Abstract
Burkholderia glumae possesses a quorum-sensing (QS) system mediated by N-octanoyl-homoserine lactone (C8-HSL) and its cognate receptor TofR. TofR/C8-HSL regulates the expression of a transcriptional regulator, qsmR. We identified one of the universal stress proteins (Usps), Usp2, from a genome-wide analysis of QS-dependent proteomes of B. glumae. In the whole genome of B. glumae BGR1, 11 usp genes (usp1 to usp11) were identified. Among the stress conditions tested, usp1 and usp2 mutants died 1 h after heat shock stress, whereas the other usp mutants and the wild-type strain survived for more than 3 h at 45°C. The expressions of all usp genes were positively regulated by QS, directly by QsmR. In addition, the expressions of usp1 and usp2 were dependent on RpoS in the stationary phase, as confirmed by the direct binding of RpoS-RNA holoenzyme to the promoter regions of the usp1 and usp2 genes. The expression of usp1 was upregulated upon a temperature shift from 37°C to either 28°C or 45°C, whereas the expression of usp2 was independent of temperature stress. This indicates that the regulation of usp1 and usp2 expression is different from what is known about Escherichia coli. Compared to the diverse roles of Usps in E. coli, Usps in B. glumae are dedicated to heat shock stress.
INTRODUCTION
Bacteria possess internal and external protection mechanisms to overcome environmental stresses such as nutrient starvation, temperature stress, oxidative stress, and toxic agents. One of the stress-responsive genes against various deleterious stresses is the universal stress protein (usp) gene (32). The usp genes were first reported for Escherichia coli (32) and are found in archaea, fungi, flies, and plants (27). In E. coli, six bona fide usp genes exist: uspA and uspC to uspG (27). These are divided into four classes based on their amino acid sequence similarities (27). UspA, UspC, and UspD belong to class I, and UspF and UspG are members of class II. UspE possesses two domains, E1 and E2, which were previously classified as class III and class IV, respectively (27). In E. coli, three isoforms of UspA have been identified in vivo, and two of them are phosphorylated on serine and threonine residues in response to stasis (13). UspG is dimeric and possesses autophosphorylation and autoadenylation activities (43). Although there have been many reports regarding the roles of Usps in defending against diverse stresses in bacteria, how Usps function biochemically is not known.
The expression of usp genes in E. coli is regulated primarily at the transcriptional level from a σ70-dependent promoter (33). The alarmone guanosine tetraphosphate (ppGpp), which is important for the regulation of many stationary-phase-induced genes, coordinately regulates four usp genes (uspA, uspC, uspD, and uspE) in E. coli (26, 27). The expression of uspA is negatively regulated by FadR, an activator or a repressor of fatty acid biosynthesis and degradation genes, respectively (9). The uspA, uspC, uspD, and uspE genes are highly expressed in the stationary phase of the carboxy domain deletion mutant of FtsK, a RecA-like double-stranded DNA (dsDNA) translocase (8). However, the induction of the UspA protein is independent of RpoS (σS) (32). In E. coli, UspA is involved in oxidative stress defense, and UspD is involved in both oxidative stress defense and iron scavenging (31). UspC, UspF, and UspG are involved mainly in motility and adhesion (31). In Pseudomonas aeruginosa, Usps are essential for survival under conditions of anaerobic energy stress and are required for pyruvate fermentation (3, 38).
Considering that usp genes of E. coli are highly expressed in the stationary phase, usp gene expression might depend on bacterial cell density. The regulation of bacterial gene expression that is dependent on cell density is called quorum sensing (QS). QS is an intercellular signaling circuit that regulates sets of genes involved in certain social behaviors of bacteria (42). QS plays important roles in physiological changes, including biofilm formation, motility, protein secretion, virulence, antibiotic production, and protection against stress defense in bacteria (7, 14, 17, 24, 41, 42). However, whether QS regulates the expression of usp genes in bacteria is not known.
In the present study, we examined QS-dependent biological phenomena of Burkholderia glumae, which is the causal agent of rice panicle blight (also called bacterial rice grain rot). The bacterium produces a phytotoxin called toxoflavin at the optimum growth temperature (37°C) in a QS-dependent manner and infects rice panicles during the flowering stage (25). Rice panicle blight has recently become widespread in most rice-growing areas and can be a serious threat, particularly when hot and humid weather conditions persist during the flowering stage (21). The bacterium possesses a LuxR-LuxI-type QS system. TofI is an N-acyl homoserine lactone synthase for the synthesis of N-octanoyl homoserine lactone (C8-HSL) that is recognized by a cognate receptor, TofR (25). A complex of TofR and C8-HSL activates the expression of qsmR, an IcIR-type transcriptional regulator (24). QsmR activates the expression of flagellum genes and type II protein secretion genes (14, 24).
From genome-wide proteome analyses of the bacterium, we found that one of the Usps is under the control of QS. Based on this finding, we decided to characterize all the usp genes in the whole genome of B. glumae BGR1. We identified 11 usp genes that are phylogenetically distinct from the 6 usp genes of E. coli. Unlike usp gene regulation in E. coli, the expression of usp genes in B. glumae depends on QS and RpoS. We show that Usp1 and Usp2 play important roles in the survival of B. glumae under conditions of heat shock stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are shown in Table 1. B. glumae BGR1 strains and all E. coli strains were grown in LB medium (1% [wt/vol] tryptone and 0.5% [wt/vol] yeast extract [pH 7.0]; USB Corp., Cleveland, OH) at 37°C or 28°C. Antibiotics were used at the following concentrations: ampicillin at 100 μg ml−1, chloramphenicol at 20 μg ml−1, kanamycin at 50 μg ml−1, nalidixic acid at 20 μg ml−1, rifampin at 100 μg ml−1, spectinomycin at 100 μg ml−1, tetracycline at 10 μg ml−1, and gentamicin at 20 μg ml−1.
