Abstract
N-Acetylglucosamine, the monomer of chitin, is a favored carbon and nitrogen source for streptomycetes. Its intracellular catabolism requires the combined actions of the N-acetylglucosamine-6-phosphate (GlcNAc-6P) deacetylase NagA and the glucosamine-6-phosphate (GlcN-6P) deaminase/isomerase NagB. GlcNAc acts as a signaling molecule in the DasR-mediated nutrient sensing system, activating development and antibiotic production under poor growth conditions (famine) and blocking these processes under rich conditions (feast). In order to understand how a single nutrient can deliver opposite information according to the nutritional context, we carried out a mutational analysis of the nag metabolic genes nagA, nagB, and nagK. Here we show that the nag genes are part of the DasR regulon in Streptomyces coelicolor, which explains their transcriptional induction by GlcNAc. Most likely as the result of the intracellular accumulation of GlcN-6P, nagB deletion mutants fail to grow in the presence of GlcNAc. This toxicity can be alleviated by the additional deletion of nagA. We recently showed that in S. coelicolor, GlcNAc is internalized as GlcNAc-6P via the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS). Considering the relevance of GlcNAc for the control of antibiotic production, improved insight into GlcNAc metabolism in Streptomyces may provide new leads toward biotechnological applications.
INTRODUCTION
Streptomycetes are Gram-positive, soil-dwelling mycelial bacteria that have a complex life cycle that starts with the germination of a spore, which then grows out to form a branched mycelium of vegetative hyphae. The multicellular mycelial lifestyle of streptomycetes makes these organisms unique among bacteria, as connected compartments are formed that are separated by cross walls. When morphological differentiation is initiated, a so-called aerial mycelium is produced, and the erected aerial hyphae eventually form chains of spores (7, 13). Streptomycetes are prolific producers of natural products, producing some 50% of all known antibiotics, as well as many anticancer, antifungal, and immunosuppressant agents. The production of these so-called secondary metabolites is part of the developmental program, roughly coinciding with growth cessation and the onset of sporulation (3, 43).
As saprophytic soil bacteria, streptomycetes utilize naturally occurring polysaccharides, such as chitin, xylan, and cellulose, as carbon sources. N-Acetylglucosamine (GlcNAc), the monomer of chitin, is a preferred carbon and nitrogen source for streptomycetes, and the related metabolite glutamate is preferred over glucose (27, 41). Chitin is one of the most abundant polysaccharides on earth and is found, among others, in the cuticles and shells of insects and crustaceans and within the cell walls of fungi; to utilize this polysaccharide, streptomycetes have an extensive chitinolytic system (8, 37). In Streptomyces coelicolor, GlcNAc is a substrate for transport via the phosphoenolpyruvate-dependent phosphotransferase system (PTS) (reviewed in reference 5). During PTS-mediated carbon source uptake, a phosphoryl group is transferred from phosphoenolpyruvate to the general phosphotransferase enzyme I (EI; encoded by ptsI), from there to HPr (encoded by ptsH), and then further onto enzyme IIA (EIIACrr; encoded by crr), enzyme IIB (NagF), and, finally, enzyme IIC (NagE2) (27, 28). NagF phosphorylates the incoming GlcNAc to form N-acetylgluco-samine-6-phosphate (GlcNAc-6P). Removal of any of the general PTS components (EI, EIIA, or HPr) blocks development (32). An alternative route for the internalization of monomeric GlcNAc exists in Streptomyces olivaceoviridis and probably in many other streptomycetes, namely, via the ATP-binding cassette (ABC) transporter NgcEFG, which imports both N-acetylglucosamine and its disaccharide, N,N′-diacetylchitobiose [(GlcNAc)2], with similar affinities (47, 49). (GlcNAc)2 is also imported via the ABC sugar transporter DasABC (10, 38).
We previously demonstrated that GlcNAc is an important signaling molecule for streptomycetes and a major decision point toward the onset of development and antibiotic production (32, 34). When GlcNAc accumulates under rich growth conditions (feast), it promotes growth, thereby blocking developmental processes, while under poor nutritional conditions (famine) the accumulation of GlcNAc promotes development and antibiotic production (34, 44). Under famine conditions, a complete signaling pathway was proposed, from GlcNAc uptake to the onset of antibiotic production, whereby the pleiotropic GntR family transcriptional regulator DasR controls the genes for GlcNAc transport and metabolism and for antibiotic synthesis (32, 34). Glucosamine-6-phosphate (GlcN-6P) is a key molecule in this signaling cascade and acts as an allosteric effector preventing DasR's DNA binding ability. This results in the loss of transcriptional repression of specific activator genes for antibiotic production, including actII-ORF4, encoding the pathway-specific activator for actinorhodin (Act) production, and redZ, encoding the response regulator RedZ that activates prodiginine (Red) production. DasR also represses the cpk gene cluster encoding the cryptic polyketide Cpk, but this effect is probably indirect (43). Under feast conditions, the molecular mechanism for GlcNAc blocking of sporulation and antibiotic production is still unknown. Nonphosphorylation of key developmental proteins by global PTS components (EI, HPr, and EIIACrr) upon GlcNAc sensing is so far the only realistic hypothesis proposed (10, 32).
As can be gleaned from extensive studies of amino sugar metabolism in other bacteria, the signaling molecule GlcN-6P is produced from N-acetylglucosamine-6-phosphate by a GlcNAc-6P deacetylase (NagA) and is then converted by a GlcN-6P deaminase (NagB) to fructose-6-phosphate (Fru-6P), which enters the glycolytic pathway (1, 30, 46). To better understand the amino sugar-mediated signaling pathway in streptomycetes, dissection of GlcNAc metabolism in these organisms is required. In this work, we analyzed the GlcNAc metabolic pathway and specifically blocked one or more steps of GlcNAc utilization via directed deletion mutations. This allowed us to provide the first assessment of how the flux of GlcNAc influences the global regulatory routes that control Streptomyces development and antibiotic production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1 and Table S1 in the supplemental material, respectively. Escherichia coli JM109 (36) and ET12567 (20) were used for routine cloning procedures and for extracting nonmethylated DNA, respectively. Streptomyces coelicolor A3(2) M145 was the parent strain for all mutants described in this work. All media and routine Streptomyces techniques are described in the Streptomyces manual of Kieser et al. (20). Cells of E. coli were grown in Luria-Bertani broth (LB) at 37°C. Phenotypic characterization of Streptomyces mutants was carried out on minimal medium (MM) and R5 agar plates (with carbon source as indicated) and in liquid NMMP or MM cultures (20) with 1% carbon sources, as indicated. Soy flour mannitol (SFM) agar plates were used to prepare spore suspensions.
Table 1.
