Abstract
The regulation of the Saccharomyces cerevisiae GAL genes in response to galactose as a source of carbon has served as a paradigm for eukaryotic transcriptional control over the last 50 years. Three proteins—a transcriptional activator (Gal4p), an inhibitor (Gal80p), and a ligand sensor (Gal3p)—control the switch between inert and active gene expression. The molecular mechanism by which the recognition of galactose within the cell is converted into a transcriptional response has been the subject of considerable debate. In this study, using a novel and powerful method of localizing active transcription factors within the nuclei of cells, we show that a short-lived complex between Gal4p, Gal80p, and Gal3p occurs soon after the addition of galactose to cells to activate GAL gene expression. Gal3p is subsequently replaced in this complex by Gal1p, and a Gal4p-Gal80p-Gal1p complex is responsible for the continued expression of the GAL genes. The transient role of the ligand sensor indicates that current models for the induction and continued expression of the yeast GAL genes need to be reevaluated.
INTRODUCTION
The Saccharomyces cerevisiae GAL genes encode the enzymes of the Leloir pathway that are required for the conversion of galactose into a metabolically useful form, glucose-6-phosphate (23). When yeast cells are grown in the absence of galactose, the GAL genes are, for the most part, transcriptionally inert. In the absence of repressing carbon sources, such as glucose, Gal4p is produced in the cell and can be found tethered upstream of the GAL genes (8). Under these conditions, the activity of DNA-bound Gal4p is inhibited by its interaction with another protein, Gal80p, which associates with the activation domain of Gal4p (20). If galactose becomes available as a carbon source, then the GAL genes are transcribed rapidly and mRNA is produced to a high level (31). Although the presence of galactose within the cell triggers the activation of Gal4p, neither Gal4p nor Gal80p functions as the galactose sensor. Instead, a ligand sensor, Gal3p, interacts with the transcriptional inhibitor Gal80p, in a galactose- and ATP-dependent manner (32). Gal3p appears to require galactose and ATP so that it can adopt the conformation required for its interaction with Gal80p (23). The net result of this interaction is that Gal4p becomes active and transcription of the GAL genes proceeds.
Understanding the interplay of the GAL-encoded regulatory proteins is the key to understanding the molecular basis of transcriptional control of these genes. Several apparently contradictory models have been proposed for the induction of GAL gene expression. It has been suggested that induction of the GAL genes occurs via the association of a tripartite complex formed between Gal4p, Gal80p, and Gal3p, resulting in a conformational change in Gal80p and thus relieving its inhibitory effects (20). In favor of this model are the observations that (i) Gal4p purified from yeast grown in either the presence or absence of galactose is associated with ScGal80p (17); (ii) artificially constructed Gal80p molecules that contain an activation domain can regulate transcription in the presence and absence of galactose (12); (iii) Gal4p, Gal80p, and Gal3p can assemble in vitro in a mobility shift assay (20); and (iv) fluorescence resonance energy transfer assays conducted in vivo indicate that Gal4p and Gal80p do not dissociate from each other in the presence or absence of galactose (4). Other evidence, however, suggests that Gal80p dissociates from Gal4p and interacts with Gal3p in the cytoplasm of yeast cells (18). The dissociation of Gal80p would result in the freeing of Gal4p from the inhibitory effects, enabling transcriptional activation to occur (19). The dissociation model is supported by data indicating that the expression of a myristoylated version of Gal3p (which is targeted to the plasma membrane of the cell) does not unduly impair the induction of the GAL genes (19). Chromatin immunoprecipitation (ChIP) experiments (19) and pulldown assays (24) both suggest that the Gal4p-Gal80p complex is somewhat weakened (although perhaps not completely dissociated) when cells are grown in the presence of galactose. Recently published data, however, suggest that nuclear-cytoplasmic shuttling of Gal80p is not required for the expression of the GAL genes and that the induction of GAL gene expression can occur with both Gal80p and Gal3p in the nucleus (5).
In this study, using a novel protein mislocalization strategy of tethering Gal4p to the inside surface of the nuclear membrane, we show that under inducing conditions, Gal80p does not dissociate from transcriptionally active Gal4p. Furthermore, we show that induction proceeds through the transient association of Gal3p with the Gal4-Gal80p complex. Later in the induction cycle, Gal3p is replaced by Gal1p. Gal1p therefore plays a regulatory role during the normal GAL gene expression induction cycle, rather than, as previously postulated, fulfilling this function only in the absence of Gal3p (2).
MATERIALS AND METHODS
Yeast strains, media, and transformation.
Saccharomyces cerevisiae strains were derived from strains FY23 (29) and JPY9 (1). A list of the yeast strains used in this study can be found in Table S1 in the supplemental material. Strains producing Gal3p-CFP, Gal80p-CFP, and Nup49-mCherry fusion proteins were constructed as described previously (28). The lithium acetate transformation procedure was used for introduction of DNA into yeast cells (7). Transformed strains were grown in yeast extract-peptone-dextrose (YPD) medium supplemented with hygromycin B (100 μg/ml) for selection of cells harboring an mCherry-tagged protein or in an appropriate synthetic nutrient dropout medium (SC) for nutrient selection. Where appropriate, raffinose or galactose was added as the sole carbon source, to a final concentration of 2% (wt/vol). In liquid culture, cells were grown at 30°C with constant agitation at 200 rpm.
Adding fluorescent tags to proteins produced from their genomic loci.
The fusion of DNA sequences to the 3′ end of an open reading frame (ORF) in its native genomic context was performed by PCR, using the tagging vectors pBS35 (Yeast Resource Center [http://depts.washington.edu/yeastrc]), carrying the gene encoding the mCherry (mCh) tag and a hygromycin B selection marker, and pKT174 (Euroscarf [http://web.uni-frankfurt.de/fb15/mikro/euroscarf]), carrying the yECFP gene and the URA3 selection marker. Briefly, GAL1-mCh was produced by PCR using oligonucleotides F7 and R7 (a list of the oligonucleotides used in this study can be found in Table S2 in the supplemental material), and GAL10-CFP was generated using primers F8 and R8. The GAL1-mCh and GAL10-CFP fusions were verified by PCR using primers F71 and F81.
