Abstract
Brucellosis is one of the major bacterial zoonoses worldwide. In the past decade, an increasing number of atypical Brucella strains and species have been described. Brucella microti in particular has attracted attention, because this species not only infects mammalian hosts but also persists in soil. An environmental reservoir may pose a new public health risk, leading to the reemergence of brucellosis. In a polyphasic approach, comprising conventional microbiological techniques and extensive biochemical and molecular techniques, all currently available Brucella microti strains were characterized. While differing in their natural habitats and host preferences, B. microti isolates were found to possess identical 16S rRNA, recA, omp2a, and omp2b gene sequences and identical multilocus sequence analysis (MLSA) profiles at 21 different genomic loci. Only highly variable microsatellite markers of multiple-locus variable-number tandem repeat (VNTR) analysis comprising 16 loci (MLVA-16) showed intraspecies discriminatory power. In contrast, biotyping demonstrated striking differences within the genetically homologous species. The majority of the mammalian isolates agglutinated only with monospecific anti-M serum, whereas soil isolates agglutinated with anti-A, anti-M, and anti-R sera. Bacteria isolated from animal sources were lysed by phages F1, F25, Tb, BK2, Iz, and Wb, whereas soil isolates usually were not. Rough strains of environmental origin were lysed only by phage R/C. B. microti exhibited high metabolic activities similar to those of closely related soil organisms, such as Ochrobactrum spp. Each strain was tested with 93 different substrates and showed an individual metabolic profile. In summary, the adaptation of Brucella microti to a specific habitat or host seems to be a matter of gene regulation rather than a matter of gene configuration.
INTRODUCTION
Brucella species are facultatively intracellular pathogens responsible for one of the world's most widespread zoonotic diseases. The bacteria may cause reproductive failure and abortion in domestic animals and a potentially debilitating multiorgan infection in humans. Like Agrobacterium and Rhizobium spp., brucellae belong to the order of Rhizobiales of the α-2 subgroup of Proteobacteria. Members of the class Alphaproteobacteria include organisms that are either mammalian or plant pathogens or symbionts (12). Within the family Brucellaceae, Ochrobactrum, a genus comprising soil-associated facultative human pathogens, contains the closest phylogenetic neighbors to Brucella. Ochrobactrum intermedium and Brucella spp. are 98.8% identical in their 16S rRNA gene sequences (14). Furthermore, Brucella species are closely related to each other (monophyletic genus), showing 98 to 99% similarity in most of the coding sequences. Despite this high genetic homology, brucellae differ widely in host tropism, phenotypic characteristics, and pathogenicity (38).
The phylogeny of Brucella species does not always match that of their nominal mammalian hosts (36). Currently, the genus Brucella consists of 10 species. With the exception of Brucella inopinata (29), at least one animal host has been described for each species. Although the host range of Brucella spp. can be variable, most species display strong host preferences. The classical Brucella spp. of terrestrial origin, i.e., Brucella melitensis, B. abortus, B. ovis, B. canis, B. suis, and B. neotomae, are best characterized as facultatively intracellular pathogens. An intracellular lifestyle has also been found for Brucella species that infect marine mammals (B. ceti and B. pinnipedialis) (20). Hence, Brucella spp. are commonly regarded as intracellular pathogens with an animal reservoir, although they share close relationships with soil organisms such as Ochrobactrum spp., with plant symbionts such as Rhizobium spp., and with phytopathogens such as Agrobacterium spp. (38). No natural reservoir outside infected mammalian hosts has been identified yet. Brucella spp. generally appear as fastidious bacteria, because their survival in the nutrient-poor phagosome requires a low overall metabolic activity.
In contrast to the classical Brucella spp., the recently described species B. microti is characterized by fast growth on standard media and exhibits remarkable metabolic capabilities (28). Indeed, the phenotype of B. microti resembles that of Ochrobactrum rather than that of Brucella. Hence, B. microti has been misidentified as Ochrobactrum intermedium by use of commercially available biochemical tests, such as the API 20 NE test (bioMérieux, Nürtingen, Germany) (17).
Initially, two strains of this novel Brucella species that had been isolated from systemically diseased common voles (Microtus arvalis) in the Czech Republic (17) were analyzed in detail (28). Subsequently, B. microti was also isolated from the mandibular lymph nodes of red foxes in a district of Lower Austria (26) and even directly from soil (27). These findings indicated that B. microti may persist in a geographical area comprising most parts of Moravia (Czech Republic) and Lower Austria. Long-term environmental persistence outside mammalian hosts and the diversity of reservoir species may play a key role in the epizootic spread of this Brucella species.
The aim of this study was to characterize all currently available B. microti isolates originating from different animal species and various geographical regions by using a selection of widely recognized classical techniques, comprehensive biotyping, and molecular analyses in direct comparison. The data generated would reveal intraspecies diversity among B. microti strains, which might help to explain their ability to survive in multiple hosts and environments.
MATERIALS AND METHODS
Bacterial strains.
A total of 11 B. microti strains, including the type strain, CCM 4915, isolated from both environmental and animal sources and originating from different geographical regions, were analyzed (Table 1). The bacteria were grown on Brucella agar for 48 h at 37°C both with and without 10% CO2.
Table 1.
