Abstract
Tertiary alcohols, such as tert-butyl alcohol (TBA) and tert-amyl alcohol (TAA) and higher homologues, are only slowly degraded microbially. The conversion of TBA seems to proceed via hydroxylation to 2-methylpropan-1,2-diol, which is further oxidized to 2-hydroxyisobutyric acid. By analogy, a branched pathway is expected for the degradation of TAA, as this molecule possesses several potential hydroxylation sites. In Aquincola tertiaricarbonis L108 and Methylibium petroleiphilum PM1, a likely candidate catalyst for hydroxylations is the putative tertiary alcohol monooxygenase MdpJ. However, by comparing metabolite accumulations in wild-type strains of L108 and PM1 and in two mdpJ knockout mutants of strain L108, we could clearly show that MdpJ is not hydroxylating TAA to diols but functions as a desaturase, resulting in the formation of the hemiterpene 2-methyl-3-buten-2-ol. The latter is further processed via the hemiterpenes prenol, prenal, and 3-methylcrotonic acid. Likewise, 3-methyl-3-pentanol is degraded via 3-methyl-1-penten-3-ol. Wild-type strain L108 and mdpJ knockout mutants formed isoamylene and isoprene from TAA and 2-methyl-3-buten-2-ol, respectively. It is likely that this dehydratase activity is catalyzed by a not-yet-characterized enzyme postulated for the isomerization of 2-methyl-3-buten-2-ol and prenol. The vitamin requirements of strain L108 growing on TAA and the occurrence of 3-methylcrotonic acid as a metabolite indicate that TAA and hemiterpene degradation are linked with the catabolic route of the amino acid leucine, including an involvement of the biotin-dependent 3-methylcrotonyl coenzyme A (3-methylcrotonyl-CoA) carboxylase LiuBD. Evolutionary aspects of favored desaturase versus hydroxylation pathways for TAA conversion and the possible role of MdpJ in the degradation of higher tertiary alcohols are discussed.
INTRODUCTION
In nature, molecules bearing tertiary alcohol groups are not unusual and can even be central metabolites, such as citric acid and mevalonic acid, which are processed by nearly all living beings. However, not much is known about the catabolism of simple tertiary alcohols not possessing additional functional groups. The homologous series starts with tert-butyl alcohol (TBA) (2-methyl-2-propanol) as the smallest molecule, with only four carbon atoms, followed by tert-amyl alcohol (TAA) (2-methyl-2-butanol). In the last years, some progress on elucidating the catabolism of these compounds has been made when research focused on the environmental fate of fuel oxygenates, since tertiary alcohols have been identified as intermediates in bacterial degradation pathways of branched-chain dialkyl ethers (10, 27, 42). Since the 1980s, methyl tert-butyl ether (MTBE) and tert-amyl methyl ether (TAME) have been used as gasoline additives in Europe. Initially introduced as an octane booster at low concentrations, oxygenated gasoline is now amended with up to 15 vol% of these ethers to improve combustion efficiency and reduce carbon monoxide emissions. Currently, the corresponding ethyl ethers, ethyl tert-butyl ether (ETBE) and tert-amyl ethyl ether (TAEE), have gained the market share due to their lower vapor pressures and higher boiling points (8). The use of ethyl tert-alkyl ethers is also promoted by legislation in some countries, as the ethyl moiety can be easily derived from bioethanol (34), thus helping to reduce carbon dioxide emissions from fossil resources. The extensive use of fuel oxygenate ethers, however, has resulted in the contamination of numerous groundwater sites in the United States and Europe due to accidental spills and leaking storage tanks (14, 24, 29, 45). Compared to other gasoline compounds, the ethers are highly water soluble and render water unfit for drinking even at concentrations in the parts-per-billion range (44). These properties often impede the removal of fuel oxygenate ethers from contaminated sites below threshold values, making fuel oxygenate ethers and also their tertiary alcohol intermediates very problematic pollutants.
