Abstract
Bacteria of the genus Dehalococcoides play an important role in the reductive dechlorination of chlorinated ethenes. A systems-level approach was taken in this study to examine the global transcriptomic and proteomic responses of exponentially growing cells of Dehalococcoides ethenogenes strain 195 to fixed nitrogen limitation (FNL), as dechlorination activity and cell yield both decrease during FNL. As expected, the nitrogen-fixing (nif) genes were differentially upregulated in the transcriptome and proteome of strain 195 during FNL. Aside from the nif operon, a putative methylglyoxal synthase-encoding gene (DET1576), the product of which is predicted to catalyze the formation of the toxic electrophile methylglyoxal and is implicated in the uncoupling of anabolism from catabolism in bacteria, was strongly upregulated in the transcriptome and could potentially play a role in the observed growth inhibition during FNL. Carbon catabolism genes were generally downregulated in response to FNL, and a number of transporters were differentially regulated in response to nitrogen limitation, with some playing apparent roles in nitrogen acquisition, while others were associated with general stress responses. A number of genes related to the functions of nucleotide synthesis, replication, transcription, translation, and posttranslational modifications were also differentially expressed. One gene coding for a putative reductive dehalogenase (DET1545) and a number of genes coding for oxidoreductases, which have implications in energy generation and redox reactions, were also differentially regulated. Interestingly, most of the genes within the multiple integrated elements were not differentially expressed. Overall, this study elucidates the molecular responses of strain 195 to FNL and identifies differentially expressed genes that are potential biomarkers to evaluate environmental cellular nitrogen status.
INTRODUCTION
Bacteria of the genus Dehalococcoides play a key role in the bioremediation of the carcinogenic and toxic chlorinated ethenes because of their ability to reductively dechlorinate tetrachloroethene (PCE) and its daughter products to the innocuous product ethene (9, 10, 29, 36, 49). While bacteria from diverse genera can dechlorinate PCE and trichloroethene (TCE) to isomers of dichloroethene (DCE), to date, dechlorination beyond DCE to vinyl chloride (VC) and ethene has been observed only for strains of Dehalococcoides (47). Through transcriptional and protein analyses, the reductive dehalogenase (RDase)- and hydrogenase-encoding genes for energy generation in Dehalococcoides isolates and enrichments have been studied extensively (6, 16, 19, 23, 27, 28, 34, 36, 39, 40, 55). However, other aspects of the physiology of Dehalococcoides are not as well understood.
Currently, the genomes of four Dehalococcoides strains have been sequenced, annotated, and published (20, 30, 45). A nitrogenase-encoding operon (nif) for the reduction of atmospheric dinitrogen to ammonia is present in the genome annotation of Dehalococcoides ethenogenes strain 195 (45) but not in the other sequenced Dehalococcoides strains (U.S. Department of Energy Joint Genome Institute Integrated Microbial Genomes [IMG] Database [http://img.jgi.doe.gov/cgi-bin/w/main.cgi]). Interestingly, genomic analyses of three recently isolated Dehalococcoides strains showed that each strain possessed a nif operon (2, 21). Furthermore, a previous study demonstrated that strain 195 is a diazotroph by showing growth in the absence of fixed nitrogen concomitant with the incorporation of labeled 15N2 into biomass (22). However, nitrogen fixation caused strain 195 to prematurely transition into the stationary phase, accompanied by reductions in the overall dechlorination activity and cell density (22).
In order to better understand the underlying reasons for the undesirable physiological responses of strain 195 to fixed nitrogen limitation (FNL), global transcriptomic and proteomic profiles are examined in this study. A DNA microarray targeting the whole genome of strain 195 was used to examine the transcriptome, while shotgun mass spectrometry-based proteomic analyses were used to query the proteome. Previous studies have demonstrated the utility of systems-level investigations for an understanding of the physiology of strain 195 (15, 17, 33) and other microbes (12, 31, 35, 48, 50, 51). The identified differentially expressed genes are also of practical use as biomarkers for diagnosing nitrogen-stressed conditions and directly assessing the physiology of Dehalococcoides (24) so that, together with geochemical measurements, they can be used to optimize contaminated-site conditions for success in bioremediation projects.
MATERIALS AND METHODS
Culture sources and growth conditions.
D. ethenogenes strain 195 was grown in defined minimal medium at 34°C with FNL and with ammonium amendment (AA), as described previously (8, 22). Nitrogen-fixing cultures of strain 195 have been growing stably with routine subculturing in our laboratory for more than 5 years.
Experimental conditions for transcriptomic and proteomic analyses.
In order to generate enough biomass for triplicate biological transcriptomic and proteomic analyses, multiple identical 100-ml cultures were prepared in 160-ml serum bottles. For AA conditions, ammonium (5.6 mM) was added to 12 bottles (4 bottles combined to represent one biological replicate) for each transcriptomic and proteomic analysis, while for FNL conditions, 42 bottles (14 bottles combined to represent one biological replicate) and 27 bottles (9 bottles combined to represent one biological replicate) were prepared for transcriptomic and proteomic analyses, respectively. Cultures were harvested when cells reached the late exponential growth phase, as determined by the amount of VC produced as described previously (22) via a gas chromatograph with a flame ionization detector (23). Cells were harvested by vacuum filtration onto 0.22-μm by 47-mm Durapore membrane filters (Millipore, Billerica, MA). For transcriptomic analysis, 200 ml of culture from two replicate bottles was applied onto each filter, and filters were immediately placed into 2-ml centrifuge tubes, flash-frozen in liquid nitrogen, and stored at −80°C until processing. This collection procedure typically took about 4 min. The samples were combined to generate biological replicates, as indicated above, after RNA extraction. For proteomic analysis, 900 ml or 400 ml of culture was collected from replicate bottles onto each filter for FNL or AA conditions, respectively. The entire cell collection process took less than 20 min prior to the freezing of the samples in liquid nitrogen. Frozen samples were stored at −80°C and later shipped in dry ice to the Oak Ridge National Laboratory for proteomic analysis.
RNA extraction.
Total RNA was extracted from frozen filters by using the RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions and processed as previously described (22). Total RNA was eluted with 45 μl of RNase-free water, and contaminating DNA was removed via the Qiagen RNase-free DNase set (Qiagen, Valencia, CA) according to the manufacturer's instructions. The mass of the total RNA was quantified with a fluorometer (Turner BioSystems, Sunnyvale, CA) using the RiboGreen RNA quantification kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Microarray sample preparation and Affymetrix GeneChips.
Five micrograms of total RNA from each biological replicate was used for each microarray analysis according to the instructions provided by the manufacturer (GeneChip Expression Analysis technical manual; Affymetrix, Santa Clara, CA). Briefly, total RNA was first reverse transcribed to cDNA, the RNA was digested, and the cDNA was fragmented and labeled with biotin. The fragmented and labeled cDNA was hybridized onto the microarray, stained, washed, and scanned by using an Affymetrix Scan300 scanner. Details of the custom-designed Affymetrix GeneChip (Santa Clara, CA) microarray targeting the whole genome of strain 195 were reported elsewhere previously (57). Gene annotations and sequence information were obtained from data described previously by Seshadri et al. (45) and from the Integrated Microbial Genomes (IMG) Database (http://img.jgi.doe.gov/cgi-bin/w/main.cgi).
Shotgun proteomic sample preparation and analysis.
Three biological replicate cell pellets under each experimental condition, each approximately 15 mg in wet weight, were quickly scraped from frozen filters and lysed, denatured, and reduced in a solution of 6 M guanidine and 10 mM dithiothreitol (DTT) in 50 mM Tris buffer (pH 7.6) with shaking overnight at 37°C. The solution was then diluted 6-fold with 50 mM Tris buffer–10 mM CaCl2 (pH 7.6), proteins were digested into peptides with a 1:100 (wt/wt) volume of sequencing-grade trypsin (Promega, Madison, WI), and insoluble cellular material was removed by centrifugation (2,000 × g for 10 min). Peptides were desalted off-line by C18 solid-phase extraction (Waters, Milford, MA), concentrated, filtered, and aliquoted as previously described (54). Two-dimensional nano-liquid chromatography (LC) tandem mass spectrometry (MS/MS) analysis of each sample was carried out with an LTQ mass spectrometer (Thermo Fisher, San Jose, CA) as described elsewhere previously (41), and a brief description can be found in the supplemental material.