Strains of Streptomyces coelicolor A3(2) described in this study
| Strain | Genotypea | Reference |
|---|---|---|
| M145 | Prototrophic SCP1− SCP2− | 20 |
| GAM1 | M145 nagA::aacC4 Aprr | This work |
| GAM2 | M145 nagB::aacC4 Aprr | This work |
| GAM3 | M145 nagK::aacC4 Aprr | This work |
| GAM4 | M145 ΔnagAIFD | This work |
| GAM5 | M145 ΔnagBIFD | This work |
| GAM6 | M145 ΔnagKIFD | This work |
| GAM7 | M145 nagKA::aacC4 Aprr | This work |
| GAM8 | M145 ΔnagKAIFD | This work |
| GAM9 | M145 ΔnagBIFDnagA::aacC4 | This work |
| GAM10 | M145 ΔnagKAIFDnagB::aacC4 | This work |
| GAM12 | GAM4/pGAM5 | This work |
| GAM13 | GAM5/pGAM6 | This work |
IFD, in-frame deletion mutant; Aprr, apramycin resistant.
Plasmids and constructs.
All constructs described in this work are listed in Table S1 in the supplemental material, and oligonucleotides are listed in Table S2.
Constructs for creating gene replacement and IFD mutants.
The strategy for creating knockout mutants was based on the unstable multicopy vector pWHM3 (45) as described previously (42). For deletion of nagA, the −1365/+6 and +1133/+2484 regions relative to the start of nagA were amplified by PCR, using primer pairs nagALF-1365 plus nagALR+6 and nagARF+1133 plus nagARR+2484, respectively. Fragments were cloned into pWHM3, and the engineered XbaI site was used for insertion of the aacC4 apramycin resistance cassette flanked by loxP sites between the flanking regions. The presence of the loxP recognition sites allows the efficient removal of the apramycin resistance cassette following the introduction of plasmid pUWLcre expressing Cre recombinase (11, 19). The constructed plasmid was called pGAM1. Using essentially the same strategy as that for pGAM1, we constructed the knockout plasmids pGAM2 and pGAM3 for the single-gene replacement of nagB and nagK, respectively. pGAM2 contained the −1185/+6 and +770/+1918 regions relative to the start of nagB, and pGAM3 contained the −1450/+6 and +963/+2570 regions relative to the start of nagK. Plasmid pGAM4 for the in-frame deletion (IFD) of nagKA contained the upstream −1450/+6 region of nagK and the downstream +1133/+2484 region of nagA, which were amplified by PCR using primer pairs nagKLF-1450 plus nagKLR+6 and nagARF+1133 plus nagARR+2484, respectively. The nagAB (GAM9) double mutant was constructed by replacing nagA with the apramycin resistance cassette in the nagB IFD mutant GAM5. GAM8 was used as the parent for the nagKAB triple mutant GAM10, following replacement of nagB by the apramycin resistance cassette.
To analyze the correctness of the mutants, PCRs were done on mycelia from liquid-grown MM cultures, using oligonucleotides nagA_FOR-198 and nagA_REV+1417 for nagA and nagBFOR-336 and nagB_REV+1098 for nagB.
For complementation of the nag mutants, the low-copy-number vector pHJL401 (22), harboring the corresponding nag gene and promoter region (nucleotides [nt] −454 to +796 relative to the start of nagB and nt −512 to +2262 relative to the start of nagK were used to complement nagK and nagA), was used.
Antibiotic production.
Quantification of prodiginines and actinorhodin was performed as described previously (20) and corrected for biomass (wet weight). Values were normalized against the parental strain M145 (set to 100%). Cultures were grown on R2YE agar plates overlaid with cellophane discs to allow separation of biomass and spent agar.
Production and purification of DasR.
Production and purification of His6-tagged DasR were based on a previously described protocol (9, 23), with several modifications. Escherichia coli BL21 Rosetta (DE3) cells carrying pFT240 (33) were grown at 37°C in 250 ml LB medium containing 100 μg/ml of ampicillin until the culture reached an optical density at 600 nm (OD600) of 0.6. Production of His6-tagged DasR was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were collected by centrifugation and ruptured by sonication in lysis buffer (100 mM phosphate buffer, pH 8, 250 mM NaCl, 30 mM imidazole, 0.1% Triton). Soluble proteins were loaded onto a preequilibrated Ni2+-nitrilotriacetic acid (NTA)-agarose column (5-ml bed volume). His6-tagged DasR eluted at around 200 mM imidazole, and fractions containing the pure protein were pooled and dialyzed overnight at 4°C against electrophoretic mobility shift assay (EMSA) buffer (see below).
EMSA.
EMSAs on double-stranded oligonucleotides were performed using Cy5-labeled dre probes (final concentration, 0.25 μM) and His6-tagged DasR (final concentration, between 1.75 and 2.5 μM) in a total reaction volume of 50 μl. All reactions were carried out in EMSA buffer (10 mM Tris-Cl, pH 7.5, 1 mM dithiothreitol [DTT], 0.25 mM CaCl2, 0.5 mM MgCl2, 50 mM KCl, and 2% glycerol) containing a 100-fold excess of nonspecific DNA (salmon sperm DNA). After 15 min of incubation at room temperature, reaction mixtures were loaded into a 1% (wt/vol) agarose gel. Bound and unbound probes were separated by gel electrophoresis at room temperature, and fluorescent DNA was visualized using a Typhoon Trio+ variable-mode imager. Oligonucleotides (see Table S2 in the supplemental material) used to generate 40-bp double-stranded DNA probes for EMSAs were taken from the upstream regions of dasA (dredasA), nagB (drenagB), and the nagKA operon (drenagKA). The cis-acting element OP1 of the gene for the Bacillus licheniformis β-lactamase repressor BlaI was used as a negative control.
RNA isolation and RT-PCR.
For transcript analysis, RNA was isolated from liquid cultures of S. coelicolor M145 or the dasR mutant. Mycelium was grown in NMMP and induced by addition of 50 mM GlcNAc at an OD of ∼0.2. Samples were taken before induction and 30 min after induction. RNA isolation and semiquantitative reverse transcription-PCR (RT-PCR) were carried out twice as described previously (4). Two hundred nanograms of RNA was used in each reaction mix (concentration was assessed using a Nanodrop spectrophotometer). Quantification was done using a Bio-Rad FX molecular imager with Quantity One software (Bio-Rad). Oligonucleotides used for RT-PCR are presented in Table S2 in the supplemental material.
Stereomicroscopy.
Strains were grown on MM and R5 agar plates supplemented with either mannitol or GlcNAc for 5 days. The mutants were imaged using a Zeiss Lumar V12 stereomicroscope.
Computer analyses.