Brucella microti strains (n = 11) isolated from different sources and of different geographical origins
| Strain | Geographical origin | Source | Yr of isolation | Reference(s) |
|---|---|---|---|---|
| CCM 4915T | South Moravia, Czech Republic | Common vole | 2000 | 17, 28 |
| CCM 4916 | South Moravia, Czech Republic | Common vole | 2000 | 17, 28 |
| IBM 284 | Gmünd, Lower Austria | Red fox | 2007 | 26 |
| IBM 257 | Gmünd, Lower Austria | Red fox | 2007 | 26 |
| FH 2208 | Horn, Lower Austria | Red fox | 2008 | This study |
| FK 21908 | Korneuburg, Lower Austria | Red fox | 2008 | This study |
| FW 70608 | Waidhofen/Thaya, Lower Austria | Red fox | 2008 | This study |
| F 303 Mi | Mistelbach, Lower Austria | Red fox | 2009 | This study |
| BMS 10 | South Moravia, Czech Republic | Soil | 2008 | This study |
| BMS 17 | South Moravia, Czech Republic | Soil | 2008 | 27 |
| BMS 20 | South Moravia, Czech Republic | Soil | 2008 | 27 |
Molecular analyses (16S rRNA [rrs], omp2a, omp2b, and recA gene sequencing, multilocus sequence typing [MLST], and multiple-locus variable-number tandem repeat analysis [MLVA]) and phenotypic characterization (biochemical profiling, agglutination, phage lysis) were carried out essentially as described previously for the B. microti type strain, CCM 4915 (28).
Molecular analysis. (i) DNA preparations.
Crude DNA was prepared by transferring a single colony of each strain from the agar plate to 200 μl 5× lysis buffer D (PCR Optimizer kit; Invitrogen, De Schelp, The Netherlands) diluted 1:5 in distilled water, supplemented with 0.5% Tween 20 (ICI America Inc., Merck, Hohenbrunn, Germany) and 2 mg/ml proteinase K (Roche Diagnostics, Mannheim, Germany). After incubation at 56°C for 1 h and inactivation at 95°C for 10 min, DNA samples were purified using the QIAamp DNA Mini kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). A total of 2 μl of the cleared lysates was used as a template in the PCR assays.
(ii) PCR analysis.
(a) bcsp31 PCR and AMOS-PCR. In a genus-specific PCR, the published B4 and B5 primers were used to amplify a 223-bp target within the bcsp31 gene encoding a 31-kDa immunogenic protein conserved among Brucella spp. (6). AMOS-PCR for the detection of B. abortus bv. 1, 2, and 4, B. melitensis, B. ovis, and B. suis bv. 1 was carried out basically as described by Bricker and Halling (7).
(b) Bruce-ladder multiplex PCR. The Bruce-ladder multiplex PCR first established by García-Yoldi and colleagues (13) was used in its modified version as described previously (21). Briefly, primer pair Bmispec_f (5′-AGATACTGGAACATAGCCCG-3′) and Bmispec_r (5′-ATACTCAGGCAGGATACCGC-3′), targeting a 12-kb genomic island specific for B. microti, was added (5, 27). The 25-μl reaction mixture contained 2.5 μl primer mix (with each primer at 2 pmol/μl), 1 μl template DNA, and 12.5 μl 2× Qiagen Multiplex PCR master mix. Thermal cycling was carried out with a model 2720 thermal cycler (Applied Biosystems, Foster City, CA). The initial denaturation step at 95°C for 15 min was followed by template denaturation at 94°C for 30 s, primer annealing at 58°C for 90 s, and a 3-min primer extension at 72°C. After a total of 25 cycles, a final extension phase of 10 min at 72°C completed the reaction, and the PCR products were analyzed using a 1.5% agarose gel.
(iii) Analysis of the 16S rRNA (rrs), recA, omp2a, and omp2b genes.
(a) 16S rRNA (rrs) and recA. The rrs (16S rRNA) and recA (recombinase A) genes were amplified and sequenced as described previously (30). Briefly, almost the complete rrs sequence was amplified using the universal primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-AAGTCGTAACAAGGTARCCG-3′). The primer pair recA-BrucOchro-f (5′-ATGTCTCAAAATTCATTGCGAC-3′) and recA-BrucOchro-r (5′-AGCATCTTCTTCCGGTCCGC-3′) generated a fragment comprising the entire recA gene (1,086 bp). The PCRs were performed in 50 μl Ready-To-Go master mix (Eppendorf GmbH, Hamburg, Germany) with the addition of 15 pmol of each primer. Amplification was carried out in a Perkin-Elmer GeneAmp 2400 thermal cycler (Perkin-Elmer, Applied Biosystems, Foster City, CA). A total of 30 cycles were conducted, each consisting of 30 s of denaturation at 94°C, 30 s of annealing at 58°C (rrs) or 65°C (recA), and elongation at 72°C for 90 s (rrs) or 60 s (recA). A final elongation step of 7 min at 72°C completed the run. Finally, PCR products were analyzed for the presence of the respective amplicons by agarose gel electrophoresis (1% [wt/vol] in Tris-acetate-EDTA [TAE] buffer).
The purified fragments of the recA and rrs sequences were sequenced with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) using the recA-BrucOchro-f/recA-BrucOchro-r primers and the internal primer set consisting of 341fw (5′-CCTACGGGAGGCAGCAG-3′), 518r (5′-ATTACCGCGGCTGCTGG-3′), and 926f (5′-AACTYAAAKGAATTGACGG-3′), respectively. Multiple sequence alignments were performed with ClustalW, version 1.8 (http://clustalw.genome.jp/).
(b) omp2a and omp2b. The omp2a and omp2b genes were amplified and sequenced as described previously (8, 9). Primers 2aA (5′-GGCTATTCAAAATTCTGGCG-3′) and 2aB (5′-ATCGATTCTCACGCTTTCGT-3′) were used to amplify the omp2a gene, and the primer set 2bA (5′-CCTTCAGCCAAATCAGAATG-3′) and 2bB (5′-GGTCAGCATAAAAAGCAAGC-3′) was used to amplify omp2b. The purified PCR products were sequenced by the dideoxy-chain termination method (25).
MLSA and MLVA comprising 16 loci (MLVA-16). (i) MLST.