GAL7-mCh was constructed by cloning PCR products into the plasmid YIplac204 (6). A region of 640 bp downstream from the GAL7 stop codon was amplified using primers F9 and R9, cut with SphI and XbaI, and cloned into the same sites of YIplac204. The GAL7 open reading frame (lacking a stop codon) was then amplified using primers F10 and R10 and cloned into the XbaI and KpnI sites. Finally, mCherry was amplified using the primers F11 and R11 and inserted into the KpnI and EcoRI sites. The resulting plasmid was linearized using XbaI and transformed into FY23::GAL10-CFP. After transformation, yeast cells were selected for the ability to grow on medium lacking tryptophan. Plasmid integration resulted in modification of the GAL7 locus such that the open reading frame was fused, at its 3′ end, to the DNA sequence encoding mCherry.
Cassettes for gene knockouts.
Gene knockouts were performed based on a previously described gene replacement strategy (22). For the knockout of GAL1, a 588-bp upstream region starting from the start codon (UP) and a 559-bp downstream region starting from the stop codon (DN) of GAL1 were generated by PCRs using primers carrying sequences for suitable restriction enzyme sites: F12 with a BamHI site and R12 with an EcoRI site for the UP region and F13 with a PstI site and R13 with a SalI site for the DN region. Both PCR products were cloned sequentially into YIplac204 (6). The resulting plasmid was linearized using BamHI and transformed into FY23::GAL3-CFP or FY23::GAL80-CFP. Transformants that were defective for growth on galactose were verified by PCR using primers F14 and R14.
The knockout of the GAL4 gene was also carried out by PCR-mediated gene replacement. The TRP1 cassette was amplified from YIPlac204 by use of oligonucleotides F15 and R15. The PCR product was transformed into strains FY23::GAL80-CFP::GAL1-mCh and FY23::GAL3-CFP::GAL1-mch. The deletion of the GAL4 gene in transformants that displayed an inability to grow on galactose as the sole carbon source was verified by PCR using primers F16 and R14 (see Table S2 in the supplemental material).
The knockout of GAL80 was performed using the same general strategy as that described above for the disruption of GAL4. Primers F17 and R17 were used for amplification of a TRP1 cassette, and the PCR product was transformed into FY23::GAL1-mCh. The GAL80 gene knockout was verified by PCR using primers F18 and R18.
Plasmids for protein production.
A list of the plasmids used in this study can be found in Table S3 in the supplemental material. pBluescript SK II(+) (Stratagene) was used for the cloning and assembly of DNA fragments, which were subsequently inserted into yeast expression vectors. Plasmids for the production of proteins in yeast were based on pRJR247, a 2μm vector containing the actin promoter (PACT1) and terminator and LEU2 and bla selection markers for yeast and bacteria, respectively. Escherichia coli strain DH5α was used as the bacterial host for plasmid propagation. The sequences of all plasmid constructs were confirmed by sequencing at the DNA sequencing facility of The University of Manchester.
pDA128 was constructed as a plasmid to produce yellow fluorescent protein (YFP) fused to ULP11–150, resulting in the tethering of YFP to the nuclear membrane. It was created by fusing DNA encoding amino acids 1 to 150 of Ulp1p (16) with residues 92 to 132 of the lambda (λ) repressor linker (15) and the YFP gene variant Ypet (14). The construct was assembled in three steps. ULP11–150 was amplified by using primer F1 to add restriction sites for HindIII and NcoI and primer R1 to add an EcoRI restriction site. The HindIII/EcoRI-digested PCR product was ligated into pBluescript SK II to generate pDA95. Second, a 123-bp λ linker was generated by PCR, using λ phage DNA as a template and primers F2 and R2, such that the amplified product contained restriction sites for EcoRI and BamHI. The amplified λ DNA fragment was inserted into the pDA95 plasmid by use of EcoRI and BamHI to produce pDA97. The ULP11–150-λ fragment was excised from the pDA97 plasmid by use of the NcoI and BamHI restriction enzymes and then ligated into pRJR247 (2μm LEU2 PACT1) to generate pDA99. Next, the gene encoding YFP was amplified by PCR, using genomic DNA from the yeast strain FY23::GAL4-Ypet (28) and primers F3 and R3. The product was digested with the restriction enzymes BamHI and SalI and cloned into the appropriate restriction sites of pDA99 to generate pDA128, which produced the Ulp1p-YFP fusion protein from the ACT1 promoter.
Full-length Gal4p fused with Ulp1 and YFP was produced using a genomic GAL4-YFP gene construct amplified by PCR from the yeast strain FY23::GAL4-Ypet (28) by use of primers F4 and R4. The amplified product was digested with BamHI and ligated into pDA99 to obtain a construct (pDA102) in which ULP11–150-λ was fused in frame to GAL4-YFP.
The major activation domain (AD) of Gal4p is located at the carboxyl-terminal end of the protein (amino acids 768 to 881) (13). A fusion of Ulp1p to the Gal4p AD and YFP was made by assembling PCR products as follows. First, DNA carrying ULP11–150 was amplified using primer F5 (to create an NcoI restriction site) and primer R5. Second, GAL4AD768–881-YFP was amplified from genomic DNA of the yeast strain FY23::GAL4-Ypet (28) by use of primers F6 and R6, creating a SalI restriction site. The separately amplified sequences were combined by a PCR using the amplified products as templates and the flanking primers F5 and R6 (see Table S2 in the supplemental material). The fused PCR product was digested with NcoI and SalI and cloned into pRJR247 to generate pDA58.
The expression of GAL4 under the control of the yeast actin promoter (PACT1) for complementation of a gal4-null mutant strain was accomplished by replacement of the HIS3 marker cassette from plasmid pRJR191 (30) with the LEU2 marker cassette and ACT1 promoter from plasmid pRJR247 (30). Both plasmids were cut with the restriction endonucleases AatII and NcoI, and the appropriate fragments were isolated and ligated together.
Live-cell imaging microscopy.
For imaging, yeast cells were mounted in synthetic dropout medium (26). A drop of culture was placed onto a Superfrost Plus glass slide (Menzel-Glaser) and left for 2 min. Excess liquid was removed by pipetting and replaced with 5 to 10 μl of growth medium (SD medium supplemented with appropriate sugar sources at a concentration of 2% [wt/vol]). A 22-mm-by-22-mm coverslip was then placed on top. For time-lapse images, 50 μl of culture was left on a glass slide for 5 min at 30°C. Excess liquid was removed, and the cells were gently washed with fresh prewarmed medium. Next, 10 μl of new prewarmed (30°C) SD medium (supplemented with an appropriate carbon source) was placed onto a slide, a coverslip was placed on top, and the sample was immediately imaged. For movies, a 15-member z stack (see Movies S1 and S2 in the supplemental material) or a 25-member z stack (see Movie S3 in the supplemental material) was constructed, with an exposure time of 1 s for the visualization of Gal3p-CFP and 0.5 s for visualization of Ulp1-Gal4AD-YFP. Images were acquired at ambient temperature (25°C).
