Abstract
Toxoplasma gondii belongs to the phylum Apicomplexa, a group of obligate intracellular parasites that rely on gliding motility to enter host cells. Drugs interfering with the actin cytoskeleton block parasite motility, host cell invasion, and egress from infected cells. Myosin A, profilin, formin 1, formin 2, and actin-depolymerizing factor have all been implicated in parasite motility, yet little is known regarding the importance of actin polymerization and other myosins for the remaining steps of the parasite lytic cycle. Here we establish that T. gondii formin 3 (TgFRM3), a newly described formin homology 2 domain (FH2)-containing protein, binds to Toxoplasma actin and nucleates rabbit actin assembly in vitro. TgFRM3 expressed as a transgene exhibits a patchy localization at several distinct structures within the parasite. Disruption of the TgFRM3 gene by double homologous recombination in a ku80-ko strain reveals no vital function for tachyzoite propagation in vitro, which is consistent with its weak level of expression in this life stage. Conditional stabilization of truncated forms of TgFRM3 suggests that different regions of the molecule contribute to distinct localizations. Moreover, expression of TgFRM3 lacking the C-terminal domain severely affects parasite growth and replication. This work provides a first insight into how this specialized formin, restricted to the group of coccidia, completes its actin-nucleating activity.
INTRODUCTION
The phylum Apicomplexa contains important protozoan parasites such as Plasmodium, Toxoplasma, Babesia, Eimeria, Neospora, Theileria, and Cryptosporidium spp., that are responsible for severe diseases in humans and farm animals. As obligate intracellular organisms, these parasites are reliant on active invasion of host cells to complete their complex life cycle. Their ability to cross host biological barriers and infect a diversity of tissues requires a unique mode of locomotion called gliding motility, which is powered by the parasite actomyosin system (reviewed in references 8 and 42). While the contributions of actin polymerization and the myosin A motor in motility and invasion are well documented, mainly in Toxoplasma and Plasmodium, the importance of actin and the function of other myosins in the biology of these parasites are still poorly understood.
Toxoplasma gondii tachyzoites replicate by endodyogeny, a process in which two daughter parasites grow within an intact, fully polarized mother parasite (44). The internal daughter cells are delimited by the inner membrane complex and associated subpellicular microtubules. Inheritance of organelles by daughter cells during parasite cell division happens in a highly synchronized fashion, starting with the centriole and Golgi apparatus, followed by the apicoplast, nucleus, and endoplasmic reticulum, and ending with the mitochondrion and de novo synthesis of the micronemes and rhoptries (32). When the daughter cells are fully mature, the maternal apical complex is disassembled and the daughters bud from the mother, adopting her plasma membrane (24). Drugs interfering with actin polymerization or actin filament stability primarily affect gliding motility and block host cell invasion (10, 49). Additionally, the turnover of mother cell organelles during daughter cell budding has also been reported to be affected upon treatment with a high concentration of actin inhibitors, leading to accumulation of organelles in residual bodies formed at the posterior ends of parasites after division (41).
Apicomplexan actin is one of the most divergent among eukaryotes and is regulated by a markedly reduced set of regulatory proteins (3, 18). Filaments of parasite actin are notoriously short and turn over rapidly, thus hampering their visualization in situ (36, 37, 40, 43). Comparative and phylogenetic analyses of apicomplexan genomes identified over 60 candidate actin-related proteins (ARPs) and revealed the presence of 10 actin-related protein groups (18). These proteins share between 17 and 60% amino acid identity with conventional actins. Seven of these ARPs, termed actin-like proteins (ALPs), are novel to apicomplexans (18), and T. gondii ALP1 (TgALP1) was implicated in the formation of daughter cell membranes during parasite division (17). Overexpression of TgALP1 interrupted the formation of the daughter cell inner membrane complex (IMC), leading to delayed intracellular growth (17). Very recently, a new actin-related protein (ARP4) was assigned as a key nuclear protein involved in chromosome segregation in T. gondii (45).
Most remarkably, the apicomplexans lack the canonical actin nucleator Arp2/3 complex and possess instead formins that act as potent nucleators of actin filaments (3, 18, 34). Formins are large proteins that are associated as dimers and implicated in many biological processes, including motility and cytokinesis (16). Typically, the formin homology 2 domain (FH2) nucleates actin assembly by binding the barbed ends of actin filaments, while the formin homology 1 domain (FH1) promotes quick assembly from the profilin-ATP/actin complex (33). In fission yeast, the cell division control protein 12 (Cdc12) is a formin that acts by joining nodes and nucleating actin filaments for the contractile ring formation during daughter cell separation (35).
The T. gondii genome encodes two formins that are well conserved in the Apicomplexa (4) and were recently shown to play important and distinct roles during parasite motility and host cell invasion (7). Additionally, T. gondii possesses a third, divergent, putative formin (3, 39).
Here, we establish that TgFRM3 acts as an actin nucleator and is a formin restricted to the group of coccidians. Biochemical studies demonstrate that the FH2 domain of TgFRM3 binds to Toxoplasma actin and can nucleate rabbit actin, processing assembly in vitro by binding the F-actin barbed ends. Intriguingly, TgFRM3 localizes to large patches predominantly at the apical and posterior ends of the parasite and also appears to associate with the mitochondrion. Disruption of the TgFRM3 gene does not negatively affect the lytic cycle of tachyzoites. Conditional stabilization of the FH2 domain alone or of TgFRM3 lacking the C-terminal part (amino acids 2181 to 2849) severely affects parasite intracellular growth, suggesting that the C-terminal region affects the regulation of TgFRM3's activity.
MATERIALS AND METHODS
Parasite culture.
T. gondii tachyzoites (RH-hxgprt-ko or RH-ku80-ko) were grown in human foreskin fibroblasts (HFF). Selection of transgenic parasites was performed with mycophenolic acid (MPA) and xanthine for hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) gene selection (11), 1 μM Shld-1 for destabilization domain (DD) fusion stabilization (21), or pyrimethamine for dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene selection (12).
Toxoplasma and Escherichia coli vectors.
The primers used in this study are listed in Table S1 in the supplemental material. For the 2854-frm3ko vector, a genomic fragment of 1,497 bp corresponding of the 5′ noncoding sequence of the TgFRM3 gene was amplified by PCR and subcloned into the KpnI and HindIII sites of 2854. The 3′ noncoding sequence of TgFRM3 was amplified from genomic DNA by PCR (1,498 bp) and cloned into the BamHI and XbaI sites of 2854. The final Tgfrm3-ko construct was linearized with EcoRV at both the 5′ and 3′ flanking sequences prior to transfection.