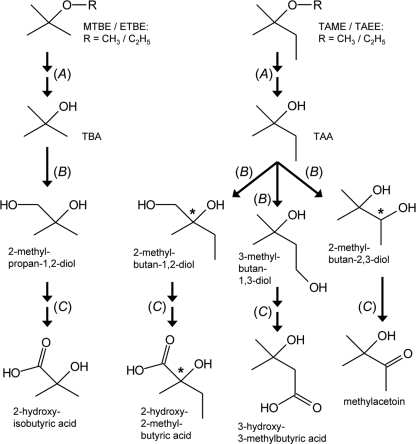
Although fuel oxygenate ethers are now banned in the United States (47) and may be phased out in other countries, they will persist at polluted sites for a long time, as they all turned out to be quite recalcitrant to biodegradation (10, 36). The main reason for this poor degradability might be the xenobiotic character of their tert-butyl and tert-amyl moieties, as the catabolism of these highly branched structures likely requires the evolution of novel enzymes linking the ether-specific pathways with common metabolic routes. In the case of the alkyl tert-butyl ethers MTBE and ETBE, there is a general agreement that aerobic degradation proceeds via several specific monooxygenase- and dehydrogenase-catalyzed steps, resulting in the formation of TBA, 2-methylpropan-1,2-diol, and 2-hydroxyisobutyric acid (Fig. 1) (27, 40, 42). 2-Hydroxyisobutyric acid is then converted into the common metabolite 3-hydroxybutyric acid in a cobalamin-dependent mutase reaction (35). The biochemistry of tert-amyl alkyl ether catabolism, on the other hand, has not been elucidated in much detail. By analogy with MTBE and ETBE, it can be proposed that TAME and TAEE are degraded via the TBA homologue TAA. However, in contrast to 2-methylpropan-1,2-diol formation from TBA, the hydroxylation of TAA would result in three possible diol products (Fig. 1), as has already been shown for TAME and TAA conversion in rats and humans (2, 43). The 1,2- and 1,3-diols could be further oxidized to 2-hydroxy-2-methyl- and 3-hydroxy-3-methylbutyric acids, respectively, while the dehydrogenation of the 2,3-diol would result in the formation of methylacetoin. Hence, it can be expected that compared to MTBE and ETBE degradation, bacterial pathways for TAME and TAEE are branched and likely involve a larger number of metabolites.
Fig 1.
Proposed pathways for the bacterial degradation of tert-butyl alkyl (MTBE and ETBE) and tert-amyl alkyl (TAME and TAEE) ethers (27, 31). (A) Initially, the methyl/ethyl groups of the ethers are hydroxylated, resulting in the formation of instable hemiacetals (not shown). The latter can spontaneously dismutate to the corresponding tertiary alcohols (TBA and TAA) and formaldehyde/acetaldehyde. Alternatively, the hemiacetals are oxidized to esters, which are hydrolyzed to the tertiary alcohols and formic/acetic acid. (B) The tertiary alcohols are hydroxylated to diols. In M. petroleiphilum PM1 and A. tertiaricarbonis L108, this step is likely catalyzed by the Rieske nonheme mononuclear iron oxygenase MdpJ (18, 39). (C) Dehydrogenation of primary and secondary alcohols of the diol intermediates to aldehyde and ketone groups. In the case of the aldehydes, further dehydrogenation to the corresponding carboxylic acids is possible. Asterisks indicate chiral carbon centers in metabolite molecules.
For the bacteria Aquincola tertiaricarbonis strain L108 and Methylibium petroleiphilum strain PM1, it has been found that the expressions of a putative Rieske nonheme mononuclear iron monooxygenase and its corresponding reductase are upregulated when cells are grown on MTBE and TBA, suggesting that these enzymes are responsible for the hydroxylation of TBA to 2-methylpropan-1,2-diol (18, 39). In strain PM1, the oxygenase and reductase are encoded by the mdpJ and mdpK genes, respectively, which show about 97% identity to the corresponding sequences found in strain L108. Recently, the importance of MdpJ in TBA metabolism has also been demonstrated by 13C metabolomic and proteomic stable isotope probing (SIP) approaches investigating oxygenate degradation in mixed cultures (3, 4). It is likely that MdpJ is also involved in TAA degradation, as strains L108 and PM1 could metabolize TAME and TAA (31, 38). However, the hydroxylation of TAA by MdpJ or other bacterial tert-alcohol monooxygenases has not been investigated so far.