DNA and protein data bank searches were performed using the BLAST server of the National Center for Biotechnology Information at the National Institutes of Health, Bethesda, MD (http://www.ncbi.nlm.nih.gov), as well as the S. coelicolor genome page services (http://strepdb.streptomyces.org.uk/). Alignments were generated with CLUSTALW, available at http://www.ebi.ac.uk/clustalw. Regulon predictions were performed using PREDetector (15). General information about the amino sugar metabolic pathway of S. coelicolor was found in the KEGG database (http://www.genome.jp/kegg-bin/show_pathway?sco00520).
RESULTS
In silico identification of GlcNAc utilization genes in S. coelicolor.
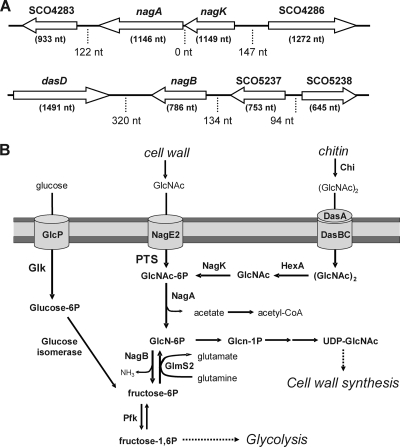
The genetic environment of the nag metabolic genes on the S. coelicolor chromosome and their predicted gene products are presented in Fig. 1A. The nagK (SCO4285) and nagA (SCO4284) genes have overlapping start and stop codons and encode the N-acetylglucosamine kinase NagK and the N-acetylgluco-samine-6P deacetylase NagA, respectively. The NagA and NagK proteins show 34.5% and 37.2% amino acid (aa) sequence identity, respectively, to the corresponding enzymes in E. coli. Downstream of the nagKA operon lies SCO4283, encoding a phosphofructokinase-like sugar kinase, and upstream lies SCO4286, which encodes an ABC-type sugar-binding protein of unknown function. The nagB gene for glucosamine-6P-deaminase (SCO5236) lies immediately downstream of the dasABCD gene cluster involved in (GlcNAc)2 uptake and degradation (10, 35) and upstream of SCO5237, encoding a likely 3-ketoacyl-(acyl carrier protein) reductase. NagB shows 44.6% amino acid sequence identity with NagB from E. coli. The GlcNAc-related metabolic pathway of S. coelicolor, based on information in the KEGG database, is shown in Fig. 1B.
Fig 1.
Genetic environment of the nag metabolic genes and GlcNAc-related primary metabolism. (A) Map of the genomic regions around the amino sugar utilization genes. nagB (SCO5236) is a single transcription unit, while nagK (SCO4285) and nagA (SCO4284) together form the nagKA operon. ORFs are indicated by open arrows. Lengths of the genes (in brackets below the genes) and intergenic regions (dotted lines) are indicated. (B) Amino sugar metabolism in S. coelicolor, with connecting pathways. Annotation is based on the KEGG database and experimental evidence. Arrows show the direction of metabolism, and the responsible enzymes are presented in bold. It is unknown whether GlcN-6P can be metabolized directly to GlcNAc-6P. Names and functions of the enzymes and the reactions they catalyze are listed in Table 2.
Table 2.
Genes involved in GlcNAc transport and metabolism in S. coelicolor A3(2)
| Gene | Locus tag | Predicted function (EC no.) | Relevant equation(s)a |
|---|---|---|---|
| crr | SCO1390 | Global PTS component EIIACrr | |
| gdhA | SCO4683 | Glutamate dehydrogenase (EC 1.4.1.4) | Glu + H2O + NADP+ ⇔ 2-OG + NH3 + NADPH + H+ |
| glmM | SCO4736 | Phosphoglucosamine mutase (EC 5.4.2.10) | GlcN-1P ⇔ GlcN-6P |
| glmS1 | SCO4740 | Glucosamine-fructose-6-phosphate aminotransferase (EC 2.6.1.16) | Gln + Fru-6P ⇔ Glu + GlcN-6P |
| glmS2 | SCO2789 | Glucosamine-fructose-6-phosphate aminotransferase (EC 2.6.1.16) | Gln + Fru-6P ⇔ Glu + GlcN-6P |
| glmU | SCO3122 | Bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase (EC 2.7.7.23) | UTP + GlcNAc-1P ⇔ 2Pi + UDP-GlcNAc (EC 2.3.1.157), AcCoA + GlcN-1P ⇔ CoA + GlcNAc-1P |
| hexA | SCO2758 | Beta-N-acetylglucosaminidase (EC 3.2.1.52) | (GlcNAc)2 + H2O ⇔ 2 GlcNAc |
| SCO2786 | Beta-N-acetylhexosaminidase (EC 3.2.1.52) | (GlcNAc)2 + H2O ⇔ 2 GlcNAc | |
| SCO2943 | Sugar hydrolase (EC 3.2.1.52) | (GlcNAc)2 + H2O ⇔ 2 GlcNAc | |
| SCO4860 | Secreted hydrolase (EC 3.2.1.52) | (GlcNAc)2 + H2O ⇔ 2 GlcNAc | |
| nagA | SCO4284 | N-Acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) | GlcNAc-6P + H2O ⇔ GlcN-6P + OAc− |
| nagB | SCO5236 | Glucosamine phosphate isomerase (EC 3.5.99.6) | GlcN-6P + H2O ⇔ Fru-6P + NH3 |
| nagE2 | SCO2907 | PTSGlcNAc transmembrane component EIIC | |
| nagF | SCO2905 | PTSGlcNAc transmembrane component EIIB | |
| nagK | SCO4285 | N-Acetylglucosamine kinase (EC 2.7.1.59) | GlcNAc + ATP ⇔ GlcNAc-6P + ADP |
| murA | SCO2949 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase (EC 2.5.1.7) | PEP + UDP-GlcNAc ⇔ UACCG + OP |
| murA2 | SCO5998 | UDP-N-acetylglucosamine transferase (EC 2.5.1.7) | PEP + UDP-GlcNAc ⇔ UACCG + OP |
| murB | SCO4643 | UDP-N-acetylenolpyruvoylglucosamine reductase (EC 1.1.1.158) | UNAM + NADP+ ⇔ UACCG + NADPH + H+ |
| murQ | SCO4307 | N-acetylmuramic acid-6-phosphate etherase (EC 4.2.-.-) | MurNAc-6P + H2O ⇔ GlcNAc-6P + lactate |
| ptsH | SCO5841 | Global PTS component HPr | |
| ptsI | SCO1391 | Global PTS component EI |
Abbreviations of metabolites: OAc−, acetate; AcCoA, acetyl coenzyme A; (GlcNAc)2, N,N′-diacetylchitobiose; 2Pi, diphosphate; Fru, fructose; Fru-6-P, fructose-6-phosphate; GlcNAc, N-acetyl-d-glucosamine; GlcNAc-1P, N-acetylglucosamine-1-phosphate; GlcNAc-6P, N-acetylglucosamine-d-6-phosphate; GlcN, glucosamine; GlcN-1P, glucosamine-1-phosphate; GlcN-6P, glucosamine-6-phosphate; Gln, glutamine; Glu, glutamate; MurNAc-6P, N-acetylmuramic acid 6-phosphate; OP, orthophosphate; 2-OG, oxoglutarate; PEP, phosphoenolpyruvate; UDP-GlcNAc, UDP N-acetyl-d-glucosamine; UACCG, UDP-N-acetyl-3-O-(1-carboxyvinyl)-d-glucosamine; UNAM, UDP-N-acetylmuramate.
