAGTACG-3′ and 5′-TGCCAATGAGGAAAGCACCT-3′; um03690, 5′-CGCTTCCACTTTGATCTGGTG-3′ and 5′-AAAGAGAGGGCGATCGAGATG-3′; um10005, 5′-ACAGCAAGACTGGCAGGATGC-3′ and 5′-CGGAAGTGAGCGAGCTGAGAG-3′; um04577, 5′-GACCAGAGACCGAGGACCACT-3′ and 5′-TCAAACGCCGTCTTTTCTTCA-3′; um03847, 5′-GACCTGAGCCGTGGTTTGTC-3′ and 5′-CATCGAGTGGATTGCCATGA-3′; um11104, 5′-CTCAGATCGCCGTTTGGAAC-3′ and 5′-CGACCCTTGTCGTCTTCACC-3′; um11105, 5′-CTTCTGGCTGCTCGTCACCT-3′ and 5′-CGAGGAAACCAGTCGTACCG-3′; um05889, 5′-GCCTACGTCATCTTGGGAACC-3′ and 5′-TCCCAGAACATCCAGGTGAGA-3′; um02625, 5′-CTGCATCCTGCTGTGGTTTTC-3′ and 5′-GCCCGACTCCAATACCAGTTC-3′; um00477, 5′-GTCTGACGTCCGGTGGAATC-3′ and 5′-GCGCTGTATACTCGCCATCC-3′; um04060, 5′-CCTGAATCTGGCCAAAGACG-3′ and 5′-CGAAAGACCTCGGAACAACG-3′; um06253, 5′-TTTTTGACCGATTGGATGCAC-3′ and 5′-GCTTGACGAGCTCCCAAAGAT-3′.
Nitrate reductase activity assay.
The maximal extractable activity of nitrate reductase in sporidia from the logarithmic growth phase was assayed as described in reference 25.
Filamentation assay.
Filamentation on a hydrophobic surface was performed as described previously (60). Sporidia were sprayed in water or in a 1 mM Gln solution on parafilm. For strains in the AB31 background, sporidia were cultivated to an OD600 of 0.3 in AM-Glc, washed once in distilled water, and transferred to an equal volume of either NM-Glc (to relieve NCR with no concomitant induction of the b locus) or NM-Ara (to relieve NCR and to induce the b locus concomitantly). After 1 h of agitation, sporidia were pelleted for 8 min at 2,500 × g, washed once in distilled water, and finally sprayed onto parafilm at an OD600 of 0.1. Filamentous growth was evaluated after 16 and 21 h at 28°C and 100% relative humidity using a Leica DMR fluorescence microscope with differential interference contrast optics. Filamentation on planta was assessed 18 h after infection. To visualize fungal material, infected leaf segments were stained for 1 min with calcofluor (fluorescence brightener 28; Sigma-Aldrich) and rinsed briefly with double-distilled water. Stained samples were immediately examined under a fluorescence microscope using a UV lamp and Leica filter cube A.
Microarray accession number.
The microarray data presented in this study are deposited at GEO under the accession number GSE28916 (http://www.ncbi.nlm.nih.gov/geo).
RESULTS
Ustilago maydis Um10417 is homologous to Nit2/AreA.
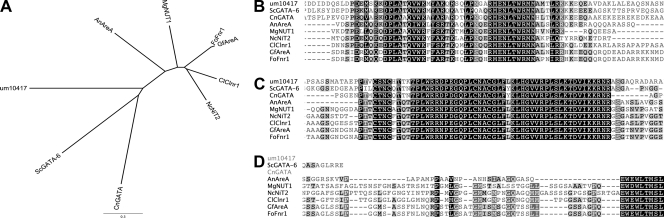
Functional Nit2/AreA proteins from ascomycete fungi contain a highly conserved GATA Cys2/Cys2-Zn finger domain followed by an adjacent basic region, which is also conserved (70). The U. maydis genome harbors 10 annotated proteins with a GATA Zn finger domain, of which only URBS1, which is a regulator of iron-siderophore biosynthesis, has been characterized (21, 84). A BLAST analysis (1) using N. crassa Nit2 and A. nidulans AreA as a query showed that Um10417 was the U. maydis GATA transcription factor with highest sequence homology to both Nit2 and AreA. Multiple sequence alignment with functionally characterized regulators of NCR (nitrogen catabolite repression) from ascomycete fungi and with Nit2 orthologs from basidiomycetes revealed that besides the highly conserved, C-terminal zinc finger domain, Um10417 contains a DUF1752 domain that is also conserved among functional Nit2/AreA proteins (Fig. 1). Nit2/AreA from ascomycete fungi also harbor a conserved sequence motif at the outermost C terminus that has been shown to participate in the direct interaction between N. crassa Nit2 and its negative regulator, Nmr1 (65). This extension is missing from Um10417 and also from the nit2 orthologs of the basidiomycetes Cryptococcus neoformans and Schizophyllum commune (Fig. 1D). The construction of a phylogenetic tree from the full protein alignment (see Fig. S1 in the supplemental material) shows that basidiomycete nit2 orthologs diverged from their ascomycete homologs (Fig. 1A). This raises the question of whether the U. maydis Nit2 serves a function similar to that of the ascomycetous orthologs in vivo.
Fig 1.
Sequence analysis of U. maydis GATA transcription factor Um10417. (A) Phylogenetic tree of characterized Nit2/AreA transcription factors from the ascomycetes Aspergillus nidulans (AnAreA), Colletotrichum lindemuthianum (ClClnr1), Fusarium oxysporum (FoFnr1), Gibberella fujikuroi (GfAreA), and Neurospora crassa (NcNit2), as well as homologous proteins from the basidiomycetes Schizophyllum commune (ScGata-6) and Cryptococcus neoformans (CnGat1). (B to D) Protein alignment showing the DUF1752 motif conserved among functional Nit2/AreA proteins (B), the Zn finger motif (C), and the C-terminal motif (D) conserved only in ascomycete Nit2/AreA proteins. Multiple sequence alignment was executed using the MUSCLE algorithm (20).
Ustilago maydis Nit2 regulates utilization of unfavorable nitrogen sources.
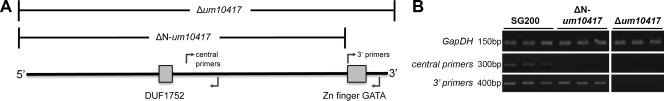
The utilization of most nonfavorable nitrogen sources is repressed in the presence of favorable nitrogen sources in fungi. Nit2/AreA proteins are necessary to release this repression in the absence of favorable nitrogen sources. To ascertain whether Um10417 regulates the utilization of unfavorable nitrogen sources, such as the Nit2 homologs in ascomycetes, we constructed two independent disruptions of Um10417 in the solopathogenic U. maydis strain SG200 (45) using a targeted gene replacement strategy as described previously (44). As the 3′ end of the um10417 gene is only 450 bp away from the start codon of the next downstream gene (um10416), we constructed two different knockout strains. The resulting strain, SG200-Δum10417, was deleted for the whole predicted CDS of um10417, also potentially affecting the promoter region of um10416, while in strain SG200-ΔN-um10417, the N-terminal 1,158 amino acids (aa) were deleted, leaving the potential promoter sequence of the downstream gene intact (Fig. 2). Both strains showed a single integration of the hygromycin resistance cassette at the um10417 locus as assessed by insert-specific PCR and DNA blot analysis (data not shown). SG200-Δum10417 completely failed to accumulate the um10417 transcript, as suggested by RT-PCR targeting different regions of the predicted full-length cDNA. In contrast, the um10417 3′ end, but not the 5′ end, could be amplified from SG200-ΔN-um10417 cDNA (Fig. 2), which suggests the accumulation of a severely truncated 3′ portion of the um10417 transcript in the latter strain.
Fig 2.