Table 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsd1 hsdR17(rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | Gibco BRL |
| C2110 | polA Nalr | 39 |
| HB101 | F−mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (Smr) supE44 λ− | Gibco BRL |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Burkholderia glumae | ||
| BGR1 | Wild type; Rifr | 21 |
| BGS2 | BGR1 tofI::Ω | 25 |
| BGS9 | BGR1 qsmR::Ω | 24 |
| BGQ8/BGSQ8/BGH8 | BGR1 usp1::Tn3-gusA20/BGS2usp1::Tn3-gusA20/BGS9 usp1::Tn3-gusA20 | This study |
| BGQ9/BGSQ9/BGH9 | BGR1 usp2::Tn3-gusA104/BGS2 usp2::Tn3-gusA104/BGS9 usp2::Tn3-gusA104 | This study |
| BGQ10/BGSQ10/BGH10 | BGR1 usp3::Tn3-gusA40/BGS2 usp3::Tn3-gusA40/BGS9 usp3::Tn3-gusA40 | This study |
| BGQ14/BGSQ14/BGH14 | BGR1 usp6::Tn3-gusA242/BGS2 usp6::Tn3-gusA242/BGS9 usp6::Tn3-gusA242 | This study |
| BGQ12/BGSQ12/BGH12 | BGR1 usp7::Tn3-gusA52/BGS2 usp7::Tn3-gusA52/BGS9 usp7::Tn3-gusA52 | This study |
| BGQ16/BGSQ16/BGH16 | BGR1 usp8::Tn3-gusA192/BGS2 usp8::Tn3-gusA192/BGS9 usp8::Tn3-gusA192 | This study |
| BGQ18/BGSQ18/BGH18 | BGR1 usp9::Tn3-gusA136/BGS2 usp9::Tn3-gusA136/BGS9 usp9::Tn3-gusA136 | This study |
| BGQ21/BGSQ21/BGH21 | BGR1 usp10::Tn3-gusA94/BGS2 usp10::Tn3-gusA94/BGS9 usp10::Tn3-gusA94 | This study |
| BGQ22/BGSQ22/BGH22 | BGR1 usp11::Tn3-gusA181/BGS2 usp11::Tn3-gusA181/BGS9 usp11::Tn3-gusA181 | This study |
| BGO6/BGSO6/BGH6 | BGQ8 rpoS::Gmr/BGSQ8 rpoS::Gmr/BGH8 rpoS::Gmr | This study |
| BGO7/BGSO7/BGH7 | BGQ9 rpoS::Gmr/BGSQ9 rpoS::Gmr/BGH9 rpoS::Gmr | This study |
| BGQ82 | BGQ9 usp1::Ω | This study |
| S70/S2S70/S9S70 | BGR1 rpoS::Tn3-gusA70/BGS2 rpoS::Tn3-gusA70/BGS9 rpoS::Tn3-gusA70 | This study |
| Plasmids | ||
| pRK2013 | Tra+; ColE1 replicon; Kmr | 12 |
| pHoKmGus | Promoterless β-glucuronidase gene; Kmr AmprtnpA | 4 |
| pSShe | Cmr | 39 |
| pLysS | Harbors the T7 lysozyme gene; Cmr | Novagen |
| pBluescript II SK(+) | Cloning vehicle, phagemid, pUC derivative; Ampr | Stratagene |
| pLAFR3 | Tra− Mob+ RK2 replicon; Tetr | 40 |
| pRK415 | Mob+lacZ Tetr | 23 |
| pBSGm | 0.8-kb DNA fragment harboring a gentamicin cassette cloned into pBluescriptII SK(+); Ampr Gmr | J.-G. Kim |
| pBGT63 | 2.2-kb DNA fragment harboring qsmR cloned into pLAFR3 | 24 |
| pQSMR-His | qsmR in pET21b; Ampr | 24 |
| pHP45Ω | Ω cassette; Spr Smr | 35 |
| pBGQ8 | 21.5-kb DNA fragment harboring usp1 in pLAFR3 | This study |
| pBGQ9 | 21.5-kb DNA fragment harboring usp2 in pLAFR3 | This study |
| pBGQ11 | 20.1-kb DNA fragment harboring usp7 in pLAFR3 | This study |
| pBGQ12 | 19.4-kb DNA fragment harboring usp3 to usp6 and usp8 to usp11 in pLAFR3 | This study |
| pRPOS | 22.8-kb DNA fragment harboring rpoS in pLAFR3 | This study |
| pRPOS1 | 0.8-kb gentamicin cassette inserted into the AfeI site in rpoS in pRK415 | This study |
| pUSP1 | 2.0-kb cassette inserted into the EcoNI site in usp1 in pRK415 | This study |
| pHS1 | 1,046-bp PCR product harboring usp1 and its promoter region cloned into pRK415 | This study |
| pHS2 | 1,649-bp EcoRI DNA fragment harboring usp2 and its promoter region cloned into pRK415 | This study |
| pRPOS-His | rpoS in pET14b; Ampr | This study |
Where more than one strain is listed in a single row, the corresponding phenotypes are separated by slashes. Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance; Rifr, rifampin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance; Tetr, tetracycline resistance; Gmr, gentamicin resistance.
Protein sample preparation for two-dimensional electrophoresis (2-DE).
Cultures of B. glumae strains grown overnight in LB medium were diluted 1:100 in LB medium and grown at 37°C with shaking for 24 h in the stationary phase. Next, 1 μM C8-HSL was added to a culture of BGS2 (tofI::Ω) cells when necessary. Cells were harvested in the stationary phase by centrifugation and washed three times with 20 mM Tris-HCl (pH 7.4), suspended in 20 mM Tris-HCl (pH 7.4), and lysed by sonication with a VCX-400 sonicator (Sonics & Materials, Newton, CT). Cellular proteins were precipitated with 10% (wt/vol) trichloroacetic acid, followed by centrifugation at 12,000 × g for 30 min at 4°C, and the protein concentration of each sample was determined by using the Bradford assay and bovine serum albumin (BSA) as a standard (5).
2-DE and MS/MS analysis.
A total of 100 μg of each protein sample was resuspended in 450 μl rehydration solution containing 8 M urea, 2% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2% (vol/vol) immobilized pH gradient (IPG) buffer (GE Healthcare), 100 mM dithiothreitol (DTT), and 0.002% (wt/vol) bromophenol blue. The proteins were focused in the first dimension by using 24-cm-long IPG strips (GE Healthcare) at pH 4 to 7. Isoelectric focusing was performed by using an Ettan IPGphor (GE Healthcare) at a constant temperature of 20°C with a total of 74,500 V/h, as follows: 80 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, and 8,000 V up to 74,500 V/h. The strips were equilibrated before the second gel was run, as described previously (15). For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), we used 12.5% T-acrylamide–bisacrylamide (37.5:1) gels to separate proteins in the 1- and 100-kDa ranges using an Ettan DALTsix Large Vertical Electrophoresis system (GE Healthcare). After electrophoresis, proteins were visualized by using a silver staining kit (GE Healthcare) as recommended by the manufacturer. 2-DE was performed several times for each strain.