Microarray and proteomic data analysis.
Microarray probe set hybridization signal intensities were calculated with the R statistical program (http://www.r-project.org) as described previously (15). The average and median coefficients of variation (CV) among biological triplicates of individual probe set signal intensities were 9.7% and 6.3%, respectively. The linear correlation (r2) of the signal intensities of the respective probe sets between biological replicates under the same conditions was greater than 0.98, and the slope ranged from 0.99 to 1.1. In the proteomic data set, in order for a protein to be considered positively present, two or more peptides had to be detected in all three biological replicates under at least one of the two experimental conditions. Of the three sampling statistics for the proteome (peptide count, spectrum count, and sequence coverage), the spectrum count has been found to have the highest reproducibility and does not saturate as rapidly (26, 58) and was therefore used in this study to identify differentially regulated proteins. The linear correlation (r2) of the spectrum count of the respective proteins between biological replicates under the same conditions was greater than 0.93, and the slope ranged from 0.89 to 1.2. The strong linear correlation (r2) between biological replicates in the respective microarray and shotgun proteomics analyses indicated reasonably high reproducibility.
In order to identify differentially expressed transcripts and proteins between the two growth conditions, a two-tailed Student t test was applied by using the “pt” function from the “multtest” package in R (7). The hybridization intensity and spectral counts were the inputs for the microarray and proteomic analyses, respectively. In the proteome, a value of 0.1 was assigned as the pseudo-spectrum count if no peptide was actually detected in a replicate. This procedure was applied to avoid running into indeterminable situations in the t-statistic calculations for proteins that had detectable peptides under one of the conditions but completely undetectable peptides under the other. The returned P values were corrected for multiple-hypothesis testing by applying a procedure described previously by Benjamini and Hochberg (1), with the “rawp2adjp” function from the multtest package in R. This procedure was used to control the false-discovery rates (FDRs) below 1% for the transcriptomic analysis and below 16% for the proteomic data (beyond this threshold, the FDRs for the next group of proteins were considerably higher). For the transcriptome, a detailed analysis was applied to those differentially expressed transcripts that had an FDR of less than 1%, an absolute hybridization signal intensity of greater than 300 under one of the two conditions, and a 2-fold or higher expression ratio, as described previously (15). Genes that changed by less than 2-fold but had an FDR of less than 1% were included in some analyses for deciphering the overall trend in the data set and interpreting how all genes within a pathway or operon were changing. For the proteome, detailed biological interpretations were drawn from those differentially expressed proteins that passed the FDR and exhibited a 1.5-fold or greater change in the spectrum count. Proteins that fell outside the FDR cutoff but exhibited an increase or decrease of 1.3-fold or greater were included only for the purpose of analyzing the overall trend in Gene Ontology (GO) terms and Clusters of Orthologous Groups (COG) distributions. Throughout this study, upregulation refers to more transcripts or peptides detected under conditions of FNL than with the AA controls, and downregulation refers to fewer transcripts or proteins detected under conditions of FNL.
Analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) algorithm to determine enriched GO terms (3, 14) and the OpWise algorithm (38) to evaluate whether genes within the same predicted operon exhibited similar changes in the microarray data were executed according to previously reported instructions. Details have been included in the supplemental material. Proteomic data showing the three sampling statistics and links to all peptide and protein identifications under each experimental condition are available online (http://compbio.ornl.gov/Dehalo195_FNL).
qPCR and RT-qPCR.
Quantitative PCR (qPCR) was used to analyze the same cDNA generated for the microarray analysis. Amplification of the diluted cDNA sample was performed with SYBR green PCR master mix (Applied Biosystems, Foster City, CA). Details of the reaction setup and primer sequences (see Table S1) can be found in the supplemental material.
Reverse transcription (RT) followed by qPCR was used to analyze the expressions of two genes (DET0138 and DET1576) over a growth cycle of strain 195. The Power SYBR green RNA-to-CT 1-Step kit (Applied Biosystems, Foster City, CA) was used, and details of the reaction setup and primer sequences (see Table S2) can be found the supplemental material. The input for the reaction was RNA collected from a previously reported experiment (22).
Microarray data accession number.
All microarray hybridization signal intensity data have been deposited in the NCBI Gene Expression Omnibus database under accession number GSE28855.
RESULTS AND DISCUSSION
Transcriptomic and proteomic analyses of nitrogen limitation.
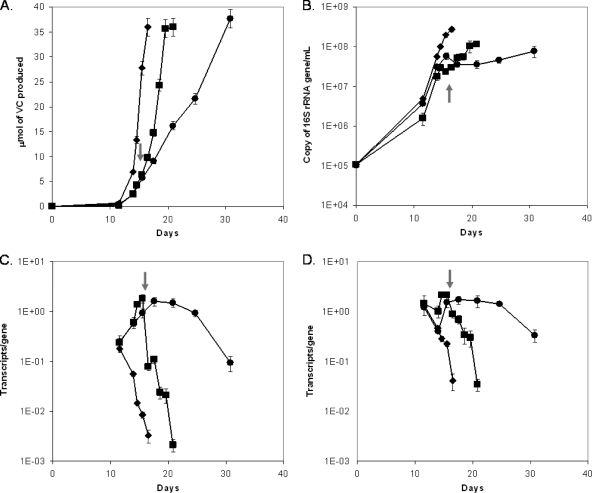
To compare the molecular responses of strain 195 grown with FNL to those of strain 195 grown without FNL, the transcriptomes and proteomes of cells collected in the late exponential phase were examined. Since cultures grown with FNL exhibited lower levels of dechlorination activity and an earlier transition into the stationary phase, two extra weeks were required to generate the same amount of VC from TCE (Fig. 1A and B).
Fig 1.
Three experimental conditions, as previously reported (22), were used to examine the gene transcription of DET1576 and DET0138 in this study. ♦ represents cultures incubated with ammonium, ● represents cultures incubated without ammonium, and ■ represents cultures incubated initially without ammonium but which later received a dose of ammonium at the time indicated by the arrow. (A) Dechlorination profile of VC production. (B) Growth characteristics. (C) DET1576 transcription (this study). (D) DET0138 transcription (this study). Transcription data are averages of triplicate RT-qPCR data from each biological duplicate, and error bars represent 1 standard deviation. (Panels A and B reprinted from reference 22.)
In the transcriptome, a total of 599 genes (see Table S3 in the supplemental material) were differentially expressed with an FDR of less than 1%. Specifically, 84 and 71 genes were differentially up- and downregulated, respectively, 2-fold or greater, with an absolute hybridization intensity of more than 300 units (see Tables S4 and S5 in the supplemental material). These 155 genes represent close to 10% of the 1,641 total predicted genes in strain 195. In the proteome, a total of 721 gene products (see Table S6 in the supplemental material) were positively identified under at least one of the two experimental conditions, representing close to 45% of the 1,590 predicted protein-coding genes in strain 195 (the shotgun proteomic technique inherently does not capture all the expressed proteins within a cell [53]). In the proteome, far more proteins were downregulated (473 proteins) than upregulated (173 proteins) in response to FNL. By applying an FDR of less than 16% and expression ratios of 1.5-fold or greater to the spectrum count, 20 and 33 proteins were found to be differentially up- and downregulated, respectively (Table 1). Some of the differentially expressed proteins showed a clear on/off response, with detectable peptides under only one of the two conditions.
Table 1.
