Abstract
The abundances of nine transcripts predicted to encode lignocellulose-modifying enzymes were measured over the course of Phanerochaete carnosa cultivation on four wood species. Profiles were consistent with sequential decay; transcripts encoding lignin-degrading peroxidases featured a significant substrate-dependent response. The chitin synthase gene was identified as the optimal internal reference gene for transcript quantification.
TEXT
Phanerochaete carnosa is a white rot fungus that has been found growing almost exclusively on coniferous wood (softwood) (3) and so might employ strategies for degradation that could be developed to reduce the recalcitrance of softwood resources to bioprocessing for the production of liquid fuel and other renewable chemicals. For example, previous transcriptome sequencing of P. carnosa suggested a principal role for manganese peroxidase (MnP) activity in lignin degradation (9). Moreover, the abundance of transcripts encoding lignin-degrading activity were much higher than those encoding carbohydrate-active enzymes, which is consistent with the selective lignin decay elicited by P. carnosa grown on softwood (10) but opposite to previous transcriptomic analyses of Phanerochaete chrysosporium grown on hardwood (12, 20). However, the transcriptome study of P. carnosa analyzed a single time point, and it is known that enzyme and transcript expression in white-rot fungi growing on lignocellulosic substrates can vary over time (2, 16, 18). Therefore, in the present study, P. carnosa was grown on four wood species, and at regular time intervals, reverse-transcription quantitative PCR (RT-qPCR) was used to determine the abundance of transcripts predicted to encode two MnPs, two lignin peroxidases (LiP), a mannanase (Man) from glycoside hydrolase family 5 (GH5; www.cazy.org), a GH6 cellobiohydrolase (Cbh), a GH10 xylanase (Xyl), an acetylxylan esterase (AXE) from carbohydrate esterase family 1 (CE1), and a CE15 glucuronoyl esterase (GE).
P. carnosa strain HHB-10118-sp was grown on 4 g of ground and sifted wood samples prepared from the softwood species balsam fir (Abies balsamea), lodgepole pine (Pinus contorta), or white spruce (Picea glauca) or the hardwood species sugar maple (Acer saccharum), each with B3 buffer as previously described (9). Cultures were inoculated with 11-mm circular plugs from the growing edge of P. carnosa on solid nutrient medium and then incubated at 27°C under stationary conditions. The central 28 mm of fungal growth was harvested from triplicate cultivations at predefined growth points (GP): 4 days (GP1), a 4-cm colony diameter (GP2), and a 6-cm colony diameter (GP3). Wood species did not significantly affect the incubation time required to reach each GP (1-way analysis of variance [ANOVA]; P > 0.1) (see Fig. S1 in the supplemental material).
The RNeasy plant minikit (Qiagen Inc., Mississauga, ON, Canada) was used to isolate total RNA from frozen samples collected at each GP. Reverse transcription was performed using RevertAID H Minus Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Fermentas Canada Inc., Burlington, ON, Canada), T25VN primer, and 30 ng of total RNA in a volume of 50 μl, while real-time qPCR was performed in 25 μl with mixtures containing 2 μl of cDNA, SYBR green JumpStart Taq ReadyMix (Sigma-Aldrich Canada Ltd., Oakville, ON, Canada), and 1 μM gene-specific primers (see Table S1 in the supplemental material). Cycles were run as follows: 1 cycle of 94°C for 2 min and 40 cycles of 94°C for 30 s, 65°C for 30 s, 72°C for 30s, and plate read, with the exception of lip-263501, lip-213241, and ge-247750 (transcript names are based on protein IDs at http://www.jgi.doe.gov/Pcarnosa), for which the annealing temperature was 63°C. Reaction products were quantified based on plasmid standard curves using the DNA Engine Opticon 2 detection system (Bio-Rad Laboratories Canada Ltd., Mississauga, ON, Canada). Plasmid standards were constructed using primers that amplified the qPCR fragments and flanking cDNA sequence (see Table S2 in the supplemental material). Graphical representations of the data and associated statistical analyses were generated using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA).
