Abstract
Ralstonia solanacearum, a major phytopathogenic bacterium, causes a bacterial wilt disease in diverse plants. Although fatty acid analyses of total membranes of R. solanacearum showed that they contain primarily palmitic (C16:0), palmitoleic (C16:1) and cis-vaccenic (C18:1) acids, little is known regarding R. solanacearum fatty acid synthesis. The R. solanacearum GMI1000 genome is unusual in that it contains four genes (fabF1, fabF2, fabF3, and fabF4) annotated as encoding 3-ketoacyl-acyl carrier protein synthase II homologues and one gene (fabB) annotated as encoding 3-ketoacyl-acyl carrier protein synthase I. We have analyzed this puzzling apparent redundancy and found that only one of these genes, fabF1, encoded a long-chain 3-ketoacyl-acyl carrier protein synthase, whereas the other homologues did not play roles in R. solanacearum fatty acid synthesis. Mutant strains lacking fabF1 are nonviable, and thus, FabF1 is essential for R. solanacearum fatty acid biosynthesis. Moreover, R. solanacearum FabF1 has the activities of both 3-ketoacyl-acyl carrier protein synthase II and 3-ketoacyl-acyl carrier protein synthase I.
INTRODUCTION
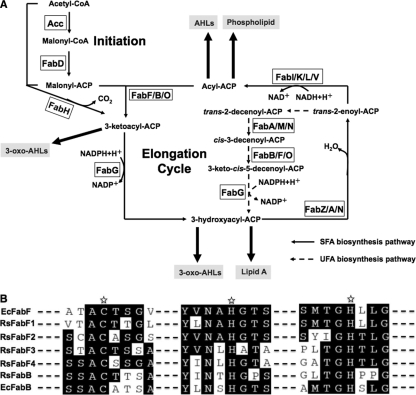
Fatty acids are not only one of the major components of bacterial cell membranes but also intermediates in the synthesis of quorum-sensing molecules, lipid A, and vitamins (19, 20) (Fig. 1A). In most bacteria, fatty acid biosynthesis is catalyzed by a series of small, soluble proteins, each encoded by a discrete gene (3, 24, 39). Four reactions, i.e., condensation, reduction, dehydration, and reduction, are required to complete each round of fatty acid elongation (Fig. 1A). The chain elongation steps in fatty acid biosynthesis consist of the condensation of acyl groups derived from acyl-acyl carrier protein (ACP) or acyl coenzyme A (acyl-CoA) substrates with malonyl-ACP in reactions catalyzed by the 3-ketoacyl-ACP synthases (KASs) (17, 26, 37). These condensing enzymes are divided into two classes (3, 37). The FabH class of condensing enzymes is responsible for the initiation of fatty acid chains, whereas the FabB-FabF classes of condensing enzymes together catalyze the elongation steps required for the synthesis of long acyl chains (17, 26). Although FabB, FabF, and FabH all share the same overall structure, the FabB-FabF class of enzymes possesses a Cys-His-His catalytic triad at the active site, whereas the FabH enzymes have a Cys-His-Asn triad (37).
Fig 1.
Fatty acid biosynthesis in bacteria and alignment of R. solanacearum 3-ketoacyl-acyl carrier protein synthase homologues with E. coli FabF and FabB. (A) Fatty acid biosynthesis in bacteria and acyl-ACPs as acyl donors in cellular metabolism. Abbreviations: ACC, acetyl-CoA carboxylase; FabD, malonyl-CoA:ACP transacylase; FabH, 3-ketoacyl ACP synthase III; FabG, 3-ketoacyl-ACP reductase; FabZ, 3-hydroxyacyl-ACP dehydratase; FabA, 3-hydroxydecanoyl-ACP dehydratase/isomerase (from E. coli); FabM, trans-2-cis-3-decenoyl-ACP isomerase (from Streptococcus pneumoniae); FabN, 3-hydroxydecanoyl-ACP dehydratase/isomerase (from Enterococcus faecalis); FabF, 3-ketoacyl-ACP synthase II; FabB, 3-ketoacyl-ACP synthase I (from E. coli); FabO, 3-ketoacyl-ACP synthase I (from E. faecalis); FabI (from E. coli), FabK (from Streptococcus pneumoniae), FabL (from Bacillus subtilis), and FabV (from Vibrio cholerae), enoyl-ACP reductase; AHLs, acylhomoserine lactones; SFA, saturated fatty acid; UFA, unsaturated fatty acid. (B) Sequence alignment of R. solanacearum 3-ketoacyl-acyl carrier protein synthase homologues with E. coli FabF and FabB. Ec, E. coli; Rs, R. solanacearum. The Cys-His-His catalytic triads are marked with stars.
Escherichia coli expresses both types of long-chain condensing enzymes (3, 26). The fabB gene is defined by a class of mutants defective in unsaturated fatty acid synthesis and encodes synthase I (KAS I) (4, 13, 14). The fabF gene encoding synthase II (KAS II) was found to be required for the elongation of cis-9-hexadecenoyl-ACP (palmitoleoyl-ACP) to cis-11-octadecenoyl-ACP (cis-vaccenoyl-ACP), a phenotype expected for a defect in chain elongation activity (5, 13, 14). Thus, although the two enzymes have overlapping substrate specificities, FabB is responsible for a condensation reaction in unsaturated fatty acid synthesis that cannot be performed by FabF (10), whereas FabF plays a role in the thermal regulation of the fatty acid composition (8).
The fabB and fabA genes, which encode the key enzymes of the classic anaerobic pathway of unsaturated fatty acid synthesis, show covariance within organisms, and fabB genes are found only in genomes that contain fabA (3). FabA and FabB homologues are encoded only in the genomes of alpha- and gammaproteobacteria (21, 24, 35, 41). In contrast, FabF homologues are found throughout the bacteria, and FabF is often considered the generic KAS of long-chain fatty acid synthesis (24, 35, 41). For several Gram-positive bacteria, FabF homologues have been shown to also have KAS I activity (29, 35, 41). Enterococcus faecalis encodes two FabF homologues. Wang and Cronan (35) first showed that one of these proteins, now called FabO, functioned as a KAS I analogous to FabB, whereas the other FabF homologue had KAS II activity. A similar picture was found for the FabF proteins of Clostridium acetobutylicum and Lactococcus lactis (29, 41).
Ralstonia solanacearum, one of the most serious phytopathogenic bacteria, causes a bacterial wilt disease that affects more than 200 plant species. Its hosts include economically important plants such as tomato (Lycopersicon esculentum), potato (Solanum tuberosum), tobacco (Nicotiana tabacum), banana (Musa acuminata), and peanut (Arachis hypogaea) (16, 28). The expression of virulence determinants in R. solanacearum is controlled by a complex regulatory network, in which PhcA, a LysR-type transcriptional regulator, plays a central role (2, 12, 28). The level of active PhcA is regulated in response to cell density by a quorum-sensing mechanism that involves the specific autoinducer molecule 3-hydroxypalmitic acid ester (11, 12). It has been demonstrated that 3-hydroxypalmitic acid methyl ester is synthesized by the gene product of phcB, a putative S-adenosyl methionine-dependent methyltransferase, which is thought to convert 3-hydroxypalmitoyl-ACP to 3-hydroxypalmitoyl methyl ester (11).