Extended multilocus sequence analysis (MLSA) was carried out, examining 21 distinct genomic fragments equating to >10.2 kb of sequence. Most of these genetic loci were housekeeping genes. Nine loci, gap, aroA, glk, dnaK, gyrB, trpE, cobQ, omp25, and int-hyp, had already been demonstrated to be useful in describing genetic relatedness among Brucella spp. (37). An additional 12 loci (ddlA, csdB, putA, fbaA, mutL, fumC, prpE, leuA, acnA, soxA, a gene encoding an acyl coenzyme A [acyl-CoA] dehydrogenase, and a gene encoding a glucose-fructose oxidoreductase precursor), part of an extended MLSA scheme designed to increase resolution (A. M. Whatmore, unpublished data), were examined in order to characterize B. microti strains. MLST was conducted essentially as described previously (37). Each allele at each locus was given a distinct arbitrary numerical designation.
(ii) MLVA-16.
MLVA was carried out as described by Le Flèche and colleagues (19) and modified by Al Dahouk and colleagues (1) using eight minisatellite markers (panel 1: bruce06, bruce08, bruce11, bruce12, bruce42, bruce43, bruce45, and bruce55) and eight microsatellite markers (panel 2). The panel 2 markers were split into two groups, panel 2A and 2B, comprising three (bruce18, bruce19, and bruce21) and five (bruce04, bruce07, bruce09, bruce16, and bruce30) markers, respectively. The most highly variable markers were included in panel 2B.
The clustering analysis was based on the categorical coefficient and the unweighted-pair group method using arithmetic averages (UPGMA). The same weight was given to a large and a small number of differences in the repeats at each locus. Three different character data sets were defined and were combined using the composite data set tool provided by Bionumerics. Different weights were assigned to the markers depending on the panel to which they belonged: individual weights of 2 for panel 1 markers, 1 for panel 2A markers, and 0.2 for panel 2B markers (1).
Analysis of phenotypic characteristics. (i) Classical microbiological methods.
All isolates were characterized using the classical microbiological methods described by Alton and colleagues, i.e., CO2 requirement, H2S production, urea hydrolysis, agglutination with monospecific sera (anti-A, anti-M, and anti-R), dye sensitivity (basic fuchsin and thionine), and phage typing (F1, F25, Tb, BK2, Iz, Wb, R/C) (4).
(ii) Biotyping by metabolic activity testing.
Metabolic activity was assessed by using a commercial biotyping system (Micronaut; Merlin Diagnostika, Bornheim-Hersel, Germany) as described previously (3) to determine if a set of phenotypic features may reveal intraspecies variability. The recently developed 96-well “Brucella identification and typing” plate tested for 29 aminopeptidases, 2 phosphatases, 4 glucosidases, 1 esterase, and the metabolism of 11 monosaccharides, 3 disaccharides, 7 sugar derivates, 15 amino acids, 11 organic acids, 1 salt, 1 amino acid derivate, 1 peptide, 1 base, and 6 classical reactions (nitrite and nitrate reduction, pyrazinamidase, Voges-Proskauer medium, urease, and H2S production). Each enzyme-substrate reaction was carried out 3 to 5 times.
Hierarchical cluster analysis was performed by Ward's linkage algorithm using binary coded data based on empirically set cutoffs, and a dendrogram was generated. Each character was considered equal to every other character within the data set.
RESULTS
Molecular analysis. (i) PCR analysis.
The genus-specific bcsp31 PCR correctly identified all B. microti strains as members of the genus Brucella. In the IS711-based AMOS-PCR, a 1,900-bp fragment was generated in all B. microti strains with the Brucella ovis-specific primers, as previously shown for the B. microti type strain, CCM 4915 (data not shown).
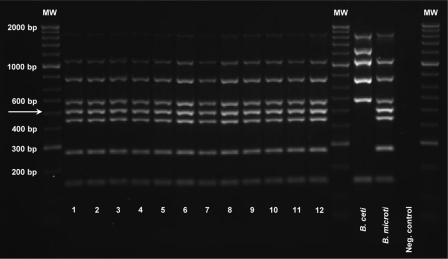
By use of the modified Bruce-ladder multiplex PCR (13, 21), the B. microti field isolates gave a consistent amplicon pattern comprising the seven fragments found in B. suis (1,682 bp, 1,071 bp, 794 bp, 587 bp, 450 bp, 272 bp, and 152 bp) and the distinguishing 510-bp amplicon of the B. microti-specific 12-kb genomic insertion (Fig. 1).
Fig 1.
Identification of Brucella microti using the Bruce-ladder multiplex PCR. The B. microti strains showed a consistent amplicon pattern comprising seven fragments also found in B. suis (1,682 bp, 1,071 bp, 794 bp, 587 bp, 450 bp, 272 bp, and 152 bp) and an additional, B. microti-specific 510-bp amplicon (indicated by the arrow). Lane 1, B. microti type strain CCM 4915; lane 2, strain CCM 4916; lane 3, field isolate IBM 257; lane 4, field isolate IBM 284; lanes 5 and 6, field isolate BMS 10, presented with a rough and a smooth phenotype, respectively; lane 7, field isolate F 303 Mi; lane 8, field isolate FK 210908; lane 9, field isolate FH 2208; lane 10, field isolate FW 70608; lane 11, field isolate BMS 17; lane 12, field isolate BMS 20. A 1-kb Plus DNA ladder (Invitrogen Ltd.) was used as molecular weight (MW) markers. The B. ceti NCTC 12891 and B. microti CCM 4915T reference strains were used as positive controls for all possible amplicons in the Bruce-ladder PCR, and distilled water was used as a negative control.
(ii) Analysis of 16S rRNA, recA, omp2a, and omp2b genes.