Analysis of the S. coelicolor genome by using the BLASTP algorithm strongly suggests that there is no redundancy of the nag metabolic genes in S. coelicolor, with single nagA, nagB, and nagK genes. Two distant homologues of nagA occur in S. coelicolor, with gene products sharing only around 20% aa identity with NagA, namely, SCO2758, encoding a secreted β-N-acetylgluco-saminidase most likely involved in glucan degradation (40), and SCO4860, encoding a GH20-type secreted β-N-acetyl-glucosaminidase. GH20-type hexosaminidases are involved in hydrolysis of the extracellular polysaccharide matrix in bacteria, such as β-1,6-linkages of PGA [poly-beta-(1,6)-N-acetylgluco-samine], and do not play a role in central metabolism. The nearest functional homologue of the NagB gene is SCO2786, which encodes the endo-β-N-acetylhexosaminidase Hex (23% aa identity) and was shown to cleave the GlcNAc oligomers (GlcNAc)3 and (GlcNAc)4 in Streptomyces plicatus (24). Based on this in silico analysis, we concluded that in S. coelicolor, PTS-transported GlcNAc-6P is metabolized solely by the gene products of open reading frames (ORFs) SCO4284 (nagA) and SCO5236 (nagB). Similarly, we also failed to identify paralogs for the GlcNAc kinase NagK in S. coelicolor. In terms of the highest end-to-end aa similarity, BLASTP analysis revealed functionally unrelated enzymes, namely, a chlorohydrolase (SCO3070; 35% aa identity to NagA), a 6-phosphogluconolactonase (SCO1939; 29% aa identity to NagB), and an FGGY family carbohydrate kinase (SCO6260; 35% aa identity to NagK) with a ROK (repressors, ORFs, kinases) family signature (39).
Expression of nagA and nagB is induced by GlcNAc and repressed by DasR.
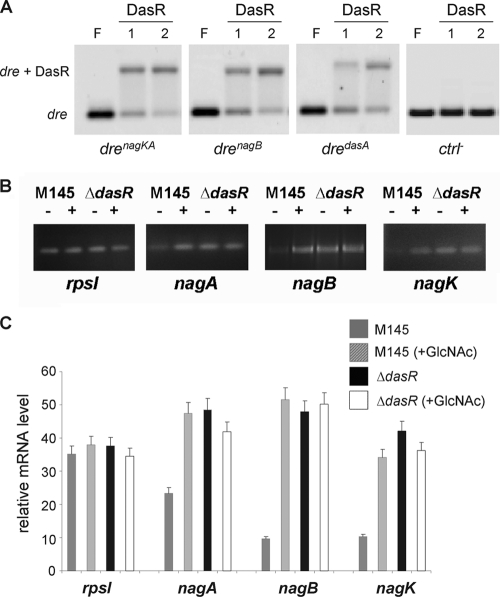
Prior to assessing the effect of inactivation of the nag genes on the GlcNAc-mediated signaling pathway in S. coelicolor, we first analyzed the GlcNAc-dependent control of the nagA, nagB, and nagK genes. DasR is a global regulator of primary metabolism and antibiotic production and is known to control, among others, the chitinolytic system in Streptomyces coelicolor (9, 26), as well as nagB (32). There is a DasR-responsive element (dre) at position −68 upstream of nagB (tGTGGTtTAGACCAaT; drenagB) with a 13-nt match to the dre consensus sequence (AGTGGTCTAGACCACT; drecons) (32). Suggestively, the program PREDetector (15) also identified a dre element (AGaGGTCTAGtCCACT; drenagKA) at position −83 upstream of the nagKA operon, with 14 of 16 nt matching the dre consensus sequence. Furthermore, the computational prediction of the DasR regulon in five other streptomycetes revealed identical or similar dre-like sequences upstream of the respective nagKA and nagB orthologs, and the very high conservation suggests that these sites are true functional cis-acting elements. In order to validate our in silico predictions, EMSAs were performed in vitro, using His6-tagged DasR and a short double-stranded oligonucleotide centered on drenagKA and drenagB. Positive specific DasR-dre interactions were observed for both dre sequences as well as for another positive control (the dre upstream of dasA), while the negative control (the cis-acting element of blaI of Bacillus licheniformis) was not bound by DasR (Fig. 2A).
Fig 2.
Direct transcriptional control of nagKA and nagB by DasR. (A) DasR binding to predicted dre upstream of nagKA and nagB. EMSAs were performed with pure DasR-His6 and a 40-bp Cy5-labeled double-stranded oligonucleotide probe centered on the DasR-responsive elements (dre) in the promoter regions of nagKA (drenagKA), nagB (drenagB), and dasA (dredasA; positive control), with short flanking sequences. The cis-acting element of blaI of Bacillus licheniformis was used as a negative control (ctrl−). Lanes: F, free probe (0.25 μM double-stranded Cy5-labeled probe); 1 and 2, DasR-dre binding reaction mixtures with 1.75 and 2.5 μM DasR-His6, respectively. (B) Induction of nag transcription by GlcNAc and control by DasR. Semiquantitative RT-PCR was carried out on RNA isolated from the dasR null mutant or its parent, S. coelicolor M145, grown in liquid mineral medium (NMMP plus mannitol) cultures immediately prior to (−) or 30 min after (+) the addition of 25 mM GlcNAc. (C) The constitutive expression of nagA, nagK, and nagB in dasR null mutants and their enhanced transcript levels in wild-type cells 30 min after induction show that the nag genes are repressed by DasR and induced by N-acetylglucosamine.
If the nag genes are repressed by DasR, it would follow that their expression should be at least partially independent of induction by GlcNAc in a dasR null mutant. To test this hypothesis, S. coelicolor M145 and its ΔdasR null mutant were grown in liquid NMMP medium until an OD600 of ∼0.2, followed by the addition of 25 mM GlcNAc. Mycelia were harvested for RNA preparation immediately before and 30 min after GlcNAc addition. Semiquantitative RT-PCR on total RNA samples from the wild-type strain S. coelicolor M145 showed induction of nagA, nagK, and nagB transcript levels of 2-, 3-, and 5-fold, respectively, following the addition of GlcNAc, while transcripts of the control rspI gene (for the ribosomal protein S9) were readily identified before and after induction and did not show significant induction (Fig. 2B and C). In contrast, expression of the nag genes was already high in the dasR null mutant before induction, with similar transcript levels before and after the addition of GlcNAc (Fig. 2B and C). This is consistent with induction of the nagA, nagB, and nagK genes by GlcNAc and repression of these genes by DasR. The induction by GlcNAc most likely acts via the allosteric inactivation of DasR by the GlcNAc-derived metabolite GlcN-6P (32, 34).
nagA and nagB gene replacement and IFD mutants.