Although fatty acid analyses of total membranes of R. solanacearum showed that they contain primarily palmitic (C16:0), palmitoleic (C16:1), and cis-vaccenic (C18:1) acids (11, 33), much remains to be elucidated regarding R. solanacearum fatty acid synthesis. To identify putative long-chain 3-ketoacyl-acyl carrier protein synthase homologues, we searched the R. solanacearum GMI1000 genome database against the E. coli fabF and fabB sequences using BLAST. Four open reading frames were identified as E. coli FabF homologues; two of these are encoded on the chromosome, whereas the others are encoded on the megaplasmid. The chromosomal RSc1054 gene is annotated as encoding 3-ketoacyl-acyl carrier protein synthase II (FabF1). The megaplasmid genes RSp0358 and RSp0361 are annotated as encoding two putative 3-ketoacyl-acyl carrier protein synthase II enzymes (FabF2 and FabF3, respectively). The megaplasmid also encodes a putative E. coli FabB homologue, RSp0357. Finally, the chromosomal RSc0427 gene encodes a putative fourth 3-ketoacyl-acyl carrier protein synthase II (FabF4). In order of their numbering, the four putative 3-ketoacyl-acyl carrier protein synthases II have 58%, 36%, 35%, and 37% identities to E. coli FabF, respectively (Fig. 1B). The FabF1 homologue is located within a gene cluster (plsX fabH fabD fabG1 acpP fabF1) that is composed of what appears to be a complete set of the genes required for saturated fatty acid synthesis, except for an enoyl-ACP reductase. The FabB, FabF2, and FabF3 homologues are located in a megaplasmid gene cluster (fabF3 RSp0360 fabG2 fabF2 fabB), whereas FabF4 is located at a distant chromosomal location. All of these long-chain 3-ketoacyl-acyl carrier protein synthase homologues possess a Cys-His-His catalytic triad (Fig. 1B).
In this report, we describe the characterization of these five R. solanacearum putative 3-ketoacyl-acyl carrier protein synthase homologues. We report that only one of these homologues, fabF1, encodes a long-chain 3-ketoacyl-acyl carrier protein synthase required for fatty acid synthesis, whereas the other homologues did not play roles in R. solanacearum fatty acid synthesis.
MATERIALS AND METHODS
The supply sources were as follows: malonyl-CoA, acetyl-CoA, fatty acids, cerulenin, NADH, NADPH, and antibiotics were obtained from Sigma; Takara Biotechnology provided molecular biology reagents; Novagen provided pET vectors; American Radiolabeled Chemicals, Inc., provided sodium [1-14C]acetate (specific activity, 50 mCi/mM); Invitrogen provided the Ni2+-agarose column; and Bio-Rad provided the Quick Start Bradford dye reagent. All other reagents were of the highest available quality.
Bacterial strains, plasmids, and growth media.
The E. coli K-12 strains, R. solanacearum strains, and plasmids used in this study are listed in Table 1. R. solanacearum strains were routinely grown at 30°C in BG broth (1% Bacto peptone, 0.1% Casamino Acids, 0.1% yeast extract, and 0.5% glucose) or BG agar (BG broth plus 1.6% agar). Luria-Bertani (LB) medium was used as the rich medium for E. coli. The phenotypes of E. coli fab strains were assessed with rich broth (RB) medium (34). Oleate neutralized with KOH was added to RB medium at a final concentration of 0.1%. M63 medium (27) supplemented with 0.1% Casamino Acids was used to screen R. solanacearum mutants, and if needed, 5% or 10% sucrose was added. Antibiotics were used at the following concentrations: sodium ampicillin at 100 μg/ml, kanamycin sulfate at 30 μg/ml, chloramphenicol at 30 μg/ml, and gentamicin at 10 μg/ml (for E. coli) or 30 μg/ml (for R. solanacearum). l-Arabinose was used at a final concentration of 0.01%. Isopropyl-β-d-thiogalactoside (IPTG) was used at a final concentration of 1 mM, and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was used at a final concentration of 20 μg/ml.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argF)U169 (ϕ80lacZΔM15) | Laboratory collection |
| MG1655 | Wild-type strain | Laboratory collection |
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal (λDE3) | Laboratory collection |
| S17-1 | F−thi pro hsdR [RP4-2 Tc::Mu Km::Tn7 (Tp Sm)] | Laboratory collection |
| CY244 | fabB15(Ts) fabF | 8 |
| CY242 | fabB15(Ts) | 34 |
| CL28 | fabF::kan of MG1655 | 23 |
| R. solanacearum | ||
| GMI1000 | Wild-type strain | ATCC |
| RSY-F1 | GMI1000 fabF1::pCJK1 | This work |
| RSY-F11 | GMI1000 fabF1::pCJK1/pV-F1Histag-Tcr | This work |
| RS-F1 | GMI1000 fabF1::Gmr/pV-F1Histag-Tcr | This work |
| RS-F2 | GMI1000 fabF2::Gmr | This work |
| RS-F3 | GMI1000 fabF3::Gmr | This work |
| RS-F4 | GMI1000 fabF4::Gmr | This work |
| RS-B | GMI1000 fabB::Gmr | This work |
| Plasmids | ||
| pMD19 | Ampr; T-vector | Takara |
| pBAD24 M | Ampr; NcoI site of expression vector pBAD24 changed to an NdeI site | |
| pET-28b | Kmr; expression vector | Novagen |
| pK18mobscaB | Kmr; sacB-based gene replacement vector | 31 |
| p34s-Gm | Ampr; Gm resistance cassette-carrying vector | 9 |
| p34s-Tc | Ampr; Tc resistance cassette-carrying vector | 9 |
| pVLT33 | Kmr; RSF1010-lacIq/Ptac hybrid-host-range expression vector; MCS of pUC18 | 7 |
| pCJ1 | Ampr; R. solanacearum fabF1 in pMD19 | This work |
| pCJ2 | Ampr; R. solanacearum fabF2 in pMD19 | This work |
| pCJ3 | Ampr; R. solanacearum fabF3 in pMD19 | This work |
| pCJ4 | Ampr; R. solanacearum fabF4 in pMD19 | This work |
| pCJ5 | Ampr; R. solanacearum fabB in pMD19 | This work |
| pCJ6 | Ampr; R. solanacearum fabH in pMD19 | This work |
| pCJ7 | Ampr; R. solanacearum fabD in pMD19 | This work |
| pCJ8 | Ampr; R. solanacearum fabZ in pMD19 | This work |
| pCJ9 | Ampr; R. solanacearum fabI in pMD19 | This work |
| pCJLF1 | Ampr; RsfabF1 in pBAD24 M (ligation of the NdeI-HindIII fragment from pCJ1 with pBAD24 M at the same sites) | This work |
| pCJLF2 | Ampr; RsfabF2 in pBAD24 M (constructed as for pCJLF1) | This work |
| pCJLF3 | Ampr; RsfabF3 in pBAD24 M (ligation of the NdeI-XbaI fragment from pCJ3 with pBAD24 M at the same sites) | This work |
| pCJLF4 | Ampr; RsfabF4 in pBAD24 M (constructed as for pCJLF1) | This work |
| pCJLB | Ampr; RsfabB in pBAD24 M (constructed as for pCJLF1) | This work |
| pCEF1 | Kmr; RsfabF1 from pCJ1 cut with NdeI and HindIII and inserted into the same sites of pET-28b | This work |
| pCEF2 | Kmr; RsfabF2 in pET-28b (constructed as for pCEF1) | This work |
| pCEF3 | Kmr; RsfabF3 from pCJ3 cut with NdeI and EcoRI and inserted into the same sites of pET-28b | This work |
| pCEF4 | Kmr; RsfabF4 in pET-28b (constructed as for pCEF1) | This work |
| pCEB | Kmr; RsfabB in pET-28b (constructed as for pCEF1) | This work |
| pCEH | Kmr; RsfabH in pET-28b (constructed as for pCEF1) | This work |
| pCED | Kmr; RsfabD in pET-28b (constructed as for pCEF1) | This work |
| pCEZ | Kmr; RsfabZ in pET-28b (constructed as for pCEF1) | This work |
| pCEI | Kmr; RsfabI in pET-28b (constructed as for pCEF1) | This work |
| pCJK1 | Kmr Gmr; pK18mobscaB-carried RsfabF1::Gmr | This work |
| pCJK2 | Kmr Gmr; pK18mobscaB-carried RsfabF2::Gmr | This work |
| pCJK3 | Kmr Gmr; pK18mobscaB-carried RsfabF3::Gmr | This work |
| pCJK4 | Kmr Gmr; pK18mobscaB-carried RsfabF4::Gmr | This work |
| pCJK5 | Kmr Gmr; pK18mobscaB-carried RsfabB::Gmr | This work |
| pV-F1His | Kmr; fabF1 DNA fragment amplified from pCEF1 digested with EcoRI and HindIII and inserted into the same sites of pVLT33 | This work |
| pV-F1His-Tc | Kmr Tcr; Tc-resistant cassette of p34s-Tc digested with PstI and cloned into the same site of pV-F1His | This work |
Recombinant DNA techniques and construction of plasmids.