The 16S rRNA gene sequences of the 11 B. microti strains were identical to the consensus sequence of the classical Brucella species. Thus, they did not reveal even the minor differences that have been observed in other atypical strains, such as BO1 (B. inopinata), BO2, and the newly described rodent strains originating from Australia (10, 33, 34).
The recA gene sequences of the B. microti strains investigated were successfully amplified by PCR with the primer set recA-BrucOchro-r/recA-BrucOchro-f, initially constructed from sequence information available for B. abortus bv. 1 strain 9-941. BLASTN analysis of the nucleotide sequences of the 1,065-bp PCR fragments generated confirmed specific amplification of the recA gene in all strains. The uniform recA sequence of the B. microti strains under study was 100% identical to the consensus sequence of Brucella spp.
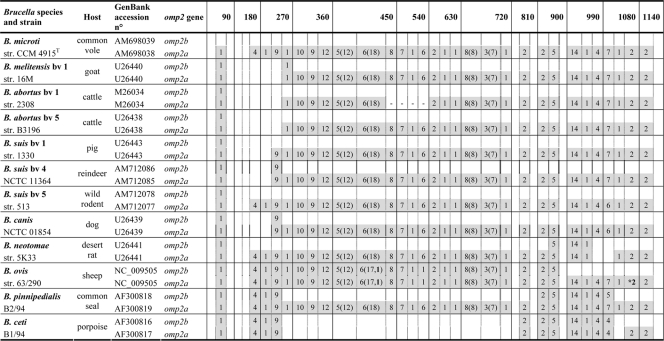
omp2 gene sequences were also 100% identical among B. microti isolates. Genetic diversity in Brucella spp. at the omp2 locus is driven mainly by gene conversion between the two omp2 gene copies, omp2a and omp2b, which are in opposite orientations at this locus and are separated by about 900 bp. Interestingly, the highest divergence (about 15%) between the two copies is found in B. microti, which appears to be ancestral to the classical Brucella species at the genomic level. In other species, omp2 gene conversion has led to exchange of either omp2a-specific or omp2b-specific motifs in the respective omp2 sequence position of each gene copy, sometimes resulting in the homogenization of the gene copies. This is, for example, the case in B. ovis, with two gene copies almost identical to the omp2a consensus, or in B. ceti, with two gene copies almost identical to the omp2b consensus (Fig. 2). The omp2 sequences closest to those of B. microti are those of B. suis bv. 5 (strain 513), another wild rodent isolate. There are indeed only minimal differences between omp2a and omp2b in B. microti CCM 4915T and B. suis bv. 5 strain 513 (1 nucleotide and 2 nucleotides, respectively). However, in terms of omp2 gene conversion, B. microti and B. suis bv. 5 are identical, as shown in Fig. 2.
Fig 2.
Schematic multiple nucleotide sequence alignment of the omp2a and omp2b genes of Brucella strains. Nucleotide sequences are represented by rectangles divided into boxes of 30 nucleotides. The B. microti omp2b gene sequence was used as a reference. B. microti omp2a-specific nucleotides are shaded. The numbers in the corresponding boxes indicate the number of omp2a-specific nucleotides present in the sequence considered. Numbers in parentheses represent insertions and deletions. Numbers in boldface indicate nucleotide differences that are not due to gene conversion. *, there is a premature stop codon in omp2a of B. ovis 63/290.
(iii) MLSA and MLVA-16.
By use of MLSA based on 21 distinct genomic fragments equating to >10.2 kb of conserved sequence, the allelic pattern identified was unique and consistent in all B. microti isolates under study. Hence, B. microti is represented by a single and specific sequence type (ST), which forms a genotypic cluster distinct from those of all other brucellae.
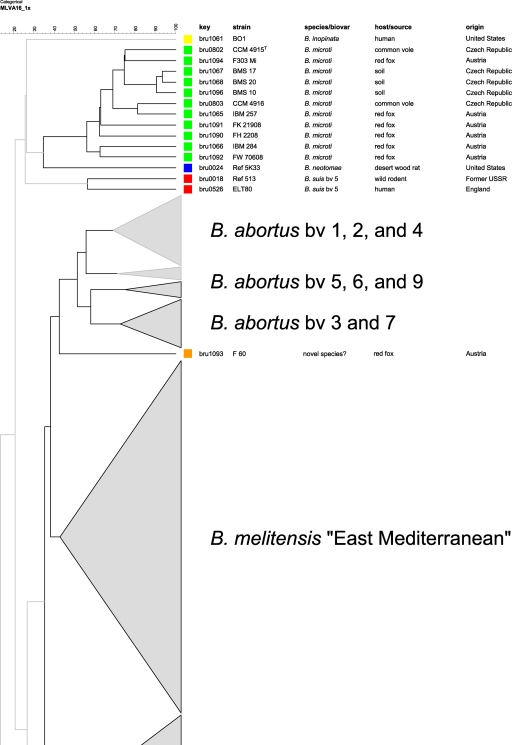
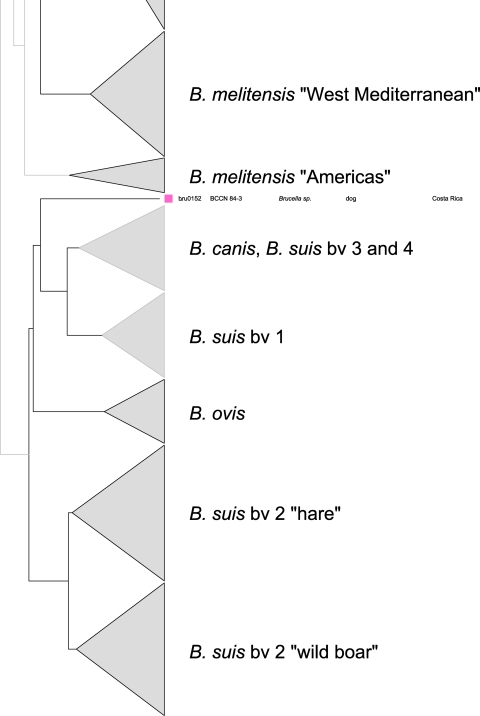
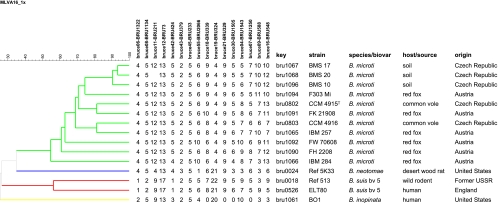
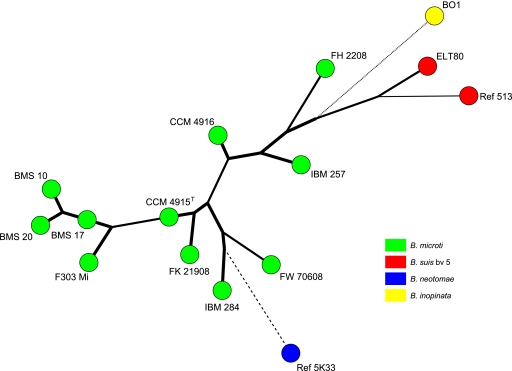