Fluorescence images were acquired using a Leica 5500B automated upright microscope system (Leica Microsystems) equipped with an oil immersion lens (100×/1.40; HCX PL APO) (Leica Microsystems). The following filter sets were used: for YFP, Chroma set 41028; and for cyan fluorescent protein (CFP) (d436/10×, 455 dcip, d480/40 M) and mCherry, Chroma set 41043. Exposure times were 1 s for Gal80p-CFP, 1 s for Gal3p-CFP, 100 ms for Nup49p-mCh, 100 ms for Gal1p-mCh, 100 ms for Gal7p-mCh, 1 s for Gal10p-CFP, 10 ms for Ulp1-YFP, 10 to 100 ms for Ulp1-Gal4AD-YFP, and 300 ms for Ulp1-Gal4p-YFP. Fluorescence was detected using a DFC360FX camera (Leica Microsystems). Images were acquired using LAS AF ver. 2.3.0 (Leica Microsystems) and ImageJ software (rsbweb.nih.gov/ij/). Apart from global changes to brightness and contrast, no other alterations were made to the images.
HA protein tagging.
To detect indirect interactions between Gal1p and its own promoter, the protein was chromosomally tagged by adding DNA encoding a 3× hemagglutinin (HA) tag to the 3′ end of the open reading frame. Briefly, a cassette containing 3 repeats of the hemagglutinin sequence and an hph selection marker was amplified using plasmid pYM24 (Euroscarf) as a template and primers F19 and R19. The cassette was transformed into yeast strains FY23 and D246 (see Table S1 in the supplemental material), and cells were cultivated in YPD medium with 50 μg/ml hygromycin B. The integration of the 3× HA tag was verified by PCR using primers F20 and R20. The growth of strains harboring Gal1p-3HA on medium containing galactose as the sole carbon source was found to be identical to that of the wild-type strain. GAL3-3HA integration was performed in the same way as that described above, using primers F25 and R25. The genomic integration was verified by PCR using primers F26 and R26 (see Table S1 in the supplemental material). Gal4p was modified with a 3× HA tag by the same procedure as that described above. A cassette was amplified using primers F21 and R21 and transformed into FY23. Tag integration was verified by PCR using primers F22 and R22.
Chromatin immunoprecipitation.
Yeast cells were grown at 30°C in YPD or YP with galactose until they reached an optical density at 600 nm (OD600) of ∼0.8 to 1.0. Chemical cross-linking was performed for 20 min with formaldehyde (final concentration, 1%) and 1.5 mM ethylene glycolbis[succinimidyl succinate]. The reaction was quenched with 125 mM glycine, and the cells were washed 3 times in cold phosphate-buffered saline (PBS). Cells were harvested and resuspended in buffer A (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail). An equal volume of glass beads was added, and the samples were vortexed for 30 min at 4°C. Lysates were collected by centrifugation and sonicated for 15 min (30 s on, 30 s off), using a Bioruptor instrument (Diagenode). After sonication, the lysate was centrifuged (16,000 × g, 10 min), and the chromatin and protein concentrations in collected supernatants were determined.
Immunoprecipitation of samples was performed overnight with protein A Dynabeads (Invitrogen) that were prewashed with bovine serum albumin (BSA) and salmon sperm DNA. ChIP-grade rabbit anti-HA antibody (Abcam) was then added and incubated for 4 h. The beads were washed sequentially with buffer B (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% sodium deoxycholate), buffer C (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA), and 1× TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and finally eluted with buffer D (20 mM Tris-HCl, pH 8.0, 1% SDS, 10 mM EDTA). Following the reversal of cross-links (65°C overnight), proteinase K (final concentration, 0.1 mg/ml) was added and incubated at room temperature for 60 min. DNA was then purified using a Qiagen PCR purification kit (Qiagen).
Real-time PCR.
Quantitative PCR was performed using an iCycler MyiQ real-time detection system (Bio-Rad) and an iScript RT-qPCR system kit with SYBR green (Bio-Rad) as a fluorescent dye for the detection of DNA. PCR was performed under the following conditions: 3 min at 95°C followed by 40 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C. For each PCR, titration of known amounts of DNA was used to calculate the ratio of immunoprecipitated DNA to input DNA (immunoprecipitation efficiency). The PCR primers F23 and R23 were used to amplify a 220-bp DNA fragment corresponding to positions −471 to −251 of the GAL1 gene (upstream activation sequence [UAS]). A 235-bp PCR fragment corresponding to the GAL1 ORF was amplified using primers F20 and R24. Five 10-fold serial dilutions of the input DNA were used to generate a standard curve for each primer pair. The PCR product was verified via analysis of the melting curves and by agarose gel electrophoresis. The ΔCT value was calculated (normalized to the input) for each sample as follows: ΔCT = [threshold cycle (CT) (sample) − CT (input)]. The ΔΔCT value was calculated as [ΔCT (UAS) − ΔCT (ORF)], and the fold difference was calculated as 2−ΔΔCT. For each sample, the amount of ChIP was computed as a percentage of the value for GAL4 occupancy in the presence of galactose. The data from three independent experiments were used to calculate the standard deviation.
Western blot analysis.
Whole-cell lysates were obtained using Cell Lytic Y cell lysis reagent (Sigma). For Western blots, polyclonal rabbit anti-HA (Abcam) and horseradish peroxidase-conjugated goat anti-rabbit (Pierce) antibodies were used as the primary and secondary antibodies, respectively. Proteins were visualized by use of an ECL detection kit (SuperSignal West Dura substrate; Thermo Scientific) and detected using a cooled charge-coupled device (CCD) camera detection system (Fluor-s MultiImager; Bio-Rad). The image exposure time was 3 s.
RESULTS
To address the nature of the interaction between Gal4p, Gal80p, and Gal3p during the galactose induction process, we set out to determine the localization and interactions of each of the regulatory proteins. We and others have previously shown that Gal4p accumulates, albeit at a low level, in the nuclei of cells and that both Gal80p and Gal3p are distributed in both the nucleus and the cytoplasm (10, 28). The low level of Gal4p within cells and its relatively diffuse localization within the nucleus (28) made it technically difficult to determine the compositions of protein complexes that form with it on the promoters of genes. We therefore set out to deliberately mislocalize Gal4p to a specific region of the nucleus. This was achieved by fusing a version of Gal4p, tagged with YFP, to the first 150 amino acids of Ulp1p, a protein that has previously been shown to localize to the inner side of the nuclear membrane (16).