2-DE gel image analysis was performed by visual inspection or by using PDQues 2-D Analysis V 8.0 software (Bio-Rad). After optical density calibration, spot volumes were normalized according to the total spot volume in each gel. The analysis of protein spots was independently repeated at least three times. Electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) of the peptides generated by in-gel digestion was performed by nano-ESI on a Q-TOF2 mass spectrometer (Micromass, Manchester, United Kingdom). The data were processed by using the MassLynx, version 3.5, Windows NT system (Micromass), and de novo sequencing from the fragmentation spectra of the peptides was performed by using PepSeq (Micromass). To assign a positive protein identification, parameters of up to 1 missed cleavage, fixed modifications of carbamidomethyl (C), a peptide tolerance of ±0.6 Da, and a peptide charge of 2+ or 3+ were used. The resulting sequences were searched against a FASTA format of the translated open reading frames (ORFs) in the B. glumae BGR1 genomic database at the KROPBASE website (http://kropbase.snu.ac.kr/). Search results were accepted when the de novo peptide sequencing data comprised at least eight matched amino acids and the matched results were below the E value inclusion threshold (E = 0.05). A homology search of each putative ORF with proteins present in the NCBI database was performed by using the BLAST program (1).
Multiple-sequence alignment and phylogenetic analysis.
All usp gene sequences were retrieved from the NCBI website (http://www.ncbi.nlm.nih.gov/) through similarity searches performed by using BLASTP (http://ncbi.nlm.nih.gov/BLAST). The retrieved sequences from E. coli and B. glumae were aligned by using the “create alignment” function in the CLC Main Workbench 5.6 program (CLCbio). The evolutionary distances between the sequences were computed by using an algorithm described previously by Jukes and Cantor (22). Phylogenetic tree calculation was performed by using the neighbor-joining method (36) in the CLC Main Workbench 5.6 program. The statistical significances of the phylogenies were tested by using bootstrap analysis (11), with each bootstrap value reflecting the confidence of each branch. Determinations of identity among Usps in B. glumae were performed by using the LALIGN program (19).
DNA construction and mutagenesis.
General and standard techniques were used for DNA manipulations, cloning, restriction digestions, and agarose gel electrophoresis (37). Mutagenesis of genes with Tn3-gusA was performed, and the insertion site and orientation of Tn3-gusA in each mutant were determined as described previously (25). The mutagenized cosmids that carried Tn3-gusA insertions were introduced individually by conjugation and then marker exchanged into wild-type strain BGR1 and strains BGS2 (tofI::Ω) and BGS9 (qsmR::Ω) as described previously (25). To construct double mutants of usp1 or usp2 and rpoS, the gentamicin cassette from pBSGm was inserted into the unique AfeI site in the 4.0-kb DNA fragment containing rpoS in pRPOS (Table 1). The resulting plasmid, pRPOS1, was introduced into strains BGQ8 (usp1::Tn3-gusA20), BGSQ8 (tofI::Ω usp1::Tn3-gusA20), and BGH8 (qsmR::Ω usp1::Tn3-gusA20) to construct double mutations of usp1 and rpoS or into strains BGQ9 (usp2::Tn3-gusA104), BGSQ9 (tofI::Ω usp2::Tn3-gusA104), and BGH9 (qsmR::Ω usp2::Tn3-gusA104) to construct double mutations of usp2 and rpoS by marker exchange. To construct double mutations of usp1 and usp2, the spectinomycin cassette from pHP45Ω was inserted into the unique EcoNI site in the 5.4-kb DNA fragment carrying usp1 in pBGQ8 (Table 1). The resulting plasmid, pUSP1, was introduced into BGQ9 (usp2::Tn3-gusA104) cells by marker exchange. All marker exchanges were confirmed by Southern hybridization analysis using each cosmid as a probe.
For usp1 complementation, we amplified a 1,046-bp PCR product carrying a putative promoter region and structural gene using primers usp1C-F (5′-AGCAGCATCCGCAGCGTGGT-3′) and usp1C-R (5′-CGATCGATCTGCAGCCCTGA-3′). The PCR product was subcloned into the SmaI site of pBluescriptII SK(+), followed by cloning into pRK415 as an XbaI-and-KpnI fragment (Table 1). The resulting plasmid, pHS1, was introduced into BGQ8 (usp1::Tn3-gusA20) by conjugation. For usp2 complementation, the 1,649-bp EcoRI fragment from pBGQ9 was subcloned into pRK415, resulting in pHS2, followed by introduction into BGQ9 (usp2::Tn3-gusA104) cells by conjugation.
RT-PCR analysis.
Total RNA was isolated by using an RNeasy minikit (Qiagen) according to the manufacturer's protocol. Isolated RNA was treated with DNase I (Qiagen) for 30 min at 37°C. A total of 1 μg of RNA was reverse transcribed into cDNA by using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) for 1 h at 42°C. PCR was performed by using a PTC-200 Thermo Cycler (MJ Research, Waltham, MA) under the following conditions: 96°C for 5 min, followed by 35 cycles of 96°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The primers used for each reverse transcription (RT)-PCR are listed in Table S1 in the supplemental material.
Overexpression and purification of B. glumae QsmR and RpoS.
QsmR was overexpressed and purified as described previously (24). To overexpress RpoS in E. coli, the coding region of rpoS was amplified by using pRPOS as the template DNA and oligonucleotide primers RPOS-F (5′-CGAGACGCATATGCCGAAAT-3′) and RPOS-R (5′-ATCGGATCCTTACAGAACGG-3′), which introduced unique NdeI and BamHI sites at the ends of the PCR product, and the amplified product was cloned into the corresponding sites of pET14b (Invitrogen), resulting in pRPOS-His (Table 1). His-RpoS was overexpressed in E. coli strain BL21(DE3), which carries pLysS, according to instructions provided by the manufacturer (Novagen). Soluble His-RpoS was purified in a buffer containing 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 1 mM DTT, 5% glycerol, and 200 mM NaCl (28), using a Ni-nitrilotriacetic acid (NTA) spin column according to the manufacturer's instructions (Qiagen).
Gel mobility shift assay.
The 340-bp regions upstream of the usp genes were amplified by PCR using primers for each usp gene (see Table S2 in the supplemental material). The fragments were eluted from an agarose gel and labeled with biotin for chemiluminescence by using a Lightshift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce). For competitor DNA, we used the 242-bp upstream region of katE, which was amplified by using primers KEN1 and KEN2 as described previously (24). Purified QsmR-His (250 nM) was incubated with 1 nM biotin-labeled DNA in binding buffer [10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 5% (vol/vol) glycerol, and 50 ng μl−1 poly(dI·dC)] for 15 min at 28°C. For the competitor DNA, a 27-fold molar excess of unlabeled target DNA was added to the reaction mixture along with the extract, before the labeled DNA target was added. The mixtures were size fractionated on a nondenaturing 4% polyacrylamide gel, followed by drying and transfer onto nitrocellulose membranes. Signals were detected by streptavidin-horseradish peroxidase (HRP) chemiluminescence for biotin-labeled probes according to the manufacturer's instructions (Pierce).