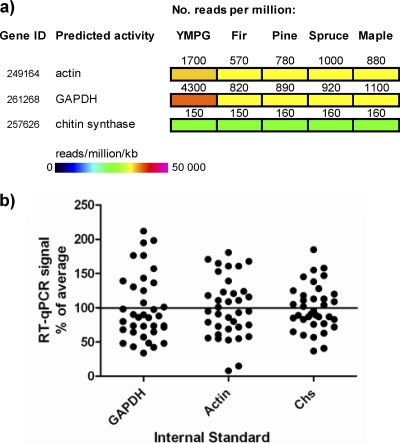
Three transcript sequences were tested as internal reference genes for RT-qPCR. Transcripts for actin and GAPDH have traditionally been used as standards in studies of P. chrysosporium (6, 14, 17). However, previous transcriptomic analysis of P. carnosa suggests that the abundance of these transcripts varies during the growth of this organism on different substrates, and that the level of transcripts encoding a chitin synthase (Chs-257626) is more stable (Fig. 1a) (9). RT-qPCR was therefore used to amplify P. carnosa transcripts encoding actin-249164, GAPDH-261268, and Chs-257626. The standard deviation of transcript abundance for each gene product in all RNA preparations indicated that the abundance of chs-257626 transcripts was less variable than that of transcripts encoding GAPDH-261268 or actin-249164 (Fig. 1b).
Fig 1.
Abundance of transcripts encoding chitin synthase (Chs) is less variable than abundance of transcripts encoding actin or GAPDH in P. carnosa. (a) Heat map representation of P. carnosa transcript abundance during growth on nutrient medium (YMPG), fir, pine, spruce, and maple from mRNA-Seq analysis (n = 1) (9). Protein IDs are from the JGI genome portal (http://www.jgi.doe.gov/Pcarnosa). (b) RT-qPCR amplification of transcripts from P. carnosa grown on fir, pine, spruce, and maple over 3 time points, plotted as the percentage of the average value for each transcript sequence (n = 36). Standard deviations for transcripts encoding Chs, actin, and GAPDH were 33, 43, and 48, respectively.
As determined by absolute quantification, the abundance of all targeted gene transcripts except for axe-248451 changed significantly over time (P < 0.05; 2-way ANOVA) (Table 1). In contrast, the source of wood fiber did not significantly affect transcript abundances (P > 0.05; 2-way ANOVA). Analyses that used chs-257626 levels as an internal reference did not significantly differ from absolute quantifications, whereas analyses that used gapdh-261268 or actin-249164 omitted time as a significant factor for two and five of the eight target transcripts that showed significance under absolute quantification, respectively (Table 1). As is often noted, it is important to test several gene transcripts as internal references to correctly interpret the abundance of queried transcripts by RT-qPCR. In this study, transcript patterns obtained using absolute quantification were consistent with those normalized to transcripts encoding chitin synthase (Table 1; also, see Fig. S2 in the supplemental material).
Table 1.
Two-way ANOVA to determine the effects of time (growth point) and wood substrate on the abundance of each target transcript in P. carnosa
| Target genea |
P valueb (with reference gene) for: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Timec |
Substrated |
|||||||
| None | chs | gapdh | actin | None | chs | gapdh | actin | |
| mnp-256991 | 0.0001 | 0.0001 | 0.0011 | 0.0198 | 0.0869 | 0.0862 | 0.5712 | 0.4424 |
| mnp-262882 | <0.0001 | <0.0001 | 0.0029 | 0.0008 | 0.2617 | 0.2594 | 0.2966 | 0.1134 |
| lip-263501 | 0.0251 | 0.0252 | 0.1332 | 0.2758 | 0.0797 | 0.0800 | 0.1951 | 0.3114 |
| lip-213241 | 0.0445 | 0.0463 | 0.0702 | 0.0291 | 0.8516 | 0.8373 | 0.8278 | 0.6076 |
| man-248589 | 0.0119 | 0.0120 | 0.0065 | 0.2418 | 0.1939 | 0.1944 | 0.2070 | 0.4782 |
| xyl-262694 | 0.0026 | 0.0025 | 0.0018 | 0.2022 | 0.6209 | 0.6098 | 0.9318 | 0.4926 |
| cbh-264060 | 0.0005 | 0.0005 | 0.0003 | 0.2467 | 0.2163 | 0.2153 | 0.1453 | 0.4329 |
| axe-248451 | 0.0885 | 0.0888 | 0.0143 | 0.1628 | 0.2295 | 0.2306 | 0.3021 | 0.5275 |
| ge-247750 | 0.0005 | 0.0005 | 0.0007 | 0.2439 | 0.2445 | 0.2428 | 0.2664 | 0.4227 |
Numbers correspond to protein IDs used by the JGI genome portal (http://www.jgi.doe.gov/Pcarnosa).