Differentially regulated genes in the proteome
| Gene | Descriptiong | Expression ratioe |
|
|---|---|---|---|
| Proteomef | Transcriptomeb | ||
| Upregulated | |||
| DET0015 | Lipoprotein, putative | 2.3a | 2.2 |
| DET0138 | Phosphate ABC transporter, phosphate-binding protein | 2.5 | 2.6 |
| DET0587 | Conserved hypothetical protein | 3.6 | 1.8 |
| DET0588 | Conserved hypothetical protein | 4.9 | 1.9 |
| DET0754 | Hypothetical protein | 1.8 | 1.3 |
| DET1123 | Glutamine synthetase, type I | 2.8 | ND |
| DET1124 | Nitrogen regulatory protein PII | 77.9 | 5.4 |
| DET1148 | Nitrogenase cofactor biosynthesis protein NifB, putative | 5.0a | 94.3 |
| DET1150 | Ferredoxin, 2Fe-2S | 6.3a | 266.6 |
| DET1154 | Nitrogenase molybdenum-iron protein, beta subunit | 17.0a | 130.6 |
| DET1155 | Nitrogenase molybdenum-iron protein alpha chain | 28.0a | 106.9 |
| DET1156 | Nitrogen regulatory protein PII | 58.3 | 72.3 |
| DET1157 | Nitrogen regulatory protein PII | 89.0a | 114.8 |
| DET1158 | Nitrogenase iron protein | 123.0a | 71.3 |
| DET1161 | Molybdenum ABC transporter, periplasmic molybdate-binding protein | 32.0a | 129.6 |
| DET1162 | Transcriptional regulator, putative | 6.0a | 38.1 |
| DET1377 | Hypothetical protein | 1.8 | 1.5 |
| DET1407 | BNR Asp-box repeat domain protein | 2.1 | ND |
| DET1503 | Ferrous iron transport protein B, putative | 4.3a | ND |
| DET1627 | Sodium hydrogen exchanger family protein | 5.3a | ND |
| Downregulated | |||
| DET0017 | Hypothetical protein | −3.0c | −1.7 |
| DET0022 | Hypothetical protein | −2.3 | ND |
| DET0146 | [Fe] hydrogenase, HymB subunit, putative | −1.8 | ND |
| DET0178 | Sensor histidine kinase | −2.7c | 1.5 |
| DET0185 | Formate dehydrogenase accessory protein FdhE, putative | −2.3 | ND |
| DET0341 | S-Adenosyl-methyltransferase MraW | −3.4 | ND |
| DET0343 | Cell division protein FtsZ | −1.6 | ND |
| DET0345 | Ribonucleotide reductase | −2.0 | 1.2 |
| DET0374 | Ribosome recycling factor | −1.7 | ND |
| DET0381 | Iron-sulfur cluster-binding protein, Rieske family | −1.9 | ND |
| DET0421 | DegV family protein | −1.6 | −1.7 |
| DET0528 | Phosphoglucomutase phosphomannomutase family protein | −2.0c | −1.3 |
| DET0545 | Amidohydrolase family protein | −2.0 | −1.4 |
| DET0614 | Hydrogenase, group 3, VhuG subunit, putative | −1.9 | 1.4 |
| DET0624 | Response regulator | −2.5 | −1.4 |
| DET0636 | Cell division protein FtsZ | −2.3 | ND |
| DET0638 | Hypothetical protein | −3.0c | ND |
| DET0645 | Nitroreductase family proteind | −1.8 | 1.4 |
| DET0655 | Histidinol-phosphate aminotransferase, putatived | −2.3c | −2.0 |
| DET0729 | [Fe] hydrogenase, HymB subunit, putative | −1.9 | ND |
| DET0730 | [Fe] hydrogenase, HymC subunit, putative | −2.4 | ND |
| DET0734 | Conserved domain protein | −2.0 | ND |
| DET0844 | Histidinol dehydrogenase | −2.9 | −2.0 |
| DET1008 | Conserved domain protein | −3.6 | ND |
| DET1127 | Conserved hypothetical protein | −1.9 | ND |
| DET1193 | Orotate phosphoribosyltransferase | −1.9 | −2.3 |
| DET1203 | Dihydroorotate dehydrogenase, electron transfer subunit | −3.4 | −2.8 |
| DET1258 | Acetylornithine aminotransferase | −3.7 | −2.1 |
| DET1301 | IS; DET4, transposase | −3.0c | 1.3 |
| DET1372 | Dihydroorotate dehydrogenase | −1.8 | ND |
| DET1434 | Hydrogenase expression formation protein HypD | −3.3c | −2.1 |
| DET1468 | Conserved hypothetical protein | −6.3c | ND |
| DET1633 | Aspartate kinase, monofunctional class | −1.7 | −1.2 |
No peptide was detected in all three biological replicates that were grown with NH4. In this case, genes are assigned a value of 1 as the pseudo-spectrum count to allow a ratio to be calculated.
ND indicates a gene that was not differentially expressed (FDR, >1%) in the transcriptome.
No peptide was detected in all three biological replicates that were grown without NH4. In this case, genes are assigned a value of 1 as the pseudo-spectrum count to allow a ratio to be calculated.
Gene is duplicated in the genome.
“−” indicates downregulation during FNL.
All proteins with an FDR of <16% and with a change of 1.5-fold or higher in the spectrum count are reported. Expression ratios were calculated by using the averaged spectrum counts from biological triplicates.
BNR, bacterial neuraminidase repeat; IS, insertion sequence.
Validation of microarray results.
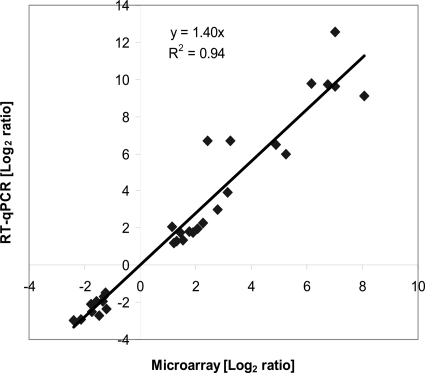
To validate the microarray results, qPCR was used to quantify changes in the transcript levels of selected genes between the two experimental conditions. The microarray and qPCR results for the 30 tested genes were strongly correlated (r2 = 0.94; slope = 1.4), and the expected trend in the expression pattern was obtained (Fig. 2). Given that the dynamic range of microarrays do not match that of qPCR (37), the quantitative agreement measured across the entire dynamic range was reasonably good. In addition to qPCR, an operon-based computational method (OpWise) (38) was used to confirm that genes within the same predicted operon showed similar changes in expression (see Fig. S1 in the supplemental material).
Fig 2.
Comparison of log2 expression ratios of 30 selected differentially regulated genes measured by using microarrays and RT-qPCR. Positive and negative log2 expression ratios represent up- and downregulation during FNL, respectively. Each data point is calculated from averages of biological triplicates.
COG distribution in the transcriptome and proteome.
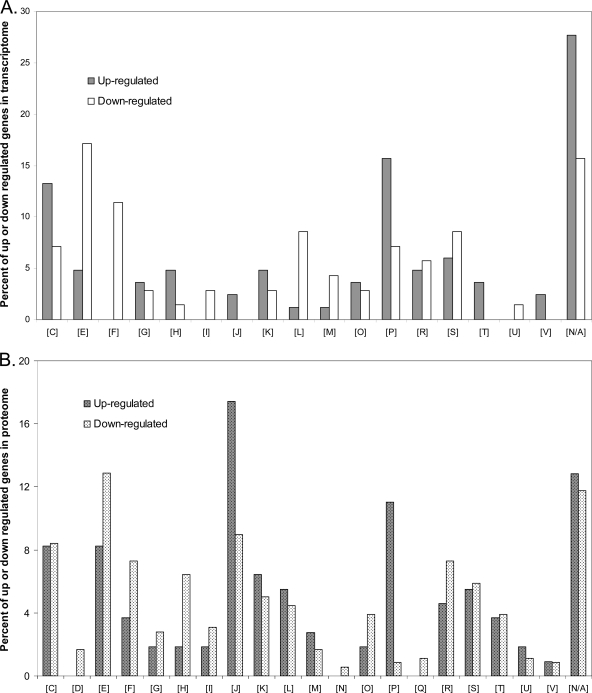
The COG distribution of genes that were up- or downregulated in the transcriptome in response to FNL is shown in Fig. 3A. Many of the differentially regulated genes are unclassified or belong to the category “function unknown, [S].” For the upregulated genes, the categories “energy production and conversion, [C],” which includes many nif genes, and “inorganic ion transport and metabolism, [P],” were the most well-represented categories in the transcriptome. For the downregulated genes, the two dominant categories in the transcriptome were “amino acid transport and metabolism, [E],” and “nucleotide transport and metabolism, [F],” suggesting that nucleic acids and amino acids are two regulated targets during nitrogen stress.