In the comparative multilocus variable-number tandem repeat (VNTR) analysis, B. microti strains also grouped together, forming a separate cluster within the genus Brucella, most closely related to the B. neotomae reference strain 5K33 (Fig. 3). B. inopinata and B. suis bv. 5 were more distantly related. By use of panel 1 VNTRs, the 11 B. microti isolates clustered into 3 different genotypes (genotypes 67, 68, and 81 [http://mlva.u-psud.fr/]) that have not yet been described in other Brucella spp., including rodent strains, i.e., B. neotomae and B. suis bv. 5. On the basis of the complete MLVA-16 data, each of the B. microti strains showed an individual genotype (Fig. 4 and 5). The B. microti isolates grouped neither according to their hosts nor according to their geographical origins. However, the soil isolates, BMS 10, BMS 17, and BMS 20 (genotype 67 with panel 1), were found in the same cluster, because they showed only minimal differences in the highly variable VNTR markers of panel 2B (Fig. 4 and 5).
Fig 3.
Condensed dendrogram of clustered MLVA-16 genotypes of Brucella spp. A total of 344 Brucella isolates revealed 340 different genotypes. The bars reflect the percentages of divergence. The cluster of the B. microti isolates and closely related species, such as B. neotomae, B. inopinata, and B. suis bv. 5, are presented in more detail. Two atypical isolates are separately marked.
Fig 4.
Dendrogram of clustered MLVA-16 genotypes (panels 1 and 2). The 11 B. microti isolates were clustered into 11 different genotypes based on the differences in the numbers of repeat units at 16 VNTR loci. Key, DNA batch.
Fig 5.
Maximum-parsimony analysis of 11 B. microti isolates and the closely related strains B. neotomae 5K33, B. inopinata BO1, and B. suis bv. 5 (513 and ELT80), based on MLVA-16 data. Species are distinguished by different colors.
Analysis of phenotypic characteristics. (i) Bacterial cultures.
All isolates could be easily cultured on Brucella agar at 28°C and 37°C without supplementary CO2. The B. microti strains were characterized by nonfastidious, rapid growth. The bacterial colonies usually appeared transparent to whitish, and were 1 to 2 mm in diameter, after 1 to 2 days of incubation. After 72 h of growth at 37°C, most of the colonies developed a markedly brownish pigmentation. However, the pigmentation intensity and rate differed between individual strains, e.g., strain IBM 284 had already developed brownish colonies after 24 h.
(ii) Differential phenotyping by classical microbiological methods and metabolic activity testing.
Except for strain IBM 257, B. microti isolates did not produce H2S. Urea was regularly hydrolyzed within 90 to 105 min. All isolates grew in the presence of thionine at dilutions of 1/25,000, 1/50,000, and 1/100,000 and in the presence of basic fuchsin at dilutions of 1/50,000 and 1/100,000. Rough (R) colonies were ruby colored after staining with crystal violet, and trypaflavine led to spontaneous agglutination of rough strains (22).
Isolates from animal sources were lysed by phages F1, F25, and Tb at 104× RTD (104 times the routine test dilution) but not at the RTD, except for the fox isolates FH 2208, FK 21908, FW 70608, and F 303 Mi, which showed plaques when phage F25 was used at the RTD. Strain F 303 Mi also showed plaques at the RTD of phage Tb. The smooth strains were all lysed by phages BK2, Iz, and Wb at both dilutions, whereas the rough strains of environmental origin were not. In contrast, the rough strains were lysed by phage R/C at 104× RTD but not at the RTD, whereas the smooth strains were not lysed at all.
The B. microti strains were M antigen dominant except for a single fox isolate. Strain FW 70608 did not agglutinate with monospecific anti-M serum but only with Brucella anti-A monospecific serum. The soil isolates (rough strains) agglutinated with anti-A and anti-M, as well as with anti-R, monospecific sera. One of the soil strains (BMS 10) revealed both smooth and rough lipopolysaccharide phenotypes. The smooth BMS 10 strain agglutinated only with monospecific anti-M serum.