To clone the R. solanacearum fab genes, genomic DNA was extracted from R. solanacearum strain GMI1000 by using the Takara DNA extraction kit. The PCR products were amplified from strain GMI1000 genomic DNA by using Pfu DNA polymerase and the primers listed in Table S1 in the supplemental material and were inserted into T-vector plasmid pMD19 to produce plasmids pCJ1 (R. solanacearum fabF1 [RsfabF1]), pCJ2 (RsfabF2), pCJ3 (RsfabF3), pCJ4 (RsfabF4), pCJ5 (RsfabB), pCJ6 (RsfabH), pCJ7 (RsfabD), pCJ8 (RsfabZ), and pCJ9 (RsfabI). The fab gene sequences were confirmed by sequencing performed by Shanhai Sangon, Inc.
To produce plasmids pCJLF1 (RsfabF1), pCJLF2 (RsfabF2), pCJLF3 (RsfabF3), pCJLF4 (RsfabF4), and pCJFB (RsfabB), or plasmids pCEF1 (RsfabF1), pCEF2 (RsfabF2), pCEF3 (RsfabF3), pCEF4 (RsfabF4), pCEB (RsfabB), pCEH(RsfabH), pCED(RsfabD), pCEZ(RsfabZ), and pCEI(RsfabI), the T-vector pMD19 fab gene plasmids were digested with NdeI and a second restriction enzyme, the site for which had been designed into the downstream primers listed in Table S1 in the supplemental material. The fragments were gel purified and ligated into pBAD24M or pET-28b digested with the same enzymes.
Disruption of the fabF and fabB genes.
To disrupt the R. solanacearum KAS genes, five suicide plasmids were constructed, as follows. Plasmids pCJ1 (RsfabF1), pCJ2 (RsfabF2), pCJ3 (RsfabF3), pCJ4 (RsfabF4), and pCJ5 (RsfabB) were digested with the restriction endonucleases SacI (RsfabF1), PstI (RsfabF2 and RsfabF3), SphI (RsfabF4), and BamHI (RsfabB), respectively, and ligated into the gentamicin-resistant cassette of p34s-Gm (9) digested with the same restriction endonuclease, to yield five intermediate plasmids. The fabF or fabB genes of the intermediate plasmids (containing the gentamicin-resistant cassette) were liberated with EcoRI and HindIII and ligated into plasmid pK18mobsacB (31) digested with the same enzymes, to produce plasmids pCJK1 (RsfabF1::Gmr), pCJK2 (RsfabF2::Gmr), pCJK3 (RsfabF3::Gmr), pCJK4 (RsfabF4::Gmr), and pCJK5 (RsfabB::Gmr), respectively.
Following the mating of derivatives of E. coli strain S17-1 carrying the disruption plasmids with R. solanacearum GMI1000 on BG plates for 24 h at 30°C, the cells were suspended in BG medium, and appropriate dilutions were spread onto BG plates containing chloramphenicol (to select against the donor strain) plus kanamycin to select for the integration of the nonreplicating plasmid into the chromosome of the recipient. Several colonies were inoculated into BG medium, and the cultures were incubated at 30°C for 24 h, after which appropriate dilutions were spread onto BG plates containing 10% sucrose. The resulting colonies were inoculated onto BG plates containing gentamicin by using sterile toothpicks. Colonies resistant to gentamicin were screened by colony PCR utilizing the primers listed in Table S1 in the supplemental material. Finally, mutant strains RS-F2 (fabF2::Gm), RS-F3 (fabF3::Gm), RS-F4 (fabF4::Gm), and RS-B (fabB::Gm) were obtained.
To obtain the conditional fabF1 mutant, plasmid pV-F1Histag-Tc was constructed as follows. The fabF1 DNA fragment, amplified by using the primers listed in Table S1 in the supplemental material and plasmid pCEF1 as a template, was digested with EcoRI and HindIII and inserted into the same sites of pVLT33 (7) to yield plasmid pV-F1Histag. Plasmid p34s-Tc (9) with a tetracycline-resistant cassette was digested with PstI and inserted into the same site of pV-F1Histag to give pV-F1Histag-Tc. Following the mating of a derivative of E. coli strain S17-1 carrying plasmid pV-F1Histag-Tc with the single-crossover integrant RSY-F1, in which plasmid pCJK1 was inserted into the RsfabF1 locus of the R. solanacearum GMI1000 chromosome, strain RSY-F11 was selected. Cells of strain RSY-F11 were grown in BG medium containing IPTG and tetracycline for 24 h, and appropriate dilutions of the culture were spread onto BG plates containing IPTG and tetracycline. Several of the resulting colonies were inoculated onto BG plates containing gentamicin or kanamycin by using sterile toothpicks. Colonies resistant to gentamicin and sensitive to kanamycin were screened by colony PCR utilizing the primers listed in Table S1 in the supplemental material. Finally, conditional fabF1 mutant strain RS-F1 was obtained.
Expression and purification of His-tagged proteins.
Plasmids pCEF1, pCEF2, pCEF3, pCEF4, and pCEB were transformed into BL21(DE3) cells. The expressions of RsfabF1, RsfabF2, RsfabF3, RsfabF4, and RsfabB were analyzed by SDS-PAGE to assay protein solubility. Based on the SDS-PAGE results, RsFabF1 was purified by its hexahistidine tag under native conditions as described previously (18). The R. solanacearum FabD, FabH, FabZ, and FabI proteins and the Vibrio harveyi AasS protein were purified by their hexahistidine tags as described previously (18, 22).
Assay of long-chain 3-ketoacyl-ACP synthase activities in vitro.
To purify E. coli holo-ACP, the PCR products harboring acpP and acpS were cloned into the NcoI and HindIII sites of pET-28b or pBAD34, in which the multiple-cloning site (MCS) of pBAD33 (15) was replaced with the MCS of pBAD24 (15), to yield plasmids pET-ACP and pBAD34-AcpS, respectively. These two plasmids were introduced into E. coli BL21(DE3) cells, and holo-ACP was expressed at high levels and purified as described previously (42). We synthesized and purified octanoyl-ACP, trans-2-decenoyl-ACP, dodecanoyl-ACP, and tetradecanoyl-ACP as described previously (42).