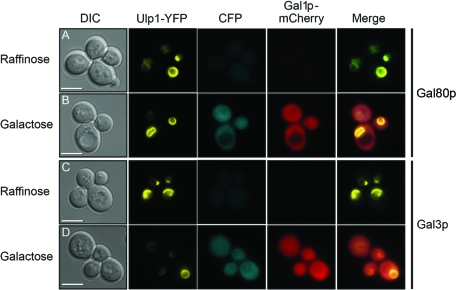
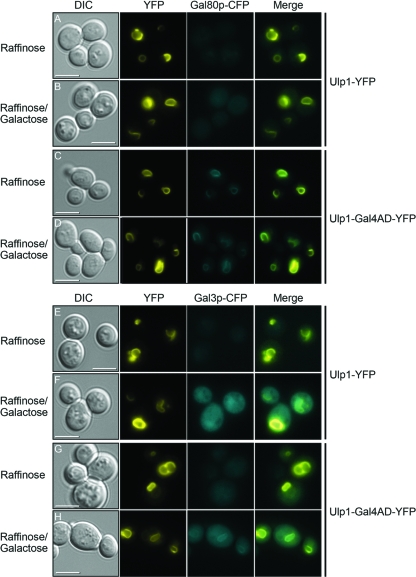
First, we showed that a fusion of Ulp1p to YFP had no effect on the localization of either Gal80p or Gal3p (Fig. 1). The Ulp1-YFP fusion protein was detected as a discrete circle indicating the position of the nuclear membrane and, by inference, the location of the nucleus itself. The localization of Ulp1-YFP was unaltered in the presence of either raffinose (a noninducing carbon source for the GAL genes) or galactose (Fig. 1, Ulp1-YFP panels). In cells expressing this protein, the localization of both Gal80p and Gal3p (each tagged with CFP) was found to be both nuclear and cytoplasmic (Fig. 1, CFP panels), as previously reported for cells not harboring an Ulp1-YFP fusion (28). We have previously shown that these Gal80p and Gal3p fusion proteins function indistinguishably from their untagged counterparts (28). Both Gal80p-CFP and Gal3p-CFP were produced from their genomic loci, and the production of both proteins was found to increase in the presence of galactose (Fig. 1B and D). However, the increase in production levels was not accompanied by an alteration in the cellular location of either protein (Fig. 1B and D). In addition, in the same cells, the production of the galactokinase Gal1p tagged with mCherry was found to increase from undetectable levels in the presence of raffinose (Fig. 1A and C) to a readily detectable signal in both the nucleus and cytoplasm in cells in the presence of galactose (Fig. 1B and D).
Fig 1.
Ulp1-YFP does not affect the localization of Gal3p-CFP or Gal80p-CFP. Yeast cells in which the genomic GAL1 locus had been modified to produce a version of Gal1p tagged at its carboxyl-terminal end with mCherry also produced Ulp1-YFP and either Gal80p-CFP (A and B) or Gal3p-CFP (C and D) from their native genomic loci. Cells were grown at 30°C in 2% raffinose (A and C) as the sole carbon source or taken from this medium, washed, resuspended in medium containing 2% galactose, and cultured for 2 h at 30°C (B and D). DIC, differential interference contrast. The merged panels are overlays of the YFP, CFP, and mCherry images. Bars = 5 μm.
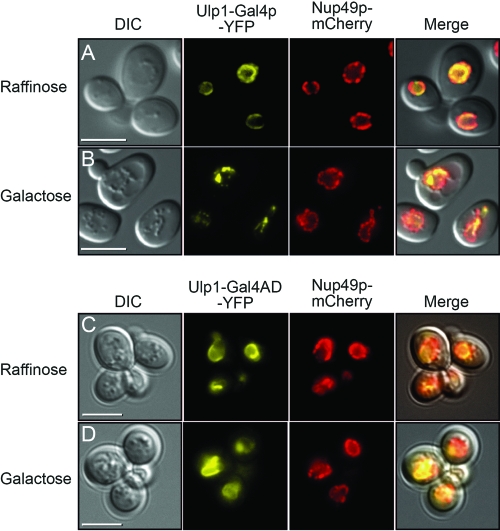
Two fusion proteins were constructed in which Gal4p sequences were tethered to the first 150 amino acids of Ulp1p and YFP. One of these fusion proteins contained just the activation domain of Gal4p (amino acids 768 to 881) fused to Ulp1p (residues 1 to 150) and YFP, while the other contained the full-length Gal4p sequence (amino acids 1 to 881). The localization of these proteins was monitored in cells by fluorescence microscopy to detect the YFP tag (Fig. 2). In the presence of raffinose, both proteins were found to be located at the nuclear membrane (Fig. 2A and C), as indicated by their colocation with the nuclear pore complex protein Nup49p, which was tagged in the same cells with mCherry. In the presence of galactose, the fusion protein containing just the activation domain of Gal4p (Ulp1-Gal4AD-YFP) (Fig. 2D) remained largely colocalized with Nup49p, while the full-length fusion protein (Ulp1-Gal4p-YFP) (Fig. 2B) adopted a more punctate appearance. We assume that the spotted manifestation was related to having a transcriptionally active protein tethered to both DNA and the nuclear periphery. Under galactose-containing conditions, the integrity of the nuclear membrane itself appears to remain intact, as the distribution of nuclear pore complexes (Fig. 2B, Nup49p-mCherry panel) was largely unaltered in these cells after switching from raffinose to galactose (Fig. 2A and B). The fusion protein containing the full-length Gal4p sequence was able to support growth of the cells on galactose when it was expressed in yeast cells that contained no other source of Gal4p (see Fig. S1 in the supplemental material), indicating that the fusion retained the ability to function as an active Gal4p molecule.
Fig 2.
Influence of Ulp1-Gal4p-YFP fusion proteins on integrity of nuclear pore complexes. Yeast cells produced a carboxyl-terminal mCherry-tagged version of Nup49p and a protein composed of the first 150 amino acids of Ulp1p fused to either full-length Gal4p and YFP (A and B) or amino acids 768 to 881 of Gal4p and YFP (C and D). In the presence of raffinose, the YFP and mCherry signals for both the full-length Gal4p and Gal4 AD fusion proteins are similar and indicate the locations of the nuclear membrane and the nuclear pore complexes, respectively. In the presence of galactose, the transcriptionally active full-length Gal4p fusion protein is punctate in appearance, but this does not affect the localization of nuclear pore complexes. The transcriptionally inactive Ulp1-Gal4AD-YFP fusion does not alter the appearance of the nuclear membrane in the presence of galactose. The merged panels are overlays of the DIC, YFP, and mCherry images. Bars = 5 μm.