Independently of the animal or environmental source and of geographical origin, all B. microti strains exhibited high biochemical and enzymatic activities (3). The key reactions typical for brucellae, i.e., Glu(pNA)-OH (ENAOH), Pyr-pNA (PYRNA) (consistently negative reactions), and H-hydroxyproline-βNA (HP) (consistently strong positive reactions), clearly identified the isolates as members of the genus Brucella. Thirteen of the 93 substrates tested in the metabolic activity assay (4 aminopeptidase reactions, 1 monosaccharide, 2 organic acids, 2 amino acids, 1 amino acid derivate, 1 peptide, and 2 classical reactions) showed differing results within the species. Despite the overall similar biochemical profiles among the B. microti strains, individual isolates differed markedly in their metabolic activities (Table 2).
Table 2.
Metabolic activities of 11 Brucella microti strainsa
| Substrate or reaction | Abbreviation | Substrate class | Activity of the following Brucella microti strain: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCM 4915T | CCM 4916 | IBM 284 | IBM 257 | FH 2208 | FK 21908 | FW 70608 | F 303 Mi | BMS 10 | BMS 17 | BMS 20 | |||
| Leu-pNA | LNA | Aminopeptidases with pNA | + | + | ++ | + | + | ++ | + | + | + | ++ | ++ |
| Ac-Gly-Lys-β | AcGK | Aminopeptidases with βNA | + | + | − | − | − | − | − | − | − | − | − |
| Ala-Phe-Pro-Ala-β | AFPA | Aminopeptidases with βNA | + | ++ | + | + | + | + | + | + | + | + | + |
| Asn-β | N | Aminopeptidases with βNA | − | + | − | − | − | − | − | − | − | − | − |
| Ac-Lys-Ala-β | AcKA | Aminopeptidases with βNA | + | + | − | − | − | − | − | − | − | − | − |
| d-Ala-d-Ala-β | dAdA | Aminopeptidases with βNA | ++ | + | + | + | + | + | ++ | + | + | + | + |
| Val-Tyr-Ser-β | VTS | Aminopeptidases with βNA | ++ | + | + | − | − | − | + | − | − | + | − |
| p-Nitrophenyl-α-d-maltoside | aMAL7 | Glucosidases | + | ++ | + | + | + | + | + | + | + | + | + |
| p-Nitrophenyl-α-d-xylopyranoside | aXYL7 | Glucosidases | + | ++ | + | + | + | + | + | + | + | + | + |
| d(−)-Ribose | d-RIB | Monosaccharides | + | ++ | ++ | ++ | ++ | + | ++ | ++ | ++ | + | ++ |
| d(−)-Arabinose | d-ARA | Monosaccharides | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| d(+)-Xylose | d-XYL | Monosaccharides | + | ++ | + | ++ | ++ | + | ++ | ++ | ++ | + | ++ |
| d-Threitol | d-TOL | Sugar derivates | ++ | ++ | + | ++ | ++ | ++ | ++ | + | ++ | + | ++ |
| dl-Lactic acid | dlLac | Organic acids | + | ++ | + | ++ | ++ | + | ++ | + | + | + | + |
| l-Asparagine | l-Asn | Amino acids | + | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ |
| l-Glutamic acid | l-Glu | Amino acids | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| d-Alanine | d-Ala | Amino acids | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | − |
| Propionic acid | Propn | Organic acids | − | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − | ++ |
| dl-β-Hydroxybutyric acid | βHBut | Organic acids | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ |
| Nα-Acetyl-l-arginine | AcArg | Amino acid derivates | ++ | ++ | − | ++ | ++ | − | ++ | − | − | − | − |
| Hippuryl-Arg | HipArg | Peptides | ++ | ++ | − | ++ | ++ | − | ++ | − | − | − | − |
| Glycine-free base | Gly | Amino acids | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
| Nitrite reduction | NTI | Classical reactions | − | + | + | + | + | ++ | ++ | + | ++ | ++ | + |
| Voges-Proskauer | VP | Classical reactions | ++ | + | + | + | ++ | ++ | ++ | ++ | ++ | + | + |
| Nitrate reduction | NTA | Classical reactions | − | + | + | + | + | ++ | ++ | + | ++ | ++ | + |
| Glu(pNA)-OH | ENAOH | Aminopeptidases with pNA | − | − | − | − | − | − | − | − | − | − | − |
| Pyr-pNA | PYRNA | Aminopeptidases with pNA | − | − | − | − | − | − | − | − | − | − | − |
| H-Hydroxyproline-βNA | HP | Aminopeptidases with βNA | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
A representative selection of the 93 substances tested in the Brucella-specific Micronaut assay has been merged (3). The quality of each biochemical reaction is presented as follows: −, no metabolic activity; +, moderate metabolic activity; ++, strong metabolic activity. Brucella-specific traits are shown in boldface, and clearly distinctive features of the isolates are shaded.
DISCUSSION
With the eradication of brucellosis in domestic animals in many parts of the developed world, the impact of wildlife brucellosis becomes increasingly relevant as a potential reservoir of the etiologic agent and a source for the reemergence of this zoonotic disease (2, 15, 16). Although rats and mice are synanthropic species living in close contact with humans and domestic animals, knowledge of the epidemiology of brucellosis in wild rodents and its impact on human health is still limited. There are some historical reports about the isolation of classical brucellae from rodents (32, 35). However, only two rodent-specific Brucella species have been described to date. In the 1960s, B. neotomae was isolated from desert wood rats (Neotoma lepida) in Utah (31), and just recently, B. microti was isolated from the common vole (Microtus arvalis) (17, 28). The significance of these findings became obvious when additional Brucella strains (non-B. microti) isolated from wild rodents in North Queensland, Australia (33), revealed notable genetic similarity with two human isolates, strains BO1 (10) and BO2 (34).