To study the effect of the accumulation of GlcNAc-related metabolites on growth, development, and antibiotic production, null mutants were constructed for nagA and nagB. For this purpose, we first replaced the entire coding region of nagA or nagB with the apramycin resistance cassette (aacC4) flanked by loxP sequences and then removed the cassette to generate the respective IFD mutants (see Materials and Methods). For each experiment, five transformants exhibiting the desired double-crossover phenotype (Aprar Thios) were selected and verified by PCR as described in Materials and Methods. The mutants were designated GAM1 (nagA::aacC4) and GAM2 (nagB::aacC4). Following removal of the aacC4 cassette by the Cre recombinase, IFD mutants were obtained for nagA (GAM4) and nagB (GAM5). For all experiments, both the apramycin-resistant and IFD mutants were analyzed. Considering that there were no phenotypic differences between them in terms of antibiotic production and development, from this point onwards we describe only the results obtained for the IFD mutants, unless stated otherwise. The nagAB double mutant GAM9 was created by replacing nagA with the apramycin resistance cassette in the nagB IFD mutant GAM5.
Effect of GlcNAc on growth of the nagA and nagB mutants in liquid-grown cultures.
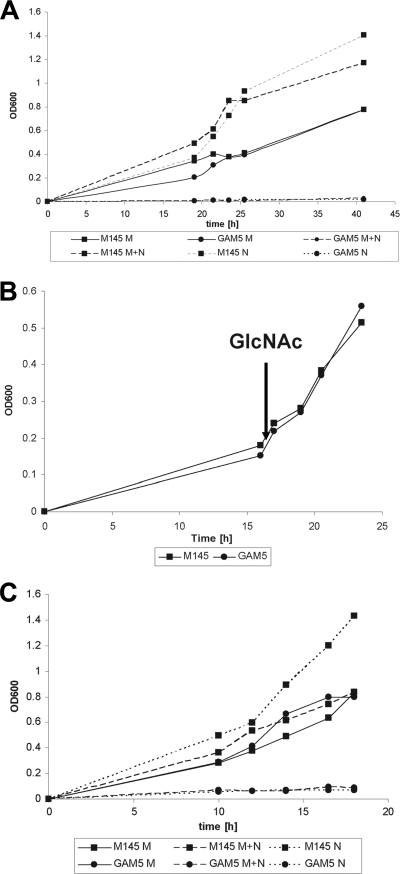
The addition of GlcNAc is expected to result in the accumulation of GlcNAc-6P in nagA mutants and GlcN-6P in nagB mutants (Fig. 1B). Growth of nagB mutants was analyzed in liquid mineral medium (NMMP) and compared to that of the parental strain S. coelicolor M145. While there was little difference in terms of growth on mannitol, nagB mutants failed to grow in the presence of GlcNAc. This is in line with the observed toxicity of the NagB substrate GlcN-6P in other bacteria (2, 29, 48). Addition of mannitol did not restore growth, strongly suggesting that rather than the inability to utilize GlcNAc, it is toxicity of a GlcNAc-derived metabolite that prevents growth of S. coelicolor in the absence of nagB (Fig. 3A). To analyze if the sensitivity to GlcNAc prevails throughout growth or occurs only during the earliest growth phases (i.e., germination and very early vegetative growth), we monitored growth in NMMP supplemented with mannitol (1% [wt/vol]), followed by the addition of 25 mM GlcNAc at an OD600 of 0.2 (early log phase) (Fig. 3B). Surprisingly, under these conditions, the mutant strain had a similar growth rate to that of the parent, indicating that GlcNAc toxicity is alleviated once a certain amount of biomass has accumulated. To rule out an effect specifically on spore germination, spores of S. coelicolor M145 and its nagB null mutant GAM5 were allowed to germinate for 8 h in 2× yeast extract-tryptone (2× YT) at 30°C and used as inocula for liquid mineral medium cultures. Despite the obvious emergence of germ tubes as well as many young vegetative hyphae, as assessed by phase-contrast microscopy, the germinated nagB mutant spores still failed to grow in the presence of GlcNAc (Fig. 3C). This illustrates that in the absence of the GlcN-6P deaminase activity of NagB, GlcNAc is toxic to young vegetative hyphae, but this is alleviated once a threshold amount of biomass has been produced.
Fig 3.
Effect of GlcNAc on growth of the nagB mutant. (A) Effects of GlcNAc on growth of S. coelicolor A3(2) M145 (■) and the nagB null mutant GAM5 (●). Strains were grown in liquid MM supplemented with either mannitol (50 mM) (M; solid lines), GlcNAc (25 mM) (N; dashed lines), or mannitol plus GlcNAc (M+N; dotted lines). (B) Effects of GlcNAc induction on growth of S. coelicolor A3(2) M145 (■) and the nagB null mutant GAM5 (●). Strains were grown in liquid NMMP supplemented with mannitol (1% [wt/vol]), followed by the addition of 25 mM GlcNAc to the cultures at an OD600 of ∼0.2. The addition of GlcNAc is indicated with an arrow. (C) Same as panel A, but with spores that were pregerminated for 8 h in 2× YT at 30°C.
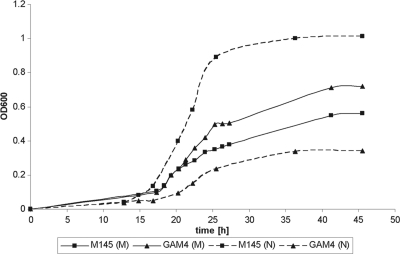
The nagA mutant GAM4 grew with a similar doubling time (Td; ∼3 h) to that of the parent strain M145 in liquid-grown MM cultures in the presence of mannitol, but it reproducibly reached higher biomass levels (Fig. 4). Conversely, in MM with GlcNAc, the doubling time of nagA mutants was much longer (∼5 h) than that of the parent strain M145 (∼2 h). This suggests that the likely accumulation of GlcNAc-6P impairs growth of nagA mutants, but the effect is not as dramatic as that of the accumulation of GlcN-6P in nagB mutants.
Fig 4.
Effect of GlcNAc on growth of the nagA mutant. The effects of GlcNAc on growth of S. coelicolor A3(2) M145 (■) and the nagA null mutant GAM4 (●) are shown. Strains were grown in liquid NMMP supplemented with either mannitol (50 mM) (M; solid lines) or N-acetylglucosamine (25 mM) (N; dashed lines). Doubling times were derived from the linear part of the growth curves.