The pTub8DD-mycHisFRM3, pTub8DD-mycHisNtF3 (F3 is the formin 3 FH2 domain), pTub8DD-mycHisF3Ct, and pTub8DD-mycHisF3 vectors were obtained by cloning of the different TgFRM3 genomic PCR fragments into the NsiI and PacI sites in the pTub8DD-myc vector. To generate the pTub8DD-mycHismCherryFRM3 construct, the mCherry sequence was amplified by PCR and cloned into the unique NsiI site in the pTub8DD-mycHisFRM3 vector. The TgMP18 gene, recently also described as TgISP3 (5), was amplified by PCR from Toxoplasma genomic DNA and cloned into the YFP/LIC vector (kindly provided by Vern Carruthers) (5, 25). A portion of the formin 3 FH2 domain was obtained by cloning the cDNA between the NcoI and EcoRI sites in pETHTB (kindly provided by Anne Houdusse).
For Tgfrm3-ki, the TgFRM3 genomic sequence was obtained from the ToxoDB database. Primers were designed to amplify 1,978 bp of the 3′ end of the open reading frame of the gene, which contains a unique AfeI restriction site. The PCR fragment was then cloned into pTUB8MycGFPPfMyoAtailTy-HX (22) between the KpnI and NsiI sites. For the Tgfrm3-ki/ko construct, primers were designed to amplify 1,534 bp upstream from the F3 domain that contain a unique NcoI site. The PCR product was then cloned into pTUB8MycGFPPfMyoAtailTy-HX as described above.
Generation of transgenic T. gondii.
Tgfrm3Ty-ki, Tgfrm3Ty-ki/ko, and 2854-Tgfrm3ko transfections were performed in the ku80-ko strain using 20 μg of vector linearized with AfeI, NcoI, and EcoRV, respectively, and subjected to MPA-xanthine selection (14, 25). DD-FRM3-expressing parasites were obtained in RH-hxgprt-ko and selected for MPA resistance. DD-myc-His-mCherry-FRM3- and MP18-YFP-expressing parasites were cotransformed with 50 μg linearized pTub8DDmycHismCherryFRM3-HX and 100 μg ISP3-YFP-DHFR.
Protein purification and analysis.
His-tagged proteins were purified on Qiagen Ni-nitrilotriacetic acid (Ni-NTA) Superflow resin (no. 30410) under native conditions (31). To detect TgFRM3 proteins, parasite lysates were fractionated on 3 to 8% Tris-acetate precast gels (Invitrogen) using the manufacturer's running buffer, and electrophoresis was continued until the 71-kDa marker reached the bottom of the gel.
Antibodies and immunoprecipitation.
Bacterial expression of a 30-kDa protein corresponding to a portion of the FH2 domain of TgFRM3 (amino acids 1480 to 1735) was used to produce truncated F3 for immunization of rabbits (Eurogentec). Anti-TgGAP45, anti-ACT, anti-myc (9E10), anti-Ty tag (BB2), and anti-MIC4 antibodies were previously described (7, 29, 34). Immunoblots were visualized using a chemiluminescent substrate (Amersham, GE Healthcare).
TgFRM3-3Ty was immunoprecipitated using mouse monoclonal anti-Ty antibodies. To achieve this, 3 × 108 parasites were lysed in buffer A (10 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, 0.5% SDS, 2% Triton X-100, 1% sodium deoxycholate). The incubation of parasite lysates for 3 h at 4°C with anti-Ty antibodies was followed by incubation with protein A beads overnight at 4°C. The beads were then washed 3 times with washing buffer (50 mM Tris-HCl [pH 8.3], 0.5% NP-40, 1 M NaCl), suspended in protein loading buffer, and analyzed by Western blotting using anti-Ty antibodies.
Indirect immunofluorescence assay (IFA) and microscopy.
Parasite-infected HFF cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, followed by 5 min of neutralization with 0.1 M glycine in PBS. Fixed cells were permeabilized with 0.2% Triton X-100 in PBS (PBS-TX) for 20 min and then blocked with 2% bovine serum albumin (BSA) in PBS-TX for 20 min. The cells were then incubated for 1 h with primary antibodies (anti-Myc, anti-Ty [1:1,000], anti-GAP45 [1:5,000], anti-Hsp70 [1:1,000], anti-MIC4 [1:1,000]), followed by goat anti-rabbit or goat anti-mouse immunoglobulin G (IgG) conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Invitrogen). Finally, glass coverslips were incubated for 10 min with 4′,6-diamidino-2-phenylindole (DAPI) in PBS and mounted in FluoromountG (Southern Biotech). Confocal images were collected with a Leica laser scanning confocal microscope (TCS-NT DM/IRB and SP2) using a 1003 Plan-Apo objective with a numerical aperture (NA) of 1.4. Single optical sections were recorded with an optimal pinhole of 1.0 (according to the Leica instructions) and 16 times averaging.
Nickel affinity pulldowns.
Freshly released parasites (3× 108 parasites) were harvested, washed once with buffer G (0.1 mM CaCl2, 5 mM Tris [pH 7.8], 0.2 mM ATP, and 1 mM dithiothreitol [DTT]), and resuspended in the same buffer containing 0.5 mM ATP and protease inhibitors. Successive rounds of freezing and thawing in liquid N2 were performed to break the cells. After ultracentrifugation at 30,000 × g, the supernatant was incubated for 2 h at 4°C with 75 μg of the bait protein (His-F1 [formin 1 FH2 domain], His-F1-IR/AA [actin binding mutant formin 1 FH2 domain], or His-F3 [formin 3 FH2 domain]), followed by incubation with 50 μl of nickel beads (Qiagen) for 1 h at 4°C. The beads were centrifuged and washed 3 times with buffer G. The binding proteins were then eluted and subjected to Western blot analysis using the anti-ACT and His monoclonal antibodies.
Polymerization assays.