The isolation of B. microti from different animal species and also from soil (26, 27) in Austria and in the Czech Republic throughout a whole decade proves the endemic persistence of the pathogen within a restricted geographical region. It is possible that enzootic transmission cycles including rodents, carnivores, and the natural environment maintain the long-term survival of B. microti in this region. Soil might be the primary reservoir of infection, but other vectors cannot be excluded. Although the pathogenicity of B. microti for livestock and humans has not yet been confirmed, new foci of Brucella infections potentially posing a public health threat have to be monitored (24). The natural environment may play a key role in the reemergence of the disease.
To gain deeper insight into the composition of the species B. microti, all currently available isolates have been characterized using a comprehensive combined molecular and classical microbiological approach.
The molecular techniques used in this study revealed that B. microti is a species with a high degree of genetic relatedness. With respect to one of its closer relatives, B. suis 1330, the genome sequence of B. microti CCM 4915T was found to be 99.84% identical in aligned regions. In contrast, the virulence and pathogenicity of B. microti are totally different, e.g., intramacrophagic replication proceeds much faster, and the mortality rate in murine models of infection is much higher, than in B. suis (18).
The 100% identity of the 16S rRNA gene sequence among the B. microti strains tested and with the consensus sequence of the genus is consistent with previous studies of the six classical Brucella species (14). 16S rRNA and recA sequence analysis, as well as MLSA (based on nine genes: gap, aroA, glk, dnaK, gyrB, trpE, cobQ, omp25, and int-hyp), also revealed a high degree of homology among the Australian rodent strains most recently described (33). However, the 16S rRNA sequence of these strains was unique in the genus Brucella (comparable to those of B. inopinata [BO1] and strain BO2), showing 99.2% identity to the consensus sequence.
The recA sequence in B. microti strains was identical to the corresponding consensus sequence of Brucella spp. In contrast, recA shows high genetic diversity in Ochrobactrum spp., which are closely related to members of the genus Brucella (30).
Interestingly, B. microti and B. suis bv. 5 (strain 513), another wild rodent strain, had almost identical omp2 genes. Hence, the position of strain 513, or, in more general terms, that of bv. 5, within the species B. suis currently appears aberrant (38).
Based on MLSA of 21 genetic loci, B. microti strains were found to represent a single clone, i.e., only a single sequence type (ST) could be identified, a pattern comparable to those of B. ovis and B. neotomae (37). However, B. microti could be clearly distinguished from the currently known Brucella species, although the global alignment of B. microti CCM 4915T and B. suis 1330 chromosomes revealed an almost perfect colinearity (5). Because of the wide ecological niche of B. microti, intraspecies polymorphisms might have been expected. For the marine mammal brucellae, isolated from different animal species, a much higher level of intragroup diversity, supporting a host-specific classification, has been described (37).
By use of MLVA-16, each B. microti strain exhibited a distinct genotype (Fig. 4), mainly due to diverging panel 2B VNTR markers. Since different Brucella isolates originating from the same outbreak usually show identical MLVA-16 genotypes (1), the B. microti strains under study have to be regarded as a heterogeneous population. IBM 284 and FW 70608 also showed aberrant panel 1 genotypes because of minor differences at the bruce55 and bruce42 loci. In both strains, these deviant genotypes were associated with unusual phenotypic characteristics. IBM 284 rapidly developed brownish colonies, and FW 70608 agglutinated with monospecific anti-A serum but not with anti-M serum, in contrast to all the other B. microti strains. A similar association of VNTR genotypes with phenotypic features has been described for fuchsin-sensitive B. melitensis strains (1).
Both molecular typing and biotyping are essential for full understanding of the population structure of the genus Brucella and the intraspecies variability of its closely related members. In the B. microti population, genotypic and phenotypic features diverged. The minimal genetic differences found in B. microti strains do not readily account for the differences in phenotype and host specificity or habitat. Furthermore, the pathogenicity of B. microti differs significantly from that of other Brucella species (18). B. microti showed an enhanced capacity for intramacrophagic replication and exhibited a highly lethal, septicemia-like course of infection in murine models. The resistance of B. microti to acidic pHs might explain its ability to survive in soil (18). In contrast to most of the other Brucella species currently known, B. microti is a fast-growing and metabolically very active microorganism (3). This metabolic activity is a feature shared by all B. microti strains with their closest phylogenetic relative, Ochrobactrum. That is the reason why brucellae, including B. microti, are commonly misidentified as Ochrobactrum species by commercially available identification systems, such as the API 20 NE system (11, 17). Mechanisms involving gene regulation and altered expression may have contributed to the adaptation of the bacteria to environmental stress, and the differing metabolic activities of the isolates may mirror such regulatory processes. The outstanding metabolic activity of B. microti compared to that of other members of the genus also justified its designation as a distinct species (3, 28).
In summary, the competence of B. microti to adapt its lifestyle so as to survive in mammals and in soil suggests that B. microti represents an ancestral Brucella species whose phenotype is more closely related to that of the soil-associated facultative human pathogen Ochrobactrum than to that of the classical Brucella species. It has been suggested that the classical Brucella species evolve intracellularly as isolated units in their preferred hosts, with recombination restricted by ecological isolation (23). In contrast, the ecological niche of B. microti in soil theoretically allows many interactions with bacteria of other genera, facilitating horizontal gene transfer. On the basis of the genetic markers investigated, however, the B. microti isolates under study revealed a clonal population structure. Since the number of B. microti isolates characterized in this study was low and their geographical distribution was rather limited, further surveillance on a global scale is urgently needed in order to obtain a deeper understanding of distribution, ecology, zoonotic potential, genomic organization, and relatedness to other rodent strains.
ACKNOWLEDGMENTS
We gratefully thank Cornelia Göllner and Peter Bahn of the Federal Institute for Risk Assessment for excellent technical assistance.
Work by G. Vergnaud, P. Le Flèche, and H. C. Scholz is part of the European Biodefence Laboratory Network (EBLN) under the auspices of the European Defense Agency (EDA).