The abilities of RsFabF1 to function in the cycle of fatty acid synthesis were assessed with reaction mixtures containing 0.1 M sodium phosphate (pH 7.0); 0.1 μg each of RsFabD, RsFabG, RsFabZ, and RsFabI; 50 μM NADH; 50 μM NADPH; 1 mM β-mercaptoethanol; 100 μM acyl-ACP; 100 μM malonyl-CoA; and 50 μM holo-ACP in a final volume of 40 μl. The reactions were initiated by the addition of RsFabF1 or E. coli FabB (EcFabB) to the mixture, followed by incubation for 1 h. The reaction products were resolved by conformationally sensitive gel electrophoresis on 15% polyacrylamide gels containing a concentration of urea optimized for separation. To test for possible inhibitory effects of cerulenin on enzyme activities, the appropriate volumes of cerulenin solution were added to the assay tubes.
Analysis of phospholipid compositions.
The cultures were grown aerobically at 30°C in BG medium overnight. Cells were harvested from 10-ml cultures and washed three times with BG medium at room temperature. The phospholipids were extracted according to a previously reported procedure. Fatty acid methyl esters were synthesized, extracted, and analyzed by gas chromatography-mass spectrometry (GC-MS) as previously described (42).
The ability of RsFabF1to restore fatty acid synthesis in vivo was tested. First, conditional mutant strain RS-F1 was grown on a BG plate containing IPTG for 2 days. The strain was then transferred onto BG plates in the absence of IPTG and grown for 4 days. The culture of the strain was collected with BG liquid medium and divided into two halves; IPTG was added to one half, and the other half was left untreated. Both halves were labeled with [1-14C]acetate in parallel, and the phospholipids were extracted and analyzed by thin-layer chromatography (TLC) and quantitated by phosphorimaging (36).
Assay of RsFabF1 expression in the RsfabF1 strain by Western blotting.
Cell cultures were conducted as described above for the [1-14C]acetate labeling experiment. The RS-F1 proteins were separated by SDS-PAGE, and FabF1 expression was assayed by Western blotting with an anti-His tag antibody according to the manufacturer's instructions.
RESULTS
R. solanacearum FabF2, FabF3, FabF4, and FabB all lack detectable 3-ketoacyl-ACP synthase activity.
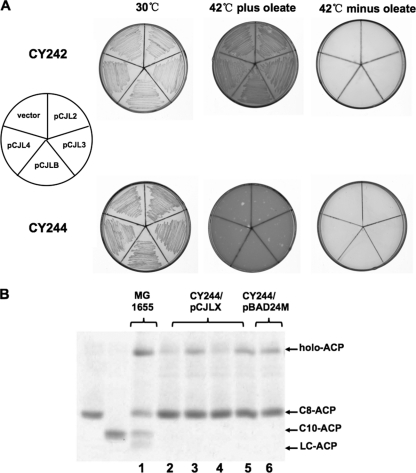
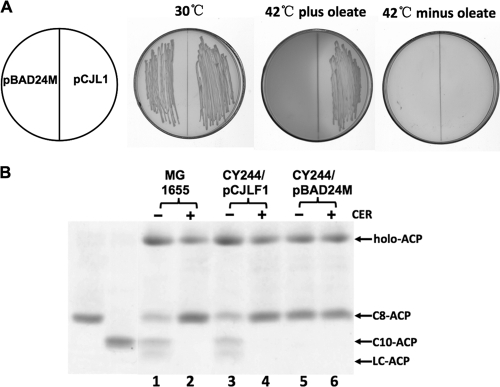
In order to confirm whether R. solanacearum fabF2, fabF3, fabF4, and fabB function in fatty acid biosynthesis, these genes were inserted into the arabinose-inducible vector pBAD24M, as described in Materials and Methods, to yield plasmids pCJL2 (Rsfabf2), pCJL3 (Rsfabf3), pCJL4 (Rsfabf4), and pCJB (RsfabB). Complementation experiments were done to test if these genes encode functional condensing enzymes. First, the resulting plasmids were introduced into E. coli fabB(Ts) mutant strain CY242 in order to investigate whether these genes encode 3-ketoacyl-ACP synthase I activity. CY242 lacks 3-ketoacyl-ACP synthase I activity at the nonpermissive temperature and is unable to grow at 42°C in the absence of oleate (3, 34). The data showed that none of derivatives of strain CY242 carrying the above-described plasmids was able to grow at 42°C in the absence of oleate (Fig. 2A). The plasmids were then introduced into E. coli fabB(Ts) fabF strain CY244 to test if these genes encode 3-ketoacyl-ACP synthase II activity. At the nonpermissive temperature, this mutant strain lacks both long-chain 3-ketoacyl-ACP synthase activities and thus is unable to grow even when the medium is supplemented with the unsaturated fatty acid oleate. The growth of the transformants at 42°C in the presence of oleate indicates a complementation of the fabF mutation, whereas the growth of the transformants in the absence of oleate indicates a complementation of the fabB mutation (34, 36). The strains carrying pCJL2, pCJL3, pCJL4, or pCJLB (harboring RsfabF2, RsfabF3, RsfabF4, or RsfabB, respectively) failed to grow in either the presence or absence of oleate (Fig. 2A). Although R. solanacearum fabF2, fabF3, fabF4, and fabB failed to complement E. coli mutant strains CY242 and CY244, it remained possible that one of these plasmids supported long-chain fatty acid synthesis but that the levels of the long-chain fatty acids synthesized were insufficient for the growth of the host strains. To test this possibility, cell extracts of CY244 derivatives were tested for long-chain 3-ketoacyl-ACP synthase activity as described in Materials and Methods. We used octanoyl-ACP as a substrate, and as expected, octanoyl-ACP was converted to decanoyl-ACP and longer-chain acyl-ACP species by an extract of E. coli MG1655 (Fig. 2B, lane 1), whereas the extracts from strain CY244 derivatives (including CY244 carrying vector pBAD24M) did not show any long-chain 3-ketoacyl-ACP synthase activity (Fig. 2B, lanes 2 to 6). We also examined [1-14C]acetate incorporation into fatty acids of CY244 derivative strains at 42°C in the absence of oleate and found no increase in the rate of fatty acid synthesis over that of CY244 carrying the pBAD24M vector by argentation thin-layer chromatographic analysis (data not shown). Thus, these results confirmed that none of the R. solanacearum proteins FabF2, FabF3, FabF4, and FabB possesses long-chain 3-ketoacyl-ACP synthase activities.
Fig 2.
Growth of transformants of E. coli mutants with plasmids carrying R. solanacearum fabF genes or fabB and analysis of the enzymatic activities of cell-free extracts from strain CY244 carrying plasmids harboring fabF or fabB. (A) Cells of E. coli strain CY242 or CY244 carrying pBAD24M-derived plasmid pCJLF2, (fabF2), pCJLF3 (fabF3), pCJLF4 (fabF4), or pCJLB (fabB) were grown at 30°C or 42°C. (B) Analysis of the activities of long-chain KAS in cell-free extracts of CY244 carrying plasmids harboring fabF genes or fabB by conformationally sensitive gel electrophoresis. The migration positions of octanoyl-ACP and decanoyl-ACP on the gel are shown. Lane 1 is the extract of wild-type strain MG1655, whereas lanes 2 to 6 show extracts of strain CY244 carrying plasmid pCJL2, pCJL3, pCJL4, pCJLB, or pBAD24M.
Properties of fabF and fabB deletion mutants of R. solanacearum.