In order to identify the role of the putative monooxygenase MdpJ in tert-amyl alkyl ether catabolism, we analyzed cultures of the bacteria A. tertiaricarbonis strain L108 and M. petroleiphilum strain PM1 for TAME- and TAEE-related metabolites. Furthermore, two mdpJ knockout mutants of A. tertiaricarbonis L108 were characterized. Surprisingly, it was shown that MdpJ is not hydroxylating the tert-amyl alkyl ether intermediate TAA and its higher homologue 3-methyl-3-pentanol but formed the corresponding desaturation products. Thus, MdpJ plays a central role in the TAME and TAEE degradation pathways by linking the conversion of TAA with the routes for catabolizing the hemiterpene 2-methyl-3-buten-2-ol and the amino acid leucine.
MATERIALS AND METHODS
Chemicals, bacterial strains, and growth medium.
The purity and supply sources of tertiary alcohols and other chemicals used in this study are listed in the supplemental material. Aquincola tertiaricarbonis strain L108, previously isolated from an MTBE-contaminated aquifer in Leuna, Germany (25, 35), was cultivated in liquid mineral salt medium (MSM) (see the supplemental material) containing MTBE at a concentration of 0.3 g liter−1. Methylibium petroleiphilum strain PM1 (32), obtained from the American Type Culture Collection (ATCC BAA-1232), was grown under the same conditions. Nitrogen-free and cobalt-free MSM was prepared by omitting NH4Cl and CoCl2 · 6H2O, respectively.
Growth and resting-cell experiments.
Cultures were incubated at 30°C on rotary shakers. Bacterial cells used in experiments were pregrown on the respective substrates in closed glass bottles in up to 1 liter of culture medium and harvested by centrifugation at 13,000 × g at 4°C for 10 min. After washing twice with MSM or nitrogen-free MSM, cells were immediately used as an inoculum for growth or resting-cell experiments. For the latter experiments, the cell concentration was adjusted to values between 1.4 and 2.2 g biomass (dry weight) per liter by dilution with MSM, whereas growth experiments were typically started with 30 to 60 mg biomass per liter. The data shown in this study represent the mean values and standard deviations (SD) of data from at least three replicate experiments.
Sampling and analytics.
Liquid and gas samples were taken as previously described (38), by puncturing the butyl rubber stoppers of incubation bottles with syringes equipped with 0.6- by 30-mm Luer Lock needles. The biomass was monitored by measuring the optical density at 700 nm (OD700), using a multiplication factor of 0.54 for calculating the dry biomass in g per liter (31). Volatile compounds (MTBE, TAME, TAEE, TBA, TAA, isoamylene, isoprene, 2-methyl-3-buten-2-ol, prenol, prenal, 3-methyl-3-pentanol, 3-methyl-1-penten-3-ol, and methylacetoin) were quantified by headspace gas chromatography (GC) using flame ionization detection (FID) (38). Compounds in samples were identified according to the retention times of pure GC standards. In addition, assignments were verified by GC mass spectrometry analysis (see Fig. S3 to S7 in the supplemental material). Diols and carboxylic acids were quantified by using high-performance liquid chromatography (HPLC) with refractive index (RI) detection as described elsewhere previously (30, 31), applying an eluent of 0.01 N sulfuric acid at 0.6 ml per min and a Nucleogel Ion 300 OA column (300 by 7.7 mm; Macherey-Nagel). In addition, carboxylic acid metabolites were identified as methyl esters by GC mass spectrometry (see the supplemental material).
Sequencing of wild-type strain L108 DNA.
Genomic DNA of A. tertiaricarbonis wild-type strain L108 was extracted by using the MasterPure DNA purification kit (Epicentre) and sequenced by Illumina HiSeq 2000 technology (GATC Biotech, Konstanz, Germany). The obtained DNA sequences were analyzed for open reading frames by using Rast (Rapid Annotation Using Subsystem Technology) (http://rast.nmpdr.org/).
Knockout mutants.