To perform an EMSA with RpoS and RpoS-RNA holoenzyme, E. coli RNA polymerase was purchased from Epicentre Technologies. EMSAs were conducted according to methods reported in a previous study (28), with some modifications; briefly, 50 nM or 100 nM core RNA polymerase was mixed with 500 nM His-RpoS in a 10-μl reaction mixture with 1× buffer A [50 mM Tris-HCl (pH 7.9), 200 mM potassium glutamate, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 100 μg ml−1 BSA, 6 ng μl−1 poly(di·dC)], incubated at 28°C for 10 min to allow RNA holoenzyme formation, and then added to 1 nM biotin-labeled DNA and incubated for 25 min on ice. For heparin challenge experiments, 0.5 μl of 1 mg ml−1 heparin was added for an additional 10 min. All samples were run on 4% polyacrylamide gels at 100 V with cold 1× Tris-borate-EDTA in a cold room and then transferred onto nitrocellulose membranes for detection by streptavidin-HRP chemiluminescence for biotin-labeled probes. The images were visualized and quantified by using Chemi Doc XRS+ with Image Lab software (Bio-Rad).
β-Glucuronidase assays.
β-Glucuronidase (GUS) activity assays were performed at least three times, as described previously (20), with modifications. The expressions of usp genes were evaluated under three different conditions. All of the BGR1 derivatives were grown in LB medium at 37°C for 12 h with shaking and then shifted to a temperature of either 28°C for 4 h or 45°C for 15 min with shaking for cold and heat stress conditions. C8-HSL was added at a final concentration of 1 μM when the cells were subcultured. The bacteria were collected by centrifugation, resuspended in GUS extraction buffer, and lysed by sonication using a VCX-400 sonicator (Sonics & Materials). The extract was used in a β-glucuronidase enzyme assay with 4-methylumbelliferyl glucuronidase as the substrate. The fluorescence was measured at 365-nm excitation and 460-nm emission wavelengths with a Hoefer DQ300 fluorometer (Hoefer Scientific Instruments, San Francisco, CA). One unit of β-glucuronidase activity was defined as the amount of enzyme required to release 1 nmol 4-methylumbelliferone per bacterium per min.
Survival tests under conditions of heat shock stress.
All B. glumae BGR1 derivatives were grown at 37°C for 12 h in LB medium and then shifted to a temperature of 45°C with shaking. Samples were taken periodically to measure CFU on LB plates at 37°C after serial dilution.
RESULTS
Identification of QS-dependent usp genes in B. glumae.
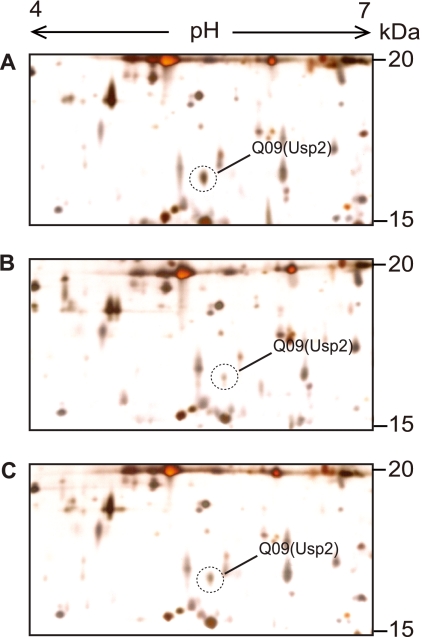
We identified one of the Usps, Usp2, from the genome-wide analysis of QS-dependent proteomes of B. glumae (Fig. 1). The estimated molecular mass and pI of Usp2 were 15.2 kDa and 5.17, respectively (Fig. 1 and see Table S3 in the supplemental material). Usp2 was expressed approximately 3-fold more in wild-type B. glumae BGR1 cells than in cells of tofI::Ω mutant strain BGS2 after 24 h of incubation in Luria-Bertani (LB) medium (Table S3). When BGS2 was supplemented with 1 μM C8-HSL, the expression of usp2 recovered to the wild-type level (Fig. 1). This indicated that the expression of usp2 is dependent on QS. Thus, we addressed questions regarding how usp genes are regulated and what the functional roles of Usps are in B. glumae BGR1.
Fig 1.
Two-dimensional gel analysis of B. glumae cell extracts. (A) BGR1 (wild type). (B) BGS2 (tofI::Ω) (N-octanoyl homoserine lactone [C8-HSL] synthase mutant). (C) BGS2 cells grown in LB medium supplemented with 1 μM C8-HSL. The cultures were harvested in the stationary phase. A total of 100 μg of protein from bacterial cells was separated and stained with silver nitrate. Gels shown are representative of three independent experiments. Spot Q09 was identified as Usp2.
Classification of Usps and their phylogenetic relationships.
B. glumae strain BGR1 has 11 Usps classified as members of the Usp family in the Pfam database (Pfam accession number PF00582). The 11 Usps, which are encoded by the genes usp1 to usp11 (thus named to avoid confusion with the uspA and uspC to uspG genes of E. coli), are similar to those of E. coli and other Burkholderia species (see Table S4 in the supplemental material).
To classify the Usps of B. glumae based on domain organization and their characteristics, we compared them to those of E. coli. The Usps of B. glumae BGR1 can be divided into three classes based on domain organization (Fig. 2A). The first class consists of Usp1, Usp2, Usp4, Usp7, Usp9, and Usp10, which contain one conserved UspA domain and are similar in size to UspA of E. coli (Fig. 2A). Usp8 and Usp11 belong to the second class, which possesses one UspA domain and a conserved domain of unknown function at the C terminus (Fig. 2A). The third class contains Usp3, Usp5, and Usp6, which possess two conserved UspA domains and are approximately twice the size of UspA of E. coli (Fig. 2A and see Table S4 in the supplemental material). The UspA domain in Usp2, Usp4, Usp8, Usp9, Usp10, and Usp11 possesses a conserved putative ATP-binding motif [G-2×-G-9×G(S/T)], whereas Usp1 and Usp7 have a similar motif, with some variation (Fig. 2B). In the third class of Usps, possessing two UspA domains, the conserved putative ATP-binding motif was not present in the first Usp domain but was found in the second domain, with some variation in Usp6 (Fig. 2B). Serine and threonine residues that are phosphorylation sites in E. coli seem to be conserved among Usps of B. glumae, except for Usp8 (see Fig. S1 in the supplemental material).
Fig 2.