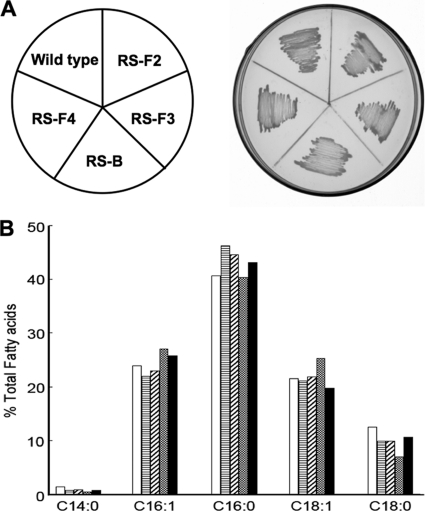
In order to identify the functions of the R. solanacearum 3-ketoacyl-acyl carrier protein synthase homologues in fatty acid biosynthesis, we attempted to inactivate each of the putative fabF and fabB genes by allelic replacement. Five pK18mobsacB-borne Gmr insertions into the genes were constructed (see Materials and Methods), and single-crossover integrants of each plasmid in the strain GMI1000 genome were selected by kanamycin resistance (see Fig. S1A in the supplemental material). Cultures grown from the integrant colonies were then plated onto a medium containing sucrose and gentamicin in order to select for the loss of the vector sequences. The successful construction of the designed mutations was assayed by PCR analysis using the primers listed in Table S1 in the supplemental material (also see Fig. S1B, S1C, and S1D in the supplemental material). Four insertion mutant strains were obtained: RS-F2 (fabF2::Gm), RS-F3 (fabF3::Gm), RS-F4 (fabF4::Gm), and RS-B (fabB::Gm) (see Fig. S1D in the supplemental material), but no fabF1 insertion mutant was obtained. The growth of the insertion mutant strains on M63 medium was tested. All mutant strains were viable at 30°C (Fig. 3A), although their generation times (RS-F2, ∼209 min; RS-B, ∼208 min; RS-F3, ∼217 min; RS-F4, ∼181 min) were significantly longer than that of the wild-type strain (∼151 min). These data showed that functional fabF2, fabF3, fabF4, and fabB genes were not essential for the growth of strain GMI1000. It was reported previously that E. coli strains with fabF null mutations are viable, although these mutants are unable to regulate their unsaturated fatty acid contents upon shifts in the growth temperature (5, 13, 14). Therefore, it is possible that fabF2, fabF3, and fabF4 could play a role in regulating the fatty acid compositions of R. solanacearum. However, the growth of mutant strain RS-B (fabB::Gm) on M63 medium without oleic acid was unexpected. Generally, the fabB mutants are auxotrophic for oleic acid, such as an E. coli fabB mutant (4) and a Pseudomonas aeruginosa fabB mutant (21). One of the explanations for this finding was that R. solanacearum FabB did not play a role in unsaturated fatty acid synthesis in this bacterium. In order to examine these hypotheses, the effects of the insertion of fabB or the fabF genes on the fatty acid composition of R. solanacearum were determined by GC-MS (see Materials and Methods). The fatty acid compositions of insertion strains RS-B (fabB::Gm), RS-F2 (fabF2::Gm), RS-F3 (fabF3::Gm), and RS-F4 (fabF4::Gm) were essentially the same as that of the wild-type strain (Fig. 3B). Thus, these results indicated that R. solanacearum FabF2, FabF3, FabF4, and FabB do not take part in fatty acid biosynthesis.
Fig 3.
Examination of growth and phospholipid fatty acid compositions of R. solanacearum fabF and fabB insertion mutants. (A) Growth of mutant strains RS-F2, RS-F3, RS-F4, and RS-B on M63 medium not containing oleate. (B) Phospholipid fatty acid compositions of mutant strains. The methyl esters were C14:0 (tetradecanoic), C16:1 (hexadecenoic), C16:0 (hexadecanoic), C18:1 (octadecenoic), and C18:0 (octadecanoic). □, strain RS-F2; ▤, strain RS-F3; ▨, strain RS-F4;  , strain RS-B; ■, wild-type GMI1000.
, strain RS-B; ■, wild-type GMI1000.
Essentiality of R. solanacearum fabF1 for fatty acid synthesis.
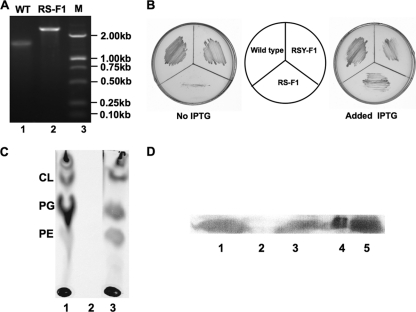
Although no R. solanacearum fabF1 insertion mutant was obtained, single-crossover integrant strain RSY-F1 was obtained by selection for gentamicin resistance following the RP4-mediated conjugation of R. solanacearum with E. coli S17-1 carrying a plasmid pK18mobsacB-borne Gmr insertion in fabF1. Since we were unable to eliminate the functional copy of fabF1 by selection for a second crossover, it seemed likely that fabF1 is an essential gene in R. solanacearum. If so, it should be possible to obtain fabF1 knockout mutants by selection for the second crossover in the presence of a functional fabF1 gene. We therefore constructed plasmid pV-F1Histag-Tc, which carried a copy of R. solanacearum GMI1000 fabF1 under the transcriptional control of the E. coli lac operon promoter, and introduced this plasmid into strain RSY-F1 as described in Materials and Methods. The conjugants were selected on medium containing kanamycin and tetracycline. Cultures grown from the conjugant colonies were then plated onto a medium containing sucrose, IPTG, gentamicin, and tetracycline in order to obtain the fabF1 insertion mutant by selection for the second crossover. Several colonies that were sensitive to kanamycin and resistant to 10% sucrose, gentamicin, and tetracycline were obtained. One of these strains, named RS-F1, was examined by colony PCR analysis using the primers listed in Table S1 in the supplemental material (Fig. 4A). As expected, the primers (fabF1 upper and fabF1 lower) amplified a 1.52-kb fabF1-containing fragment from wild-type GMI1000 DNA. In strain RS-F1, this fragment increased to 2.40 kb, as expected from the insertion of the Gmr cassette plus fabF1 sequences (Fig. 4A). The strains were further verified by the sequencing of the 2.40-kb fragment, which validated the construction.
Fig 4.
Characterization of R. solanacearum conditional mutant strain RS-F1. (A) PCR analysis of genomic DNA from mutant strain RS-F1. Lane 1, PCR product amplified from wild-type (WT) strain GMI1000; lane 2, PCR product amplified from mutant strain RS-F1. (B) Growth of R. solanacearum conditional mutant strain RS-F1 under conditions of IPTG induction. Wild type, R. solanacearum GMI1000; RS-F1, GMI1000 fabF1::Gmr strain carrying plasmid pV-F1His-Tc, in which the fabF1 gene of R. solanacearum GMI1000 was cloned into vector pVL33 and fabF1 expression was controlled by IPTG; RSY-F1, a fabF1 merodiploid, where one copy of fabF1 was normal, whereas the other copy had a Gmr cassette insertion. (C) Culture of strain RS-F1 labeled with [1-14C]acetate, followed by extraction of the cellular phospholipids and analysis by TLC (see Materials and Methods). Lane 1, phospholipids from wild-type strain GMI1000; lane 2, phospholipids from cultures of strain RS-F1 without IPTG induction; lane 3, phospholipids from cultures of strain RS-F1 with IPTG induction. CL, cardiolipin; PG, phosphatidylglycerol, PE, phosphatidylethanolamine. (D) Analysis of RsFabF1 expression in the RS-F1 mutant by Western blotting (see Materials and Methods). Lanes 1 and 3, RsFabF1 expression in strain RS-F1 under conditions of IPTG induction; lane 2, RsFabF1 expression in strain RS-F1 under conditions of no IPTG induction; lane 4, RsFabF1 protein purified with native nickel chelate chromatography; lane 5, expression of RsFabF1 in E. coli strain BL21(DE3) carrying plasmid pCEF1.