In order to prove the enzymatic function of MdpJ in tertiary alcohol degradation, we generated knockout mutants of A. tertiaricarbonis strain L108. Site-directed mutagenesis by the homologous recombination of the designed modified target gene mdpJ::tet (our unpublished data) out of diverse special knockout vectors failed. However, mutants were successfully obtained by using the high-transposable but unspecific 1.2-kb small linear EZ-Tn5<KAN-2> Tnp transposome (Epicentre Biotechnologies). We transformed 1 μl of the transposome into 70 μl highly competent mid-exponential-phase bacterial cells by electroporation (MicroPulser; Bio-Rad) in 0.1-cm cuvettes at 600 Ω and 1.8 kV for about 5 ms. Transformed cells were rescued in 5 ml MSM amended with 10 mM fructose as a carbon source, incubated for 6 h at 30°C and 150 rpm. Dilutions were then plated onto MSM fructose agar containing 50 μg ml−1 kanamycin as a selection marker and incubated for 2 days at 30°C. As the transposome integrates randomly into the DNA, all colonies obtained had to be analyzed for the loss of their capability to grow on MSM TBA agar (containing 0.5 g liter−1 TBA). In addition, copies of the colonies were maintained on MSM fructose agar for further analysis. About 5,000 colonies were screened in this way. Colonies with restricted or even lost TBA degradation potential were transferred again onto MSM TBA agar plates. The mutants that failed to grow were analyzed further.
The exact integration site of the transposome into the genomic DNA was determined by direct sequencing with flanking KAN-2 primers (Epicentre) out of 1 μg μl−1 high-concentrated genomic DNA (Master Pure DNA purification kit; Epicentre Biotechnologies) according to the following protocol. Sequencing forward primer KAN-2 FP-1 (5′-ACCTACAACAAAGCTCTCATCAACC-3′) and reverse primer KAN-2 RP-1 (5′-GCAATGTAACATCAGAGATTTTGAG-3′) were used. Direct sequencing conditions were 4 min at 95°C and 60 cycles of 30 s at 95°C and 4 min at 60°C. The products were cleaned with Centri-Sep columns (Applied Biosystems) and sequenced according to a method described previously by Sanger et al. (37), using an ABI Prism 3100 genetic analyzer and the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems). The resulting sequences were analyzed with BLAST (1) and aligned via Sequencher, version 5.0, sequence analysis software (Gene Codes Corporation, Ann Arbor, MI).
Nucleotide sequence accession numbers.
Sequences of the mdpJK gene environment and the liu operon obtained by the sequencing of genomic DNA of A. tertiaricarbonis strain L108 have been deposited in the GenBank/EMBL/DDBJ database under accession numbers JQ062962 and JQ001939.
RESULTS
TAA formation in cultures grown on TAME and TAEE.
When grown on TAME, cells of M. petroleiphilum strain PM1 temporarily accumulated TAA in significant amounts (see Fig. S1 in the supplemental material), proving that TAA is a central metabolite of the TAME degradation pathway. TAEE did not support the growth of strain PM1. In contrast, A. tertiaricarbonis L108 metabolized TAME and also TAEE without an accumulation of TAA (see Fig. S1 and S2 in the supplemental material), indicating that TAA is more efficiently degraded by this strain.
mdpJ gene environment and knockout mutants.
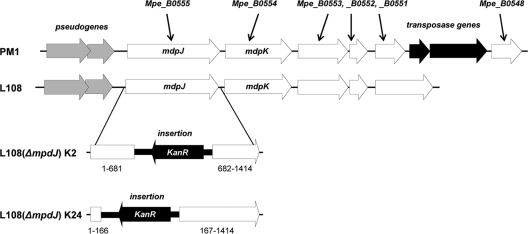
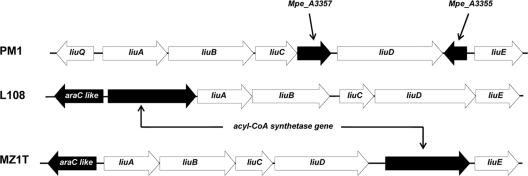
By the sequencing of genomic DNA from strain L108, a 6.2-kb fragment containing the mdpJ and mdpK genes was obtained. An alignment with the corresponding gene region of strain PM1 showed a very high level of similarity (Fig. 2). Both the gene and intergenic sequences possess >97% identity. The only substantial difference that could be found were two transposase genes present in strain PM1 and interrupting Mpe_B0551 and Mpe_B0548. At the corresponding position in strain L108, only a single gene encoding a hypothetical protein with 290 amino acids could be identified.
Fig 2.
Comparison of the mdpJ gene regions in M. petroleiphilum PM1 and A. tertiaricarbonis L108. The location of transposome integration into mdpJ found in the two knockout mutants of strain L108 resulting in a complete loss of the capability to grow on TBA and TAA is indicated.