Actin was purified from rabbit muscle acetone powder and isolated in monomeric form by gel filtration on Superdex-200 in buffer G. Spontaneous assembly of actin was monitored using the enhancement of the fluorescence of 5% pyrenyl-labeled actin in a Safas Xenius spectrofluorimeter. Conditions were as follows: 2.5 μM MgATP–G-actin, 5 mM Tris Cl (pH 7.8), 0.2 mM ATP, 1 mM DTT, 0.1 mM CaCl2, 0.25 mM EGTA, 1 mM MgCl2, and 0.1 M KCl. Seeded actin assembly assays were performed similarly using spectrin-actin seeds, 2.5 μM pyrenyl-labeled G-actin, and various amounts of F3. The amount of spectrin-actin seeds was large enough (0.11 nM) to provide more barbed ends than nucleation by F3 in the range of F3 concentrations investigated (0 to 9 nM). The initial rate of growth measured without seeds in parallel control samples was subtracted from the initial rate measured with both F3 and seeds. The dependence of the corrected rate ΔV on F3 concentration expressed the saturation of barbed ends by F3, as described by the equation (ΔV0 − ΔV)/(ΔV0 − ΔV∞) = [F3]/([F3] + KF), where [F3] is the concentration of F3, KF is the equilibrium dissociation constant for binding of F3 to barbed ends, and ΔV0 and ΔV∞ represent the corrected rates in the absence and presence of a saturating concentration of F3.
Dilution-induced depolymerization assays were performed by diluting 40-fold a solution of 2.5 μM F-actin (50% pyrenyl labeled) in polymerization buffer containing the desired concentrations of formins. The initial rate of fluorescence decrease was measured. The decrease in rate expressed the binding of F3 to barbed ends. Data were analyzed as described above to derive the equilibrium dissociation constant for binding of F3 to ADP-bound depolymerizing barbed ends.
Plaque assay.
Monolayers of HFF on circular coverslips were infected with parasites in presence or absence of 1 μM Shld-1 for 7 days. Fixation, staining, and visualization were performed as previously described (34).
Intracellular growth.
Parasites were treated for 48 h with or without 1 μM Shld-1, collected promptly after egress, and inoculated onto new HFF monolayers. Twenty-four hours later, the culture was fixed with paraformaldehyde (PFA) and stained with anti-TgGAP45. The numbers of parasites per vacuole were counted for more than 100 vacuoles for each condition.
RESULTS
Formin 3 is restricted to the coccidians.
A bioinformatics search of the T. gondii genome revealed the existence of a third gene coding for an FH2 domain-containing protein (3, 39). TgFRM3 has a predicted molecular mass of 299 kDa (see Fig. S1A in the supplemental material). Atypically, the FH2 domain is located in the second half of the protein sequence between amino acids residues 1480 and 2181 and shows weak homology to the FH2 domains of TgFRM1 and TgFRM2 (see Fig. S1A and S1B in the supplemental material). A survey of EuPathDB revealed that a gene related to TgFRM3 can be found only in the close relative Neospora caninum and in Eimeria tenella, for which only a partial sequence is currently available (see Fig. S1C in the supplemental material). A canonical FH1 domain is not recognizable, although TgFRM3 possesses short stretches of proline residues upstream of its FH2 domain, which could serve to recruit and bind to profilin (see Fig. S1A in the supplemental material). As for the other apicomplexan formins, TgFRM3 lacks the diaphanous autoregulatory domain (DAD) and diaphanous inhibitory domain (DID) (23), and no other obvious domains can be identified (see Fig. S1A in the supplemental material).
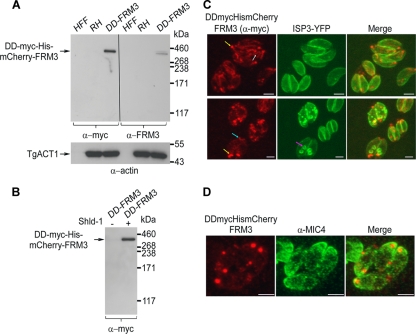
Specific antibodies to TgFRM3 were obtained by immunizing rabbits with a recombinant portion of the FH2 domain purified from E. coli. This reagent failed to detect endogenous TgFRM3 in the tachyzoite stage despite the presence of mRNA coding for the TgFRM3 gene (Fig. 1A; see Fig. S2 in the supplemental material). In contrast, the same antibodies detected the fusion DD-myc-His-mCherry-FRM3, stably expressed at the predicted size of ca. 340 kDa, in transgenic parasites cultivated for 48 h in the presence of Shld-1. Antiactin antibodies were used to detect TgACT1 as loading control (Fig. 1A). Intriguingly, attempts to constitutively express the cDNA of TgFRM3 under the control of the tubulin promoter failed, whereas conditional expression as a fusion with the destabilization domain (DD) allowed expression of DD-myc-His-mCherry-FRM3 and hence circumvented the deleterious effect likely caused by the overexpression of this formin. The DD-TgFRM3 fusion led to tight control of expression at the protein level in the presence of Shld-1 (Fig. 1B), as previously reported for other proteins (21).
Fig 1.
Controlled expression and localization of TgFRM3 in tachyzoites. (A) Western blot analysis of HFF cells, parasite lysates (RHΔhxgprt, a type I virulent strain), and parasite lysates expressing the DD-myc-His-mCherry-FRM3 protein (DD-FRM3) in the presence of Shld-1 for 48 h, probed with either anti-myc or rabbit antiserum specific to TgFRM3. T. gondii actin (TgACT1) served as a loading control. (B) Western blot analysis of parasites grown in presence or absence of Shld-1 for 48 h, using anti-myc antibodies. (C) Immunofluorescence assays performed on transgenic parasites expressing both DD-myc-His-mCherry-FRM3 and ISP3-YFP. Parasites were grown in the presence of Shld-1 for 24 h before fixation. White, blue, and yellow arrows represent a tubular structure and posterior and apical end labeling, respectively. Anti-myc antibodies were used to visualize the DD-myc-His-mCherry-FRM3 protein by immunofluorescence. Scale bars represent 2 μm. (D) Immunofluorescence assays performed on transgenic parasites expressing DD-myc-His-mCherry-FRM3, in the presence of Shld-1. Parasites were grown in the presence of Shld-1 for 24 h before fixation. The endogenous mCherry fluorescence is shown. T. gondii MIC4 was used as a apical microneme organelle marker. Scale bars represent 2 μm.
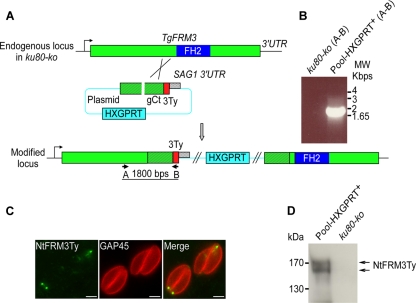
In an attempt to detect expression of endogenous TgFRM3, a parasite line was generated in which a triple Ty-1 tag was inserted at the carboxy terminus of TgFRM3 by recombination at the endogenous locus in a ku80-ko background strain (see Fig. S3A in the supplemental material). The ku80-ko recipient strain facilitates the recovery of homologous recombination events (14, 25). Reverse transcription-PCR (RT-PCR) analysis of the transgenic parasites and sequencing of the product confirmed an in-frame integration of the 3 Ty tags; however, no signal was obtained either by IFA or by Western blotting, in agreement with the absence of detection in tachyzoites with specific antibodies (see Fig. S3B in the supplemental material and data not shown). Despite a very low expression level, we were able to detect TgFRM3-3Ty by Western blotting after concentration by immunoprecipitation from a large number of parasites (3 × 108) using anti-Ty antibodies (see Fig. S3C in the supplemental material).