The growth of strain RS-F1 on M63 medium was then tested. Strain RS-F1 grew normally in the presence of IPTG, but upon the removal of IPTG, cell growth was significantly attenuated (Fig. 4B). In order to confirm that fabF1 was an essential gene for fatty acid synthesis in R. solanacearum, the incorporation of [1-14C]acetate into the phospholipids of strain RS-F1 was examined when FabF1 expression was induced by the addition of IPTG. In the presence of IPTG, strain RS-F1 incorporated [1-14C]acetate into phospholipids (Fig. 4C, lane 3), whereas cultures grown in the absence of IPTG failed to incorporate [1-14C]acetate into phospholipids (Fig. 4C, lane 2). The effect of IPTG on the cellular FabF1 content was confirmed directly by the separation of cell extracts by denaturing gel electrophoresis followed by immunodetection (Fig. 4D). In the absence of the inducer, no FabF1 could be detected in strain RS-F1 (Fig. 4D, lane 2), whereas in the presence of IPTG, increased levels of FabF1 were observed (Fig. 4D, lanes 1 and 3). We also determined the fatty acid composition of strain RS-F1 under conditions of IPTG induction (Table 2). Although there were the same fatty acid species in RS-F1 as those in the wild-type strain, the ratio of unsaturated fatty acids to saturated fatty acids in RS-F1 was higher than that in wild-type strain GMI1000. These experiments demonstrated an essential role for the fabF1 gene product in the fatty acid synthesis of R. solanacearum.
Table 2.
Fatty acid composition of R. solanacearum strainsa
| Fatty acid | Composition (%) |
|
|---|---|---|
| RS-F1 | GMI1000 | |
| Tetradecanoic C14:0 | 1 | 0.7 |
| 9-Hexadecenoic C16:1 | 33.70 | 25.7 |
| Hexadecanoic C16:0 | 40.30 | 43.2 |
| 11-Octadecenoic C18:1 | 18.70 | 19.80 |
| Octadecanoic C18:0 | 6.40 | 10.6 |
The ratios of unsaturated fatty acids to saturated fatty acids were 1.1 for strain RS-F1 and 0.83 for strain GMI1000.
R. solanacearum fabF1 can functionally replace E. coli FabF in vivo and in vitro.
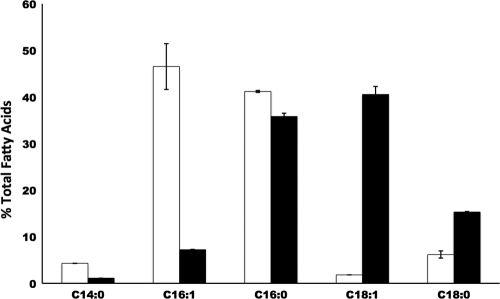
We inserted the R. solanacearum fabF1 gene into vector pBAD24M to yield pCJL1. The resulting plasmid was also introduced into E. coli strain CY244. CY244 carrying pCJL1 encoding FabF1 grew at 42°C in the presence of oleate, whereas this strain failed to grow in the absence of oleate (Fig. 5A). We then examined the activity of long-chain 3-ketoacyl-ACP synthase in cell-free extracts of this strain and found that octanoyl-ACP was converted to decanoyl-ACP and longer-chain acyl-ACPs (Fig. 5B, lane 3), as seen for the extract of wild-type E. coli MG1655 (Fig. 5B, lane 1). The addition of cerulenin, a known specific inhibitor of long-chain 3-ketoacyl-ACP synthases, blocked the synthesis of long-chain acyl-ACP species (Fig. 5B, lane 4). Therefore, R. solanacearum fabF1 complemented the E. coli fabF mutation, and FabF1 has long-chain 3-ketoacyl-ACP synthase activity. Plasmid pCJL1 was transformed into E. coli fabF mutant strain CL28, which lacks 3-ketoacyl-ACP synthase II and is unable to convert the C16 unsaturated fatty acid species to C18 species (23), and the fatty acids of the transformants were examined by GC-MS. For strain CL28 carrying vector pBAD24M, the main unsaturated fatty acid was palmitoleic acid, whereas for strain CL28 carrying pCJL1, the main unsaturated fatty acid was cis-vaccenic acid (Fig. 6). Therefore, FabF1 functionally replaced E. coli FabF and allowed the elongation of palmitoleoyl-ACP acid to cis-vaccenoyl-ACP.
Fig 5.
Expression of R. solanacearum fabF1 restores growth of CY244 on oleate-containing medium. Shown are data from analyses of the enzymatic activities of cell extracts from strain CY244 carrying plasmids harboring fabF1. (A) Transformants of strain CY244 were grown at 30°C or 42°C. (B) Analysis the activities of long-chain KAS in cell-free extracts of CY244 carrying plasmids harboring fabF1. Lanes 1 and 2 are the extracts of the wild-type strain MG1655, and lanes 3 and 4 are the extracts of strain CY244 carrying plasmid pCJLF1, whereas lanes 5 and 6 are the extracts of strain CY244 carrying the pBAD24M vector. Fatty acid synthesis was allowed to proceed for 10 min in the presence or absence of cerulenin (CER) (40 μM), as shown.
Fig 6.
Fatty acid compositions of E. coli fabF strain CL28 transformed with R. solanacearum fabF1. White bars, strain CL28 transformed with plasmid pBAD24M; black bars, strain CL28 transformed with plasmid pCJLF1 harboring R. solanacearum fabF1. All cultures were grown at 37°C, and the fatty acid compositions were determined by GC-MS as described in Materials and Methods.
In order to directly assay the long-chain 3-ketoacyl-ACP synthase activities of FabF1, the protein was expressed in E. coli. SDS-gel electrophoresis showed that FabF1 was expressed well and had a molecular weight in good agreement with that expected for the His6-tagged protein (see Fig. S2A in the supplemental material). The N-terminally His6-tagged version of R. solanacearum FabF1 was purified by nickel chelate chromatography to give a preparation that gave a single band upon SDS-gel electrophoresis (see Fig. S2A in the supplemental material). In order to reconstitute fatty acid synthesis in vitro, the N-terminally His6-tagged versions of R. solanacearum FabG, FabD, FabH, FabZ, and FabI, Vibrio harveyi AasS, and E. coli FabB were purified by nickel chelate chromatography (data not shown). E. coli holo-ACP was also expressed and purified (see Materials and Methods). First, malonyl-ACP, octanoyl-ACP, and trans-2-decenoyl-ACP were synthesized (see Materials and Methods). When only RsFabZ and RsFabG were added to the mixture of malonyl-ACP and octanoyl-ACP, no new bands appeared (see Fig. S2B, lane 3, in the supplemental material). However, upon the addition of either EcFabB or RsFabF1 to the mixture of malonyl-ACP and octanoyl-ACP, a trans-2-decenoyl-ACP band appeared (see Fig. S2B, lanes 1 and 2, in the supplemental material). Meanwhile, we tested the sensitivity of RsFabF1 to cerulenin and found that like EcFabB, RsFabF1 was sensitive to cerulenin (see Fig. S2C in the supplemental material). We also investigated the elongation of the long-chain acyl-ACP by RsFabF1; the data showed that RsFabF1 could elongate these substrates (see Fig. S2D in the supplemental material). Thus, FabF1, like E. coli FabF, is able to catalyze all of the elongation reactions required for the synthesis of saturated fatty acids.