For precisely studying the role of MdpJ in the bacterial degradation of tertiary alcohols, we tried to create mdpJ knockout mutants of strain L108 by a site-specific mutagenesis approach. However, even after several trials, corresponding knockout mutants were not obtained. Unspecific transposon mutagenesis, on the other hand, and screening for a loss of the capability to grow on TBA resulted in two mdpJ knockout mutants, which possessed insertions at different positions in the wild-type mdpJ gene (Fig. 2). In contrast to wild-type strain L108, both mutant strains were not able to grow on the tertiary alcohols TBA and TAA, whereas 2-methylpropan-1,2-diol, the putative product of TBA hydroxylation by MdpJ catalysis (Fig. 1), could still be used as the sole source of energy and carbon. In addition, the postulated 2-methylpropan-1,2-diol intermediate 2-hydroxyisobutyric acid was metabolized by wild-type and mutant strains at the same rates, indicating that only the tertiary alcohol-attacking enzymatic step was affected by the mdpJ knockout.
Accumulation of TAA metabolites by resting cells.
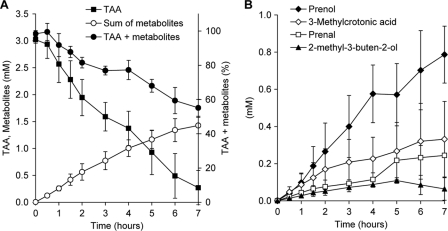
When shifting TBA-grown cells of A. tertiaricarbonis wild-type strain L108 to TAA and concomitantly inhibiting protein synthesis by chloramphenicol, significant amounts of various metabolites accumulated (Fig. 3). Surprisingly, not the expected diol compounds and derived carboxylic acids (Fig. 1) but several hemiterpenes were detected. About 30% of the TAA was converted to the primary alcohol prenol (3-methyl-2-buten-1-ol), while the unsaturated tertiary alcohol 2-methyl-3-buten-2-ol and the aldehyde prenal (3-methyl-2-butenal) corresponded to less than 5% of the metabolized substrate. In addition, significant amounts of 3-methylcrotonic acid accumulated. However, in contrast to the previously found alkene formation by TAA-grown cells (38), isoamylene was not produced from TAA in significant amounts by TBA-grown cells. Interestingly, not pure isoamylene but a mixture of this alkene and isoprene was formed, indicating the enzymatic dehydration of the tertiary alcohols TAA and 2-methyl-3-buten-2-ol (see also the next section). Both dehydration products represented about 1.6% of TAA conversion (data not shown). In the first hours of the experiment, cumulative concentrations of the substrate and all identified metabolites resulted in a nearly complete recovery of the carbon initially added. Toward the end of the experiment, this value steadily decreased. However, even after the complete degradation of TAA, still about 60% of the substrate was recovered as the metabolites listed in Fig. 3. This final analytical gap of about 40% may be attributed to common metabolites produced from 3-methylcrotonic acid, such as acetoacetate and acetyl coenzyme A (acetyl-CoA), which are readily processed via constitutive pathways. Overall, the observed accumulation of metabolites indicates that the degradation of TAA proceeds dominantly via hemiterpenes and that the enzymes required for catabolizing them are not well induced in TBA-grown cells of A. tertiaricarbonis L108. In contrast, TBA-grown cells not incubated in the presence of the translation inhibitor chloramphenicol accumulated 2-methyl-3-buten-2-ol, prenol, and the corresponding aldehyde only temporarily in the first 2 h of the experiment (data not shown). The concentration of metabolites then decreased steadily, indicating the induction of enzymes involved in the TAA- and hemiterpene-specific degradation pathway.
Fig 3.
Accumulation of metabolites in resting-cell experiments with TBA-grown wild-type A. tertiaricarbonis L108 cells incubated on TAA in nitrogen-free MSM in the presence of chloramphenicol (2 mM). (A) Concentrations of TAA, the sum of quantified metabolites, and the percentages of all TAA-derived compounds compared to the initial TAA concentration. (B) Concentrations of the metabolites 2-methyl-3-buten-2-ol, prenol, prenal, and 3-methylcrotonic acid.