Phylogenetic relationships among Usps of E. coli and B. glumae and conserved ATP-binding motifs. (A) Classification of 11 Usps of B. glumae (Pfam accession number PF00582) based on domain organization. CD denotes a conserved domain of unknown function. (B) Conservation of the putative ATP-binding motif of the MJ0577 (Methanocaldococcus jannaschii DSM 2661) Usp in the B. glumae Usps. The corresponding region of UspA in E. coli is shown for comparison. Conserved amino acid residues are in red. (C) Phylogenetic relationships among Usps of B. glumae using amino acid sequences. (D) Similarities of Usps in B. glumae and E. coli at the amino acid level. The phylogenetic tree calculation was performed by using the CLC Main Workbench 5.6 program, based on a sequence distance method that utilizes the neighbor-joining algorithm. Values at branches indicate the percent bootstrap support for 100 replicates.
We analyzed phylogenetic relationships using the amino acid sequences of the Usps of B. glumae. The Usps grouped into four separate clusters (Fig. 2C). Usp2 was distinct from the other Usps and belonged to cluster I, and Usp10 and Usp11 belonged to cluster II. Usp1, Usp3, Usp5, Usp6, Usp7, and Usp9 belonged to cluster III, and Usp4 and Usp8 belonged to cluster IV (Fig. 2C). The level of identity among the Usps ranged from 14.3% to 48% (see Table S5 in the supplemental material). When similarities among the Usps of B. glumae and E. coli were analyzed at the amino acid level, the Usps grouped into three clades (Fig. 2D). Usp3, Usp5, and Usp6 of B. glumae belonged to clade I, and all E. coli Usps and B. glumae Usp1 and Usp7 belonged to clade II. Usp2, Usp4, Usp8, Usp9, Usp10, and Usp11 of B. glumae formed clade III (Fig. 2D).
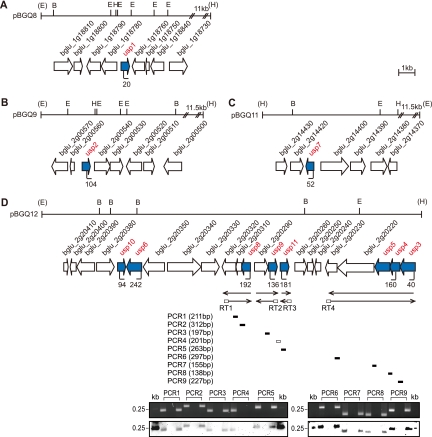
Genomic organization of the usp genes of B. glumae.
The usp1 gene was located in chromosome 1, and the other usp genes were located in chromosome 2. The usp1, usp2, and usp7 genes were not clustered and were possibly expressed monocistronically (Fig. 3A to C), and the other usp genes were clustered in one region present in a cosmid, pBGQ12 (Fig. 3D). To determine whether any usp genes are cotranscribed with other genes, we performed reverse transcription (RT)-PCR using mRNA isolated from B. glumae strain BGR1 and the designated primers. The usp3, usp4, and usp5 genes were cotranscribed with a histidine kinase gene (bglu_2g20220) and a two-component transcriptional regulator gene (bglu_2g20230) (Fig. 3D). The usp8 gene was part of an operon with a hypothetical protein (bglu_2g20310) and a cyclic nucleotide-binding protein (bglu_2g20320) (Fig. 3D). The usp9 gene was transcribed together with an acetoacetyl coenzyme A (acetoacetyl-CoA) reductase (bglu_2g20290) (Fig. 3D).
Fig 3.
Genetic organization of usp genes in B. glumae. (A to C) Schematic organization of the usp1, usp2, and usp7 genes. (D) Schematic organization of the usp3, usp4, usp5, usp6, usp8, usp9, usp10, and usp11 genes. Arrows below transcript arrows represent the direction and extent of cDNA. The short thick bars below the reverse transcription (RT) arrows indicate the nine PCR products from the corresponding RT reactions. The expected sizes of the PCR products are indicated in parentheses for each PCR. Shown are data for agarose gel analysis (top) and Southern hybridization analysis (bottom) of the RT-PCR products. Southern hybridization was performed by using pBGQ12 as probe DNA. The first lane used chromosomal DNA, the second lane used total RNA, and the third lane used cDNA as a template for each PCR. Vertical bars in the map denote the positions and orientations of the Tn3-gusA insertions. The restriction enzyme sites are indicated as follows: E, EcoRI; B, BamHI; H, HindIII.
Phenotypes of usp mutants under stress conditions.
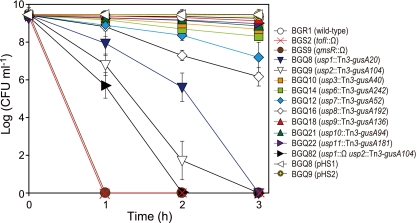
To generate usp mutants and to determine how the expression of usp genes is regulated at the transcriptional level under different stress conditions, we generated Tn3-gusA gene fusions to each usp gene in wild-type strain BGR1, tofI::Ω mutant strain BGS2, and qmsR::Ω mutant strain BGS9 (Fig. 4 and Table 2). Using these mutants, we determined the functional roles of each Usp under different stress conditions, such as hydrogen peroxide, ternary butyl hydroperoxide (t-BOOH), cold and heat stress, mitomycin C, UV light, pH stress, osmotic shock with 1 M NaCl, and 4% ethanol. No increased sensitivity or resistance was detected in any of the usp mutants, except for heat shock, compared to the wild-type strain (Fig. 4 and see Fig. S2 in the supplemental material). Under heat shock stress conditions, the populations of the usp1 and usp2 mutants rapidly decreased after 2 h and completely died after 3 h, whereas that of wild-type strain BGR1 was maintained at approximately 1 × 109 CFU ml−1 (Fig. 4). The populations of the usp7 and usp8 mutants also decreased to 1 × 107 CFU ml−1 and 2 × 106 CFU ml−1, respectively, but both mutants were far less sensitive to heat shock stress than the usp1 and usp2 mutants (Fig. 4). The survival rates for the other usp mutants were similar to that of the wild-type strain (Fig. 4). The death of the usp1 usp2 double mutant cells under conditions of heat stress was more rapid than the death of cells of each single mutant (Fig. 4). Thus, usp genes may not be involved in oxidative stress defense or swarming and swimming motilities in B. glumae (see Fig. S2C, S2I, and S3 in the supplemental material).
Fig 4.
Survival of usp mutants after exposure to heat shock in the stationary phase. All B. glumae BGR1 derivatives were cultured at 37°C and then shifted to 45°C to expose cells to heat shock stress. All experiments were performed by using three independent cultures; error bars represent the standard deviations.
Table 2.