(A) Domain structure of Um10417. Deleted regions in the respective strains and primer binding sites for the verification of transcript abundance via RT-PCR are indicated. (B) RT-PCR analysis of Um10417 deletion strains. The um10417-specific primer pairs correspond to the ones shown in panel A. The glyceraldehyde-3-phosphate dehydrogenase gene (GapDH) was used as a positive control. Contamination by genomic DNA was excluded beforehand with genomic DNA-specific primers.
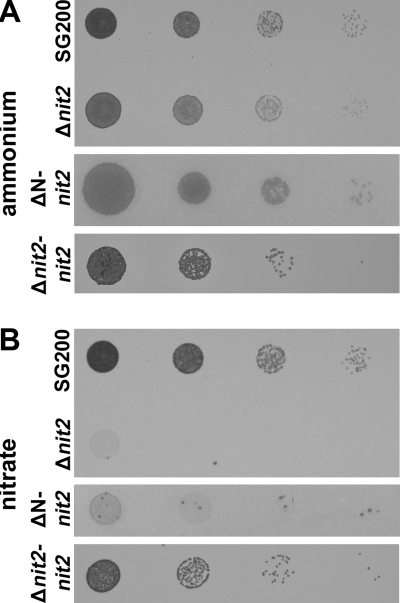
Ustilago maydis has the genetic setup to synthesize every proteinogenic amino acid (59) and is capable of utilizing a plethora of nitrogen sources, e.g., inorganic N, such as nitrate and ammonium, as well as organic nitrogen sources, such as biogenic amino acids and nucleobases (34). We assessed the role of um10417 in nitrogen utilization by cultivating SG200, SG200-Δum10417, and SG200-ΔN-um10417 sporidia in the presence of different inorganic and organic nitrogen sources. Neither mutant strain showed a growth phenotype when cultured on complex media, and both grew as well as SG200 on minimal medium supplemented with ammonium as the sole nitrogen source (Fig. 3A). When grown on nitrate minimal medium, however, both mutant strains were unable to grow (Fig. 3B). The growth of the SG200-Δum10417 deletion strain also was assessed on minimal medium supplemented with organic nitrogen sources. While SG200 grew on all nitrogen sources tested except spermine, SG200-Δum10417 sporidia exhibited growth similar to that of SG200 only when Gln, Glu, Asn, or putrescine was provided. Growth on Tyr was slightly retarded, and no growth of the Δum10417 strain was observed on all other nitrogen sources tested (Table 2). All defects in the utilization of nonfavorable nitrogen sources could be complemented in Δum10417 sporidia by transformation with a wild-type um10417 copy (Table 2). These results suggest that Um10417 is indeed a functional homolog of Nit2/AreA and subsequently is termed Nit2.
Fig 3.
Growth of U. maydis sporidia on minimal medium plates supplemented with 0.3% (wt/vol) ammonium (AMM) (A) or 0.3% (wt/vol) nitrate (NMM) (B). Dilution series (OD600 of 1 to 10−4 in 10−1 steps from left to right) of SG200, Δnit2, ΔN-Nit2, and Δnit2 complemented with Nit2 (Δnit2-Nit2) were plated onto the respective medium, and growth was assessed after incubation at 28°C for 1 day.
Table 2.
Summary of observed growth of SG200, Δnit2, and Δnit2-Nit2 sporidia on minimal medium plates supplemented with a single nitrogen sourcea
| Supplement | Growth observed for: |
||
|---|---|---|---|
| SG200 | Δnit2 | Δnit2-Nit2 | |
| Ammonium | + | + | + |
| Nitrate | + | −− | + |
| Ala | + | − | + |
| Arg | + | − | + |
| Asn | + | + | + |
| Asp | + | − | + |
| Citrullin | + | −− | + |
| Cys | + | −− | + |
| Gln | + | + | + |
| Glu | + | + | + |
| Gly | + | −− | + |
| His | + | −− | + |
| Ile | + | −− | + |
| Leu | + | −− | + |
| Lys | + | − | + |
| Met | + | −− | + |
| Ornithine | + | − | + |
| Phe | + | −− | + |
| Pro | + | − | + |
| Ser | + | −− | + |
| Thr | + | −− | + |
| Trp | + | − | + |
| Tyr | + | +/− | + |
| Uracil | + | − | + |
| Val | + | −− | + |
| Adenine | + | −− | + |
| Cytosine | + | −− | + |
| Guanine | + | − | + |
| Thymine | + | −− | + |
| Putrescine | + | + | + |
| Spermidine | + | − | + |
| Spermine | −− | −− | −− |
| Proline-chlorateb | − | + | − |
Sporidial dilution series of each genotype were plated, and growth properties were rated 1 day after plating as normal growth (+), reduced growth (+/−), strongly reduced growth (−), and no growth (−−). ND, not determined. Resistance to chlorate was assessed 7 days after plating in the presence of proline.
Growth characteristics in the chlorate assay are not compared to growth on media without chlorate.
In general, Nit2/AreA-deficient ascomycetes were unable to induce nitrate reductase (NR) activity in the absence of favorable nitrogen sources (58). To test whether U. maydis SG200-Δnit2 strains also lacked the inducibility of NR activity, sporidia were grown on chlorate plates supplemented with proline, a nonrepressing nitrogen source (Table 2). While SG200 showed only slight growth, quickly followed by death due to the NR-catalyzed reduction of chlorate to toxic chlorite, the two nit2 deletion strains were viable on chlorate plates but showed strongly reduced growth compared to that of SG200 grown on proline due to its inability to efficiently use this nitrogen source.
Nitrate reductase activity was completely repressed in SG200 sporidia grown in liquid culture with ammonium, but it was induced by 2 h after transfer from ammonium to nitrate minimal medium (6.1 ± 0.8 nmol min−1 per OD unit) or to minimal medium without a nitrogen source (11.8 ± 1.3 nmol min−1 per OD unit). Δnit2 sporidia failed to induce NR activity upon transfer to nonrepressive media, which explains their resistance to chlorate.
Transcriptional regulation of nitrate assimilation by Nit2.
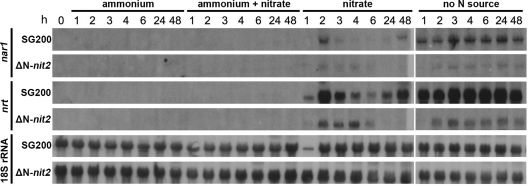
Nitrate reductase expression is induced at the transcriptional level by Nit2/AreA in filamentous fungi in the absence of favorable nitrogen sources only in the presence of nitrate (24). Furthermore, NR has been shown to be subject to transcriptional regulation in U. maydis (4), and the induction of the nitrate reductase promoter by nitrate was demonstrated to be tightly controlled (10). Nevertheless, the underlying regulatory mechanisms have remained elusive. The genes coding for nitrate reductase (nar1), nitrite reductase (nir1), and a putative nitrate transporter (nrt) are located in one gene cluster in U. maydis (59), suggesting their coordinated regulation. Thus, the transcriptional response of nar1 and nrt in SG200 and Nit2-deficient SG200 was assessed in response to different nitrogen regimens by RNA blot analysis. To this end, sporidia were cultured in medium with ammonium as the sole nitrogen source and transferred to medium containing ammonium, ammonium plus nitrate, nitrate only, or no nitrogen source at all. Samples were taken at different time points after transfer.
With ammonium as the sole nitrogen source or with both ammonium and nitrate in the medium, no nar1 and nrt transcripts were detectable in SG200 or the nit2 deletion strain, showing that the presence of a favorable nitrogen source completely inhibits the induction of the nitrate assimilation pathway at the transcriptional level (Fig. 4). Upon transfer to nitrate medium, SG200 sporidia showed a strong induction of nar1 and nrt, reaching peak transcript levels 2 h after transfer. The transcript abundance of both genes decreased up to 6 h after transfer but increased again after 24 to 48 h. The similar transcript accumulation patterns of nar1 and nrt in response to changing nitrogen availability further suggest that these two nitrate assimilatory genes are regulated in concert.
Fig 4.