Effect of GlcNAc on development and antibiotic production of nagA and nagB mutants in solid-grown cultures.
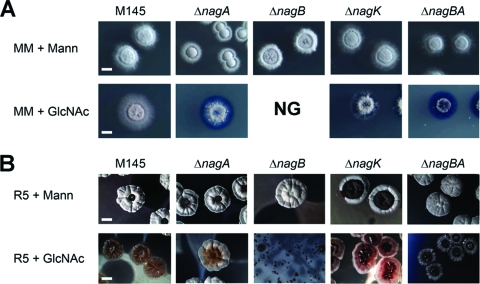
The nagA and nagB mutants and the nagAB double mutant were similar to the parental strain in terms of colony morphology on SFM, R5, or MM agar plates supplemented with glucose, fructose, galactose, glycerol, maltose, mannitol, mannose, or xylose as the sole carbon source (see Fig. S1 in the supplemental material). However, major differences were observed when GlcNAc was added to the media. The toxicity of GlcNAc to nagB mutants in liquid-grown cultures was also observed on solid media, with nagB null mutants failing to grow on MM agar plates supplemented with 25 mM GlcNAc (Fig. 5A), while very poor growth was observed on R5 agar plates with GlcNAc (Fig. 5B). nagA mutants showed normal growth and development on MM agar plates with mannitol, while GlcNAc resulted in impaired growth, although some aerial hyphae were formed. Interestingly, while GlcNAc was highly toxic to nagB null mutants on R5 agar plates supplemented with GlcNAc, the nagA null mutant GAM4 was able to enter development under these conditions (Fig. 5B), a phenotype that is similar to that of mutants lacking the GlcNAc transporter gene nagE2 (28). In line with the idea that GlcN-6P accumulation causes toxicity to nagB null mutants grown on GlcNAc, the double mutant GAM9 (M145 ΔnagAB) had a phenotype that was very similar to that of nagA mutants, with impaired growth on MM with GlcNAc and normal development on R5 agar plates with GlcNAc (Fig. 5). This most likely was due to the fact that the absence of nagA prevents the accumulation of GlcN-6P, and thus the toxicity of GlcNAc to nagB mutants.
Fig 5.
Phenotypic analysis of nag mutants. Phenotypes of S. coelicolor M145 and its nagA, nagB, nagK, and nagAB mutants (designated GAM4, GAM5, GAM6, and GAM9, respectively) were analyzed by stereomicroscopy. Colonies were grown for 5 days on MM (A) or R5 (B) agar plates with either mannitol (top) or GlcNAc (bottom). For phenotypes of the strains on other media, see Fig. S1 in the supplemental material. Note that nagB mutants failed to grow (NG) on MM supplemented with GlcNAc, while on R5 agar plates with GlcNAc, nagB mutants produced only a very thin layer of biomass. For complementation of the nagA and nagB mutants, see Fig. S3 in the supplemental material.
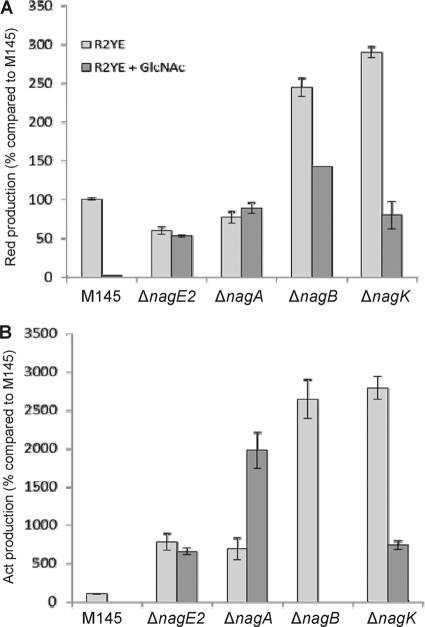
The ability of the mutants to produce the pigmented antibiotics prodiginine (Red) and actinorhodin (Act) was assessed in detail on R2YE agar plates. Since Red production is switched on earlier than Act production (3), antibiotic production was measured at different time points, namely, after 42 h of growth for Red production just prior to Act biosynthesis and after 48 and 120 h for early and late Act production, respectively. Deletion of nagA caused only a minor reduction of Red production (about 25%), while deletion of nagB resulted in a 2.5-fold increase of prodiginine biosynthesis (Fig. 6A). Both deletions had a more drastic effect on Act production, resulting in earlier and higher production, with 7- and 25-fold increases of Act biosynthesis after 48 h of growth for nagA and nagB mutants, respectively (Fig. 6B). After 120 h of growth, all mutants showed similar excesses of Act biosynthesis compared to the parental strain S. coelicolor M145 (increases of around 13-fold) (see Fig. S2 in the supplemental material). In contrast, nagB null mutant GAM5 formed extremely small colonies, with around 40% reduction of Red production and complete inhibition of extracellular Act production, in the presence of GlcNAc (Fig. 6). After prolonged incubation (120 h), the nagB null mutant GAM5 accumulated large amounts of intracellular Act on R5 agar plates supplemented with GlcNAc (see Fig. S2).
Fig 6.
Quantification of antibiotic production. Production of prodiginines (top) and actinorhodin (bottom) was quantified relative to their production by the parental strain S. coelicolor M145 (which was set to 100%). Cultures were grown for 42 h (Red) or 48 h (Act) on R2YE agar plates with (dark bars) or without (light bars) GlcNAc. For data obtained at 120 h, see Fig. S2 in the supplemental material.
In a control experiment, the nagA and nagB gene replacement and IFD mutants were complemented with the low-copy-number vector pHJL401 (22) harboring the corresponding nag gene and its native promoter. nagA mutants GAM1 and GAM4 were complemented with pGAM5 (pHJL401/nagKA), and nagB mutants GAM2 and GAM5 were complemented with pGAM6 (pHJL401/nagB). The nagKA operon was used for complementation of nagA mutants to allow expression from the operon promoter. Growth on MM and R5 agar plates with and without GlcNAc demonstrated that the complemented mutants had normal growth and GlcNAc sensing, showing that the defects in growth and development in response to GlcNAc were indeed due specifically to the deletion of the respective nag genes (see Fig. S3 in the supplemental material). Specifically, toxicity of GlcNAc to nagB mutants was relieved on both media, and GlcNAc sensing was restored to both nagA and nagB mutants (e.g., bald phenotype and blocked antibiotic production on R5 agar plates with GlcNAc).
Effect of deletion of nagK.