Expression of usp genes in B. glumae
| Tn3-gusA fusion | Mean sp act of β-glucuronidase (10−11 U CFU−1 min−1) ± SDa |
||||
|---|---|---|---|---|---|
| BGR1 (wild type) | BGS2 (tofI::Ω) | BGS9 (qsmR::Ω) | BGS2 + 1μM C8-HSL | BGS9 (pBGT63) | |
| usp1::Tn3-gusA20 | 137.6 ± 10.0 | 34.6 ± 7.2 | 30.7 ± 4.5 | 112.3 ± 5.6 | 76.5 ± 9.2 |
| usp2::Tn3-gusA104 | 364.8 ± 9.8 | 24.0 ± 3.3 | 55.5 ± 7.8 | 185.4 ± 6.4 | 152.0 ± 8.7 |
| usp3::Tn3-gusA40 | 48.4 ± 3.0 | 16.1 ± 0.4 | 17.3 ± 0.6 | 50.8 ± 5.4 | 40.0 ± 2.8 |
| usp6::Tn3-gusA242 | 35.1 ± 1.1 | 9.18 ± 0.2 | 12.4 ± 0.8 | 39.6 ± 2.3 | 25.8 ± 1.7 |
| usp7::Tn3-gusA52 | 35.2 ± 0.9 | 7.0 ± 0.1 | 9.59 ± 0.9 | 34.8 ± 1.4 | 40.4 ± 1.9 |
| usp8::Tn3-gusA192 | 61.9 ± 5.8 | 19.5 ± 0.7 | 12.7 ± 0.8 | 81.7 ± 3.1 | 48.0 ± 2.9 |
| usp9::Tn3-gusA136 | 61.1 ± 0.9 | 22.3 ± 1.1 | 30.0 ± 1.0 | 65.1 ± 4.9 | 50.3 ± 3.7 |
| usp10::Tn3-gusA94 | 54.4 ± 7.2 | 17.1 ± 0.5 | 9.65 ± 0.6 | 65.0 ± 3.7 | 49.2 ± 4.2 |
| usp11::Tn3-gusA181 | 52.1 ± 4.1 | 16.8 ± 0.8 | 12.7 ± 0.8 | 65.5 ± 5.1 | 52.3 ± 4.3 |
Bacterial cells were grown for 12 h at 37°C. One unit of β-glucuronidase was defined as 1 nmol 4-methylumbelliferone released per bacterium per min. All values are means ± standard deviations of values from triplicate experiments.
Expression of usp genes is regulated by QsmR.
When the expression levels of each usp gene were measured with Tn3-gusA gene fusions to each usp gene in LB medium, the expressions of all usp genes were 3- to 15-fold higher in wild-type strain BGR1 than in tofI::Ω mutant strain BGS2 and in qmsR::Ω mutant strain BGS9 (Table 2). The expression levels of usp1 and usp2 were higher than those of the other usp genes (Table 2). When 1 μM C8-HSL was exogenously added to the cultures of each strain carrying the usp::Tn3-gusA fusion in the tofI::Ω mutant background, the expression levels were recovered to wild-type levels (Table 2). The lower level of expression of each of the usp genes in qsmR::Ω mutant strain BGS9 recovered to the level of the wild-type strain by providing pBGT63 carrying the qsmR gene in trans to each strain (Table 2). This indicated that the expression of usp genes is regulated by QS and, particularly, by QsmR.
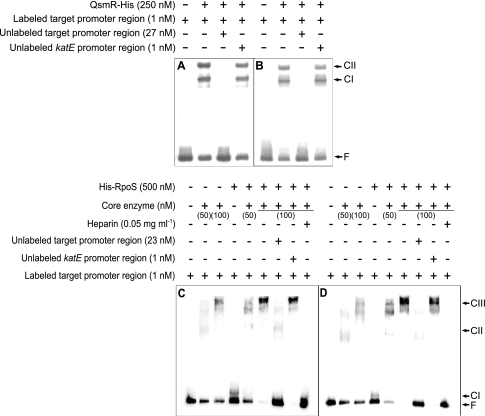
To determine whether the expression of usp genes is regulated directly by QsmR, we performed electrophoretic mobility shift assays (EMSAs) with purified QsmR-His and the promoter regions of the usp genes. As shown in Fig. 5A and B and Fig. S4 in the supplemental material, QsmR-His specifically bound to the promoter regions of all usp genes, proving that the expression of the usp genes is activated directly by QsmR.
Fig 5.
Gel mobility shift assay of the usp1 and usp2 regulatory regions using QsmR-His, His-RpoS, and E. coli RNA core enzyme. (A and B) Binding of QsmR-His to the usp1 (A) and usp2 (B) regulatory regions. (C and D) Binding of RpoS, core enzyme, or RpoS-RNA holoenzyme to the usp1 (C) and usp2 (C) regulatory regions. CI, CII, and CIII indicate DNA-protein complexes I, II, and III, respectively, and F denotes free DNA.
Expression of usp1 and usp2 is dependent on RpoS.
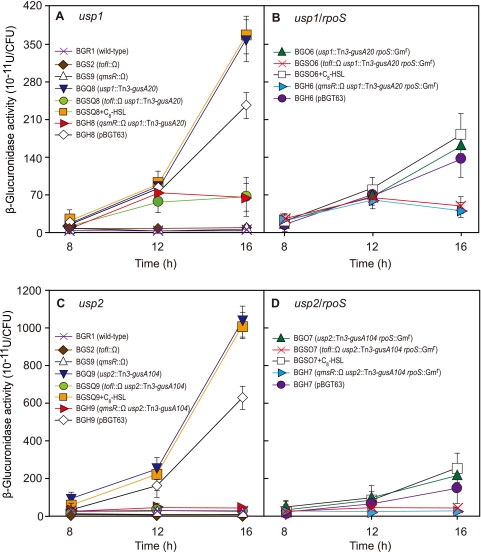
Because the usp1 and usp2 genes play important roles in the response to heat shock stress, we investigated the expression levels of these genes at different growth stages and under heat shock stress conditions in LB medium. The expression levels of both genes increased in a QsmR-dependent manner as cells entered the stationary phase (Fig. 6A and C). Because both genes were highly expressed in the stationary phase, we determined whether their expressions were dependent on RpoS. The expression level of usp1 was approximately 2-fold lower in strain BGO6 (usp1::Tn3-gusA20 rpoS::Gmr) than in strain BGQ8 (usp1::Tn3-gusA20) (Fig. 6B), whereas that of usp2 was approximately 5-fold lower in strain BGO7 (usp2::Tn3-gusA104 rpoS::Gmr) than in strain BGQ9 (usp2::Tn3-gusA104) (Fig. 6D). This indicated that the expressions of both genes are dependent on RpoS.
Fig 6.
Expression of the usp1 and usp2 genes at different growth phases and in different genetic backgrounds. (A and B) The expression level of usp1 was elevated in the stationary phase and was dependent on QsmR and RpoS. (C and D) The usp2 gene was highly expressed in the stationary phase and in a QsmR- and RpoS-dependent manner.