Expression analysis of Nar1 and Nrt in U. maydis sporidia. Transcript abundance was determined using specific probes against Nar1 (top) and Nrt (middle) at 1, 2, 3, 4, 6, 24, and 48 h upon the transfer of SG200 and ΔN-Nit2 sporidia from ammonium minimal medium to minimal medium containing ammonium, ammonium and nitrate, nitrate, or no nitrogen source. Ammonium and nitrate concentrations in the media were 0.3% (wt/vol). As a loading control, abundance of 18S rRNA was determined (bottom).
Although the deletion of nit2 completely inhibited the activity of NR on nitrate-containing medium, transcripts of nar1 and nrt were still detectable in the Nit2-deficient strain, although much less so than in SG200 (Fig. 4). The induction kinetics also differed in the Nit2-deficient strain compared to those for SG200, with peak transcript levels at 6 h and no detectable transcript 24 and 48 h after transfer. In medium without nitrogen, nar1 and nrt were strongly and immediately induced in SG200, while induction was less pronounced and was delayed in Nit2 mutant sporidia.
These data suggest that nar1 and nrt are subject to NCR in U. maydis but that Nit2 is not the sole transcriptional activator of these two NCR target genes. However, no NR activity was detected in the Nit2-deprived mutant despite nar1 and nrt transcript accumulation in the absence of favorable nitrogen sources, indicating that NR is additionally regulated at the translational or posttranslational level.
Nit2-dependent and Nit2-independent regulation of nitrogen-responsive genes.
As the transcriptional analysis of the nitrate assimilation pathway revealed that Nit2 is not the sole master regulator of nitrogen utilization in U. maydis, microarray analyses were performed to identify nitrogen-responsive genes that are regulated in an Nit2-dependent and -independent fashion.
Transcriptome analysis of Nit2-deficient sporidia cultivated either on nitrate or without a nitrogen source revealed that the transcript pattern of the deletion strain was very similar in these two N-deficient conditions, with only 5 genes being significantly regulated more than 3-fold in sporidia cultivated on nitrate or without an N source (data not shown). Based on this similarity, further transcriptome analyses with SG200, ΔN-Nit2, and Δnit2 were performed with sporidia that had been transferred from ammonium minimal medium to fresh minimal medium containing ammonium (control) or to fresh medium without any nitrogen source in two completely independent experiments. Samples were taken 2 h after transfer, which was the time point of strongest nar1 and nrt induction in the RNA blot experiments (Fig. 4).
Forty-six genes were upregulated under N starvation conditions in both ΔN-nit2 and Δnit2 strains compared to their expression in SG200, while 72 genes were commonly downregulated (see Table S1 in the supplemental material). Classification into functional categories (FunCats) according to the online tool available on the MUMDB homepage (http://mips.helmholtz-muenchen.de/genre/proj/ustilago/) showed that regulated genes were significantly enriched in the categories metabolism, energy, and transport (see Table S2 in the supplemental material).
To verify the microarray data and to compare individual transcript accumulation after the transfer of sporidia from ammonium to either nitrogen starvation medium or nitrate medium, the transcript abundance of selected genes was quantified in sporidia of SG200 and Δnit2 using qRT-PCR (Table 3). As already shown by RNA blot analysis (Fig. 4), the transcriptional induction of the nitrate assimilatory gene cluster consisting of nar, nir, and nrt was only partially Nit2 dependent in response to nitrate or N starvation. However, the induction of a putative purine transporter (um01756.2) and a putative purine permease (um03690) was entirely Nit2 dependent, since no induction was observed when Nit2-deficient sporidia were transferred to nitrate or N starvation medium. A high-affinity ammonium transporter (ump2 or um05889), which was previously discovered as a nitrogen starvation-induced gene (32), was also strongly induced under nitrogen starvation in our assay. The induction of ump2 was partially Nit2 dependent (Table 3).
Table 3.
Verification of transcript accumulation of selected genes by qRT-PCRa
| Gene name | Annotation | Change in transcript accumulation by strain and medium |
Category | |||
|---|---|---|---|---|---|---|
| SG200 |
Δnit2 |
|||||
| −N vs AMM | NMM vs AMM | −N vs AMM | NMM vs AMM | |||
| um01756.2 | Purine transporter | 12 ± 5 | 1.7 ± 0.2 | 0.7 ± 0.1 | 0.45 ± 0.01 | Nit2-dependent induction |
| um03690 | Purine permease | 671 ± 161 | 39 ± 7 | 2.9 ± 0.5 | 1.9 ± 0.2 | Nit2-dependent induction |
| um10005 | Gal4-TF (ton1) | 12 ± 2 | 3.1 ± 0.4 | 1.6 ± 0.5 | 1.9 ± 0.2 | Nit2-dependent induction |
| um04577 | Urea permease | 1,236 ± 530* | 702 ± 114* | 7 ± 2 | 12 ± 5 | Nit2-dependent induction |
| um03847 | nar1 | 131 ± 32 | 71 ± 8 | 13 ± 3 | 23 ± 3 | Partial Nit2-dependent induction |
| um11104 | nir1 | 2,484 ± 787* | 2,013 ± 393* | 154 ± 60 | 294 ± 90 | Partial Nit2-dependent induction |
| um11105 | nrt | 3,281 ± 863* | 3,604 ± 703* | 232 ± 70 | 559 ± 16 | Partial Nit2-dependent induction |
| um05889 | ump2 | 996 ± 73* | 423 ± 28 | 177 ± 21 | 152 ± 24 | Partial Nit2-dependent induction |
| um02625 | Urea permease | 0.0027 ± 0.0004 | 10 ± 2 | 0.021 ± 0.007 | 0.045 ± 0.008 | Nit2-dependent induction on NMM only |
| um00477 | mct1 | 28 ± 8 | 1.2 ± 0.8 | 153 ± 42 | 60 ± 2 | Nit2-dependent repression |
| um04060 | dic1 | 65 ± 10 | 17 ± 3 | 52 ± 12 | 50 ± 9 | Nit2 dependent repression on NMM |
| um06253 | Urea permease | NR | NR | NR | NR | Not regulated |
SG200 and Δnit2 sporidia were transferred to nitrate minimal medium (NMM), nitrogen starvation (−N), or ammonium minimal medium (AMM) and harvested 2 h after transfer. Shown is the absolute fold change and standard errors of the respective genes in −N or NMM compared to levels for the control determined from three experimental replicates with three technical replicates each. Values marked with an asterisk show strong induction due to a basically absent transcript on AMM and have to be considered qualitative, not quantitative.
The expression of three genes with strong homology to urea permeases showed distinct expression patterns. While um04577 induction was predominantly Nit2 dependent in both nitrate and N starvation medium, um02625 was induced in an Nit2-dependent fashion only when sporidia were transferred to nitrate-containing medium. Under nitrogen starvation, this gene was repressed. The third urea permease, um06253, was not transcriptionally regulated in either SG200 or in Δnit2 under our experimental conditions. Another level of complexity in the response of U. maydis to changing nitrogen regimens is added by the observation that a putative monocarboxylate transporter (um00477) is repressed on nitrate and N starvation medium, and a putative dicarboxylate transporter (um04060) is repressed on nitrate, but not under N starvation, in a Nit2-dependent fashion.
The Gal4-like transcription factor Ton1 is a target of Nit2 but is not involved in regulating nitrogen metabolism.
The only transcription factor, besides the deleted Nit2 itself, that was commonly downregulated in both Nit2 deletion strains was the Gal4-type transcription factor um10005 (see Table S1 in the supplemental material). The predicted protein sequence harbors an N-terminal Zn2Cys6-type DNA binding domain and does not show any sequence homology to transcription factors from ascomycete fungi outside this DNA-binding domain. Its closest homologs can be found in the basidiomycete C. neoformans (accession no. XP_774034.1; expect value of 2e−16), so this transcription factor might represent a novel class of transcription factors specific to basidiomycetes.