R. solanacearum FabF1 has modest 3-ketoacyl-ACP synthase I activity.
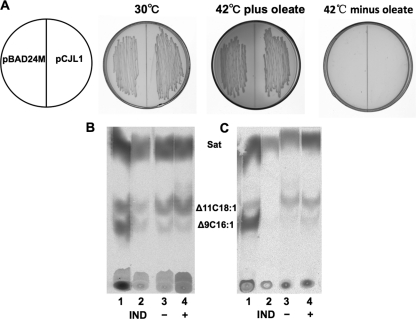
It was reported previously that in several Gram-positive bacteria, FabF homologues possess the activities of 3-ketoacyl-acyl carrier protein synthase I and thereby play a role in unsaturated fatty acid synthesis (29, 35, 41). Given that the expression of the putative R. solanacearum fabB gene failed to complement an E. coli fabB strain, it seemed possible that R. solanacearum FabF1 might also have 3-ketoacyl-ACP synthase I activity. In order to test this possibility, plasmid pCJL1 harboring fabF1 was introduced into E. coli CY242 cells, and the resulting transformants were tested for growth in the absence of oleate supplementation. No growth of the CY242 transformants was found at 42°C in the absence of oleate (Fig. 7A). Although R. solanacearum fabF1 failed to complement the E. coli mutation, it remained possible that plasmid pCJL1 supported unsaturated fatty acid synthesis but that the levels of unsaturated fatty acids synthesized were insufficient for the growth of strain CY242. To test this possibility, strain CY242 carrying pCJL1 was grown at 30°C to an optical density at 600 nm (OD600) of 0.6 with arabinose induction and then transferred to 42°C; after incubation for 2 h, the culture was labeled with [1-14C]acetate. The cellular phospholipids were extracted, and the fatty acid moieties were converted to methyl esters, which were analyzed by argentation thin-layer chromatography (which resolves each of the unsaturated species from the saturated species and from one another). We found that strain CY242 carrying the pBAD24M vector produced trace levels of unsaturated fatty acids (Fig. 7B, lane 2), whereas the strain carrying pCJL1 synthesized more unsaturated fatty acids in both the presence and absence of arabinose induction (Fig. 7B, lanes 3 and 4). Moreover, for strain CY242 carrying pCJL1, the levels of the unsaturated C18 fatty acids were higher than the levels of the unsaturated C16 species. These data indicate that R. solanacearum FabF1 has a low level of 3-ketoacyl-ACP synthase I activity. We also tested E. coli fabB(Ts) fabF strain CY244 carrying pCJL1 and obtained similar results (Fig. 7C, lanes 3 and 4).
Fig 7.
Growth of E. coli mutant strain CY242 carrying R. solanacearum fabF1 and argentation thin-layer chromatographic analysis of [1-14C]acetate-labeled E. coli CY242 or CY244 carrying plasmids harboring R. solanacearum fabF1. (A) Cells of strain CY242 carrying plasmid pCJL1 (harboring fabF1) were grown at 30°C and 42°C. (B and C) Argentation thin-layer chromatographic analysis of [1-14C]acetate-labeled E. coli CY242 (B) or CY244 (C) carrying plasmid pCJL1 harboring R. solanacearum fabF1. The migration positions of the methyl esters of the fatty acid species are shown. Sat, saturated fatty acid esters; Δ9C16:1, methyl ester of cis-9-hexadecenoic acid; Δ11C18:1, methyl ester of cis-11-octadecenoic acid. IND denotes cell cultures uninduced (−) or induced (+) with arabinose. Lane 1 is the methyl esters of wild-type E. coli strain MG1655, lane 2 is the methyl esters of E. coli mutant strains carrying pBAD24M, and lanes 3 and 4 are the methyl esters of E. coli mutant strains carrying pCJL1.
DISCUSSION
The known elongation enzymes that catalyze the formation of long fatty acid chains in bacteria are 3-ketoacyl-acyl carrier protein synthases I (FabB) and II (FabF) (3, 37). R. solanacearum is a betaproteobacterium, and its genome contains four genes (fabF1, fabF2, fabF3, and fabF4) annotated as encoding 3-ketoacyl-acyl carrier protein synthase II plus one gene (fabB) annotated as encoding 3-ketoacyl-acyl carrier protein synthase I (30). However, we report that only one of these homologues, fabF1, encodes a long-chain 3-ketoacyl-acyl carrier protein synthase that is active in fatty acid synthesis, whereas the remaining homologues are not essential for growth and were unable to replace the cognate E. coli genes. Thus, although the genomic databases provide some functional clues to the function of an unknown gene, biochemical and genetic assays are required to establish functions. Although fabF2, fabF3, fabF4, and fabB did not encode 3-ketoacyl-acyl carrier protein synthases that are active in fatty acid synthesis, and the deletion of these genes did not affect cellular fatty acid compositions, the generation times of all these mutant strains were significantly longer than that of the wild-type strain. These data indicated that these genes function in R. solanacearum and impart a growth advantage. This raises the question of what functions these proteins perform. The synthesis of polyketide species is a strong possibility (6). There is little doubt that polyketide synthesis is an evolutionary descendant of fatty acid synthesis, and this is reflected in the sequences of polyketide synthetic proteins, which are very similar to those of fatty acid synthesis proteins. Roles in the synthesis of other products are also possible. For example, PqsD, a homologue of the short-chain 3-ketoacyl-acyl carrier protein synthase, is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa (38).
FabF1 is an essential protein for the fatty acid biosynthesis of R. solanacearum, and mutants deficient in fabF1 are nonviable. Previously, this situation was observed only for Gram-positive bacteria such as Bacillus subtilis (32). In other Gram-negative bacteria, such as E. coli, fabF null mutations are viable, although these strains are unable to regulate their unsaturated fatty acid contents upon shifts in the growth temperature (3, 8, 34). Since fabF1 is the only gene encoding long-chain 3-ketoacyl-acyl carrier protein synthase in the R. solanacearum genome, its inactivation should mimic the loss of both FabF and FabB activities in E. coli (8, 34), and this is the case.
R. solanacearum synthesizes the same unsaturated fatty acid species as E. coli (33). Thus, it seemed likely that R. solanacearum uses a similar mechanism to produce unsaturated fatty acids. However, the fabB homologue gene of R. solanacearum failed to complement an E. coli fabB mutant strain, and an R. solanacearum strain having a defective fabB gene was viable in M63 medium in the absence of oleate and synthesized unsaturated fatty acids. Thus, this indicates that the R. solanacearum FabB homologue does not play a role in unsaturated fatty acid synthesis. However, in some bacteria that lack a FabB homologue, such as Enterococcus faecalis, Clostridium acetobutylicum, and Lactococcus lactis, a FabF homologue provides FabB functions (29, 35, 41). Although there are four fabF homologues in the R. solanacearum genome, only fabF1 encoded a 3-ketoacyl-acyl carrier protein synthase II activity. R. solanacearum FabF1 also possesses a low level of 3-ketoacyl-acyl carrier protein synthase I activity. Whether or not this activity is physiologically relevant remains to be seen. It is unclear how R. solanacearum introduces the cis double bonds of long-chain acyl-ACPs. Three pathways are known to introduce a cis double bond into a 10-carbon intermediate of acyl-ACPs under anaerobic conditions. The classical pathway is E. coli FabA, which is a bifunctional dehydratase/isomerase (10). A second pathway is catalyzed by FabN (E. faecalis), another bifunctional dehydratase/isomerase (35), which is a FabZ-like enzyme, and a third is catalyzed by FabM (Streptococcus pneumoniae), an isomerase that lacks dehydratase activity (25). However, the R. solanacearum genome encodes none of these proteins (30). We have tested the only FabZ homologue of R. solanacearum and found that this FabZ protein possesses only dehydratase activity and lacks isomerase activity (data not shown). We also tested RSp0360, which is in a cluster with fabB, fabF2, and fabF3 on the R. solanacearum megaplasmid. An RSp0360 homologue, Bamb3609, is found in a similar gene cluster in the Burkholderia ambifaria genome and was annotated as encoding 3-hydroxyacyl-ACP dehydratase (or the FabA/FabZ protein). We tested the RSp0360 gene for the functional complementation of an E. coli fabA mutant strain, and it failed to restore growth (data not shown).