To determine whether RpoS is involved directly in the expressions of the usp1 and usp2 genes, we performed EMSAs using the promoter regions of the genes. We observed DNA band shifts with 500 nM His-RpoS (complex I [CI]) and 50 nM or 100 nM E. coli RNA core enzyme (CII), which indicates that they bind to each 340-bp promoter region of the usp1 and usp2 genes (Fig. 5C and D). His-RpoS at 500 nM together with 50 nM E. coli RNA core polymerase bound to the same region and appeared at positions similar to those of samples treated with 50 nM or 100 nM E. coli RNA core polymerase alone (Fig. 5C and D). However, shifted bands appeared in a higher position (CIII) when samples were treated with 500 nM His-RpoS and 100 nM E. coli RNA core enzyme, and heparin disturbed the direct binding of RpoS-RNA holoenzyme (Fig. 5C and D). This indicates that His-RpoS-RNA holoenzyme specifically binds to each 340-bp promoter region of the usp1 and usp2 genes.
To determine whether QS regulates rpoS expression, we measured rpoS expression levels in strains S70 (rpoS::Tn3-gusA70), S2S70 (tofI::Ω rpoS::Tn3-gusA70), and S9S70 (qsmR::Ω rpoS::Tn3-gusA70). The expression levels in strains S2S70 and S9S70 were no different than that in the wild-type strain (see Fig. S5 in the supplemental material), indicating that rpoS expression is independent of QS.
Expression of usp1 is upregulated by a temperature shift.
To determine whether the expressions of usp1 and usp2 are regulated by temperature stress, we measured the changes in the expression levels of the two genes after cold and heat shock stresses at 28°C and 45°C, respectively. The expression level of the usp1 gene was consistently about 2- or 3-fold higher under cold and heat shock conditions than under normal growth conditions at 37°C (Fig. 7A). In addition, the upregulation of usp1 by temperature stress was dependent on QsmR, whereas that of usp2 was independent of temperature stress (Fig. 7A and B).
Fig 7.
Expression of the usp1 and usp2 genes under conditions of a temperature shift. (A) Expression of usp1 was activated by QsmR and upregulated by cold and heat shock stresses. (B) Expression of usp2 was dependent on QsmR, and the expression pattern was not influenced by a temperature shift.
Mutations of usp1 and usp2 retain full virulence.
To determine whether Usp1 and Usp2 play any role in the virulence of B. glumae, we inoculated the usp1 or usp2 mutant strain into rice panicles. Both mutants were highly virulent to the level of the wild type (see Fig. S6 in the supplemental material), indicating that neither protein affects the virulence of B. glumae.
DISCUSSION
The numbers of usp genes in Gram-negative bacteria are diverse, ranging from 1 to 12 (3, 6, 18, 30, 34), indicating that some functional redundancies of Usps exist or that each Usp might have a different role. We characterized the 11 usp genes identified in B. glumae BGR1. The genes exhibited relatively low levels of identity among each other; however, low levels of identity among known Usps at the amino acid level seem to be common. This indicates that predictions of Usp paralogs or orthologs based on similarities of amino acid sequences from various bacterial species may not be a preferred method of characterizing usp genes. In addition, comparisons of the Usps of B. glumae and E. coli provided no valuable information for predicting the possible functions of Usps. Thus, a more thorough characterization of these Usps is required for predicting which gene is a bona fide usp gene and for avoiding the misclassification of these genes, as happened with the “uspB” gene in E. coli, which was subsequently found not to be a bona fide usp gene (16). Thus, we cannot rule out that some of the annotated usp genes in B. glumae may not be bona fide usp genes.
There are many different types of Usp family members in bacteria, archaea, and plants (27). The Usps of bacteria are relatively small and simple compared to those of archaea, cyanobacteria, and plants (34). The domain organization of the 11 Usps identified in B. glumae in the present study is similar to that of Usps of E. coli and Mycobacterium tuberculosis (34). Three Usps possess two Usp domains in tandem, but the other eight have one Usp domain. However, unlike the four Usps that have two Usp domains carrying two ATP-binding motifs in M. tuberculosis, only one ATP-binding motif was found for one of the two Usp domains of Usp3, Usp5, and Usp6 in B. glumae. The relationship between the organization of the Usp domain and the slight variation in the ATP-binding motif in the Usp domain and its effect on the biochemical functions of Usps remain unclear, because the biochemical properties of Usps are limited. From the limited information on the biochemical properties of Usps, posttranslational modification seems to be common (13). The phosphorylation of UspA and UspG was not unexpected because of the presence of the conserved ATP-binding motif. It has been known that serine or threonine of UspA is phosphorylated in response to stasis (13). Interestingly, we identified one conserved serine or threonine residue from the alignment of amino acid residues of UspA and all 11 Usps of B. glumae (see Fig. S1 in the supplemental material). This residue would be a strong candidate for a phosphorylation site of Usps.
Some aspects of the functional roles of Usps have been relatively well characterized for E. coli compared to other organisms. However, information regarding the functions of Usps in other bacteria is very limited. The Usps of E. coli have individual roles, with some overlap, whereas those of B. glumae (except for Usp1 and Usp2) have no apparent major role under diverse stress conditions. In E. coli, the sensitivities of the uspA mutant and the wild type during a temperature shift from 28°C to 50°C were indistinguishable (33). In some cases, wild-type cells preadapted to a nonlethal temperature of 42°C for 15 min were somewhat more resistant than preadapted uspA mutant cells (33). However, the sensitivities of the usp1 and usp2 mutants to heat shock are clearly distinguishable in B. glumae. This indicates that Usp1 and Usp2 play major roles in protection against heat shock stress in B. glumae. The finding that the usp7 and usp8 mutant populations of B. glumae decreased approximately 100-fold 3 h after heat treatment indicates that Usp7 and Usp8 might be associated with the Usp1- and Usp2-mediated heat shock response or may play minor roles in the response to heat stress.
The biochemical mechanisms through which Usp1 and Usp2 confer resistance to heat shock stress in B. glumae are not clear. One plausible mechanism is that they might interact with GroEL to form proper protein conformations, thereby preventing unwanted protein aggregation under conditions of heat shock stress, similarly to how UspG in E. coli forms complexes with GroEL (2). However, this does not explain the biochemical properties of Usp1 and Usp2.