In order to gather some indirect information regarding the localization of TgFRM3, IFAs were performed on transgenic parasites expressing DD-myc-His-mCherry-FRM3 following treatment with Shld-1. DD-myc-His-mCherry-FRM3 was observed at the apical and posterior ends of parasites as well as along a tubular structure reminiscent of the mitochondrion (Fig. 1C, yellow, blue, and white arrows, respectively). No specific association was observed with the inner membrane complex (IMC) of nascent daughter cells (Fig. 1C, pink and yellow arrows), which was detected as a YFP signal fused to ISP3 (IMC subcompartment protein 3) (5). The large dots formed by DD-myc-His-mCherry-FRM3 were found to cluster at the apical rather than posterior pole of the parasites based on costaining with the apical microneme protein MIC4 (Fig. 1D).
F3 binds to TgACT1 and acts as nucleator of skeletal muscle actin in vitro.
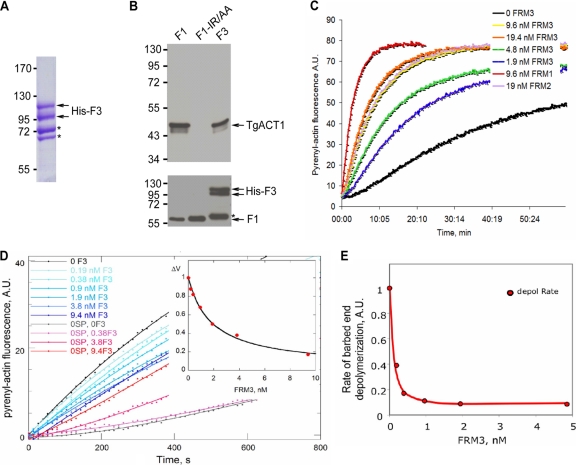
The FH2 domains of formins usually carry the actin-nucleating activity, although they lack the regulatory domains. To demonstrate that the FH2 domain of TgFRM3 (F3) can nucleate actin filaments, its boundaries first had to be delineated using a comparative alignment with the FH2 domains of Toxoplasma formin 1 and formin 2 (see Fig. S1B in the supplemental material). Recombinant F3 was produced and purified from E. coli, and two bands were visualized by SDS-PAGE after His column purification from bacterial extracts (Fig. 2A, arrows). An Ni-NTA-Sepharose bead pulldown assay demonstrated that both F1 (formin1 FH2 domain), used as positive control, and F3 bind to TgACT1 when incubated with parasite lysates (Fig. 2B, upper panel). In contrast, binding to actin was completely abolished when the recombinant actin binding mutant protein F1-IR/AA was used (7) (Fig. 2B, upper panel). As a control, all recombinant FH2 proteins were shown to bind quantitatively to the nickel column (Fig. 2B, lower panel).
Fig 2.
The recombinant TgFRM3 FH2 domain (F3) binds to TgACT1 and polymerizes rabbit actin. (A) Separation of the His-tagged F3 domain expressed in E. coli (BL21) and purified on a nickel column. The F3 protein was loaded on an 8% SDS-polyacrylamide gel, followed by Coomassie blue staining. Asterisks indicate bacterial histidine-rich proteins or chaperone binding to nickel beads, which are eluted with the F3 domain under native conditions. (B) Nickel affinity pulldown assay, measuring the abilities of the TgFRM1 FH2 domain (F1), the F1 actin binding domain mutant (F1-IR/AA), and the F3 domain fused to His to bind to TgACT1. The amounts of TgACT1 and FH2 domains were determined by Western blot analysis using antiactin and anti-His antibodies. The asterisk represents F3 domain degradation. (C) F3 nucleates actin assembly in vitro. Spontaneous actin assembly was recorded in the presence of F3 at the indicated concentrations. (D) F3 inhibits barbed-end growth of actin filaments. The initial rate of barbed end growth was measured at different concentrations of F3 in the presence (blue curves) and in the absence (red curves) of 0.11 nM spectrin-actin (SP) seeds. The normalized difference between the rates measured with and without seeds is plotted versus the concentration of F3 (inset). The calculated curve fits the data with an affinity of 0.55 nM−1 for binding of F3 to barbed ends, leading to 85% ± 5% inhibition of barbed-end growth. (E) F3 caps barbed ends and inhibits barbed-end depolymerization with high affinity. The rate of dilution-induced depolymerization was about 90% inhibited by F3 binding to the barbed ends, with an affinity of 10 nM−1.
To determine if F3 is capable of nucleating actin assembly in vitro, the recombinant protein was examined in a pyrenyl-actin fluorescence assay (Fig. 2C). From the fluorescence intensity data, F3 was three times less efficient in nucleating actin assembly than F1 but was more efficient than F2 (formin 2 FH2 domain) (Fig. 2C). However, the pyrenyl-actin fluorescence spontaneous assembly assay does not discriminate effects on elongation and nucleation of F-actin, and hence it does not provide direct information on the number of nucleated filaments or on the respective rates of growth at barbed and pointed ends in the presence of F3. We verified that the observed polymerization occurs in large part at barbed ends by measuring that it was 60% inhibited by a saturating amount (100 nM) of cytochalasin D (CD), a drug that blocks barbed-end growth specifically. However, CD routinely inhibits filament growth by 90% when the two ends are free. The fact that CD inhibition was limited to 60% hinted that barbed-end growth of filaments nucleated by F3 is slowed approximately 10-fold by F3 binding to barbed ends. This point was further addressed by quantitative analysis of the effect of F3 on barbed-end growth initiated by spectrin-actin seeds, using a large enough quantity of seeds to compete with the nuclei formed by F3. In this assay, F3 inhibited barbed-end growth up to about 80% at extrapolated saturation and at an affinity of 0.55 nM−1 (Fig. 2D). This result further confirms the qualitative evidence derived from the CD data. Furthermore, in dilution-induced depolymerization of filaments, F3 behaved as a strong barbed-end capper that blocked depolymerization, with an affinity of 10 nM−1 (Fig. 2E). In summary, F3 behaves very similarly to F1, which was also shown to block depolymerization of ADP-bound barbed ends. Moreover, the general properties of F3 are similar to those of Cdc12 and DAAM formins, which likewise greatly slow barbed-end growth and depolymerization (2, 26).