P values are shown for absolute transcript abundances (no reference gene) and for transcript abundances normalized using reference genes (chs, gapdh, and actin). Values of <0.05 are in bold.
Null hypothesis: the values of the means from each of the three time points (GPs), without considering the wood substrates, are the same.
Null hypothesis: the values of the means from each of the four wood substrates, without considering the GP, are the same.
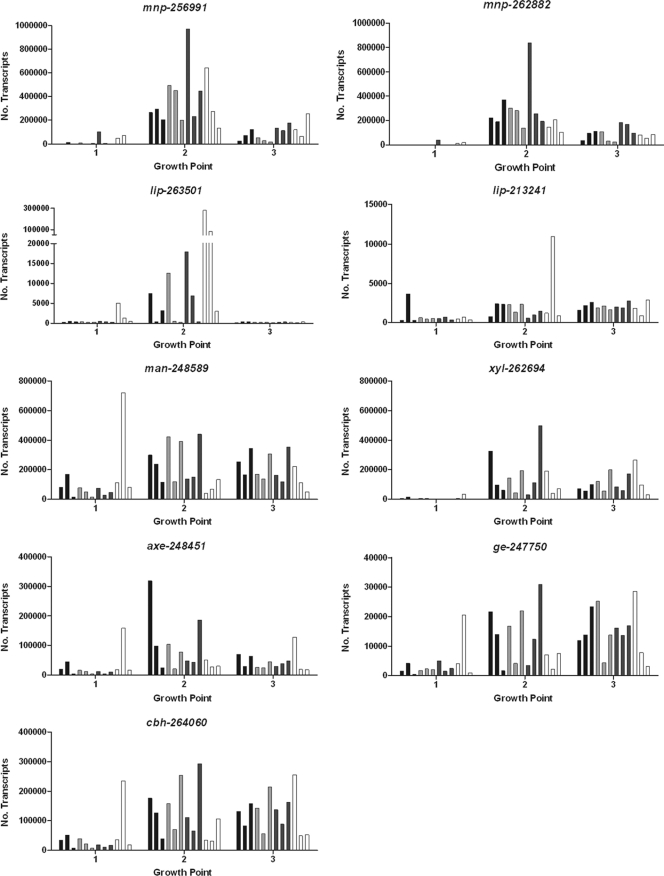
The particular sequences used to monitor the expression of genes that encode lignocellulose-degrading activities were selected based on their being among the most abundant in P. carnosa during growth on wood substrates (9). Manganese peroxidase (MnP) and lignin peroxidase (LiP) are oxidative enzymes that promote the degradation of lignin. In the current study, the abundance of mnp-256991, mnp-262882, and lip-263501 transcripts increased from GP1 to GP2 and then decreased at GP3 during growth on each of the wood substrates, whereas the abundance of lip-213241 remained more similar between GP2 and GP3 (Fig. 2; also, see Data Set S1 in the supplemental material). Overall, the abundance of the mnp transcripts was greater than that of the lip transcripts at GP2 and GP3. In fact, at GP2 the average abundance of transcript mnp-256991 during growth on all wood samples combined was significantly higher than that of any of the other target transcripts except for mnp-262882 (Bonferroni's posttest following repeated-measures ANOVA) (see Table S3 in the supplemental material). At GP2, both mnp transcripts had highest abundance in P. carnosa grown on spruce, and the transcript abundance of mnp-262882 was lowest in P. carnosa grown on maple (Fig. 2). The difference in mnp-262882 abundance between growth on spruce and maple at GP2 was determined to be significant by Bonferroni's multiple comparison posttest following 2-way ANOVA (P < 0.05). In contrast, at GP2 the abundance of lip-263501 was significantly higher in P. carnosa grown on maple than in P. carnosa grown on each of the three softwoods (P < 0.01). Bonferroni's posttest is a conservative estimate of significance because it reduces the chance of obtaining a higher number of false positives as the number of comparisons is increased (13).