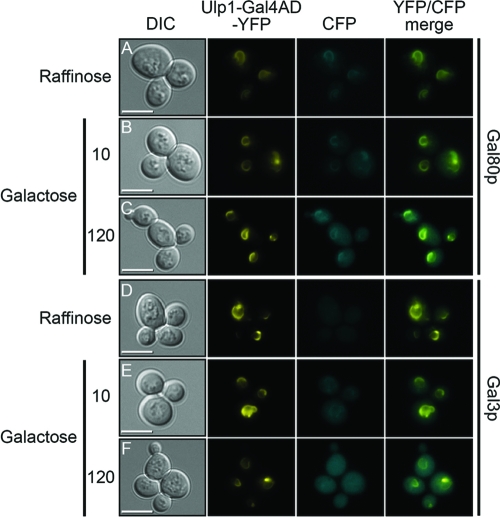
The nuclear membrane-tethered fusion protein containing the Gal4p activation domain was found to localize to the nuclear membrane (Fig. 3, Ulp1-Gal4AD-YFP panel) in a manner similar to that of Ulp1-YFP (Fig. 1), but this protein was able to colocalize Gal80p-CFP to the nuclear membrane (Fig. 3A to C). The localization of Gal80p-CFP to the nuclear membrane by Ulp1-Gal4AD-YFP was observed in both raffinose- and galactose-containing media, suggesting that the complex between Gal4p and Gal80p does not dissociate substantially in the switch between uninduced and induced GAL gene expression. A comparison of the localization of Gal3p-CFP in a yeast strain expressing Ulp1-Gal4AD-YFP showed little difference in the nuclear and cytoplasmic localization of Gal3p in cells grown in the presence of raffinose or in cells that had been grown for a substantial period (>2 h) in galactose-containing medium (Fig. 3D and F). However, at times shortly after the addition of galactose to the cells (∼10 min), we observed a brief localization of Gal3p-CFP at the nuclear membrane (Fig. 3E). At later times in galactose-containing medium, this localization dissipated (Fig. 3F). A time-lapse movie showing the transient association of Gal3p with nuclear membrane-tethered Gal4p is shown in Movie S1 in the supplemental material.
Fig 3.
Influence of Ulp1-Gal4AD-YFP on localization of Gal3p-CFP and Gal80p-CFP. Cells producing a carboxyl-terminally CFP-tagged version of either Gal80p (A to C) or Gal3p (D to F) also produced a fusion protein composed of the first 150 amino acids of Ulp1p, amino acids 768 to 881 of Gal4p, and YFP (Ulp1-Gal4AD-YFP). Cells were grown in 2% raffinose (A and D) or washed from this medium and resuspended in 1% raffinose and 1% galactose (B, C, E, and F). For cells in the presence of galactose, images were collected either 10 min after the addition of the fresh medium (B and E) or after 120 min (C and F). The merged panels are overlays of the YFP and CFP images. Bars = 5 μm.
A similar pattern of Gal80p and Gal3p localization in both raffinose- and galactose-containing media to that observed in Fig. 3 was also observed for a fusion protein composed of full-length Gal4p fused to Ulp1 and YFP (see Fig. S2 in the supplemental material). In raffinose, the Ulp1-Gal4p-YFP fusion protein was localized as a relatively discrete circle with similar characteristics to those of the Ulp1-Gal4AD-YFP protein shown in Fig. 3, and Gal80p-CFP colocalized with this protein under these conditions (see Fig. S2A in the supplemental material). In galactose, the Ulp1-Gal4p-YFP fusion protein adopted a more punctate appearance, but mirroring our results with Ulp1-Gal4AD-YFP (Fig. 3), the colocalization of Gal3p (albeit briefly) and Gal80p was still observed (see Fig. S2B, C, E, and F in the supplemental material).
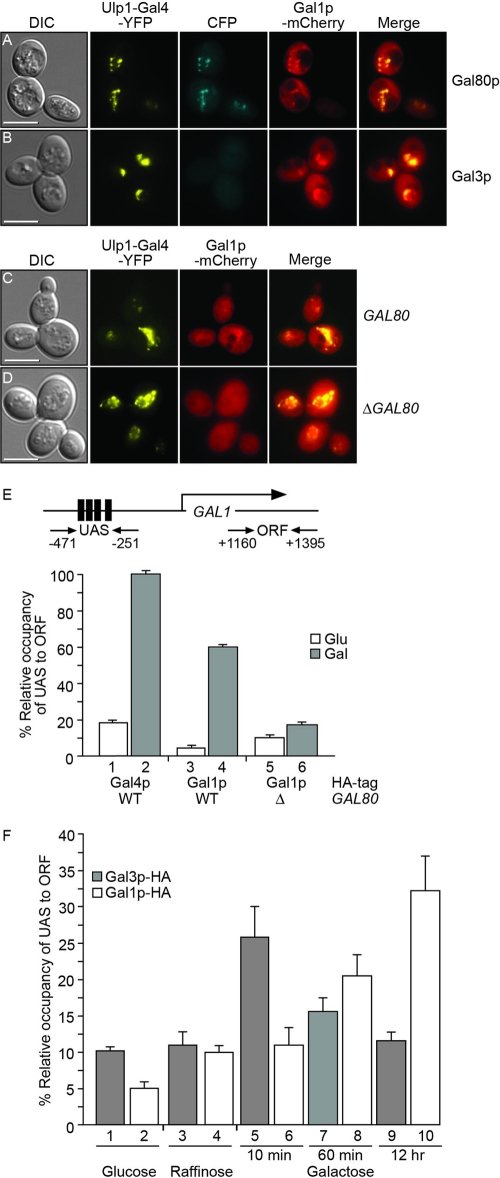
The brief association of Gal3p with a complex of Gal4p and Gal80p may suggest that, for example, Gal80p is altered in some way by the transient association with Gal3p to reduce its inhibitory effect on the transcriptional activity of Gal4p. Evidence for the posttranscriptional modification of Gal80p is very limited. Therefore, we sought to understand how a transient association with a ligand sensor could result in continued and long-term activation of the GAL genes. We have already shown that the galactokinase (Gal1p) is located relatively uniformly in both the nuclei and cytoplasm of yeast cells that are grown in the presence of galactose (Fig. 1B and D). We next asked whether mislocalization of Gal4p was able to influence the localization of Gal1p (Fig. 4). In cells producing Ulp1-Gal4p-YFP as the sole source of Gal4p and grown in galactose for 2 h, we observed colocalization between the YFP signal and Gal80p-CFP (Fig. 4A) but not Gal3p-CFP (Fig. 4B). However, in the same cells, we observed colocalization between the YFP signal and Gal1p-mCherry (Fig. 4A to C). The localization of Gal1p by Ulp1-Gal4p-YFP was not replicated by either Gal7p or Gal10p under similar conditions (see Fig. S3 in the supplemental material). In addition, the deletion of GAL80 abrogated the localization of Gal1p by Ulp1-Gal4p-YFP (Fig. 4D), indicating that the localization of the galactokinase occurred through Gal80p tethered at the nuclear membrane by Gal4p.