TgFRM3 is dispensable for survival of Toxoplasma gondii tachyzoites.
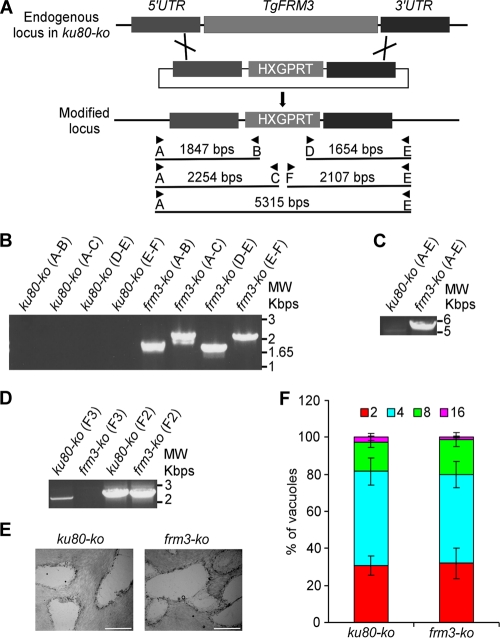
The presence of TgFRM3 at the apical and posterior ends of the parasite suggests a possible role during endodyogeny and/or organelle inheritance (41). To determine the importance of this weakly expressed protein in the lytic cycle of tachyzoites, we generated a vector that would disrupt the gene by integrating within the open reading frame, upstream from the FH2 domain (Fig. 3A). Homologous recombination in the ku80-ko strain was achieved, as demonstrated by genomic PCR analysis on the isolated frm3-ki/ko clone (Fig. 3B). IFA and Western blot analysis confirmed the disruption of the TgFRM3 gene and additionally revealed that a low level of the truncated protein with the expected size was detectable using monoclonal anti-Ty antibodies (Fig. 3C and D). The fact that the truncated form of TgFRM3 is detectable suggests that the carboxy-terminal region confers a much lower level of expression to the full protein. We then checked by semiquantitative PCR the impact of TgFRM3 3′ untranslated region (UTR) exchange on its level of expression at the transcriptional level. We observed no obvious changes in the level of expression of TgFRM3 mRNAs when the SAG1 3′ UTR was used (see Fig. S2 in the supplemental material). Interestingly, a dot-like staining pattern detected at the apical pole of the parasites was consistent with one of the localizations observed when full-length FRM3 fused to DD-myc-His-mCherry was stabilized in the presence of Shld-1 (Fig. 1C and Fig. 3C). These results provide evidence that TgFRM3 is not vital for survival of tachyzoites. Dispensability was definitively confirmed by complete deletion of the gene (Fig. 4A). Genomic PCR analysis confirmed the replacement of the open reading frame of TgFRM3 with the HXGPRT resistance cassette in frm3-ko parasites but not in the parental ku80-ko cells (Fig. 4A, B, and C). RT-PCR confirmed that the TgFRM3 transcript was absent in the frm3-ko strain but that the TgFRM2 mRNAs level was unchanged (Fig. 4D).
Fig 3.
Truncation of TgFRM3 by a knock-in strategy. (A) Insertion of three Ty epitope tags upstream from the FH2 domain of TgFRM3 by single homologous recombination (knock-in). The schematic representation shows the TgFRM3 locus, the gene-targeting construct for gene disruption by single homologous recombination, and the locus resulting from integration of the knock-in construct. The HXGPRT resistance cassette is in blue, the TgFRM3 gene in green, the genomic region coding for TgFRM3 C terminus is in hatched green, the FH2 domain in violet, and the 3 Ty tags in red. The primers used for PCR analysis are indicated. (B) PCR analysis performed on pool-HXGPRT+, showing that single homologous recombination occurred. Genomic DNA from ku80-ko parasites was used as negative control. (C) Immunofluorescence assays performed on transgenic parasites using the monoclonal anti-Ty antibodies. GAP45 was used as a peripheral marker. Scale bars represent 2 μm. (D) Western blot analysis performed on transgenic or ku80-ko parasite lysates probed with anti-Ty antibodies. A doublet at around 170 kDa was detected, corresponding to the truncated NtFRM3Ty.
Fig 4.
Generation of a TgFRM3 knockout. (A) Schematic representation of the strategy used to replace the TgFRM3 open reading frame with the HXGPRT resistance cassette. The knockout plasmid for TgFRM3 contains 1,497 bp of the 5′ untranslated region (UTR) of TgFRM3 (dark gray), the HXGPRT gene (light gray), and 1,498 bp of the 3′ untranslated region (UTR) of TgFRM3 (dark gray). Black arrows represent the primers used for PCR analysis, and the lengths of the PCR products generated are indicated. (B and C) PCR analysis performed on the frm3-ko strain, showing that double homologous recombination occurred. Genomic DNA from ku80-ko parasites was used as a control. (D) Endogenous-locus PCR analysis of ku80-ko and frm3-ko parasites, which shows that TgFRM3 open reading frame mRNA was undetectable in frm3-ko parasites. The TgFRM2 FH2 domain (F2) served as a loading control. (E) Plaque assay performed on an HFF monolayer infected with ku80-ko or frm3-ko parasites. After 7 days, the HFF were stained with Giemsa stain. The scale bar represents 1 mm. (F) Intracellular growth of ku80-ko and frm3-ko strains cultivated in parallel for 48 h. Freshly released parasites from both strains invade new HFF cells. Numbers of parasites per vacuole were counted at 24 h after inoculation. The percentage of vacuoles containing a given number of parasites is represented on the y axis. Values are means ± standard deviations (SD) for three independent experiments.
The phenotypic consequence of TgFRM3 disruption was first examined by plaque assay, which consists of scoring the size of plaques of lysis formed in a monolayer of HFF that recapitulates multiple lytic cycles over 7 days. The frm3-ko parasites formed normal lysis plaques compared with the ku80-ko parental strain (Fig. 4E). Further analysis confirmed that the absence of TgFRM3 did not affect parasite growth and replication (Fig. 4F).
Stabilization of truncated forms of FRM3 severely affects parasite growth and replication.