Fig 2.
Time-dependent abundance of transcript sequences in triplicate cultivations of P. carnosa grown on fir (black), pine (light gray), spruce (dark gray), and maple (white). Absolute transcript abundances are indicated.
The pattern of mnp and lip transcript abundances in P. carnosa suggests that MnP might be particularly important for the degradation of softwood guaiacyl (G) lignin compared to the guaiacyl-syringyl (GS) lignin of hardwood. Notably, eucalyptus (hardwood) treated with MnP from Ceriporiopsis subvermispora shows a greater percent reduction in G than syringyl (S) lignin units (4), whereas P. chrysosporium, which produces more lip than mnp transcripts under ligninolytic conditions (5, 12, 19), depolymerizes natural and synthetic GS lignins more quickly than natural and synthetic G lignins (7, 11).
Mannanases (Man) and xylanases (Xyl) hydrolyze glycosidic bonds that link backbone sugars of the most abundant hemicelluloses in higher plants. In contrast to mnp and lip transcript abundances, which in most cases peaked at GP2, both man-248589 and xyl-262694 retained higher relative abundance through GP2 and GP3 (Fig. 2; also, see Data Set S1 in the supplemental material). When all wood species were considered together at GP3, the abundance of man-248589 transcripts was significantly higher than the abundance of all other target transcripts, including xyl-262694 (see Table S3 in the supplemental material). This observation is consistent with P. carnosa adaptation to hemicelluloses that predominate in coniferous wood fiber, where glucomannans are 2 to 4 times more abundant than xylan (15). Notably, when maple cultivations were assessed alone, the abundances of man-248589 and xyl-262694 transcripts were deemed more similar. This subtle difference in expression of predicted hemicellulases was not statistically significant but is consistent with the higher xylan content in the maple cultivations.
Cellobiohydrolases (Cbh) degrade cellulose by releasing cellobiose from either the reducing or nonreducing end of cellulose molecules. Overall, the abundance of the cbh-264060 transcript increased from GP1 to GP2 and remained at similar levels from GP2 to GP3.
Acetylxylan esterase (AXE) and glucuronoyl esterase (GE) catalyze the hydrolysis of acetyl groups from acetylated hemicelluloses and ester linkages between hydroxyl groups of lignin and glucuronic acid residues of glucuronoxylans, respectively. Given the partial substitution of the backbone sugars, the levels of transcripts encoding AXE and GE were expected to be lower than those encoding the main-chain glycoside hydrolases. Indeed, at GP2 and GP3, the levels of axe-248451 and ge-247750 transcripts were lower than those of transcripts encoding mannanase and xylanase activities (Fig. 2; also, see Data Set S1 in the supplemental material). Similar to man-248589 and xyl-262694, axe-248451 and ge-247750 had higher relative abundances at GP2 and GP3.
The higher abundance of transcripts encoding MnPs than carbohydrate-active enzymes at GP2, and the prolonged abundance of transcripts encoding carbohydrate-active enzymes through to GP3, is consistent with a sequential mode of decay where lignin is degraded to some extent before the carbohydrate components (1). Indeed, Fourier transform infrared spectroscopy (FT-IR) analysis of the wood substrates used in this study confirmed that lignin degradation most clearly distinguished untreated and fungal treated wood fiber (10).