Fig 3.
COG distribution of differentially up- and downregulated genes in the transcriptome (A) and proteome (B). Transcripts that changed by 2-fold or greater and that had an FDR of less than 1% are included in the transcriptome, and proteins that changed by 1.3-fold or greater are included in the proteome. The percentages represent the number of genes that are in a category relative to the respective total number of genes that were up- or downregulated. The COG abbreviations are as follows: [C], energy production and conversion; [D], cell cycle control, cell division, and chromosome partitioning; [E], amino acid transport and metabolism; [F], nucleotide transport and metabolism; [G], carbohydrate transport and metabolism; [H], coenzyme transport and metabolism; [I], lipid transport and metabolism; [J], translation, ribosomal structure, and biogenesis; [K], transcription; [L], replication, recombination, and repair; [M], cell wall/membrane/envelope biogenesis; [N], cell motility; [O], posttranslational modification, protein turnover, and chaperones; [P], inorganic ion transport and metabolism; [Q], secondary metabolite biosynthesis, transport, and catabolism; [R], general function prediction only; [S], function unknown; [T], signal transduction mechanisms; [U], intracellular trafficking, secretion, and vesicular transport; [V] defense mechanisms; [N/A], unclassified.
Similarly to the transcriptome, proteins with no COG assignment or those with unknown function represented a significant fraction of the differentially expressed members of the proteome (Fig. 3B). Among the upregulated proteins and consistent with the transcriptome, the categories “inorganic ion transport and metabolism, [P],” and “energy production and conversion, [C],” were well-represented COG categories, while the category “translation, ribosomal structure, and biogenesis, [J],” was overrepresented in the proteome compared to the transcriptome. For proteins that were downregulated in the proteome, most were in the categories “amino acid transport and metabolism, [E],” and “translation, ribosomal structure, and biogenesis, [J].” Furthermore, the DAVID algorithm (3, 14) was used to identify GO terms that were significantly enriched in the transcriptome and proteome during FNL, and the results obtained were similar to those obtained by the COG analysis (see Tables S7 and S8 in the supplemental material).
Correlation between the transcriptome and the proteome.
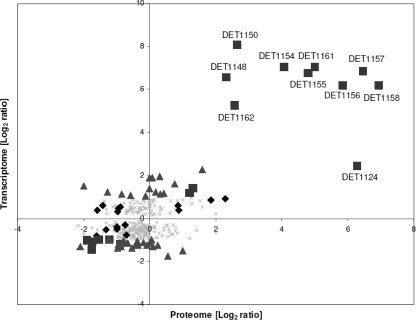
In order to quantitatively assess similarities in transcript and protein regulatory patterns, the expression ratios of 316 genes were plotted (Fig. 4). Overall, the transcriptome and proteome showed similar trends in expression, with up- or downregulated proteins corresponding to transcripts. Among the 53 differentially regulated proteins, 18 had corresponding transcripts differentially regulated by more than 2-fold (Table 1). These proteins and transcripts were mostly genes within the nif operon, and they were all regulated in the same direction. Genes with proteins and transcripts regulated in the opposite direction (Fig. 4, top left and bottom right quadrants) generally had proteins that were not differentially expressed and/or transcripts that were regulated by less than 2-fold. These genes represented a relatively small fraction, and interestingly, most of them were upregulated at the transcript levels but downregulated at the protein levels (Fig. 4, top left quadrant). A previous study examining oxygen exposure in Desulfovibrio vulgaris Hildenborough also showed that the transcripts and proteins of some genes were not expressed in the same direction (35).
Fig 4.
Plot of log2 expression ratios of selected genes from transcriptomic data against proteomic data. In cases where no peptides were detected in the proteome in all three biological replicates, a value of 1 is assigned as the pseudo-spectrum count to avoid infinity and allow a ratio to be calculated. ■ represents genes that were differentially regulated in the proteome and in the transcriptome by 2-fold or greater, ♦ represents genes that were differentially regulated in the proteome and in the transcriptome but by less than 2-fold, ▲ represents genes that were differentially regulated by 2-fold or greater in the transcriptome but were not differentially regulated in the proteome, and × represents genes that were differentially regulated by less than 2-fold in the transcriptome and were not differentially regulated in the proteome. Genes that are related to nitrogen fixation are labeled with their locus tags.
Nitrogen metabolism.
Both the transcripts and proteins of the nif operon (DET1151 to DET1158) were highly upregulated in response to FNL (Tables 1 and 2). The transcripts of all nif genes were among the most strongly upregulated in the transcriptome, and increases of as much as 281-fold were measured (Table 2). Transcripts and proteins of genes encoding the catalytic units of nitrogenases (nifD, nifK, and nifH) and the two PII nitrogen regulatory proteins that control nitrogenase activity (25) were all differentially upregulated (Tables 1 and 2). No peptides were detected for the majority of the nif genes when cells were grown with AA (Table 1), strongly showing that the expressions of these proteins are indeed modulated by the available fixed nitrogen. This stringency in the regulation of the nif operon is not surprising, given the high energy cost of nitrogen fixation and the regulation of nif expression in other organisms (4).
Table 2.
Selected differentially upregulated genes in the transcriptome
| Genea | Descriptiond | Expression ratio | Log2 ratio |
|---|---|---|---|
| DET0096 | FeoA family protein | 4.2 | 2.1 |
| DET0097 | Iron-dependent repressor, putative | 2.6 | 1.4 |
| DET0100 | Membrane protein, putative | 2.1 | 1.1 |
| DET0101 | Molybdopterin oxidoreductase | 2.3 | 1.2 |
| DET0102 | Molybdopterin oxidoreductase, membrane subunit, putative | 2.9 | 1.5 |
| DET0103 | Molybdopterin oxidoreductase, iron-sulfur-binding subunit, putative | 2.8 | 1.5 |
| DET0138 | Phosphate ABC transporter, phosphate-binding protein | 2.6 | 1.4 |
| DET0139 | Phosphate ABC transporter, permease protein | 4.9 | 2.3 |
| DET0140 | Phosphate ABC transporter, permease protein | 4.8 | 2.3 |
| DET0141 | Phosphate ABC transporter, ATP-binding protein | 3.1 | 1.6 |
| DET0142 | Phosphate transport system regulatory protein PhoU | 2.2 | 1.2 |
| DET0143 | Arsenate reductase | 2.2 | 1.1 |
| DET0198 | Glutaredoxin family protein | 2.1 | 1.1 |
| DET0253 | DNA primase domain proteinb,c | 2.6 | 1.4 |
| DET0271 | Hypothetical proteinb,c | 2.2 | 1.1 |
| DET0275 | Hypothetical proteinb | 2.3 | 1.2 |