We termed it Ton1 (for target of Nit2) and confirmed the Nit2-dependent induction on nitrate and N starvation medium by qRT-PCR (Table 3). We constructed a deletion strain, termed Δton1, in the SG200 background and tested the ability of this strain to utilize complex nitrogen sources as described for the Δnit2 strains. The deletion of ton1 did not affect growth on any nitrogen source tested compared to that of SG200 sporidia (data not shown), so a major role of Ton1 in nitrogen utilization is unlikely.
To more precisely unravel the function of ton1, a transcriptome analysis was performed with the Δton1 strain. To this end, sporidia were grown on ammonium minimal medium and transferred to ammonium or N starvation medium, and the global expression pattern was compared to the expression pattern of the Nit2 deletion strains. Sixty-five genes were commonly upregulated in the Δnit2 strains as well as in the Δton1 strain under nitrogen-limiting conditions, while 21 genes were synonymously downregulated in these three genotypes (see Table S3 in the supplemental material), indicating that this gene set is regulated in the Nit2-deficient strains due to ton1 downregulation. The classification of these genes into FunCats showed that the majority of regulated genes are involved in metabolism and in DNA, RNA, and protein metabolism, but no category was significantly enriched (see Table S4 in the supplemental material). Interestingly, rrm4, an RNA-binding protein involved in cell polarity and also required for full pathogenicity (6, 7), is strongly repressed (>1,000-fold) in Δton1 and slightly repressed in Δnit2 (2.3-fold). The deletion of ton1 in the solopathogenic SG200 strain, however, did not affect filamentous growth on charcoal plates, as was shown for rrm4 mutants (see Fig. S2 in the supplemental material).
Ustilagic acid biosynthesis is not controlled by Nit2.
One facet of the nitrogen starvation response of U. maydis is the production and secretion of glycolipids, mainly ustilagic acid (cellobiose lipid) and ustilipid (mannosylerythritol lipid), which act as biosurfactants (31). The biosynthetic genes are organized in gene clusters that have already been characterized (78). Recently, the transcriptional activator Rua1 has been shown to control the upregulation of the ustilagic acid gene cluster under nitrogen-limiting conditions (79). We also observed the formation of needle-shaped crystals that are reminiscent of ustilagic acid production after keeping both SG200 and Nit2-deficient sporidia for 4 days on nitrogen starvation medium (Fig. 5A, white arrowheads). Furthermore, the analysis of secreted glycolipids isolated from these cultures revealed that the composition was the same for SG200 or Nit2-deficient cultures (see Fig. S3 in the supplemental material).
Fig 5.
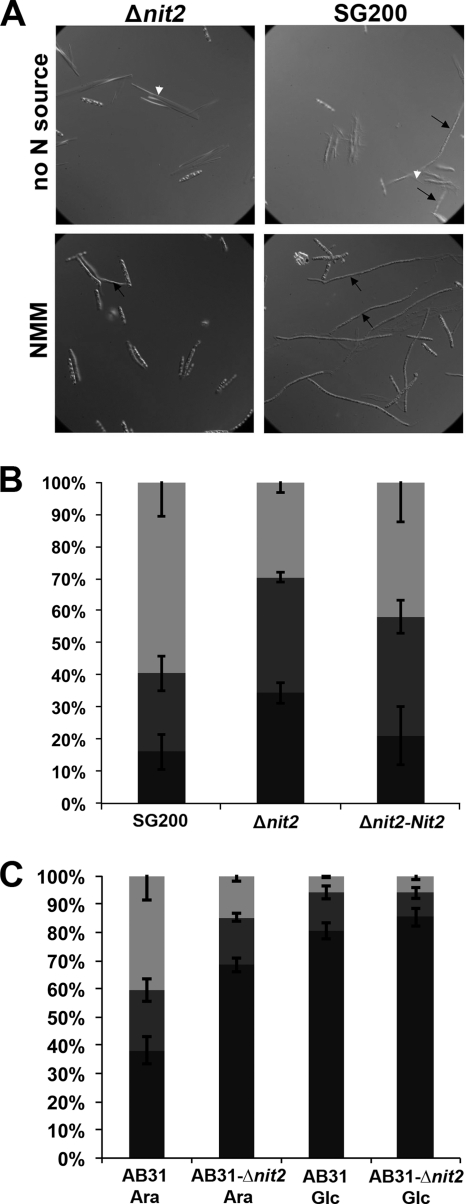
Filamentation of U. maydis sporidia. (A) Sporidia were cultivated for 4 days in nitrogen starvation medium (no N source; top panel) or nitrate minimal medium (NMM; bottom panel). Filamentous growth was assessed using a microscope at ×400 magnification and differential interference contrast optics. Black arrows point to sporidia that had switched to filamentous growth. White arrowheads mark ustilagic acid crystals. (B) Filamentous growth on planta. Filamentation of strains SG200 (control), Δnit2, and Δnit2-Nit2 was assessed 18 h after infection by the calcofluor white staining of fungal structures at the injection site. The difference between SG200 and Δnit2 strains was statistically significant in a Student's t test (P < 0.05). (C) Filamentous growth in vitro. Filamentous growth of strains AB31 and AB31-Δnit2 was assessed 18 h after spraying the sporidia in water onto parafilm. Previously, the b locus had been induced with arabinose (Ara) or sporidia were mock treated with glucose (Glc) for 1 h before spraying. Four replicates were assessed per genotype, and 100 to 200 sporidia per replicate were classified into three groups: no visible filaments (black), filaments shorter than sporidium length (dark gray), and filaments longer than sporidium length (light gray). Error bars represent the standard errors (n = 4). The difference between AB31 and AB31-Δnit2 after Ara treatment was statistically significant in a Student's t test (P < 0.05).
Nit2 is required for full pathogenicity.
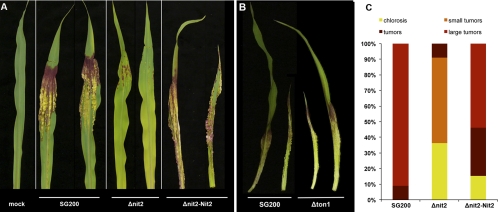
Homologs of Nit2/AreA have been shown to be required for full pathogenicity in some, but not all, plant pathogenic ascomycete fungi, as their corresponding null mutants showed reduced virulence on planta (summarized in reference 9). To test if the deletion of nit2 affects pathogenicity in U. maydis, maize seedlings were infected with Δnit2 strains and the infection phenotype was scored 10 dpi (Fig. 6C). While SG200 induced the formation of large tumors, leaves infected with Nit2 deletion strains displayed strongly reduced tumor formation and tumor size (Fig. 6A and C). The complementation of the Nit2 deletion strains with endogenous nit2 restored virulence on maize seedlings to nearly, but not completely, the SG200 level (Fig. 6A and C). However, transcript amounts of Nit2 in several independent complementation strains were higher than that in SG200 (data not shown), suggesting posttranscriptional regulation of Nit2 activity or suboptimal gene dosage effects. The disease index scored at 14 and 20 dpi was quite identical to the score obtained at 10 dpi (results not shown), indicating that host colonization is impeded in Δnit2, and not just delayed, compared to that of the wild type.
Fig 6.
Pathogenicity of U. maydis strains on maize leaves. (A) Maize leaves 10 days postinfection with SG200, Δnit2, or Δnit2 complemented with a genomic promoter-gene fragment of nit2. (B) Maize leaves 8 days postinfection with SG200 or Δton1. Two representative leaves per strain are shown. In total, between 12 and 20 infected seedlings per strain were analyzed. (C) Disease index for SG200, Δnit2, or Δnit2-Nit2 scored at 10 dpi. Maize seedlings were infected by injecting sporidia suspensions (OD600 of 1.0) of the respective strain into the leaf canal with a syringe. The incidence of the indicated symptoms was scored as described in Materials and Methods (n = 12 to 20). Results for one representative out of four experiments are displayed.