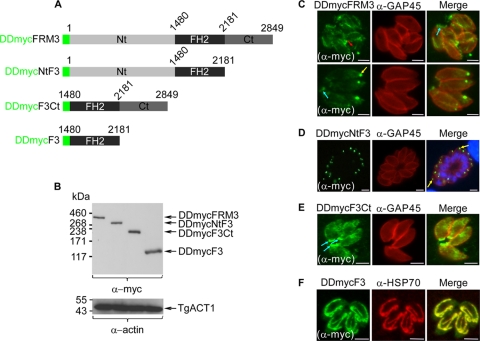
To gain insight into the regulation and localization of TgFRM3, truncated forms of TgFRM3 were fused to DD-myc and stably expressed in parasites (Fig. 5A). The three deletions made corresponded to Nt-F3 (lacking amino acids 2181 to 2849), F3-Ct (lacking amino acids 1 to 1480), or F3 (FH2 domain only). Upon treatment with Shld-1, DDmycFRM3, DDmycNtF3, DDmycF3Ct, and DDmycF3 were expressed and migrated at their expected molecular masses by Western blot analysis (Fig. 5B upper panel). TgACT1 protein was used as a loading control (Fig. 5B, lower panel). In the absence of Shld-1, the products of these transgenes were not detectable (data not shown). DDmycFRM3 exhibited the same localizations to multiple structures, including possibly the mitochondrion (red arrow), as observed with DD-myc-His-mCherry-FRM3 (Fig. 1C and 5C). When the C-terminal region was deleted, the truncated protein appeared to be targeted mainly to the apical part, of the parasites (Fig. 5D, yellow arrows). The same apical localization was observed when the N-terminal part was expressed without the FH2 domain and in the knock-in experiment described in Fig. 3C. In contrast, the DDmycF3Ct was sorted primarily to the posterior ends of the parasites (Fig. 5E, blue arrows). Expression of DDmycF3 alone was unambiguously targeted to the mitochondrion based on colocalization with HSP70 protein (Fig. 5F). Taken together, these results reveal that the three major parts of TgFRM3 possess information for localization and that some of the determinants dominate over the others in the context of the full protein.
Fig 5.
Conditional expression and localization of deletion mutants of TgFRM3 in parasites. (A) Green represents the DD-myc (the destabilization domain and the myc tag), light gray the N-terminal part of TgFRM3 (amino acids 1 to 1479), black the FH2 domain (amino acids 1480 to 2180), and dark gray the C-terminal part of TgFRM3 (amino acids 2181 to 2849). (B) Western blot analysis of parasite lysates expressing the DDmycFRM3 protein, DDmycNtF3, DDmycF3Ct, or DDmycF3 in the presence of Shld-1 for 48 h, probed with anti-myc antibodies. T. gondii actin (TgACT1) served as a loading control. (C, D, E, and F) Immunofluorescence assays performed on transgenic parasites expressing DDmycFRM3, DDmycNtFRM3, DDmycF3Ct, or DDmycF3 using monoclonal anti-myc antibodies. Parasites were grown in the presence of Shld-1 for 24 h prior to fixation. Red, blue, and yellow arrows represent tubular structure and posterior and apical end labeling, respectively. TgGAP45 and putative TgHSP70 were used as pellicle and mitochondrion markers, respectively. Scale bars represent 2 μm.
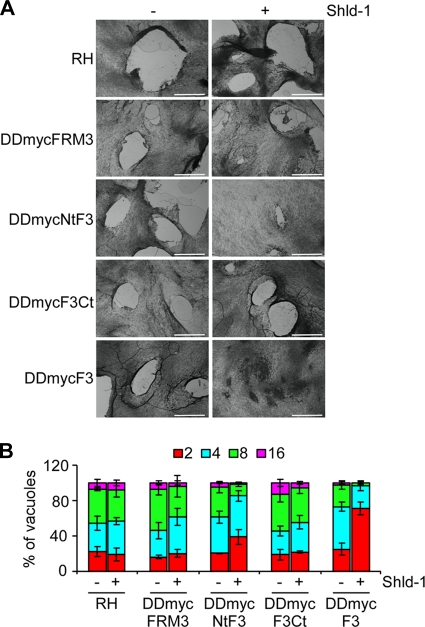
Stabilization of DDmycNtF3 and DDmycF3 in the presence of Shld-1 severely impaired parasite survival as shown by plaque assay (Fig. 6A). Quantification of the number of parasites per vacuole at 24 h postinvasion revealed that these two mutants are considerably impaired in intracellular growth and replication (Fig. 6B). In contrast, expression of DDmycFRM3 and DDmycF3Ct showed no significant defect compared to the parental RH strain by either plaque assay or intracellular replication assay, indicating that their overexpression was not detrimental to parasite growth (Fig. 6A and B).
Fig 6.
Phenotypic analysis of parasites expressing DDmycFRM3, DDmycNtF3, DDmycF3Ct, or DDmycF3. (A) Plaque assays were performed on the RH parental line and on parasites expressing DDmycFRM3, DDmycNtF3, DDmycF3Ct, or DDmycF3. HFF monolayers were infected with parasites in the presence or absence of Shld-1, fixed after 7 days, and stained with Giemsa stain. The scale bar represents 1 mm. (B) Intracellular growth/replication assay of the parasite strains cited above. Parasites were grown in the presence or absence of Shld-1 for 72 h. The percentage of vacuoles containing various numbers of parasites is represented on the y axis. Values are means ± SD for three independent experiments.
DISCUSSION
The elaborate cytoskeleton of apicomplexan parasites provides the framework for organellar partitioning, maintenance of parasite shape, and anchoring of the motor to support motility. The dynamics of actin filaments have been associated primarily with parasite motility and invasion. A role for actin in other steps of the parasite lytic cycle can be envisioned, however, based on the existence of a fairly large repertoire of 11 divergent myosin motors encompassing 5 distinct classes (13). Previous work assessed the role of actin in Toxoplasma division using drugs interfering with F-actin assembly and disassembly (41). As monitored by electron microscopy, CD treatment did not prevent parasite replication but resulted in large residual bodies containing various organelles, suggesting a role of actin in organelle turnover and/or positioning (41).
In T. gondii, actin polymerization and depolymerization are governed by a rather limited set of regulatory proteins, including profilin (34), two formins (4, 7), and an actin-depolymerizing factor (28). Here, the characterization of a new FH2 domain-containing protein (TgFRM3) establishes the existence of a third active formin in T. gondii. TgFRM3 produced in E. coli functions as a nucleator on rabbit actin assembly in spontaneous actin polymerization assays. Interestingly, the His-F3 recombinant protein exhibited a strong barbed-end nucleator activity in filament barbed-end growth assays using spectrin-actin seeds. That result suggests that the region of TgFRM3 retained in F3 possesses processing activity that is in agreement with the postulated conformational change of the FH2–barbed-end complex. This conformational change allows processive elongation of actin filaments. In Saccharomyces pombe, barbed-end elongation with G-actin is inhibited ∼90% by Cdc12p-FH2. However, elongation with profilin-actin is not inhibited by Cdc12-FH2 if the FH1 domain is present. Even more surprisingly, profilin counteracts inhibition of barbed-end depolymerization by Cdc12p-FH2 (26). The TgFRM3 FH2 domain is very divergent from the classical FH2 domains described in the literature. For example, it lacks the conserved W residue found at the beginning of canonical FH2 domains and does not possess the R or K residue known to bind to actin subunits during processive assembly at the barbed ends. Despite the absence of such key residues, TgFRM3 is capable of binding to TgACT1 in vitro and hence qualifies as a formin.