| DET0299 | Transcriptional regulator, Crp Fnr family | 2.3 | 1.2 |
| DET0801 | Pyrroline-5-carboxylate reductase, putative | 3.8 | 1.9 |
| DET0802 | Aspartate 1-decarboxylase | 3.4 | 1.8 |
| DET0803 | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | 3.7 | 1.9 |
| DET0804 | Pantoate–beta-alanine ligase | 4.5 | 2.2 |
| DET0813 | ABC transporter, ATP-binding protein | 2.2 | 1.2 |
| DET0814 | ABC transporter, permease protein | 2.4 | 1.2 |
| DET0871 | Transcriptional regulator, MarR family | 2.0 | 1.0 |
| DET0908 | Arsenical pump membrane protein, putative | 2.7 | 1.4 |
| DET1005 | Transcriptional regulator, ArsR family | 2.0 | 1.0 |
| DET1057 | Conserved hypothetical protein | 8.9 | 3.1 |
| DET1101 | Conserved hypothetical proteinc | 4.0 | 2.0 |
| DET1102 | Conserved hypothetical proteinc | 3.2 | 1.7 |
| DET1103 | Hypothetical proteinc | 2.9 | 1.5 |
| DET1124 | Nitrogen regulatory protein PII | 5.4 | 2.4 |
| DET1125 | Ammonium transporter | 9.4 | 3.2 |
| DET1146 | Hypothetical protein | 2.1 | 1.0 |
| DET1147 | Transcriptional regulator, Fur family | 5.4 | 2.4 |
| DET1148 | Nitrogenase cofactor biosynthesis protein NifB, putative | 94.3 | 6.6 |
| DET1149 | Acetyltransferase, GNAT family | 118.5 | 6.9 |
| DET1150 | Ferredoxin, 2Fe-2S | 266.6 | 8.1 |
| DET1151 | Dinitrogenase iron-molybdenum cofactor NifBYX family protein | 281.4 | 8.1 |
| DET1152 | Nitrogenase molybdenum-iron protein, beta subunit, putative | 122.0 | 6.9 |
| DET1153 | Nitrogenase MoFe cofactor biosynthesis protein NifE | 136.1 | 7.1 |
| DET1154 | Nitrogenase molybdenum-iron protein, beta subunit | 130.6 | 7.0 |
| DET1155 | Nitrogenase molybdenum-iron protein alpha chain | 106.9 | 6.7 |
| DET1156 | Nitrogen-regulatory protein PII | 72.3 | 6.2 |
| DET1157 | Nitrogen-regulatory protein PII | 114.8 | 6.8 |
| DET1158 | Nitrogenase iron protein | 71.3 | 6.2 |
| DET1159 | Molybdenum ABC transporter, ATP-binding protein | 228.4 | 7.8 |
| DET1160 | Molybdenum ABC transporter, permease protein | 103.6 | 6.7 |
| DET1161 | Molybdenum ABC transporter, periplasmic molybdate-binding protein | 129.6 | 7.0 |
| DET1162 | Transcriptional regulator, putative | 38.1 | 5.3 |
| DET1241 | Peptide methionine sulfoxide reductase MsrA | 2.9 | 1.5 |
| DET1253 | Hypothetical protein | 7.0 | 2.8 |
| DET1286 | Serine protease, DegP HtrA family | 2.3 | 1.2 |
| DET1324 | Conserved hypothetical protein | 5.4 | 2.4 |
| DET1454 | Ribosomal 5S rRNA E-loop-binding protein Ctc L25 TL5 | 2.0 | 1.0 |
| DET1459 | Ribosomal subunit interface protein, putative | 2.3 | 1.2 |
| DET1460 | Methylated-DNA-protein-cysteine methyltransferase | 2.7 | 1.4 |
| DET1532 | Dinitrogenase iron-molybdenum cofactor family protein | 2.2 | 1.1 |
| DET1545 | Reductive dehalogenase, putative | 2.0 | 1.0 |
| DET1575 | Hydrogenase, HycC subunit, putative | 2.0 | 1.0 |
| DET1576 | Methylglyoxal synthase, putative | 29.6 | 4.9 |
| DET1580 | Transcription regulator, TetR family | 2.5 | 1.3 |
| DET1616 | Transcriptional regulator, AbrB family | 2.3 | 1.2 |
Selected genes with an FDR of <1% and with expression levels changed by 2-fold or greater are reported. Expression ratios were calculated by using averages from biological triplicates, and positive values indicate upregulation during FNL.
Gene is duplicated in the genome.
Gene is within integrated elements.
GNAT, Gcn5-related N-acetyltransferase.
The primary nitrogen carriers in cells are glutamate and glutamine, which serve to donate nitrogen for amino acid and nucleic acid synthesis and other reactions (42). During nitrogen fixation, the nif genes convert dinitrogen to ammonium, and the glutamine synthetase-glutamate synthase (GS-GOGAT) pathway (42) assimilates ammonium. DET1123 encodes a glutamine synthetase to assimilate ammonium, which is then transferred by glutamate synthase (DET1127, DET1128, DET1130, and DET0038) to α-ketoglutarate to form glutamate. In the transcriptome, none of the genes involved in ammonium assimilation were differentially regulated by more than 2-fold. In the proteome, glutamine synthetase was differentially upregulated, but DET1127, which is predicted to encode a subunit of glutamate synthase, was downregulated (Table 1).
Three homologues of PII proteins are present in the genome of strain 195. Two of them (DET1156 and DET1157) are within the nif operon, are cotranscribed with the nif genes, and regulate nitrogenase activity in response to ammonia (25), and one (DET1124) is located next to the ammonium transporter and colocated with genes for the GS-GOGAT pathway. Both the proteins and transcripts were differentially upregulated for DET1124 during nitrogen fixation (Tables 1 and 2). This PII homolog is expected to regulate the ammonium transporter (DET1125) and the GS-GOGAT pathway (25). The upregulation of all three PII proteins should allow cells to respond immediately to changes in ammonium concentrations, and the differential upregulation of PII proteins is commonly observed for the Bacteria and Archaea under conditions of nitrogen stress (31, 51, 52).
Transporters.
A number of transporters were differentially regulated during FNL. Genes predicted to encode an ABC transporter for molybdenum (Mo) (DET1159 to DET1161), which are located next to the nif operon, are absent from other Dehalococcoides genomes, and are transcribed in the same direction (http://img.jgi.doe.gov/cgi-bin/w/main.cgi), were differentially upregulated at both the transcript and protein levels (Tables 1 and 2). Two proposed transporters for Fe2+ (DET0095 to DET0097 and DET1503 and DET1504) were also differentially upregulated at the transcript and protein levels, while transcripts for a predicted ABC transporter for Fe3+ (DET1174 to DET1176) were differentially downregulated (Tables 1 to 3). The influxes of Mo and iron are likely to support the synthesis of FeS clusters and FeMo cofactors of nitrogenases (4). As expected, transcripts of the ammonium transporter (DET1125) were differentially upregulated (Table 2). However, unexpectedly, the differential upregulation of a predicted ABC transporter for phosphate (DET0138 to DET0142) was identified in the transcriptome, and the phosphate-binding protein (DET0138) was also upregulated in the proteome (Tables 1 and 2). This finding is surprising, as a phosphate concentration of 1.5 mM in the medium should not be limiting and therefore should not trigger differential expression between the two growth conditions. In addition, two predicted antiporters for ions (DET0908 and DET1627) were differentially upregulated at either the transcript or protein level, and a major facilitator family protein (DET1502) for small-solute transport was differentially downregulated (Tables 1 to 3).
Table 3.