Since plants infected with Δton1 showed wild-type-like infection symptoms (Fig. 6B), the reduced virulence of Δnit2 cannot be explained by a secondary effect of ton1 downregulation in Δnit2 mutants.
Initiation of U. maydis filamentous growth is disturbed in Δnit2.
Haploid U. maydis sporidia proliferate by budding under favorable conditions, but filamentous growth has been observed under nitrogen limitation (46). Filaments also formed when sporidia were kept on low-ammonium medium, and this filamentation has been shown to be dependent on the high-affinity ammonium permease Ump2 (73). The induction of ump2 during nitrogen starvation was less pronounced in the Nit2 deletion strains than in SG200 (Table 3), and furthermore, we also observed that some SG200 sporidia initiated filamentous growth when grown overnight in liquid medium containing nitrate or no nitrogen source, while Δnit2 sporidia did not (Fig. 5A). Thus, we hypothesized that Nit2 is required to execute the dimorphic switch from budding to filamentous growth.
To further investigate if Nit2 is involved in the initiation of filamentous growth, we quantified filament formation on planta and on an artificial hydrophobic surface capable of inducing this morphological differentiation (60). After 16 h of incubation in water on parafilm, 21% ± 2% of the SG200 sporidia had started to produce filaments, while only 15% ± 2% of Δnit2 sporidia had done so. After treatment with 1 mM Gln, only 5% ± 1% of the SG200 sporidia had initiated filamentous growth. On planta, 89% of the SG200 sporidia had produced filaments 16 h postinfection (hpi), the majority of which had already exceeded the length of sporidia. In contrast, only 68% of Δnit2 sporidia had produced filaments at 16 hpi, most of which were shorter than those formed by SG200 (Fig. 5B).
Similarly to the situation in S. cerevisiae, where invasive filamentous-like growth is initiated under nitrogen limitation (27), filaments were not formed when high concentrations of the favorable nitrogen source ammonium were provided to U. maydis sporidia (data not shown). Furthermore, the addition of Gln, another favored nitrogen source for U. maydis, to SG200 sporidia resulted in reduced filamentation in the filamentation assay on parafilm compared to that of SG200 sporidia kept in water only.
To test whether Nit2 acts downstream of the bE/bW heterodimer, we introduced the nit2 knockout into the haploid strain AB31, which carries the bE and bW genes under the control of an arabinose-inducible promoter (10). Without arabinose induction, the occurrence of filaments is restricted in both AB31 and AB31-Δnit2 (Fig. 5C). After the arabinose-triggered induction of b locus-dependent genes, only 40% of AB31 sporidia had not formed filaments at 18 hpi, while 70% of AB31-Δnit2 sporidia failed to form filaments at this time point (Fig. 5C). Likewise, the fraction of sporidia that had formed long filaments had doubled upon the induction of the b locus in the AB31-Δnit2 strain, while it had increased 7-fold in the AB31 strain (Fig. 5C). This suggests that Nit2 does not act independently of bE/bW during filament induction and that the b locus is the major player in the induction of filamentous growth.
Taken together, these data indicate that Nit2 is a major positive regulator of nitrogen utilization and a molecular player that mediates the dimorphic switch in U. maydis growth downstream of bE/bW.
DISCUSSION
Fungi are capable of utilizing a plethora of nitrogen sources but show preference for nitrogen compounds that are easy to metabolize from the energetic point of view. The regulatory circuit behind this is called nitrogen catabolite repression (NCR) and has been extensively studied in S. cerevisiae and in the filamentous ascomycete fungi N. crassa and A. nidulans, where GATA-type transcription factors act as positive master regulators of nitrogen utilization (reviewed in references 15 and 58). GAT1 is required for full virulence in the opportunistic human pathogen Candida albicans (53) and Nit2 homologs in some, but not all, plant pathogenic ascomycete fungi (summarized in reference 9). Little is known about the regulatory mechanisms of NCR in basidiomycetes. It has been demonstrated that basidiomycetes possess genes coding for GATA factors homologous to Nit2/AreA but lack orthologs of the negative acting factors known from yeast and filamentous fungi (32, 59, 87).
This study demonstrates that the U. maydis GATA factor Um10417 is a functional homolog of Nit2/AreA and that U. maydis Nit2 is a positive regulator of unfavorable nitrogen source catabolism. We further show that Nit2 is required for the initiation of filamentous growth under unfavorable nitrogen conditions and for full virulence on maize leaves.
Nit2 is a master regulator of nitrogen utilization.
Ustilago maydis Nit2 showed higher homology to Nit2/AreA from filamentous fungi than to Gat1p from S. cerevisiae and had the same conserved Zn finger domain and RMENLTWRMM motif in the N-terminal part, a motif that is not conserved in S. cerevisiae Gat1p. However, Nit2 and Nit2 homologs from other basidiomycetes exhibit a long N-terminal extension with no homology to any known proteins, and basidiomycete Nit2 homologs are missing the conserved Nmr1 interaction site at the outermost C terminus that is found in ascomycete Nit2/AreA proteins (65). Although it was previously reported that the U. maydis genome does not harbor an Nmr1 homolog (87), we could identify a gene coding for a protein with an NmrA multidomain. The gene um11107 showed 35% sequence identity to the gene encoding Tar1, a functional Nmr1 homolog of C. neoformans (40). Interestingly, a um11107 knockout mutant did not exhibit the expression of nar1 in the presence of favorable nitrogen sources (unpublished results), indicating that Um11107 does not directly inhibit Nit2 activity. In contrast to A. nidulans areA, U. maydis nit2 also did not show changes in transcript accumulation under different nitrogen regimens (unpublished observations), suggesting that it is subject to regulatory mechanisms completely different from the previously described master regulators of nitrogen metabolism.
We generated two independent nit2 deletion strains and tested their ability to utilize different nitrogen sources. Sporidia of the deletion strains were unable to grow on most nitrogen compounds tested, as only ammonium, Gln, Asn, Glu, putrescine, and Tyr supported the proliferation of Δnit2. While only ammonium and Gln or ammonium, Gln, and Glu can be utilized in a Nit2/AreA-independent manner by A. nidulans or N. crassa, respectively (28), several complex nitrogen sources still could be used by nit2 knockout mutants of the ascomycete phytopathogens C. lindemuthianum, M. grisea, and C. fulvum (23, 66, 67). We therefore hypothesize that the large set of favored nitrogen sources of U. maydis is not basidiomycete specific but reflects an adaptation to the pathogenic lifestyle.
Taken together, these observations showed that U. maydis harbors a functional Nit2 homolog, and that it acts as a derepressor of nitrogen utilization in U. maydis.
Transcript analysis reveals Nit2-dependent and -independent gene expression in response to the nitrogen source.
The inability of Δnit2 sporidia to utilize nitrate was due to a lack of NR activity. NR activity is regulated by the available nitrogen source (51), which is known to be mediated on the transcriptional level (4, 10). As it was shown that the induction of the nitrate assimilatory genes in N. crassa and A. nidulans strictly depend on the global derepressor Nit2/AreA and pathway-specific induction (13, 24, 41), we analyzed the induction kinetics of nar1 and the nitrate uptake transporter nrt under different nitrogen regimens in SG200 and Δnit2. Ammonium completely repressed transcript accumulation of nar1 and nrt, even in the presence of nitrate, while the removal of the favored nitrogen source ammonium led to a swift induction of these genes. For A. nidulans nitrate reductase (NiaD) it was shown that niaD transcript accumulation is highly dependent on transcript stability, which is mediated by the nitrogen source (12). Based on our data, we cannot exclude that U. maydis nar1 is regulated at the posttranscriptional level as well.
In contrast to the regulation during N starvation in N. crassa and A. nidulans, we could detect fast and strong induction of nar1, nrt, and other Nit2-regulated genes involved in nitrogen metabolism in the absence of any nitrogen source. The pathway-independent induction of nitrogen-metabolizing enzymes was also observed in the pythopathogenic ascomycetes F. fujikoroi, F. oxysporum, and M. griseae (17, 19, 71) and the mycorrhizal basidiomycete Hebeloma cylindrosporum (38, 39) and may, therefore, represent a general adaptation to the pathogenic lifestyle of these organisms. However, a putative urea permease (um02625) showed induction in the presence of nitrate but not under N starvation. This indicates that pathway-specific induction is necessary for some, but not all, genes.