The uspA genes in many bacteria are coordinately expressed by ppGpp, FadR, FtsK, RecA, CspC, and CspE under different conditions (27). However, the regulation of usp genes in B. glumae is very different from that in E. coli. First, we showed that the expression of all usp genes in B. glumae is positively regulated by QS, specifically by QsmR. This is the first report of such QS regulation. Second, we demonstrated that the expression of usp1 and usp2 is dependent on RpoS, suggesting that RpoS plays a role in usp gene expression at entry into the stationary phase. In contrast, in E. coli, the expression of uspA is dependent on σ70 (33), and that of uspB during ethanol stress is dependent on σS (10); we excluded uspB because it is not a bona fide usp gene (16). Third, the expression of usp1 was affected by a temperature shift in a QS-dependent manner in LB medium, which has not been reported for any usp gene of E. coli. Finally, unlike the regulation of uspA by ppGpp in E. coli, ppGpp may not influence usp gene expression in B. glumae. This is consistent with our observation that mutations in usp genes in B. glumae under starvation conditions do not show phenotypes similar to those observed for E. coli. These findings provide new data on usp gene regulation, which may help elucidate how bacteria coordinate gene expression to cope with stress conditions.
UspA of Salmonella spp. was reported previously to be important for virulence in mice and survival in host cells (29). However, usp1 and usp2 may not play important roles in the virulence of B. glumae, considering that Usp1 and Usp2 appear to play a role in overcoming heat shock stress but not other stresses.
In conclusion, Usps in B. glumae play very limited roles in the responses to various stresses, compared to the roles of Usps in E. coli. This suggests that usp gene regulation is diverse in different bacteria. If Usps possess no enzymatic functions and are regulatory proteins, Usps may interact with other proteins in bacteria, because Usps do not possess DNA-binding motifs. This suggests that Usp-mediated networks might be present and would be important for an understanding of how Usps function to overcome various stresses. It would be very interesting to determine whether QS is involved in usp gene regulation in other bacteria. To understand additional biological roles of Usps, the biochemical properties of Usps should be further investigated.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Creative Research Initiatives Programs (2010-0018280) of the National Research Foundation of Korea.
Footnotes
Published ahead of print 16 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bochkareva ES, Girshovich AS, Bibi E. 2002. Identification and characterization of the Escherichia coli stress protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 269:3032–3040 [DOI] [PubMed] [Google Scholar]
- 3. Boes N, Schreiber K, Härtig E, Jaensch L, Schobert M. 2006. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J. Bacteriol. 188:6529–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonas U, Stall RE, Staskawicz BJ. 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218:127–136 [DOI] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Chen W, Honma K, Sharma A, Kuramitsu HK. 2006. A universal stress protein of Porphyromonas gingivalis is involved in stress responses and biofilm formation. FEMS Microbiol. Lett. 264:15–21 [DOI] [PubMed] [Google Scholar]
- 7. Chun H, et al. 2009. The quorum sensing-dependent gene katG of Burkholderia glumae is important for protection from visible light. J. Bacteriol. 191:4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diez AA, Farewell A, Nannmark U, Nystrom T. 1997. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J. Bacteriol. 179:5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farewell A, Diez AA, DiRusso CC, Nystrom T. 1996. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J. Bacteriol. 178:6443–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farewell A, Kvint K, Nystrom T. 1998. uspB, a new sigmaS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 180:6140–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 12. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freestone P, Nystrom T, Trinei M, Norris V. 1997. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274:318–324 [DOI] [PubMed] [Google Scholar]
- 14. Goo E, Kang Y, Kim H, Hwang I. 2010. Proteomic analysis of quorum sensing-dependent proteins in Burkholderia glumae. J. Proteome Res. 9:3184–3199 [DOI] [PubMed] [Google Scholar]
- 15. Gorg A, Boguth G, Obermaier C, Posch A, Weiss W. 1995. Two-dimensional polyacrylamide gel electrophoresis with immobilized pH gradients in the first dimension (IPG-Dalt): the state of the art and the controversy of vertical versus horizontal systems. Electrophoresis 16:1079–1086 [DOI] [PubMed] [Google Scholar]
- 16. Gustavsson N, Diez A, Nystrom T. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107–117 [DOI] [PubMed] [Google Scholar]
- 17. Hassett DJ, et al. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082–1093 [DOI] [PubMed] [Google Scholar]
- 18. Hingley-Weilson SM, Lougheed KEA, Ferguson K, Leiva S, Williams HD. 2010. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis 90:236–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang X, Miller W. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337–357 [Google Scholar]
- 20. Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive versatile gene fusion marker in higher plants. EMBO J. 6:3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeong Y, et al. 2003. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 87:890–895 [DOI] [PubMed] [Google Scholar]
- 22. Jukes TH, Cantor CR. 1969. Evolution of protein molecules, p 21–132 In Munro HN. (ed), Mammalian protein metabolism. Academic Press, New York, NY [Google Scholar]
- 23. Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmid for DNA cloning in gram negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 24. Kim J, et al. 2007. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol. Microbiol. 64:165–179 [DOI] [PubMed] [Google Scholar]
- 25. Kim J, et al. 2004. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54:921–934 [DOI] [PubMed] [Google Scholar]
- 26. Kvint K, Hosbond C, Farewell A, Nybroe O, Nystrom T. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35:435–443 [DOI] [PubMed] [Google Scholar]
- 27. Kvint K, Nachin L, Diez A, Nystrom T. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140–145 [DOI] [PubMed] [Google Scholar]
- 28. Lee S, Gralla J. 2001. Sigma38 (rpoS) RNA polymerase promoter engagement via −10 region nucleotides. J. Biol. Chem. 276:30064–30071 [DOI] [PubMed] [Google Scholar]
- 29. Liu WT, et al. 2007. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 42:2–10 [DOI] [PubMed] [Google Scholar]
- 30. Mot R, Schoofs G, Nagy I. 2007. Proteome analysis of Streptomyces coelicolor mutants affected in the proteasome system reveals changes in stress-responsive proteins. Arch. Microbiol. 188:257–271 [DOI] [PubMed] [Google Scholar]
- 31. Nachin L, Nannmark U, Nystrom T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nystrom T, Neidhardt FC. 1992. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6:3187–3198 [DOI] [PubMed] [Google Scholar]
- 33. Nystrom T, Neidhardt FC. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537–544 [DOI] [PubMed] [Google Scholar]
- 34. O'Toole R, Williams HD. 2003. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 154:387–392 [DOI] [PubMed] [Google Scholar]
- 35. Prentki BS, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 9:303–313 [DOI] [PubMed] [Google Scholar]
- 36. Saitou N, Nei M. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Schreiber K, et al. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stachel SE, An G, Flores C, Nester EW. 1985. A Tn3-lacZ transposon for the random generation of β-galactosidase gene fusion: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Staskawicz B, Dahlbeck D, Keen N, Napoli C. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. syringae. J. Bacteriol. 169:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. von Bodman SB, Bauer WD, Coplin DL. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455–482 [DOI] [PubMed] [Google Scholar]
- 42. Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 43. Weber A, Jung K. 2006. Biochemical properties of UspG, a universal stress protein of Escherichia coli. Biochemistry 45:1620–1628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.