Selected differentially downregulated genes in the transcriptome
| Genea | Description | Expression ratio | Log2 ratio |
|---|---|---|---|
| DET0114 | ABC transporter, substrate-binding protein, putative | −2.4 | −1.3 |
| DET0115 | ABC transporter, permease protein, putative | −2.3 | −1.2 |
| DET0183 | ATP-dependent RNA helicase, DEAD DEAH-box family | −3.4 | −1.8 |
| DET0370 | Membrane-associated zinc metalloprotease, putative | −2.3 | −1.2 |
| DET0371 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | −2.0 | −1.0 |
| DET0372 | Phosphatidate cytidylyltransferase | −2.1 | −1.1 |
| DET0417 | Amino acid ABC transporter, ATP-binding protein | −2.2 | −1.1 |
| DET0418 | Amino acid ABC transporter, permease protein, His Glu Gln Arg opine family | −2.1 | −1.1 |
| DET0608 | Ribosomal protein-alanine acetyltransferase | −2.3 | −1.2 |
| DET0654 | Cobalamin biosynthesis protein CobDb | −2.1 | −1.0 |
| DET0655 | Histidinol-phosphate aminotransferase, putativeb | −2.0 | −1.0 |
| DET0716 | Tyrosine recombinase XerC | −2.3 | −1.2 |
| DET0770 | Transcription elongation factor GreA | −2.1 | −1.1 |
| DET0784 | V-type H(+)-translocating pyrophosphatase | −2.0 | −1.0 |
| DET0838 | Phosphoribosylamine-glycine ligase | −2.3 | −1.2 |
| DET0840 | Adenylosuccinate lyase | −2.2 | −1.1 |
| DET0841 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | −2.6 | −1.4 |
| DET0844 | Histidinol dehydrogenase | −2.0 | −1.0 |
| DET0964 | Single-stranded-DNA-specific exonuclease RecJ | −2.4 | −1.3 |
| DET1173 | FwdE family protein | −5.2 | −2.4 |
| DET1174 | Fec-type ABC transporter, periplasmic iron-binding protein | −4.4 | −2.1 |
| DET1175 | Fec-type ABC transporter, permease protein | −4.0 | −2.0 |
| DET1176 | Fec-type ABC transporter, ATP-binding protein | −3.0 | −1.6 |
| DET1192 | Signal peptidase I | −3.0 | −1.6 |
| DET1193 | Orotate phosphoribosyltransferase | −2.3 | −1.2 |
| DET1194 | Conserved hypothetical protein | −2.8 | −1.5 |
| DET1196 | ATP-dependent DNA helicase PcrA | −2.5 | −1.3 |
| DET1199 | Aspartate carbamoyltransferase | −2.3 | −1.2 |
| DET1200 | Dihydroorotase, multifunctional complex type | −2.2 | −1.1 |
| DET1202 | Carbamoyl-phosphate synthase, large subunit | −2.3 | −1.2 |
| DET1203 | Dihydroorotate dehydrogenase, electron transfer subunit | −2.8 | −1.5 |
| DET1258 | Acetylornithine aminotransferase | −2.1 | −1.0 |
| DET1259 | Hypothetical protein | −3.8 | −1.9 |
| DET1389 | Formamidopyrimidine-DNA glycosylase | −2.0 | −1.0 |
| DET1422 | Phosphoglycerate mutase family protein | −2.2 | −1.1 |
| DET1434 | Hydrogenase expression formation protein HypD | −2.1 | −1.1 |
| DET1435 | Hydrogenase expression formation protein HypE | −2.6 | −1.4 |
| DET1461 | Lipoprotein, putative | −3.0 | −1.6 |
| DET1462 | DNA repair protein, RadC family | −2.2 | −1.2 |
| DET1490 | Peptide ABC transporter, ATP-binding protein | −2.5 | −1.3 |
| DET1491 | Peptide ABC transporter, ATP-binding protein | −3.3 | −1.7 |
| DET1492 | Peptide ABC transporter, permease protein | −3.1 | −1.6 |
| DET1493 | Peptide ABC transporter, permease protein | −2.5 | −1.3 |
| DET1494 | Peptide ABC transporter, periplasmic peptide-binding protein | −3.4 | −1.8 |
| DET1495 | Conserved hypothetical protein | −2.8 | −1.5 |
| DET1502 | Major facilitator family protein | −2.4 | −1.3 |
| DET1515 | Conserved hypothetical protein | −2.7 | −1.4 |
| DET1557 | Lipoprotein, putativec | −2.8 | −1.5 |
| DET1558 | Reductive dehalogenase-anchoring protein, putativec | −2.0 | −1.0 |
| DET1568 | Hypothetical protein | −5.5 | −2.5 |
Selected genes with an FDR of <1% and with an expression level changed by 2-fold or greater are reported. Expression ratios were calculated by using averages from biological triplicates, and negative values indicate downregulation during FNL.
Gene is duplicated in the genome.
Gene is within integrated elements.
Moreover, in the transcriptome, predicted ABC transporters for peptides (DET1490 to DET1494) and polar amino acids (DET0417 to DET0419) were both differentially downregulated (Table 3) in response to FNL. Peptide transporters are typically activated by ammonium (32), and its absence should trigger downregulation. A transmembrane signal peptidase for protein export (DET1192) was differentially downregulated in the transcriptome (Table 3).
Carbon metabolism.
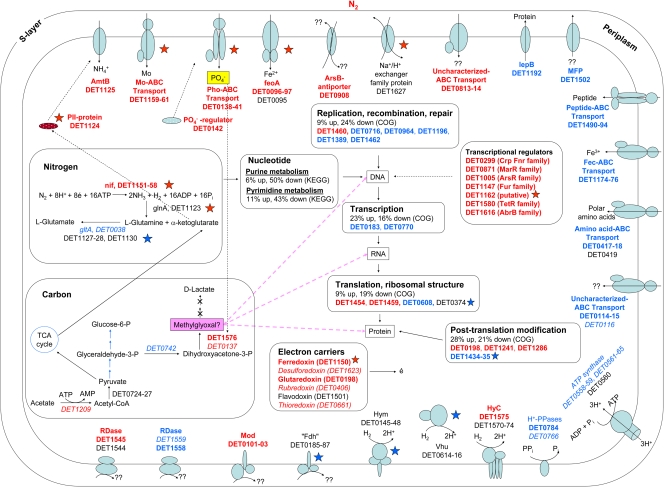
Aside from genes related to nitrogen fixation, a putative methylglyoxal synthase-encoding gene (DET1576) was the most differentially upregulated gene in the transcriptome, with a 30-fold signal ratio (Table 2), but no peptide was measured. Strain 195 contains a second copy of the methylglyoxal synthase gene (DET0137) located next to an ABC-phosphate transporter gene, and although this gene exhibited increased detection in both the transcriptome and proteome, the increase in the transcriptome was far lower, and its proteome FDR exceeded the cutoff value (see Tables S3 and S6 in the supplemental material). The upregulation of the methylglyoxal synthase gene is physiologically interesting, as the characterized methylglyoxal synthase is known to catalyze the conversion of dihydroxyacetone-3-phosphate (P) to methylglyoxal, a toxic electrophile that can react with macromolecules such as DNA, RNA, and proteins to inhibit growth and cause cell death (5). The loss of balance between carbon and nitrogen was previously shown to result in methylglyoxal accumulation and cell death in the rumen bacterium Prevotella ruminicola (44) and to result in uncoupled anabolism and catabolism in Escherichia coli (5, 56). Here, in strain 195, even though methylglyoxal was not analytically detected in the spent medium (which may be due to a low cell density), cell growth was stunted by nitrogen limitation (Fig. 1B), and the differential downregulation of the two cell division proteins FtsZ1 and FtsZ2 (DET0343 and DET0636) in the proteome (Table 1) supports the physiological observation.
In strain 195, dihydroxyacetone-3-P, the precursor to methylglyoxal, is associated with gluconeogenesis as part of the central carbon metabolism following acetate assimilation (see Fig. S2 in the supplemental material). In some organisms, methylglyoxal synthesis is initiated to consume excess phosphorylated sugars in order to avoid the growth inhibition caused by the sugar phosphates (18), uncoupling anabolism from catabolism (5), but paradoxically, methylglyoxal itself is a toxin. Some organisms are known to contain detoxification pathways of methylglyoxal via the production of d-lactate (5), but strain 195 does not. However, transcripts and proteins of genes along the gluconeogenesis and tricarboxylic acid pathways were found to be either differentially downregulated or unchanged in response to FNL (see Fig. S2 in the supplemental material). The downregulation of carbon pathways suggests that cells were attempting to repress carbon catabolism when insufficient nitrogen was present.
Transcription of the phosphate transporter and methylglyoxal synthase genes.