In summary, transcript profiles revealed comparatively high abundances of transcripts encoding MnP and LiP at early stages of cultivation followed by comparatively high abundances of carbohydrate-active enzyme transcripts at later stages of cultivation. Substrate-dependent patterns of transcript abundance were most evident for transcripts encoding lignin-degrading activity. The relative abundance of transcripts encoding hemicellulase activity at GP3 was mannanase > xylanase > glucuronoyl esterase, which reflects the typical abundance and composition of softwood hemicellulose. However, the P. carnosa genome contains at least two mannanase- and four xylanase-encoding genes (http://www.jgi.doe.gov/Pcarnosa), and only one of each was analyzed in the current study. Finally, an analysis of three internal standards for RT-qPCR revealed that transcript patterns obtained using absolute quantification were consistent with those normalized to transcripts encoding chitin synthase.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Hitoshi Suzuki for providing the cbh qPCR primer.
This work was supported by grants from the Natural Sciences and Engineering Research Council to E.R.M.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Blanchette RA, Otjen L, Effland MJ, Eslyn WE. 1985. Changes in structural and chemical-components of wood delignified by fungi. Wood Sci. Technol. 19:35–46 [Google Scholar]
- 2. Bogan BW, Schoenike B, Lamar RT, Cullen D. 1996. Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:3697–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burdsall HH. 1985. A contribution to the taxonomy of the genus Phanerochaete. Mycologia Memoirs 10:1–165 [Google Scholar]
- 4. Cunha GGS, Masarin F, Norambuena M, Freer J, Ferraz A. 2010. Linoleic acid peroxidation and lignin degradation by enzymes produced by Ceriporiopsis subvermispora grown on wood or in submerged liquid cultures. Enzyme Microb. Technol. 46:262–267 [Google Scholar]
- 5. Doddapaneni H, Chakraborty R, Yadav JS. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doddapaneni H, Yadav J. 2004. Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white-rot fungus Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 65:559–565 [DOI] [PubMed] [Google Scholar]
- 7. Faix O, Mozuch MD, Kirk TK. 1985. Degradation of gymnosperm (guaiacyl) vs angiosperm (syringyl guaiacyl) lignins by Phanerochaete chrysosporium. Holzforschung 39:203–208 [Google Scholar]
- 8. Reference deleted.
- 9. MacDonald J, et al. 2011. Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl. Environ. Microbiol. 77:3211–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahajan S, Jeremic D, Goacher RE, Master ER. 2011. Mode of coniferous wood decay by the white rot fungus Phanerochaete carnosa as elucidated by FTIR and ToF-SIMS. Appl. Microbiol. Biotech. doi:10.1007/s00253-011-3830-1 [DOI] [PubMed] [Google Scholar]
- 11. Otjen L, Blanchette RA, Leatham GF. 1988. Lignin distribution in wood delignified by white-rot fungi: X-ray microanalysis of decayed wood treated with bromine. Holzforschung 42:281–288 [Google Scholar]
- 12. Sato S, Feltus FA, Iyer P, Tien M. 2009. The first genome-level transcriptome of the wood-degrading fungus Phanerochaete chrysosporium grown on red oak. Curr. Genet. 55:273–286 [DOI] [PubMed] [Google Scholar]
- 13. Shaffer JP. 1995. Multiple hypothesis testing. Annu. Rev. Psychol. 46:561–584 [Google Scholar]
- 14. Shary S, et al. 2008. Differential expression in Phanerochaete chrysosporium of membrane-associated proteins relevant to lignin degradation. Appl. Environ. Microbiol. 74:7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sjöström E. 1993. Wood chemistry: fundamentals and applications. Academic Press, San Diego, CA [Google Scholar]
- 16. Suzuki H, Igarashi K, Samejima M. 2009. Quantitative transcriptional analysis of the genes encoding glycoside hydrolase family 7 cellulase isozymes in the basidiomycete Phanerochaete chrysosporium. FEMS Microbiol. Lett. 299:159–165 [DOI] [PubMed] [Google Scholar]
- 17. Suzuki H, Igarashi K, Samejima M. 2010. Cellotriose and cellotetraose as inducers of the genes encoding cellobiohydrolases in the basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:6164–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vallim MA, Janse BJH, Gaskell J, Pizzirani-Kleiner AA, Cullen D. 1998. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl. Environ. Microbiol. 64:1924–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanden Wymelenberg A, et al. 2009. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl. Environ. Microbiol. 75:4058–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanden Wymelenberg A, et al. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.