In G. fujikuroi, many of the nitrogen starvation-induced genes were only partially dependent on AreA (71). We also observed this in U. maydis. While a subset of genes that are controlled by the nitrogen source are strictly dependent on Nit2, a subset of genes was identified whose induction under N starvation was only partially dependent on Nit2. The nitrate assimilation cluster is induced in Δnit2, although at a lower level than that in SG200. Since it was shown that the U. maydis NR protein is subject to high turnover (51), the low nar1 transcript levels in Δnit2 do not seem to be sufficient to drive NR activity to detectable levels. In turn, this indicates that there are additional positive regulators that are involved in the transcriptional activation of nar1 and other Nit2 target genes identified in this study. Alternatively, nar1 expression might be regulated at the posttranscriptional level, preventing the generation of NR protein from the detectable nar1 transcripts.
Ustilago maydis produces and secretes glycolipids under nitrogen-limiting conditions (31). The activation of the biosynthetic gene cluster has been shown to be mediated by the transcription factor Rua1, and it has been postulated that Rua1 is under the control of Nit2 (79). Our data suggest that glycolipid production is not under the control of Nit2 and may, therefore, be controlled by additional transcriptional inducers. Recently, it has been shown that the bikaverin biosynthetic cluster in G. fujikuroi is regulated mainly in an AreA-independent manner by the bZIP factor MeaB (86), while gibberellic acid biosynthesis is under the strict control of AreA (61), suggesting that biosynthetic genes of different secondary metabolites are regulated independently, although they are concertedly induced under N starvation.
Taken together, these data suggest that regulatory circuits differ in U. maydis from those known to control nitrogen utilization in filamentous fungi, and that at least a second positively acting regulator is present that controls the expression of nitrogen-metabolizing genes. As there is no Gln3p homolog in U. maydis, these additional regulators remain elusive.
Among the genes that are regulated in a Nit2-dependent manner are genes involved in carbon metabolism, specifically those encoding the putative carboxylate transporters Mct1 and Dic1. These are upregulated under nitrogen starvation but seem to be repressed by Nit2. While the closest yeast homolog of Mct1 is the riboflavin transporter Mch5p (68), yeast Dic1p represents a mitochondrial carboxylate transporter (43) that is thought to be required for providing carbon backbones to the mitochondrial matrix (64). The fact that Asp and Glu supplementation restores growth on ethanol or acetate in a yeast dic1 deletion strains indicates potential cross-talk between carbon and nitrogen metabolism via UmDic1 (64).
Nit2 is required for filamentous growth and full virulence.
The deletion of nit2 led to strongly decreased virulence on maize leaves and strongly reduced tumor size and number. Similar effects on pathogenicity have been described for Nit2/AreA deletion strains of several plant pathogenic ascomycetes (17, 47, 66). The reduced growth of F. verticillioides AreA mutants on mature maize kernels was attributed to its inability to adapt to the low nitrogen conditions, since the growth of the mutants was not impaired on kernel blisters that contain high concentrations of free amino acids (47). A similar explanation was found for the reduced virulence of C. lindemuthianium clnr1 mutants on the leaves of the common bean (Phaseolus vulgaris), where the initial penetration of the leaf epidermis and the biotrophic phase were not affected but where the mutant was arrested in the early necrotrophic stage (66). The authors of this study hypothesized that in the biotrophic stage, amino acids are readily available to the pathogen but are limiting growth in the early necrotrophic stage.
Fusarium oxysporum invades plants via growth within the xylem vessels, which is an environment generally low in favored nitrogen sources. Divon et al. therefore concluded that the loss of the AreA ortholog Fnr1 decreases the fitness of the pathogen in this environment (17). More recently, F. oxysporum MeaB was shown to be an inhibitor of virulence functions under favorable nitrogen conditions, and it was demonstrated that MeaB acts independently of Fnr1 (54). While the invasive growth of F. oxysporum is generally inhibited in the presence of ammonium, the deletion of MeaB resulted in invasive growth even in the presence of ammonium (54). While U. maydis does not possess a meaB ortholog, it has been described that nitrogen starvation induces the filamentous growth of sporidia (46), similarly to the pseudohyphal growth of yeast cells observed under nitrogen starvation (27). Furthermore, C. albicans Δgat1 and Δgln3 strains showed impaired filamentous growth under certain nitrogen conditions (16, 52). In U. maydis, the initiation of filamentous growth represents a crucial morphological switch to start the pathogenic program (42), and the activity of the active bE/bW heterodimer is necessary and sufficient to trigger pathogenic development (8). Nitrogen limitation has been shown to trigger the dimorphic switch, but this cue still requires the activity of bE/bW (5). Using a temperature-sensitive bE allele, Wahl and coworkers have shown that not all b-dependent genes in planta are regulated upon b locus induction in axenic cultures (85). The authors suggested that other regulators integrate plant-derived environmental cues into the b locus-mediated cascade. We observed reduced filamentous growth of Δnit2 sporidia in both in vitro assays and on planta. Furthermore, the inhibition of the filament formation of SG200 sporidia could be achieved by supplementing the favored nitrogen source, Gln. The pheromone response factor (prf1) is a central regulator of the dimorphic transition in U. maydis, as prf1 mutants cannot initiate filamentous growth and are sterile and nonpathogenic (29). Prf1 integrates environmental signals, such as the availability of carbon sources, and relays these signals to the activity of the b locus (30). Neither prf1 itself nor any of the known proteins regulating Prf1 activity (reviewed in reference 11) are differentially regulated in Δnit2 sporidia, suggesting that Nit2 acts independently or downstream of Prf1 in the haploid pathogenic strain used in this study. In the absence of Nit2, filament formation could be restored only partially by inducing the expression of bE/bW in haploid sporidia, suggesting that nitrogen-induced filamentation via Nit2 integrates directly into the b locus-triggered signaling cascade. Nit2 did not influence filament induction by bE/bW, indicating that filamentation is predominantly controlled by the b locus. Consistently with this finding, it has been demonstrated previously that the induction of filamentous growth by nitrogen limitation is dependent on b locus-triggered gene expression (4). We conclude that Nit2 is one regulator of filamentous growth under low nitrogen conditions that acts downstream of bE/bW. Nevertheless, nitrogen limitation on the plant surface appears to be a cue to induce the dimorphic switch from budding to filamentous growth.
The Gal4-like transcription factor Ton1, which is regulated in a Nit2-dependent manner, is involved in inducing the expression of rrm4 under nitrogen starvation. While rrm4 mutants were impaired in filamentous growth and pathogenicity (7), the deletion of ton1 did not affect either. Most likely, there is residual Rrm4 activity in Δton1 sporidia that is able to promote filamentous growth.
Ho et al. found the ammonium permease Ump2 to be upregulated under nitrogen starvation (32). We observed the same in this study and showed that this upregulation is dependent on Nit2. Ump2 is a close homolog of S. cerevisiae Mep2p, which is involved in sensing low nitrogen conditions, eventually leading to pseudohyphal growth (55). Yeast Δmep2 mutants did not form pseudohyphae on low ammonium, a defect that could be complemented by U. maydis Ump2 (73). The deletion of ump2 from U. maydis led to a defect in the filament formation of haploid cells, but no effect on the pathogenicity of these mutants has been reported (73). Similarly, the deletion of the Mep2p homolog Amt2 in C. neoformans also led to impaired invasive growth but had no effect on virulence in two different cryptococcus assays (69). While the reduced induction of ump2 in Δnit2 sporidia under low N conditions may contribute to the impaired filamentation, this reduction alone does not explain the reduced virulence of Δnit2 on planta.