Since a previous study carried out with Escherichia coli indicated that methylglyoxal synthase is susceptible to inhibition by inorganic phosphate (13), the differential upregulation of the predicted ABC transporter genes for phosphate observed in this study might be related to the methylglyoxal synthase genes in strain 195. In order to further investigate their potential roles, the transcriptions of the putative methylglyoxal synthase gene (DET1576) and the gene encoding the phosphate-binding component (DET0138) of the ABC-phosphate transporter were quantified over a growth cycle under three experimental conditions (Fig. 1): FNL, AA, and initial growth with FNL but with ammonium later abruptly amended prior to the stationary phase. When cultures were grown with FNL, the methylglyoxal synthase gene (DET1576) transcripts were at relatively high levels throughout the growth cycle, increasing initially and then decreasing as TCE conversion to the final daughter product of VC neared completion (Fig. 1C). In contrast, the AA culture exhibited rapid decreases in levels of methylglyoxal synthase transcripts throughout. The culture that was initially under conditions of FNL and then abruptly amended with ammonium exhibited an expression pattern with levels that initially increased and then decreased rapidly in response to the amendment, and growth subsequently resumed. The phosphate transporter gene (DET0138) followed a pattern similar to that of the methylglyoxal synthase gene, responding to the abrupt ammonium amendment with rapid downregulation (Fig. 1D). These results suggest that the transcriptions of the phosphate transporter and methylglyoxal synthase genes are tightly coupled in response to nitrogen stress, and their gene products might be of physiological importance.
Nucleotide metabolism, transcription, translation, and posttranslational modification.
In response to FNL and in the late exponential growth phase, a significant downregulation of genes responsible for purine and pyrimidine synthesis was measured in the transcriptome. In fact, 50% and 43% of the genes that are in the KEGG pathway of purine and pyrimidine metabolism, respectively, were differentially downregulated, with a number of them changing by more than 2-fold. For example, the gene for orotate phosphoribosyltransferase (DET1193), which is involved in pyrimidine synthesis, was differentially downregulated in both proteins and transcripts (Tables 1 and 3). Figures S3 and S4 in the supplemental material show how genes participating in the early part of the nucleotide synthesis pathways were consistently downregulated. In addition to restricting the synthesis of nucleotides, a gene encoding a methylated-DNA-protein-cysteine methyltransferase (DET1460) for repairing damage to DNA was differentially upregulated in the transcriptome in response to FNL (Table 2). Furthermore, 24% of all genes that belong to the COG category of replication, recombination, and repair were downregulated in the transcriptome in response to FNL (Table 3), suggesting that the cell replication function is limited during nitrogen stress.
Under conditions of FNL, cells also systematically regulated their transcription, translation, and posttranslation modification functions (Fig. 5). For example, seven transcriptional regulators (DET0299, DET0871, DET1005, DET1147, DET1162, DET1580, and DET1616) and genes encoding ribosomal proteins (DET1454 and DET1459) were differentially upregulated in the transcriptome (Table 2).
Fig 5.
Conceptual model of the response of strain 195 to FNL. Red text and blue text indicate differential upregulation and downregulation in the transcriptome, respectively, and black indicates insignificant differential expression (FDR of >1%). Boldface and italic fonts represent expression ratios of 2-fold or greater and less than 2-fold, respectively. Red and blue stars indicate proteins that are differentially up- and downregulated by 1.5-fold or greater, respectively. Red and blue arrows and lines indicate metabolic pathways or cycles that are up- and downregulated, respectively. All genes in selected COG categories and KEGG pathways were examined to determine their direction of change in the transcriptome, and the indicated percentages are the relative fractions that were differentially up- or downregulated with an FDR of less than 1%. Two homologs of the Fe2+ transporter are present in strain 195 (DET0095 to DET0097 and DET1503 and DET1504), and analyses of the transcriptome and proteome have separately identified these two homologs as being differentially regulated. TCA, tricarboxylic acid; CoA, coenzyme A.
Integrated elements.
The genome of strain 195 contains ∼200 kb of integrated elements (IEs) with phage- and transposon-like genes and a few reductive dehalogenase (RDase)-encoding genes located within them (45). In a previous study examining the transcriptome of strain 195 during different growth phases, a large number of genes located within the IEs were differentially regulated as cells transitioned into the stationary phase (15). This includes all genes that are within a 22-gene transposon that is present in three identical copies (IEs III, IV, and VI) in the genome and genes (IE VII) that encode components of a temperate bacteriophage. In contrast, in this study, only seven genes (including duplicated ones) within the IEs were differentially regulated by more than 2-fold in the transcriptome: four encode hypothetical proteins and one encodes an RDase anchor protein (DET1558) (Tables 2 and 3). The striking differences in these transcriptomic results suggest that cells of strain 195 respond differently to growth-related stress and unfavorable environmental conditions.
Reductive dehalogenases.
Seventeen intact RDase genes are present in the genome of strain 195 (45). Thus far, the tceA (DET0079) and pceA (DET0318) genes are the only ones associated with confirmed dehalogenation functions, and some of the substrates of their gene products have been identified (6, 27, 28). In a previous transcriptomic analysis of strain 195 at different growth phases, four putative RDase genes (DET0173, DET0180, DET1535, and DET1545) were differentially upregulated as cells transitioned into the stationary phase (15). In this study, only one RDase-encoding gene (DET1545) was found to be differentially upregulated by more than 2-fold in response to FNL (Table 2), while the functionally important tceA gene, for catalyzing TCE to VC, was not differentially expressed. In contrast, DET1559 was differentially downregulated albeit by only 1.9-fold (see Table S3 in the supplemental material), and its associated membrane anchor gene (DET1558) was differentially downregulated by 2-fold (Table 3). It was demonstrated previously that the level of expression of DET1545 increases during the latter part of the dechlorination cycle and when the dechlorination rate is low (40). Together, these results suggest that DET1545 might be important when dechlorination conditions are suboptimal and that its expression might be a stress response to maintain energy generation.
Oxidoreductases.
Dehalococcoides spp. are obligate hydrogenotrophs, and this is reflected in the five different multisubunit putative hydrogenases encoded in the genome of strain 195 (45). Four of the hydrogenases (Hup, Ech, Hyc, and Hym) are membrane bound, while one (Vhu) is located in the cytoplasm (20, 34, 45). The Hup, Hym, and Vhu hydrogenase proteins were shown to be highly expressed in a previous study of a hydrogen-grown Dehalococcoides culture, suggesting that they are important for electron transport in dehalogenation, while the Ech and Hyc hydrogenases are possibly important for developing low-potential electrons for biosynthetic reactions (34).
In the current investigation, the Hym (DET0145 to DET0148) and Vhu (DET0614 to DET0616) hydrogenases were differentially downregulated in the proteome in response to FNL (Table 1). Furthermore, in the transcriptome and proteome, genes that encode proteins for posttranslational hydrogenase maturation (DET1434 and DET1435) were differentially downregulated (Tables 1 and 3). It was demonstrated previously for Rhizobium leguminosarum that these maturation proteins are necessary for hydrogenase activity (43). The downregulation of the Hym and Vhu hydrogenases and the maturation proteins seems to correspond with the observed decline in levels of dechlorination activity under conditions of FNL (Fig. 1A).
The Hyc hydrogenase (DET1570 to DET1575) was differentially upregulated in the transcriptome during FNL (Table 2). This upregulation might serve to generate low-potential electrons for nitrogenase, which typically uses ferredoxin as an electron donor, and this can be tied to the upregulation at the transcript and protein levels of genes that encode potential cytoplasmic electron carriers (DET1150 and DET0198) (Tables 1 and 2). DET1150, located contiguously with the nif operon and transcribed in the same direction, encodes a ferredoxin that likely supplies electrons to reduce the Fe protein of nitrogenases. It was proposed previously that the cytoplasmic electron carriers in strain 195 play a role in sensing the cellular energy level through changes in the proton motive force or redox potential of the electron transport chain (45). Under conditions of FNL, this role might become critical to ensure that the energy and redox status is maintained for biosynthesis.
In addition to the hydrogenases, the differential upregulation of the molybdopterin-containing oxidoreductase genes (DET101 to DET103) in the transcriptome is of interest (Table 2). The functional role of this oxidoreductase has yet to be determined, and it was speculated that it might be involved in the reduction of nonhalogenated electron acceptors (45). The molybdopterin-containing oxidoreductase genes were previously observed to be upregulated when strain 195 was grown in a mixed culture relative to a pure culture (34). The upregulation of this gene under stress conditions suggests that the protein might indeed function to maintain the cellular redox status by directing electrons to alternative electron acceptors.