None of the apicomplexan formins characterized to date appear to carry a canonical FH1 domain. The three T. gondii formins harbor regions rich in very short stretches of proline residues directly upstream of the FH2 domain; however, their potential to act as binding sites for profilin is uncertain (7).
Triple epitope tagging at the C terminus of endogenous TgFRM3 revealed that the product of the gene is weakly expressed and not readily detectable by IFA in tachyzoites. Nevertheless TgFRM3 cDNA could be amplified in this stage, and additional epigenomic evidence indicated that the TgFRM3 gene is indeed transcribed (15). Moreover, the presence of specific peptides by mass spectrometry (12 peptides according to Einstein et al. [6] and 11 phosphopeptides according to Treeck and Sanders [47]) (http://toxodb.org/toxo/) indicates that the protein is produced in tachyzoites, and TgFRM3-3Ty was successfully immunoprecipitated with monoclonal mouse anti-Ty antibodies (6, 47). To gain information about its localization, TgFRM3 was expressed in a conditional fashion to eliminate the deleterious effect of overexpression. Unlike TgFRM1 and TgFRM2, which are confined to the pellicle of the parasite (7), TgFRM3 is distributed around the mitochondrion, at the posterior end, and in patches at the apical pole of the parasite. Expression of subdomains of TgFRM3 recapitulates these three localizations, with F3 associated with the mitochondrion, NtF3 with apical patches, and F3Ct mainly at the posterior pole. Given the way the data are generated, information about localization needs to be considered with some level of caution, as truncated proteins expressed via an exogenous promoter may not recapitulate the localization of the full-length, endogenous protein. However, the knock-in experiment with insertion of a tag after the N-terminal region confirmed the apical staining observed with the NtF3 transgene and proved that TgFRM3 is produced at a detectable level in tachyzoites. Assignment of the distinct localizations to different domains of the protein suggests the existence of several interacting partners that might selectively guide the formin to its sites of action. A connection between formins and mitochondria has previously been established, with the diaphanous formin mDia1 affecting the distribution of mitochondria through the regulation of F-actin concentration (30).
Intriguingly, during early endodyogeny, TgFRM3 was found predominantly at the posterior ends of the mother cells. It is well established that the basal complex is assembled at the beginning of daughter parasite assembly and is involved in the initiation of the parasite cortical cytoskeleton. T. gondii MORN1, Centrin2, and DLC1 have been described to be located at the extreme basal end of the parasite (19, 24). Disruption of TgMORN1 altered the structure of the parasite posterior end and affected cytokinesis and apicoplast segregation (20, 27). However, the absence of TgFRM3 at the basal ends of daughter cells distinguishes it from having a MORN1-related role in parasite division. TgFRM3 is conserved and present exclusively in coccidian parasites. This finding is relevant considering the fact that three class XIV myosins (TgMyoD, TgMyoB/C, and TgMyoG) are also restricted to coccidians (13). The alternatively spliced MyoB/C was also reported to locate to the posterior polar ring, and overexpression of the tail caused a substantial replication defect with a severe loss of pellicle integrity (9). Given their partial colocalization, it is possible that TgFRM3 contributes to actin polymerization at the posterior ring and hence assists MyoC function.
The presence of TgFRM3 at discrete sites within the parasite suggests a role in actin assembly at those locations in coccidian parasites. However, the generation of a frm3-ko strain showed no significant impact on parasite growth and survival, suggesting a more prominent role at another life stage of the parasite. Deletion of the TgFRM3 gene was performed in the virulent strain RH, and when assessed in mice, the frm3-ko strain showed no difference in virulence from the ku80-ko parental cell line (data not shown).
As an alternative approach to learn more about the possible mode of regulation of TgFRM3, truncated forms of the formin 3 protein were conditionally expressed in parasites and shown to exhibit differential phenotypes. Deletion of the carboxy-terminal region in both NtF3 and F3 led to a dramatic alteration of parasite growth and replication. In contrast, stabilization of full-length or N-terminal deletion F3Ct caused no defect. These results strongly point toward an important regulatory function of the C-terminal region of the protein even if it does not possess the typical modular domain architectures recently described for formin families (38). Despite the absence of recognizable domains, there is an apparent mechanistic similarity with the autoregulatory domain (DAD), which is also positioned at the C terminus of Dia1 and interacts with the N-terminal FH3 domain and hence leads to autoinhibition of the formin. As for TgFRM3, the truncation of the DAD in Dia1 produces a constitutively active formin variant, which exhibits a strong phenotype of actin cytoskeleton reorganization (1). C-terminal phosphorylation of two formins (FHOD1 and Bni1p) by two different kinases (ROCK and Prk1p, respectively) was reported to disrupt the autoinhibitory interaction between DAD and DID domains, rendering these formins active (46, 48). Interestingly, publicly available phosphoproteomic data have revealed the presence of several phosphorylated residues in the N-terminal region upstream of the FH2 of TgFRM1 and TgFRM3 (http://toxodb.org/toxo/) (47). TgFRM2 and to a less extent TgFRM3 also appear to be phosphorylated in the C-terminal region. These findings suggest a possible role of phosphorylation in the control of the apicomplexan formins, as described for mammalian or yeast formins (46, 48). Further investigations of these technically challenging large proteins would be needed to assess the biological significance of this posttranslational modification. The conditional stabilization of truncated forms of FRM3 revealed here that NtF3 and F3 cause considerable defects in parasite growth and replication without an apparent impact on organelle biogenesis. The presence of the C-terminal domain prevented such a deleterious effect, stressing its importance in controlling TgFRM3 activity.
Given that the presence of FRM3 is restricted to the coccidians, we cannot exclude that this formin performs a more prominent function in other stages of the parasite. We also cannot exclude a functional redundancy with TgFRM1 or TgFRM2.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the Swiss National Foundation (FN3100A0-116722). W.D. was supported by the HHMI. N.K. is supported by the Faculty of Medicine. M.F.C. acknowledges support from an ERC advanced grant 249982 “Forcefulactin,” EU FP7 grant 241548, and the Ligue Nationale contre le Cancer.