Some bacteria (e.g., Bacillus subtilis and Pseudomonas aeruginosa) have an oxygen-dependent desaturase pathway for unsaturated fatty acid synthesis (1, 40). We searched the R. solanacearum GMI1000 genome database and found three open reading frames (RSc2450, RSp1029, and Rsc2025) annotated as fatty acid desaturases. However, the facts that R. solanacearum grows anaerobically and that the membrane phospholipids of anaerobically grown cells of R. solanacearum contain palmitoleic and cis-vaccenic acids argue that desaturation is not responsible for unsaturated synthesis in this bacterium. It therefore seems possible that a new pathway for unsaturated fatty acid synthesis in R. solanacearum remains to be discovered.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (30870036/C010201) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (20104404110).
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aguilar PS, Cronan JE, Jr, de Mendoza D. 1998. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 180:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brencic A, Winans SC. 2005. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69:155–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell JW, Cronan JE., Jr 2001. Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu. Rev. Microbiol. 55:305–332 [DOI] [PubMed] [Google Scholar]
- 4. Cronan JE, Jr, Birge CH, Vagelos PR. 1969. Evidence for two genes specifically involved in unsaturated fatty acid biosynthesis in Escherichia coli. J. Bacteriol. 100:601–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cronan JE, Jr, Godson GN. 1972. Mutants of Escherichia coli with temperature-sensitive lesions in membrane phospholipid synthesis: genetic analysis of glycerol-3-phosphate acyltransferase mutants. Mol. Gen. Genet. 116:199–210 [DOI] [PubMed] [Google Scholar]
- 6. Cronan JE, Thomas J. 2009. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459:395–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Lorenzo V, Eltis L, Kessler B, Timmis KN. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17–24 [DOI] [PubMed] [Google Scholar]
- 8. de Mendoza D, Klages Ulrich A, Cronan JE., Jr 1983. Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of beta-ketoacyl-acyl carrier protein synthase I. J. Biol. Chem. 258:2098–2101 [PubMed] [Google Scholar]
- 9. Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng Y, Cronan JE. 2009. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284:29526–29535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flavier AB, Clough SJ, Schell MA, Denny TP. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251–259 [DOI] [PubMed] [Google Scholar]
- 12. Flavier AB, Ganova-Raeva LM, Schell MA, Denny TP. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:7089–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garwin JL, Klages AL, Cronan JE., Jr 1980. Beta-ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem. 255:3263–3265 [PubMed] [Google Scholar]
- 14. Garwin JL, Klages AL, Cronan JE., Jr 1980. Structural, enzymatic, and genetic studies of beta-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J. Biol. Chem. 255:11949–11956 [PubMed] [Google Scholar]
- 15. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayward AC. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65–87 [DOI] [PubMed] [Google Scholar]
- 17. Heath RJ, Rock CO. 2002. The Claisen condensation in biology. Nat. Prod. Rep. 19:581–596 [DOI] [PubMed] [Google Scholar]
- 18. Heath RJ, Rock CO. 1996. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271:27795–27801 [DOI] [PubMed] [Google Scholar]
- 19. Heath RJ, White SW, Rock CO. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58:695–703 [DOI] [PubMed] [Google Scholar]
- 20. Heath RJ, White SW, Rock CO. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467–497 [DOI] [PubMed] [Google Scholar]
- 21. Hoang TT, Schweizer HP. 1997. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding beta-hydroxyacyl-acyl carrier protein dehydratase (FabA) and beta-ketoacyl-acyl carrier protein synthase I (FabB). J. Bacteriol. 179:5326–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Y, Chan CH, Cronan JE. 2006. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45:10008–10019 [DOI] [PubMed] [Google Scholar]
- 23. Lai CY, Cronan JE. 2004. Isolation and characterization of beta-ketoacyl-acyl carrier protein reductase (fabG) mutants of Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu YJ, Zhang YM, Rock CO. 2004. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol. 82:145–155 [DOI] [PubMed] [Google Scholar]
- 25. Marrakchi H, Choi KH, Rock CO. 2002. A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J. Biol. Chem. 277:44809–44816 [DOI] [PubMed] [Google Scholar]
- 26. Marrakchi H, Zhang YM, Rock CO. 2002. Mechanistic diversity and regulation of type II fatty acid synthesis. Biochem. Soc. Trans. 30:1050–1055 [DOI] [PubMed] [Google Scholar]
- 27. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 28. Mole BM, Baltrus DA, Dangl JL, Grant SR. 2007. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15:363–371 [DOI] [PubMed] [Google Scholar]
- 29. Morgan-Kiss RM, Cronan JE. 2008. The Lactococcus lactis FabF fatty acid synthetic enzyme can functionally replace both the FabB and FabF proteins of Escherichia coli and the FabH protein of Lactococcus lactis. Arch. Microbiol. 190:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salanoubat M, et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497–502 [DOI] [PubMed] [Google Scholar]
- 31. Schäfer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 32. Schujman GE, Choi KH, Altabe S, Rock CO, de Mendoza D. 2001. Response of Bacillus subtilis to cerulenin and acquisition of resistance. J. Bacteriol. 183:3032–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stead DE. 1992. Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int. J. Syst. Bacteriol. 42:15 [Google Scholar]
- 34. Ulrich AK, de Mendoza D, Garwin JL, Cronan JE., Jr 1983. Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature-dependent regulation of membrane lipid composition. J. Bacteriol. 154:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H, Cronan JE. 2004. Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J. Biol. Chem. 279:34489–34495 [DOI] [PubMed] [Google Scholar]
- 36. Wang H, Cronan JE. 2003. Haemophilus influenzae Rd lacks a stringently conserved fatty acid biosynthetic enzyme and thermal control of membrane lipid composition. J. Bacteriol. 185:4930–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. White SW, Zheng J, Zhang YM, Rock CO. 2005. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74:791–831 [DOI] [PubMed] [Google Scholar]
- 38. Zhang YM, Frank MW, Zhu K, Mayasundari A, Rock CO. 2008. PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa. J. Biol. Chem. 283:28788–28794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]
- 40. Zhu K, Choi KH, Schweizer HP, Rock CO, Zhang YM. 2006. Two aerobic pathways for the formation of unsaturated fatty acids in Pseudomonas aeruginosa. Mol. Microbiol. 60:260–273 [DOI] [PubMed] [Google Scholar]
- 41. Zhu L, et al. 2009. Functions of the Clostridium acetobutylicium [sic] FabF and FabZ proteins in unsaturated fatty acid biosynthesis. BMC Microbiol. 9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu L, Lin J, Ma J, Cronan JE, Wang H. 2010. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 54:689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.