The genome of U. maydis harbors clusters of genes coding for small, secreted peptides that are required for full virulence on maize leaves (45). In contrast to the situation in C. fulvum and M. griseae, where the expression of a subset of effector genes is induced under N limitation (19, 77, 83), the transcriptome analysis performed in this study did not reveal the upregulation of any putative effector genes under nitrogen starvation and no altered expression of such genes in Δnit2 sporidia. Since these transcriptome analyses were not performed on planta, however, it cannot be excluded that some putative effectors are dependent on Nit2 at later infection stages. Nevertheless, we hypothesize that the reduced virulence of U. maydis Δnit2 is unlikely to be a consequence of altered effector expression but rather is a consequence of impaired filament formation.
At this stage, we cannot rule out that the impaired utilization of complex nitrogen sources in later infection stages also contributes to the overall reduced virulence on maize. As U. maydis-induced tumors constitute strong sinks for organic nitrogen and amino acids to accumulate to high concentrations in the vicinity of fungal hyphae (36), it appears unlikely that Nit2 function in the utilization of complex nitrogen sources plays an important role for U. maydis pathogenicity at late infection stages.
Our study provides insight into the regulatory mechanisms of nitrogen catabolite repression in a basidiomycete phytopathogen. We showed that the U. maydis Nit2 homolog is a central, but not the only, regulator of NCR, that this transcription factor integrates nitrogen metabolism into the induction of filamentous growth, and that Nit2 therefore is required for full pathogenicity.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Deutsche Forschungsgemeinschaft via the priority program FOR 666, project SO 300 11-2.
We thank Sabine Karpeles (FAU Erlangen-Nuremberg) for technical assistance, Stephen Reid (FAU Erlangen-Nuremberg) for microarray hybridizations, Marlis Dahl and Christian Koch (FAU Erlangen-Nuremberg) for their advisory support on microbiological, molecular, and genetic work with U. maydis, Jörg Kämper (KIT, Karlsruhe, Germany) for the provision of U. maydis strains SG200 and AB31, and Regine Kahmann (Max-Planck Institute for Terrestrial Microbiology, Germany) for the provision of vectors used in this study.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Amon J, Titgemeyer F, Burkovski A. 2010. Common patterns–unique features: nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol. Rev. 34:588–605 [DOI] [PubMed] [Google Scholar]
- 3. Andrianopoulos A, Kourambas S, Sharp JA, Davis MA, Hynes MJ. 1998. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180:1973–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banks GR, Shelton PA, Kanuga N, Holden DW, Spanos A. 1993. The Ustilago maydis nar 1 gene encoding nitrate reductase activity: sequence and transcriptional regulation. Gene 131:69–78 [DOI] [PubMed] [Google Scholar]
- 5. Banuett F, Herskowitz I. 1994. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp. Mycol. 18:247–266 [Google Scholar]
- 6. Becht P, Konig J, Feldbrugge M. 2006. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119:4964–4973 [DOI] [PubMed] [Google Scholar]
- 7. Becht P, Vollmeister E, Feldbrugge M. 2005. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot. Cell 4:121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boelker M, Genin S, Lehmler C, Kahmann R. 1995. Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73:320–325 [Google Scholar]
- 9. Bolton MD, Thomma BPHJ. 2008. The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72:104–110 [Google Scholar]
- 10. Brachmann A, Weinzierl G, Kamper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063 [DOI] [PubMed] [Google Scholar]
- 11. Brefort T, et al. 2009. Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47:423–445 [DOI] [PubMed] [Google Scholar]
- 12. Caddick MX, et al. 2006. Opposing signals differentially regulate transcript stability in Aspergillus nidulans. Mol. Microbiol. 62:509–519 [DOI] [PubMed] [Google Scholar]
- 13. Chiang T, Marzluf G. 1995. Binding affinity and functional significance of NIT2 and NIT4 binding sites in the promoter of the highly regulated nit-3 gene, which encodes nitrate reductase in Neurospora crassa. J. Bacteriol. 177:6093–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 15. Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26:223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dabas N, Morschhauser J. 2007. Control of ammonium permease expression and filamentous growth by the GATA transcription factors GLN3 and GAT1 in Candida albicans. Eukaryot. Cell 6:875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Divon HH, Ziv C, Davydov O, Yarden O, Fluhr R. 2006. The global nitrogen regulator, FNR1, regulates fungal nutrition-genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 7:485–497 [DOI] [PubMed] [Google Scholar]
- 18. Doehlemann G, et al. 2008. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56:181–195 [DOI] [PubMed] [Google Scholar]
- 19. Donofrio NM, et al. 2006. Global gene expression during nitrogen starvation in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol. 43:605–617 [DOI] [PubMed] [Google Scholar]
- 20. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eichhorn H, et al. 2006. A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18:3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng B, Marzluf GA. 1998. Interaction between major nitrogen regulatory protein NIT2 and pathway-specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa. Mol. Cell. Biol. 18:3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Froeliger EH, Carpenter BE. 1996. NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol. Gen. Genet. 251:647–656 [DOI] [PubMed] [Google Scholar]
- 24. Fu YH, Marzluf GA. 1987. Molecular cloning and analysis of the regulation of NIT-3, the structural gene for nitrate reductase in Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 84:8243–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gibon Y, et al. 2004. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16:3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gillissen B, et al. 1992. A 2-component regulatory system for self non-self recognition in Ustilago maydis. Cell 68:647–657 [DOI] [PubMed] [Google Scholar]
- 27. Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth–regulation by starvation and Ras. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 28. Griffin DH. 1994. Fungal physiology. Wiley-Liss, New York, NY [Google Scholar]
- 29. Hartmann HA, Kahmann R, Bolker M. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632–1641 [PMC free article] [PubMed] [Google Scholar]
- 30. Hartmann HA, Kruger J, Lottspeich F, Kahmann R. 1999. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator prf1. Plant Cell 11:1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hewald S, Josephs K, Bolker M. 2005. Genetic analysis of biosurfactant production in Ustilago maydis. Appl. Environ. Microbiol. 71:3033–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho EC, Cahill MJ, Saville BJ. 2007. Gene discovery and transcript analyses in the corn smut pathogen Ustilago maydis: expressed sequence tag and genome sequence comparison. BMC Genomics 8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hofman-Bang J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12:35–73 [DOI] [PubMed] [Google Scholar]
- 34. Holliday R. 1961. Genetics of Ustilago maydis. Genet. Res. 2:204–230 [Google Scholar]
- 35. Holliday R. 1974. Ustilago maydis, p. 575–595 In King R. C. Handbook of genetics, vol. 1. Bacteria, bacteriophages, and fungi. Plenum Press, New York, NY [Google Scholar]
- 36. Horst RJ, et al. 2010. Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol. 152:293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horst RJ, Engelsdorf T, Sonnewald U, Voll LM. 2008. Infection of maize leaves with Ustilago maydis prevents establishment of C-4 photosynthesis. J. Plant Physiol. 165:19–28 [DOI] [PubMed] [Google Scholar]
- 38. Jargeat P, Gay G, Debaud JC, Marmeisse R. 2000. Transcription of a nitrate reductase gene isolated from the symbiotic basidiomycete fungus Hebeloma cylindrosporum does not require induction by nitrate. Mol. Gen. Genet. 263:948–956 [DOI] [PubMed] [Google Scholar]