Fig 4.
Localization of Gal1p to Gal4p. Yeast cells in which the genomic GAL1 locus had been modified to produce a version of Gal1p tagged at its carboxyl-terminal end with mCherry also produced Ulp1-Gal4p-YFP. In addition, cells were modified to either produce Gal80p-CFP (A) or Gal3p-CFP (B) from the native genomic locus. Yeasts producing Gal1p-mCherry and Ulp1-Gal4p-YFP were also analyzed in a wild-type GAL80 background (C) or in a background in which the GAL80 gene had been deleted (D). Cells were grown at 30°C for 2 h in a galactose-containing medium. The merged panels in panels A and B are overlays of the YFP, CFP, and mCherry images, and those in panels C and D are overlays of the YFP and mCherry images. Bars = 5 μm. (E) Differential protein occupancy by the tagged protein at the GAL1 promoter, calculated as the ratio of binding to the UAS in comparison to that to the ORF, as indicated in the diagram above, was analyzed by chromatin immunoprecipitation. Relative occupancy was calculated using the 2−ΔΔCT method, and the standard deviation was calculated for at least three independent immunoprecipitation experiments. Yeast cells contained either a wild-type GAL80 gene (bars 1 to 4) or a deletion of the gene (bars 5 and 6) and produced either HA-tagged Gal4p (bars 1 and 2) or HA-tagged Gal1p (bars 3 to 6). Cells were grown in the presence of glucose or galactose, as indicated. The occupancy of Gal4p in the presence of galactose was set at 100%, and the error bars indicate standard deviations. (F) Replacement of Gal3p and the GAL1 promoter by Gal1p. The GAL1 promoter occupancies by Gal3p-HA and Gal1p-HA in cells grown in the presence of glucose (bars 1 and 2) or raffinose (bars 3 and 4) or after exposure to galactose for 10 min, 60 min, or 12 h (bars 5 to 10) was calculated by chromatin immunoprecipitation as described for panel E. The occupancy of Gal4p at the UAS in the presence of galactose was set at 100%, and the error bars indicate standard deviations (n = 3).
The association of the galactokinase with Gal80p also occurred at transcriptionally active promoters (Fig. 4E). Chromatin immunoprecipitation of either HA-tagged Gal4p (Fig. 4E, bars 1 and 2) or Gal1p (Fig. 4E, bars 3 and 4) showed a clear increase in the accumulation of the tagged protein at the GAL1 promoter when cells were grown in the presence of galactose. For both Gal4p (9) and Gal1p, protein levels in cells grown under noninducing conditions were considerably lower than those in cells grown in the presence of galactose (see Fig. S4 in the supplemental material). However, in the absence of Gal80p, galactokinase levels were high in both the presence and absence of galactose (see Fig. S4 in the supplemental material), but the lack of Gal80p means that Gal1p did not become localized to active GAL promoters (Fig. 4E, bars 5 and 6). We then used chromatin immunoprecipitation to investigate the timing of the interaction of Gal3p with the GAL1 promoter (Fig. 4F). HA-tagged Gal3p was found to associate with the GAL1 promoter at low levels when the cells were grown in medium containing either glucose or raffinose as a carbon source (Fig. 4F, bars 1 and 3). However, when the cells were switched to medium containing galactose, a relatively strong but short-lived association of Gal3p with the GAL1 promoter occurred (Fig. 4F, bars 5, 7, and 9). This interaction could be detected around 10 min after the addition of galactose to cultures (Fig. 4F, bar 5) but not after the cells were incubated for 60 min or longer (Fig. 4F, bar 7). Over this time frame, the association of Gal1p with its own promoter increased (Fig. 4F, bars 6, 8, and 10), suggesting that the brief association of Gal3p with the transcriptionally active DNA-tethered Gal4p-Gal80p complex is replaced by Gal1p shortly after induction.
Therefore, it appears that the transient interaction between Gal4p, Gal80p, and Gal3p that occurs early after the addition of galactose to yeast cells is replaced by a complex between Gal4p, Gal80p, and Gal1p at later times. A prediction of this observation would be that in the absence of Gal1p, the complex between Gal4p, Gal80p, and Gal3p should be more stable following the switch of sugar source from raffinose to galactose. The results of such an experiment are shown in Fig. 5. In the absence of Gal1p, the interaction between Ulp1-Gal4AD-YFP and Gal80p-CFP was observed in the presence of both raffinose and galactose (Fig. 5C and D). A fusion protein composed of just Ulp1p-YFP was unable to drive the localization of Gal80p-CFP to the nuclear membrane (Fig. 5A and B). However, the absence of Gal1p stabilized the association between Ulp1-Gal4AD-YFP and Gal3p-CFP, such that the association between the two proteins at the nuclear membrane was now observed constitutively in the presence of galactose (Fig. 5H), even when cells were exposed to the presence of galactose for extended periods (12 h). Again, an Ulp1p-YFP fusion protein lacking Gal4p sequences was unable to colocalize Gal3p-CFP to the nuclear membrane (Fig. 5E and F). Movie S2 in the supplemental material shows the formation of a stable complex of Gal3p-CFP with nuclear membrane-tethered Gal4p following the addition of galactose to cells lacking Gal1p. In the absence of Gal1p, the interaction between Gal3p and the Gal4p-Gal80p complex is reversible. The removal of galactose from the medium resulted in the loss of Gal3p localization within 60 min (see Movie S3 in the supplemental material). A stabilization of the interaction between Gal3p and full-length Gal4p in the absence of Gal1p was also noted (see Fig. S5 in the supplemental material).