The sugar kinase NagK catalyzes the phosphorylation of GlcNAc to GlcNAc-6P, and the chromosomal deletion of nagK was performed as described for the nagA and nagB mutants (see Materials and Methods for details). We previously showed that transport of GlcNAc in S. coelicolor is mediated via the PTS (28). If GlcNAc is transported exclusively via the PTS, then the only internal source of GlcNAc is the dismantling of the cell wall during programmed cell death (PCD) or chitin degradation providing chitooligosaccharides [(GlcNAc)n, e.g., (GlcNAc)2 and (GlcNAc)3] (Fig. 1B). When grown on rich medium agar plates, the nagK mutant presented ∼27-fold-increased Red and Act production compared to the parental strain M145. Interestingly, on R2YE agar plates with GlcNAc, the nagK mutant showed partial loss of the GlcNAc repression effect, which we quantified to be about 75% of the wild-type GlcNAc-mediated repression. This suggests that GlcNAc may be internalized in a PTS-independent manner under these growth conditions. As expected, the additional deletion of nagK did not significantly alter the morphology of the nagA, nagB, or nagAB mutant under any of the conditions tested (data not shown).
DISCUSSION
The amino sugar N-acetylglucosamine (GlcNAc) is a major carbon and nitrogen source for Streptomyces which contributes to both cell wall synthesis and primary metabolism but also plays an important role as a developmental signaling molecule (27, 34). The signaling cascade from nutrient sensing to development and antibiotic production is a highly complex process, based on cooperative interaction between signaling molecules, the phosphorylation state of the PTS, and global and pathway-specific regulators and their effector molecules. We previously demonstrated that GlcNAc acts as a signaling molecule that controls the onset of development and antibiotic production (32, 34). Higher concentrations of GlcNAc outside the cell lock Streptomyces coelicolor in the vegetative growth phase under rich growth conditions (feast) but activate development and antibiotic production under poor conditions (famine). This principle has been applied successfully to enhance the expression of antibiotic biosynthetic gene clusters, including the clusters for Act, Red, and the cryptic polyketide Cpk (34). The GntR regulator DasR acts as a responsive nutrient sensor which has a pleiotropic regulon and represses, among others, nag metabolic genes, chitinolytic genes, and the pathway-specific activator genes for antibiotic production (9, 26, 32, 34). Since the repressing activity of DasR is relieved allosterically by the effector molecule GlcN-6P (32), it follows that controlling the flux of GlcNAc may be a promising approach for metabolic engineering relating to improved screening of novel antimicrobial compounds.
From analysis of amino sugar metabolism and connected pathways (Fig. 1B), it follows that during growth on GlcNAc, nagA null mutants likely accumulate GlcNAc-6P and nagB null mutants likely accumulate GlcN-6P. Indeed, in line with the requirement of NagA for the production of the signaling molecule GlcN-6P, nagA null mutants had lost GlcNAc sensing. This underpins the current model of a direct signaling pathway from external GlcNAc to the accumulation of GlcN-6P, which then acts as an allosteric effector of the developmental regulator DasR. However, during growth on GlcNAc, nagA null mutants likely accumulate large amounts of GlcNAc-6P, which in E. coli has two important regulatory roles, namely, as an allosteric activator of NagB (6) and an effector molecule for NagC, the repressor of the nag regulon. In E. coli, inactivation of nagA (and hence accumulation of GlcNAc-6P) thus evokes derepression of the nag regulon (29, 31). An additional effect is cell lysis, suggesting a moderate toxicity of the accumulation of GlcNAc-6P in E. coli (48). Similar observations were made for nagA mutants of Staphylococcus aureus or Gluconacetobacter xylinus, which showed strongly reduced growth rates in the presence of GlcNAc (21, 50). Indeed, we also found growth reduction for nagA null mutants of S. coelicolor in liquid minimal medium with GlcNAc, and the negative effect of GlcNAc on growth of the nagA mutant was shown by microscopy.
The accumulation of GlcN-6P is toxic to S. coelicolor, as demonstrated by the inability of nagB mutants to grow on GlcNAc. This toxicity was relieved by the additional deletion of nagA or the complete nagKA operon (ΔnagABK) and could also be complemented by reintroducing nagB on a low-copy-number vector. Interestingly, exponentially growing cells of nagB mutants had overcome the sensitivity to GlcNAc, as addition of GlcNAc during early logarithmic growth (OD600 of ∼0.2) did not lead to growth inhibition. In contrast, prolonged germination of the spores, which also resulted in the formation of many young vegetative hyphae, did not relieve the sensitivity. This strongly suggests that young vegetative hyphae in particular are sensitive to the accumulation of GlcN-6P, perhaps because alternative ways to reduce the GlcN-6P pool are not yet active enough. GlcN-6P occupies a central position between cell wall synthesis and glycolysis, and besides being converted to Fru-6P by NagB, it is also incorporated into murein following its conversion to UDP-GlcNAc by the action of GlmM (phosphoglucosamine mutase) and GlmU (N-acetyl-glucosamine-1-phosphate uridyltransferase) (Fig. 1B) (18, 25). In the absence of NagB, the major route to deplete GlcN-6P is incorporation into murein. Cell wall synthesis takes place exclusively at the apical sites (12, 14), and with very few growing tips during early growth, there may not be sufficient cell wall synthesis to convert sufficient amounts of GlcN-6P, with the net accumulation of large amounts of this toxic metabolite. An alternative explanation would be lower enzymatic activity of GlmM during early growth. In E. coli, GlmM is activated by phosphorylation, either by autophosphorylation or by the Ser/Thr kinase StkP (16, 17). It is still unknown how GlmM is activated in streptomycetes.
Preliminary work investigating the role of the GlcNAc kinase NagK revealed that the addition of GlcNAc has a less profound effect on development and antibiotic production in rich media when the nagK gene is deleted. The effect of NagK on the DasR-mediated GlcNAc nutrient sensing system suggests the accumulation of its substrate, GlcNAc. GlcNAc is internalized via the PTS in S. coelicolor (27, 28), but the existence of another import system that may be active under specific growth conditions cannot be ruled out. One candidate is the ABC transporter encoded by SCO6005-6007 in S. coelicolor, which is orthologous to the GlcNAc/chitobiose transport operon ngcEFG in Streptomyces olivaceoviridis, although the sequence similarity is low (the various gene products share 30 to 40% aa identity). Alternatively, the accumulated GlcNAc-6P (which is a labile molecule) may be hydrolyzed to GlcNAc, which is not toxic. In both cases, NagK would be required to produce GlcNAc-6P in order to allow formation of the DasR effector molecule GlcN-6P.