- 39. Jargeat P, et al. 2003. Characterisation and expression analysis of a nitrate transporter and nitrite reductase genes, two members of a gene cluster for nitrate assimilation from the symbiotic basidiomycete Hebeloma cylindrosporum. Curr. Genet. 43:199–205 [DOI] [PubMed] [Google Scholar]
- 40. Jiang N, et al. 2009. Negative roles of a novel nitrogen metabolite repression-related gene, TAR1, in laccase production and nitrate utilization by the basidiomycete Cryptococcus neoformans. Appl. Environ. Microbiol. 75:6777–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnstone IL, et al. 1990. Isolation and characterization of the CrnA-NiiA-NiaD gene-cluster for nitrate assimilation in Aspergillus nidulans. Gene 90:181–192 [DOI] [PubMed] [Google Scholar]
- 42. Kahmann R, Kämper J. 2004. Ustilago maydis: how its biology relates to pathogenic development. New Phytol. 164:31–42 [DOI] [PubMed] [Google Scholar]
- 43. Kakhniashvili D, Mayor JA, Gremse DA, Xu Y, Kaplan RS. 1997. Identification of a novel gene encoding the yeast mitochondrial dicarboxylate transport protein via overexpression, purification, and characterization of its protein product. J. Biol. Chem. 272:4516–4521 [DOI] [PubMed] [Google Scholar]
- 44. Kämper J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271:103–110 [DOI] [PubMed] [Google Scholar]
- 45. Kämper J, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101 [DOI] [PubMed] [Google Scholar]
- 46. Kernkamp MF. 1939. Genetic and environmental factors affecting growth types of Ustilago zeae. Phytopathology 29:473–484 [Google Scholar]
- 47. Kim H, Woloshuk CP. 2008. Role of AREA, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 45:947–953 [DOI] [PubMed] [Google Scholar]
- 48. Kmetzsch L, et al. 2011. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet. Biol. 48:192–199 [DOI] [PubMed] [Google Scholar]
- 49. Langdon T, et al. 1995. Mutational analysis reveals dispensability of the N-terminal region of the Aspergillus transcription factor mediating nitrogen metabolite repression. Mol. Microbiol. 17:877–888 [DOI] [PubMed] [Google Scholar]
- 50. Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349–377 [DOI] [PubMed] [Google Scholar]
- 51. Lewis CM, Fincham JRS. 1970. Regulation of nitrate reductase in basidiomycete Ustilago maydis. J. Bacteriol. 103:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao WL, Ramon AM, Fonzi WA. 2008. GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans. Fungal Genet. Biol. 45:514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Limjindaporn T, Khalaf RA, Fonzi WA. 2003. Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50:993–1004 [DOI] [PubMed] [Google Scholar]
- 54. Lopez-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A. 2010. A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22:2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lorenz MC, Heitman J. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marzluf GA. 1993. Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu. Rev. Microbiol. 47:31–55 [DOI] [PubMed] [Google Scholar]
- 58. Marzluf GA. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCann MP, Snetselaar KM. 2008. A genome-based analysis of amino acid metabolism in the biotrophic plant pathogen Ustilago maydis. Fungal Genet. Biol. 45:S77–S87 [DOI] [PubMed] [Google Scholar]
- 60. Mendoza-Mendoza A, et al. 2009. Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71:895–911 [DOI] [PubMed] [Google Scholar]
- 61. Mihlan M, Homann V, Liu TWD, Tudzynski B. 2003. AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol. Microbiol. 47:975–991 [DOI] [PubMed] [Google Scholar]
- 62. Morozov IY, Galbis-Martinez M, Jones MG, Caddick MX. 2001. Characterization of nitrogen metabolite signalling in Aspergillus via the regulated degradation of areA mRNA. Mol. Microbiol. 42:269–277 [DOI] [PubMed] [Google Scholar]
- 63. Morozov IY, Martinez MG, Jones MG, Caddick MX. 2000. A defined sequence within the 3′ UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol. Microbiol. 37:1248–1257 [DOI] [PubMed] [Google Scholar]
- 64. Palmieri L, et al. 1999. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol. Microbiol. 31:569–577 [DOI] [PubMed] [Google Scholar]
- 65. Pan H, Feng B, Marzluf GA. 1997. Two distinct protein-protein interactions between the NIT2 and NMR regulatory proteins are required to establish nitrogen metabolite repression in Neurospora crassa. Mol. Microbiol. 26:721–729 [DOI] [PubMed] [Google Scholar]
- 66. Pellier AL, Lauge R, Veneault-Fourrey C, Langin T. 2003. CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48:639–655 [DOI] [PubMed] [Google Scholar]
- 67. Perez-Garcia A, Snoeijers SS, Joosten M, Goosen T, De Wit P. 2001. Expression of the avirulence gene Avr9 of the fungal tomato pathogen Cladosporium fulvum is regulated by the global nitrogen response factor NRF1. Mol. Plant Microbe Interact. 14:316–325 [DOI] [PubMed] [Google Scholar]
- 68. Reihl P, Stolz J. 2005. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 280:39809–39817 [DOI] [PubMed] [Google Scholar]
- 69. Rutherford JC, Lin XR, Nielsen K, Heitman J. 2008. Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot. Cell 7:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Scazzocchio C. 2000. The fungal GATA factors. Curr. Opin. Microbiol. 3:126–131 [DOI] [PubMed] [Google Scholar]
- 71. Schönig B, Brown DW, Oeser B, Tudzynski B. 2008. Cross-species hybridization with Fusarium verticillioides microarrays reveals new insights into Fusarium fujikuroi nitrogen regulation and the role of AreA and NMR. Eukaryot. Cell 7:1831–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schulz B, et al. 1990. The B-alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295–306 [DOI] [PubMed] [Google Scholar]
- 73. Smith DG, Garcia-Pedrajas MD, Gold SE, Perlin MH. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50:259–275 [DOI] [PubMed] [Google Scholar]
- 74. Snoeijers SS, Perez-Garcia A, Joosten MHAJ, De Wit PJGM. 2000. The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 106:493–506 [Google Scholar]
- 75. Spellig T, Bottin A, Kahmann R. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503–509 [DOI] [PubMed] [Google Scholar]
- 76. Stergiopoulos I, de Wit PJGM. 2009. Fungal effector proteins. Annu. Rev. Phytopathol. 47:233–263 [DOI] [PubMed] [Google Scholar]
- 77. Talbot NJ, Ebbole DJ, Hamer JE. 1993. Identification and characterization of Mpg1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Teichmann B, Linne U, Hewald S, Marahiel MA, Bolker M. 2007. A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis. Mol. Microbiol. 66:525–533 [DOI] [PubMed] [Google Scholar]
- 79. Teichmann B, Liu LD, Schink KO, Bolker M. 2010. Activation of the ustilagic acid biosynthesis gene cluster in Ustilago maydis by the C2H2 zinc finger transcription factor Rua1. Appl. Environ. Microbiol. 76:2633–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Todd RB, Fraser JA, Wong KH, Davis MA, Hynes MJ. 2005. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot. Cell 4:1646–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tsukuda T, Carleton S, Fotheringham S, Holloman WK. 1988. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8:3703–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tudzynski B, Homann V, Feng B, Marzluf GA. 1999. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol. Gen. Genet. 261:106–114 [DOI] [PubMed] [Google Scholar]
- 83. Van den Ackerveken GF, et al. 1994. Nitrogen limitation induces expression of the avirulence gene Avr9 in the tomato pathogen Cladosporium fulvum. Mol. Gen. Genet. 243:277–285 [DOI] [PubMed] [Google Scholar]
- 84. Voisard C, Wang J, McEvoy JL, Xu P, Leong SA. 1993. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13:7091–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wahl R, Zahiri A, Kaemper J. 2010. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol. Microbiol. 75:208–220 [DOI] [PubMed] [Google Scholar]
- 86. Wiemann P, et al. 2009. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol. Microbiol. 72:931–946 [DOI] [PubMed] [Google Scholar]
- 87. Wong KH, Hynes MJ, Davis MA. 2008. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 7:917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xiao XD, Fu YH, Marzluf GA. 1995. The negative-acting Nmr regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein Nit2. Biochemistry 34:8861–8868 [DOI] [PubMed] [Google Scholar]
- 89. Young JL, Jarai G, Fu YH, Marzluf GA. 1990. Nucleotide sequence and analysis of Nmr, a negative-acting regulatory gene in the nitrogen circuit of Neurospora crassa. Mol. Gen. Genet. 222:120–128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.