The formate dehydrogenase (Fdh)-homologous genes (DET0185 to DET0187) were previously observed to be highly expressed in both the transcriptome and proteome of strain 195 during active dechlorination (15, 34). However, strain 195 is not able to use formate as an electron donor or carbon source (34), suggesting that “Fdh” has a novel function. Under conditions of nitrogen limitation, Fdh was differentially downregulated in the proteome (Table 1).
Conceptual model and environmental implications.
A global transcriptomic and proteomic approach was used in this study to query the molecular responses of strain 195 under conditions of FNL. By incorporating the transcriptomic and proteomic data, a conceptual model that summarizes key differentially regulated genes and the interactions between different cellular components was developed and is illustrated in Fig. 5.
The problem of FNL is of environmental relevance, since ammonium, a much more energetically favorable nitrogen source than dinitrogen, is not always readily available at chlorinated ethene-contaminated sites (46). Furthermore, members of the Geobacteraceae were previously shown to express their nitrogen-fixing genes in subsurface environments contaminated with petroleum (11). Under conditions of FNL, strain 195 prematurely transitions into the stationary phase and reduces its dechlorination activity (22). According to the transcriptomic data, a possible clue to the limited growth may be related to the differential upregulation of the methylglyoxal synthase gene, whose product catalyzes the formation of methylglyoxal, a toxic electrophile that can lead to growth inhibition. Although not all Dehalococcoides strains contain a nif operon to acquire dinitrogen, all four sequenced strains have homologues of both putative methylglyoxal synthase genes (http://img.jgi.doe.gov/cgi-bin/w/main.cgi). In order to monitor the nitrogen status of Dehalococcoides spp. in the environment, genes that have been identified to be differentially regulated in this study are potential biomarkers. These genes include the nif (for strains that contain this operon), ammonium transporter, PII nitrogen regulatory protein, and methylglyoxal synthase genes and a number of general stress response genes. Monitoring the expressions of these genes at the transcript and/or protein level could provide direct information about cellular physiology and complement geochemical measurements to guide field-scale manipulations (e.g., whether ammonium should be added as a supplement) to potentially improve treatment outcomes.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIEHS Superfund basic research project ES04705-19 and SERDP grant ER 1567. The Oak Ridge National Laboratory part of this research was sponsored in part by the U.S. Department of Energy under contract DE-AC05-00OR22725 with Oak Ridge National Laboratory, managed and operated by UT-Battelle, LLC.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 2. Cheng D, He J. 2009. Isolation and characterization of “Dehalococcoides” sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl. Environ. Microbiol. 75:5910–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dennis G, Jr, et al. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:R60. [PubMed] [Google Scholar]
- 4. Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631 [DOI] [PubMed] [Google Scholar]
- 5. Ferguson GP, Tötemeyer S, MacLean MJ, Booth IR. 1998. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170:209–219 [DOI] [PubMed] [Google Scholar]
- 6. Fung JM, Morris RM, Adrian L, Zinder SH. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73:4439–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He J, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65 [DOI] [PubMed] [Google Scholar]
- 10. He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442–1450 [DOI] [PubMed] [Google Scholar]
- 11. Holmes DE, Nevin KP, Lovley DR. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holmes DE, et al. 2009. Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 3:216–230 [DOI] [PubMed] [Google Scholar]
- 13. Hopper DJ, Cooper RA. 1972. The purification and properties of Escherichia coli methylglyoxal synthase. Biochem. J. 128:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang DW, et al. 2007. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35:W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson DR, et al. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson DR, Lee PKH, Holmes VF, Fortin AC, Alvarez-Cohen L. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294:198–206 [DOI] [PubMed] [Google Scholar]
- 18. Kadner RJ, Murphy GP, Stephens CM. 1992. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J. Gen. Microbiol. 138:2007–2014 [DOI] [PubMed] [Google Scholar]
- 19. Krajmalnik-Brown R, et al. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kube M, et al. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269–1273 [DOI] [PubMed] [Google Scholar]
- 21. Lee PKH, et al. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee PKH, He J, Zinder SH, Alvarez-Cohen L. 2009. Evidence for nitrogen fixation by “Dehalococcoides ethenogenes” strain 195. Appl. Environ. Microbiol. 75:7551–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72:6161–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee PKH, Macbeth TW, Sorenson KS, Jr, Deeb RA, Alvarez-Cohen L. 2008. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74:2728–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349–377 [DOI] [PubMed] [Google Scholar]
- 26. Liu H, Sadygov RG, Yates JR., III 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193–4201 [DOI] [PubMed] [Google Scholar]
- 27. Magnuson JK, Romine MF, Burris DR, Kingsley MT. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571 [DOI] [PubMed] [Google Scholar]
- 30. McMurdie PJ, et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Methé BA, Webster J, Nevin K, Butler J, Lovley DR. 2005. DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:2530–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Monnet V. 2003. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 60:2100–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morris RM, et al. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73:320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris RM, Sowell S, Barofsky D, Zinder S, Richardson R. 2006. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ. Microbiol. 8:1499–1509 [DOI] [PubMed] [Google Scholar]
- 35. Mukhopadhyay A, et al. 2007. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189:5996–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Müller JA, et al. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park PJ, et al. 2004. Current issues for DNA microarrays: platform comparison, double linear amplification, and universal RNA reference. J. Biotechnol. 112:225–245 [DOI] [PubMed] [Google Scholar]
- 38. Price MN, Arkin AP, Alm EJ. 2006. OpWise: operons aid the identification of differentially expressed genes in bacterial microarray experiments. BMC Bioinformatics 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rahm BG, Morris RM, Richardson RE. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72:5486–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rahm BG, Richardson RE. 2008. Dehalococcoides ' gene transcripts as quantitative bioindicators of tetrachloroethene, trichloroethene, and cis-1,2-dichloroethene dehalorespiration rates. Environ. Sci. Technol. 42:5099–5105 [DOI] [PubMed] [Google Scholar]
- 41. Ram RJ, et al. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920 [PubMed] [Google Scholar]
- 42. Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155–176 [DOI] [PubMed] [Google Scholar]
- 43. Rey L, et al. 1993. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar viciae. Mol. Microbiol. 8:471–481 [DOI] [PubMed] [Google Scholar]
- 44. Russell JB. 1993. Glucose toxicity in Prevotella ruminicola: methylglyoxal accumulation and its effect on membrane physiology. Appl. Environ. Microbiol. 59:2844–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seshadri R, et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108 [DOI] [PubMed] [Google Scholar]
- 46. Skubal KL, Barcelona MJ, Adriaens P. 2001. An assessment of natural biotransformation of petroleum hydrocarbons and chlorinated solvents at an aquifer plume transect. J. Contam. Hydrol. 49:151–169 [DOI] [PubMed] [Google Scholar]
- 47. Smidt H, de Vos WM. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43–73 [DOI] [PubMed] [Google Scholar]
- 48. Stolyar S, et al. 2007. Response of Desulfovibrio vulgaris to alkaline stress. J. Bacteriol. 189:8944–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson MR, et al. 2007. Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J. Proteome Res. 6:1745–1757 [DOI] [PubMed] [Google Scholar]
- 51. Tolonen AC, et al. 2006. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol. Syst. Biol. 2:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Veit K, et al. 2006. Global transcriptional analysis of Methanosarcina mazei strain Gö1 under different nitrogen availabilities. Mol. Genet. Genomics 276:41–55 [DOI] [PubMed] [Google Scholar]
- 53. VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF. 2009. Systems biology: functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 7:196–205 [DOI] [PubMed] [Google Scholar]
- 54. VerBerkmoes NC, et al. 2006. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J. Proteome Res. 5:287–298 [DOI] [PubMed] [Google Scholar]
- 55. Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257–8264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weber J, Kayser A, Rinas U. 2005. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. II. Dynamic response to famine and feast, activation of the methylglyoxal pathway and oscillatory behaviour. Microbiology 151:707–716 [DOI] [PubMed] [Google Scholar]
- 57. West KA, et al. 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Appl. Environ. Microbiol. 74:3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang B, et al. 2006. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5:2909–2918 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.