Interestingly, when the nagB null mutant was challenged with GlcNAc, we readily obtained suppressor mutants of this mutant. This is similar to observations in other bacterial systems, where spontaneous second-site mutations relieved the toxicity of accumulated sugar phosphates in nagB mutants (29, 48). Interestingly, when the nagB mutant spores were plated at a high density on MM agar plates containing GlcNAc, some colonies emerged, with a frequency of around 1:105, suggesting that they had undergone spontaneous suppressor mutations to render the colonies insensitive to the toxicity of GlcNAc. Likely candidates for suppressor mutations include the GlcNAc-specific transporter genes nagE2 and nagF and the metabolic genes nagA and nagK, but analysis of 16 independent suppressor mutants did not reveal changes in these genes or their promoter regions. We are currently analyzing the genomes of a series of the obtained suppressor mutants, and following recreation of the mutations in nagB mutants, this will reveal which genes have undergone mutations, which will shed important new light on the metabolism of GlcNAc in Streptomyces.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Andriy Luzhetskyy for providing pUWLcre and for discussions and to Fritz Titgemeyer for discussions.
S.R. is a research associate of the FRS-FNRS. This work was supported by a FRIA fellowship from the FRS-FNRS to E.T. and by grant R.CFRA.1237 from the Fonds Spéciaux of the University of Liège and a VICI grant from the Netherlands Applied Research Council (STW) to G.P.V.W.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alvarez-Anorve LI, Calcagno ML, Plumbridge J. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 187:2974–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernheim NJ, Dobrogosz WJ. 1970. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J. Bacteriol. 101:384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 [DOI] [PubMed] [Google Scholar]
- 4. Birko Z, et al. 2009. Lack of A-factor production induces the expression of nutrient scavenging and stress-related proteins in Streptomyces griseus. Mol. Cell. Proteomics 8:2396–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruckner R, Titgemeyer F. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141–148 [DOI] [PubMed] [Google Scholar]
- 6. Calcagno M, Campos PJ, Mulliert G, Suastegui J. 1984. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from Escherichia coli. Biochim. Biophys. Acta 787:165–173 [DOI] [PubMed] [Google Scholar]
- 7. Chater KF, Losick R. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p 149–182 In Shapiro J. A., Dworkin M. (ed), Bacteria as multicellular organisms. Oxford University Press, New York, NY [Google Scholar]
- 8. Cohen-Kupiec R, Chet I. 1998. The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9:270–277 [DOI] [PubMed] [Google Scholar]
- 9. Colson S, et al. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J. Mol. Microbiol. Biotechnol. 12:60–66 [DOI] [PubMed] [Google Scholar]
- 10. Colson S, et al. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382 [DOI] [PubMed] [Google Scholar]
- 11. Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A. 2008. Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 78:1065–1070 [DOI] [PubMed] [Google Scholar]
- 12. Flärdh K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564–571 [DOI] [PubMed] [Google Scholar]
- 13. Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 14. Gray DI, Gooday GW, Prosser JI. 1990. Apical hyphal extension in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 136:1077–1084 [DOI] [PubMed] [Google Scholar]
- 15. Hiard S, et al. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357:861–864 [DOI] [PubMed] [Google Scholar]
- 16. Jolly L, et al. 1999. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur. J. Biochem. 262:202–210 [DOI] [PubMed] [Google Scholar]
- 17. Jolly L, Pompeo F, van Heijenoort J, Fassy F, Mengin-Lecreulx D. 2000. Autophosphorylation of phosphoglucosamine mutase from Escherichia coli. J. Bacteriol. 182:1280–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jolly L, et al. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J. Bacteriol. 179:5321–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khodakaramian G, et al. 2006. Expression of Cre recombinase during transient phage infection permits efficient marker removal in Streptomyces. Nucleic Acids Res. 34:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 21. Komatsuzawa H, et al. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221–1231 [DOI] [PubMed] [Google Scholar]
- 22. Larson JL, Hershberger CL. 1986. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid 15:199–209 [DOI] [PubMed] [Google Scholar]
- 23. Mahr K, et al. 2000. Glucose kinase of Streptomyces coelicolor A3(2): large-scale purification and biochemical analysis. Antonie Van Leeuwenhoek 78:253–261 [DOI] [PubMed] [Google Scholar]
- 24. Mark BL, et al. 1998. Structural and functional characterization of Streptomyces plicatus beta-N-acetylhexosaminidase by comparative molecular modeling and site-directed mutagenesis. J. Biol. Chem. 273:19618–19624 [DOI] [PubMed] [Google Scholar]
- 25. Mengin-Lecreulx D, van Heijenoort J. 1994. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 176:5788–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nazari B, et al. 2011. High expression levels of chitinase genes in Streptomyces coelicolor A3(2) grown in soil. FEMS Microbiol. Ecol. 77:623–635 [DOI] [PubMed] [Google Scholar]
- 27. Nothaft H, et al. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nothaft H, et al. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 75:1133–1144 [DOI] [PubMed] [Google Scholar]
- 29. Plumbridge J. 2009. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J. Bacteriol. 191:5641–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Plumbridge J, Vimr E. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plumbridge JA. 1991. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053–2062 [DOI] [PubMed] [Google Scholar]
- 32. Rigali S, et al. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237–1251 [DOI] [PubMed] [Google Scholar]
- 33. Rigali S, et al. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 32:3418–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rigali S, et al. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saito A, et al. 2007. The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N′-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 73:3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Schrempf H. 2001. Recognition and degradation of chitin by streptomycetes. Antonie Van Leeuwenhoek 79:285–289 [DOI] [PubMed] [Google Scholar]
- 38. Seo JW, Ohnishi Y, Hirata A, Horinouchi S. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol. 184:91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Titgemeyer F, Reizer J, Reizer A, Saier MH., Jr 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349–2354 [DOI] [PubMed] [Google Scholar]
- 40. Tsujibo H, et al. 1998. Cloning, characterization and expression of beta-N-acetylglucosaminidase gene from Streptomyces thermoviolaceus OPC-520(1). Biochim. Biophys. Acta 1425:437–440 [DOI] [PubMed] [Google Scholar]
- 41. van Wezel GP, et al. 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 72:5283–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Wezel GP, et al. 2005. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2). Mol. Microbiol. 55:624–636 [DOI] [PubMed] [Google Scholar]
- 43. van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28:1311–1333 [DOI] [PubMed] [Google Scholar]
- 44. van Wezel GP, McKenzie NL, Nodwell JR. 2009. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458:117–141 [DOI] [PubMed] [Google Scholar]
- 45. Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogler AP, Lengeler JW. 1989. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol. Gen. Genet. 219:97–105 [DOI] [PubMed] [Google Scholar]
- 47. Wang F, Xiao X, Saito A, Schrempf H. 2002. Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol. Genet. Genomics 268:344–351 [DOI] [PubMed] [Google Scholar]
- 48. White RJ. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao X, et al. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol. Genet. Genomics 267:429–439 [DOI] [PubMed] [Google Scholar]
- 50. Yadav V, et al. 2011. N-Acetylglucosamine 6-phosphate deacetylase (NagA) is required for N-acetylglucosamine assimilation in Gluconacetobacter xylinus. PLoS One 6:e18099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.