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Al Dahouk S, et al. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69:137–145 [DOI] [PubMed] [Google Scholar]
- 2. Al Dahouk S, et al. 2005. Seroprevalence of brucellosis, tularemia, and yersiniosis in wild boars (Sus scrofa) from north-eastern Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 52:444–455 [DOI] [PubMed] [Google Scholar]
- 3. Al Dahouk S, et al. 2010. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol. 10:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alton GG, Jones LM, Angus RD, Verger JM. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France [Google Scholar]
- 5. Audic S, Lescot M, Claverie JM, Scholz HC. 2009. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics 10:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baily GG, Krahn JB, Drasar BS, Stoker NG. 1992. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J. Trop. Med. Hyg. 95:271–275 [PubMed] [Google Scholar]
- 7. Bricker BJ, Halling SM. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cloeckaert A, Verger JM, Grayon M, Grépinet O. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 141:2111–2121 [DOI] [PubMed] [Google Scholar]
- 9. Cloeckaert A, et al. 2001. Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3:729–738 [DOI] [PubMed] [Google Scholar]
- 10. De BK, et al. 2008. Novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J. Clin. Microbiol. 46:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elsaghir AA, James EA. 2003. Misidentification of Brucella melitensis as Ochrobactrum anthropi by API 20 NE. J. Med. Microbiol. 52:441–442 [DOI] [PubMed] [Google Scholar]
- 12. Ficht T. 2010. Brucella taxonomy and evolution. Future Microbiol. 5:859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. García-Yoldi D, et al. 2006. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin. Chem. 52:779–781 [DOI] [PubMed] [Google Scholar]
- 14. Gee JE, et al. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 42:3649–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godfroid J, et al. 2005. From the discovery of the Malta fever's agent to the discovery of a marine mammal reservoir, brucellosis has continuously been a re-emerging zoonosis. Vet. Res. 36:313–326 [DOI] [PubMed] [Google Scholar]
- 16. Godfroid J. 2002. Brucellosis in wildlife. Rev. Sci. Tech. 21:277–286 [DOI] [PubMed] [Google Scholar]
- 17. Hubálek Z, et al. 2007. Brucellosis of the common vole (Microtus arvalis). Vector Borne Zoonotic Dis. 7:679–687 [DOI] [PubMed] [Google Scholar]
- 18. Jiménez de Bagüés MP, et al. 2010. The new species Brucella microti replicates in macrophages and causes death in murine models of infection. J. Infect. Dis. 202:3–10 [DOI] [PubMed] [Google Scholar]
- 19. Le Flèche P, et al. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maquart M, Zygmunt MS, Cloeckaert A. 2009. Marine mammal Brucella isolates with different genomic characteristics display a differential response when infecting human macrophages in culture. Microbes Infect. 11:361–366 [DOI] [PubMed] [Google Scholar]
- 21. Mayer-Scholl A, Draeger A, Göllner C, Scholz HC, Nöckler K. 2010. Advancement of a multiplex PCR for the differentiation of all currently described Brucella species. J. Microbiol. Methods 80:112–114 [DOI] [PubMed] [Google Scholar]
- 22. Meyn A, Schmid DO, Schrinner EJ. 1957. Contribution to the understanding and characterization of the dissociation forms of Brucella abortus. Zentralbl. Veterinarmed. 4:933–944 [Google Scholar]
- 23. Moreno E, Cloeckaert A, Moriyón I. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90:209–227 [DOI] [PubMed] [Google Scholar]
- 24. Morris JG, Jr, Southwick FS. 2010. Brucella, voles, and emerging pathogens. J. Infect. Dis. 202:1–2 [DOI] [PubMed] [Google Scholar]
- 25. Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scholz HC, et al. 2009. Isolation of Brucella microti from mandibular lymph nodes of red foxes, Vulpes vulpes, in lower Austria. Vector Borne Zoonotic Dis. 9:153–156 [DOI] [PubMed] [Google Scholar]
- 27. Scholz HC, et al. 2008. Isolation of Brucella microti from soil. Emerg. Infect. Dis. 14:1316–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scholz HC, et al. 2008. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58:375–382 [DOI] [PubMed] [Google Scholar]
- 29. Scholz HC, et al. 2010. Brucella inopinata sp. nov., isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 60:801–808 [DOI] [PubMed] [Google Scholar]
- 30. Scholz HC, et al. 2006. Genotyping of Ochrobactrum anthropi by recA-based comparative sequence, PCR-RFLP, and 16S rRNA gene analysis. FEMS Microbiol. Lett. 257:7–16 [DOI] [PubMed] [Google Scholar]
- 31. Stoenner HG, Lackmann DB. 1957. A preliminary report on a Brucella isolated from the desert wood rat, Neotoma lepida Thomas. J. Am. Vet. Med. Assoc. 130:411–412 [PubMed] [Google Scholar]
- 32. Stoll L, Manz D. 1971. Isolation of Brucella from wild mammals. Dtsch. Tierarztl. Wochenschr. 78:193–195 [PubMed] [Google Scholar]
- 33. Tiller RV, et al. 2010. Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl. Environ. Microbiol. 76:5837–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiller RV, et al. 2010. Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol. 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vershilova PA, Liamkin GI, Malikov VE, Dranovskaya EA, Taran IF. 1983. Brucella strains from mouselike rodents in Southwestern USSR. Int. J. Syst. Bacteriol. 33:399–400 [Google Scholar]
- 36. Wattam AR, et al. 2009. Analysis of ten Brucella genomes reveals evidence for horizontal gene transfer despite a preferred intracellular lifestyle. J. Bacteriol. 191:3569–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whatmore AM, Perrett LL, MacMillan AP. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whatmore AM. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9:1168–1184 [DOI] [PubMed] [Google Scholar]