Fig 5.
Localization of Gal3p in cells lacking Gal1p and producing a nuclear membrane-tethered version of Gal4p. ΔGAL1 yeast cells producing either Gal80p-CFP (A to D) or Gal3p-CFP (E to H) and either Ulp1-YFP (A, B, E, and F) or Ulp1-Gal4AD-YFP (C, D, G, and H) were grown in medium containing either raffinose as the sole source of carbon (A, C, E, and G) or a mixture of raffinose and galactose (B, D, F, and H). Images were collected 12 h following the change of medium. The merged panels are overlays of the YFP and CFP images. Bars = 5 μm.
DISCUSSION
For many years, the yeast GAL genetic switch has been viewed as a paradigm for the control of gene expression in eukaryotic cells. Despite intensive efforts, the precise molecular mechanism by which the recognition of galactose as the preferred source of carbon available to the cell is converted into a suitable transcriptional output has remained obscure. A number of somewhat contradictory models have been proposed to explain a wide range of experimental data. Here we have uncovered a regulatory mechanism that largely explains previously published data. Our observations were built upon the premise of observing protein colocalization in yeast strains where a fluorescently tagged version of Gal4p was mislocalized within the nucleus. It has previously been shown that a fluorescently tagged version of Gal4p produced from its own promoter is technically difficult to detect in yeast cells due to its low level of accumulation (28). We therefore produced our fusion protein from a heterologous promoter and localized it to a specific part of the nucleus to aid in the detection process. The nuclear membrane-tethered version of Gal4p-YFP remains fully functional as a transcription factor for the GAL genes (see Fig. S1 in the supplemental material) and is able to drive the localization of Gal80p from both the nucleus and cytoplasm to the nuclear membrane under both induced and noninduced conditions (Fig. 3). This suggests that Gal4p and Gal80p do not substantially dissociate during GAL induction or that, if dissociation occurs, the proteins are capable of reassociation. During the switch between uninduced and induced GAL gene expression, Gal3p associates with the nuclear membrane only transiently in the presence of tethered Gal4p, and then the protein dissipates across both the nucleus and cytoplasm (Fig. 3; see Movie S1 in the supplemental material). Our results are consistent with recently published data suggesting that under inducing conditions, Gal3p transiently associates with a Gal4p-Gal80p complex (5). We go on to show, however, that the dissipation of Gal3p is coincident with the recruitment of the galactokinase (Gal1p) to Gal4p (Fig. 4). Moreover, we show that Gal1p is recruited to the promoter of its own gene under inducing conditions (Fig. 4E). This recruitment is dependent upon the presence of Gal80p, and hence a complex between Gal4p, Gal80p, and Gal1p is required for the sustained activation of the yeast GAL genes.
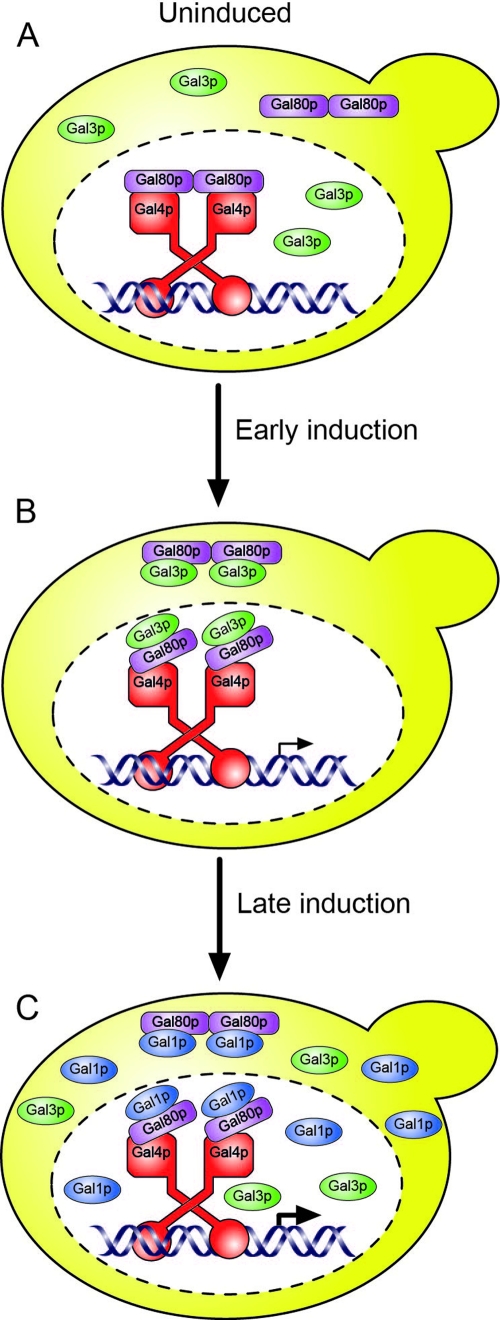
Taken together, our data suggest a model for the induction of the GAL genes that involves the formation of two distinct complexes of proteins. In the uninduced state, the transcriptional activity of DNA-bound Gal4p is inhibited by the physical association with Gal80p (Fig. 6A). Under these circumstances, Gal4p is present in the nucleus, while both Gal80p and Gal3p are found in both the nucleus and cytoplasm of cells. A proportion of Gal80p is presumed to be associated with Gal4p in order to inhibit its transcriptional activation properties. The addition of galactose to the cells is detected by the association of the sugar, in the presence of ATP, with Gal3p. In this form, Gal3p is able to form a complex with Gal80p (Fig. 6B). This results in relief of the inhibitory effect of Gal80p on the ability of Gal4p to activate transcription but does not result in the dissociation of Gal80p from Gal4p. The net result of the relief of inhibition is the transcription of the GAL genes and, in particular, the production of Gal1p. The high-level accumulation of Gal1p within the cell results, we suggest, in the replacement of Gal3p in the Gal4p-Gal80p-Gal3p complex with Gal1p (Fig. 6C). Gal3p shows a high degree of homology with Gal1p, with the two proteins sharing ∼70% amino acid identity and >90% similarity over their entire length. Despite this similarity, Gal3p has no galactokinase activity itself. However, it can be imparted with galactokinase activity, albeit a weak one, by the addition of two amino acids (a serine and an alanine immediately following amino acid 164) which are not normally present in Gal3p (21). This underscores the functional similarities between the proteins. The transient nature of the interaction between the ligand sensor and the Gal4p-Gal80p complex and its replacement by a Gal4p-Gal80p-Gal1p complex suggest that current mathematical models for the activation of the GAL genes need to be reassessed.