We are thankful to Louise Kemp for critical reading of the manuscript. We are grateful to Vern Carruthers for sharing the ΔKu80hxgprt strain and the YFP/LIC vector and to Markus Meissner for generously providing us with the DDFKBP system prior to publication. The vector pETHTB was kindly provided by Anne Houdusse. We acknowledge the publicly available genome databases (EuPathDB, Wellcome Trust, and Sanger Institute) and Fiona Tomley, Arnab Pain, and Adam Reid.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alberts AS. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824–2830 [DOI] [PubMed] [Google Scholar]
- 2. Barko S, et al. 2010. Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J. Biol. Chem. 285:13154–13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. 2006. Regulation of apicomplexan actin-based motility. Nat. Rev. Microbiol. 4:621–628 [DOI] [PubMed] [Google Scholar]
- 4. Baum J, et al. 2008. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe 3:188–198 [DOI] [PubMed] [Google Scholar]
- 5. Beck JR, et al. 2010. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 6:e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Che FY, et al. 2011. Comprehensive proteomic analysis of membrane proteins in Toxoplasma gondii. Mol. Cell Proteomics 10:M110000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daher W, Plattner F, Carlier MF, Soldati-Favre D. 2010. Concerted action of two formins in gliding motility and host cell invasion by Toxoplasma gondii. PLoS Pathog. 6:e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daher W, Soldati-Favre D. 2009. Mechanisms controlling glideosome function in apicomplexans. Curr. Opin. Microbiol. 12:408–414 [DOI] [PubMed] [Google Scholar]
- 9. Delbac F, et al. 2001. Toxoplasma gondii myosins B/C: one gene, two tails, two localizations, and a role in parasite division. J. Cell Biol. 155:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobrowolski JM, Sibley LD. 1996. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84:933–939 [DOI] [PubMed] [Google Scholar]
- 11. Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 [DOI] [PubMed] [Google Scholar]
- 12. Donald RG, Roos DS. 1993. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl. Acad. Sci. U. S. A. 90:11703–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foth BJ, Goedecke MC, Soldati D. 2006. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U. S. A. 103:3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. 2009. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot. Cell 8:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goode BL, Eck MJ. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76:593–627 [DOI] [PubMed] [Google Scholar]
- 17. Gordon JL, Beatty WL, Sibley LD. 2008. A novel actin-related protein is associated with daughter cell formation in Toxoplasma gondii. Eukaryot. Cell 7:1500–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordon JL, Sibley LD. 2005. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genomics 6:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. 2006. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J. Cell Sci. 119:2236–2245 [DOI] [PubMed] [Google Scholar]
- 20. Heaslip AT, Dzierszinski F, Stein B, Hu K. 2010. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathog. 6:e1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herm-Gotz A, et al. 2007. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nat. Methods 4:1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herm-Gotz A, et al. 2002. Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. EMBO J. 21:2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgs HN. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353 [DOI] [PubMed] [Google Scholar]
- 24. Hu K, et al. 2006. Cytoskeletal components of an invasion machine—the apical complex of Toxoplasma gondii. PLoS Pathog. 2:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huynh MH, Carruthers VB. 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8:530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kovar DR, Kuhn JR, Tichy AL, Pollard TD. 2003. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161:875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lorestani A, et al. 2010. A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS One 5:e12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehta S, Sibley LD. 2011. Actin depolymerizing factor controls actin turnover and gliding motility in Toxoplasma gondii. Mol. Biol. Cell 22:1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meissner M, et al. 2002. A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J. Cell Sci. 115:563–574 [DOI] [PubMed] [Google Scholar]
- 30. Minin AA, et al. 2006. Regulation of mitochondria distribution by RhoA and formins. J. Cell Sci. 119:659–670 [DOI] [PubMed] [Google Scholar]
- 31. Moseley JB, Maiti S, Goode BL. 2006. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 406:215–234 [DOI] [PubMed] [Google Scholar]
- 32. Nishi M, Hu K, Murray JM, Roos DS. 2008. Organellar dynamics during the cell cycle of Toxoplasma gondii. J. Cell Sci. 121:1559–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paul AS, Pollard TD. 2009. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66:606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plattner F, et al. 2008. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3:77–87 [DOI] [PubMed] [Google Scholar]
- 35. Pollard TD. 2010. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. 2006. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol. Biol. Cell 17:895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitz S, et al. 2005. Malaria parasite actin filaments are very short. J. Mol. Biol. 349:113–125 [DOI] [PubMed] [Google Scholar]
- 38. Schonichen A, Geyer M. 2010. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803:152–163 [DOI] [PubMed] [Google Scholar]
- 39. Schuler H, Matuschewski K. 2006. Regulation of apicomplexan microfilament dynamics by a minimal set of actin-binding proteins. Traffic 7:1433–1439 [DOI] [PubMed] [Google Scholar]
- 40. Schuler H, Mueller AK, Matuschewski K. 2005. Unusual properties of Plasmodium falciparum actin: new insights into microfilament dynamics of apicomplexan parasites. FEBS Lett. 579:655–660 [DOI] [PubMed] [Google Scholar]
- 41. Shaw MK, Compton HL, Roos DS, Tilney LG. 2000. Microtubules, but not actin filaments, drive daughter cell budding and cell division in Toxoplasma gondii. J. Cell Sci. 113:1241–1254 [DOI] [PubMed] [Google Scholar]
- 42. Sibley LD. 2010. How apicomplexan parasites move in and out of cells. Curr. Opin. Biotechnol. 21:592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skillman KM, et al. 2011. Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites. PLoS Pathog. 7:e1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Striepen B, Jordan CN, Reiff S, van Dooren GG. 2007. Building the perfect parasite: cell division in apicomplexa. PLoS Pathog. 3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suvorova ES, Lehmann MM, Kratzer S, White MW. 2012. Nuclear actin-related protein is required for chromosome segregation in Toxoplasma gondii. Mol. Biochem. Parasitol. 181:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takeya R, Taniguchi K, Narumiya S, Sumimoto H. 2008. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 27:618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Treeck M, Sanders JL, Elias JE, Boothroyd JC. 2011. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10:410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang J, Neo SP, Cai M. 2009. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic 10:528–535 [DOI] [PubMed] [Google Scholar]
- 49. Wetzel DM, Hakansson S, Hu K, Roos D, Sibley LD. 2003. Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol. Biol. Cell 14:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.