Fig 6.
Model for the induction of yeast GAL genes. (A) In the absence of galactose, Gal3p and Gal80p are produced at relatively low levels in the cell, and both proteins may be found in the nucleus and cytoplasm. (B) Galactose entering the cell promotes an interaction between Gal3p and Gal80p. This association occurs, at least in part, at the location of Gal4p and results in the expression of the GAL genes. (C) As Gal1p is produced and accumulates to a high level, the complex between Gal3p and Gal80p is disrupted and replaced by a complex between Gal1p and Gal80p. The Gal1p-Gal80p interaction occurs at the location of Gal4p.
The purpose of or advantage to S. cerevisiae using two different ligand sensor proteins, Gal3p and Gal1p, to regulate GAL gene expression is perhaps not obvious. In terms of the initial response to galactose, it would appear to be of benefit to the cell to have a ligand sensor that does not possess galactokinase activity. The formation of galactose-1-phosphate before that of the protein (Gal10p) that is capable of further processing this metabolite is likely to be toxic to the cell (3). Therefore, the constitutive production of Gal3p provides a mechanism for rapidly inducing all of the genes required for the metabolism of galactose in a concerted fashion. We assume, however, that the massive expression, production, and accumulation of Gal1p that occur in the presence of galactose are sufficient to outcompete Gal3p for the Gal4p-Gal80p complex. This results in the effective removal of Gal3p from the transcriptionally active complex and its replacement by Gal1p. Further experimentation is required to determine whether the activity of the Gal4p-Gal80p-Gal1p complex is substantially different from that of the Gal4p-Gal80p-Gal3p complex.
To date, high-resolution three-dimensional structures are available for Gal80p (26) and for complexes between Gal80p and peptides representing the activation domain of Gal4p (11, 25). In addition, the structure of Gal1p has been solved (27). However, in order to understand the basis of transcriptional control, it will be necessary to understand the interaction, at the molecular level, between Gal80p and the ligand sensor. The data presented here suggest that the interaction between Gal80p and Gal1p represents a tractable target for such analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the BBSRC and The Wellcome Trust.
We thank Ray Wightman and all members of the Reece lab for their help and assistance in the execution and interpretation of the work presented here.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Barberis A, et al. 1995. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell 81:359–368 [DOI] [PubMed] [Google Scholar]
- 2.Bhat PJ, Hopper JE. 1991. The mechanism of inducer formation in gal3 mutants of the yeast galactose system is independent of normal galactose metabolism and mitochondrial respiratory function. Genetics 128:233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat PJ, Hopper JE. 1992. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: evidence for a new model of induction for the yeast GAL/MEL regulon. Mol. Cell. Biol. 12:2701–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik SR, Raha T, Aiello DP, Green MR. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egriboz O, Jiang F, Hopper JE. 2011. Rapid GAL gene switch of Saccharomyces cerevisiae depends on nuclear Gal3, not nucleocytoplasmic trafficking of Gal3 and Gal80. Genetics 189:825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 7.Gietz RD, Woods RA. 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313:107–120 [DOI] [PubMed] [Google Scholar]
- 8.Giniger E, Varnum SM, Ptashne M. 1985. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40:767–774 [DOI] [PubMed] [Google Scholar]
- 9.Griggs DW, Johnston M. 1991. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. U. S. A. 88:8597–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE. 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol. Cell. Biol. 29:5604–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L. 2008. NADP regulates the yeast GAL induction system. Science 319:1090–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leuther KK, Johnston SA. 1992. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science 256:1333–1335 [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Ptashne M. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847–853 [DOI] [PubMed] [Google Scholar]
- 14.Nguyen AW, Daugherty PS. 2005. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23:355–360 [DOI] [PubMed] [Google Scholar]
- 15.Ohashi Y, Brickman JM, Furman E, Middleton B, Carey M. 1994. Modulating the potency of an activator in a yeast in vitro transcription system. Mol. Cell. Biol. 13:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panse VG, Kuster B, Gerstberger T, Hurt E. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21–27 [DOI] [PubMed] [Google Scholar]
- 17.Parthun MR, Jaehning JA. 1992. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol. Cell. Biol. 12:4981–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng G, Hopper JE. 2000. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5140–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng G, Hopper JE. 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. U. S. A. 99:8548–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt A, Reece RJ. 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p/Gal80p/Gal3p complex. EMBO J. 17:4086–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt A, Ross HC, Hankin S, Reece RJ. 2000. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc. Natl. Acad. Sci. U. S. A. 97:3154–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph H, Koenig-Rauseo I, Hinnen A. 1985. One-step gene replacement in yeast by cotransformation. Gene 36:87–95 [DOI] [PubMed] [Google Scholar]
- 23.Sellick CA, Reece RJ. 2005. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 30:405–412 [DOI] [PubMed] [Google Scholar]
- 24.Sil AK, et al. 1999. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol. Cell. Biol. 19:7828–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoden JB, Ryan LA, Reece RJ, Holden HM. 2008. The interaction between an acidic transcriptional activator and its inhibitor: the molecular basis of Gal4p recognition by Gal80p. J. Biol. Chem. 283:30266–30272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoden JB, Sellick CA, Reece RJ, Holden HM. 2007. Understanding a transcriptional paradigm at the molecular level: the structure of yeast Gal80p. J. Biol. Chem. 282:1534–1538 [DOI] [PubMed] [Google Scholar]
- 27.Thoden JB, Sellick CA, Timson DJ, Reece RJ, Holden HM. 2005. Molecular structure of Saccharomyces cerevisiae Gal1p—a bifunctional galactokinase and transcriptional inducer. J. Biol. Chem. 280:36905–36911 [DOI] [PubMed] [Google Scholar]
- 28.Wightman R, Bell R, Reece RJ. 2008. Localization and interaction of the proteins constituting the GAL genetic switch in Saccharomyces cerevisiae. Eukaryot. Cell 7:2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winston F, Dollard C, Ricupero-Hovasse SL. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55 [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Reece RJ, Ptashne M. 1996. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 15:3951–3963 [PMC free article] [PubMed] [Google Scholar]
- 31.Yarger JG, Halvorson HO, Hopper JE. 1984. Regulation of galactokinase (GAL1) enzyme accumulation in Saccharomyces cerevisiae. Mol. Cell. Biochem. 61:173–182 [DOI] [PubMed] [Google Scholar]
- 32.Zenke FT, et al. 1996. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272:1662–1665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.