Resting cells pregrown on TAA, on the other hand, did not show a significant accumulation of metabolites. When these cells were incubated with or without chloramphenicol on TAA, the latter was rapidly degraded, and neither prenol nor prenal or 3-methylcrotonic acid was detectable (data not shown). Only small amounts of 2-methyl-3-buten-2-ol were produced, reaching maximal concentrations of less than 0.1 mM, which represented about 2% of the TAA metabolized. Interestingly, resting cells pregrown on 2-methyl-3-buten-2-ol gave similar results (Fig. 4), suggesting that all enzymes necessary for efficient TAA degradation were induced under these conditions. Like L108 wild-type and mutant strains, strain PM1 was able to grow on 2-methyl-3-buten-2-ol as the sole source of carbon and energy (data not shown), indicating that this hemiterpene could be metabolized in case it was also occurring as a TAA metabolite in this strain.
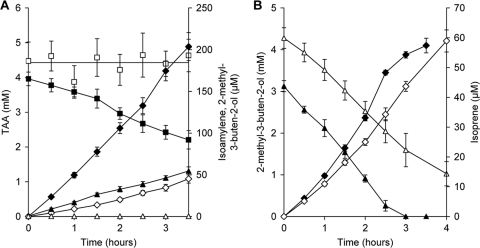
Fig 4.
Degradation of TAA and 2-methyl-3-buten-2-ol by resting cells of wild-type strain L108 (closed symbols) and the L108(ΔmdpJ) K2 knockout mutant strain (open symbols). (A) Isoamylene, as the sum of gamma- and beta-isomers (diamonds), was formed from TAA (squares). In all cases, mainly beta-isoamylene was emitted, representing 96 to 97% of the alkenes produced (data not shown). In addition, the accumulation of small amounts of 2-methyl-3-buten-2-ol (triangles) from TAA with the wild-type strain but not with the mutant was observed. (B) Isoprene (diamonds) was formed from 2-methyl-3-buten-2-ol (triangles). For a direct comparison with alcohol conversion, values of dehydration products refer to concentrations in the liquid phase, although they were found exclusively in the gas phase of the closed incubation bottles.
Dehydration of TAA and 2-methyl-3-buten-2-ol.
As the mutant strains of L108 were not able to grow on TAA but could still use 2-methyl-3-buten-2-ol as a growth substrate, resting cells pregrown on this hemiterpene were used to test whether the mutant strains accumulated metabolites from TAA. As expected, and in contrast to the wild-type strain, with both mutant strains, a formation of 2-methyl-3-buten-2-ol or other hemiterpene compounds was not observed. However, as was described previously for cells of the wild-type strain grown on TAA (38) and now for TBA-grown cells (see the section above), the alkenes gamma- and beta-isoamylene were still formed from TAA by resting cells of both mutant strains, as shown exemplarily for the L108(ΔmdpJ) K2 strain in Fig. 4A, proving that this dehydration reaction is not catalyzed by MdpJ. Interestingly, the level of alkene formation from TAA by the wild-type strain was about 5 times higher, indicating that a complete metabolization of the alcohol may be beneficial for a high level of dehydration activity. Accordingly, isoprene was emitted from the metabolizable tertiary alcohol 2-methyl-3-buten-2-ol by mutant and wild-type strains at similar rates (Fig. 4B), representing less than 2% of the substrate turnover.
Degradation of 3-methyl-3-pentanol.
Wild-type strain L108 and the mdpJ knockout mutant strains were also tested for their capabilities to metabolize the tertiary alcohol 3-methyl-3-pentanol, a C6 homologue of TBA and TAA. In analogy with 2-methyl-3-buten-2-ol accumulation from TAA, resting cells of the wild-type strain pregrown on the tertiary hemiterpene alcohol produced the unsaturated tertiary alcohol 3-methyl-1-buten-3-ol. However, compared to hemiterpene accumulation from TAA, the formation of 3-methyl-1-buten-3-ol was more pronounced, temporarily amounting to up to more than 30% of the substrate conversion (Fig. 5). In line with the assumption that MdpJ is responsible for tertiary alcohol degradation, the mutant strains neither produced 3-methyl-1-buten-3-ol from 3-methyl-3-pentanol nor converted the substrate at all (Fig. 5). The wild-type strain not only metabolized the C6 tertiary alcohol in short-term experiments but also could grow on it as the sole source of carbon and energy (data not shown). However, with generation times of about 40 h, the growth of strain L108 on 3-methyl-3-pentanol was significantly slower than that on TAA (31).
Fig 5.
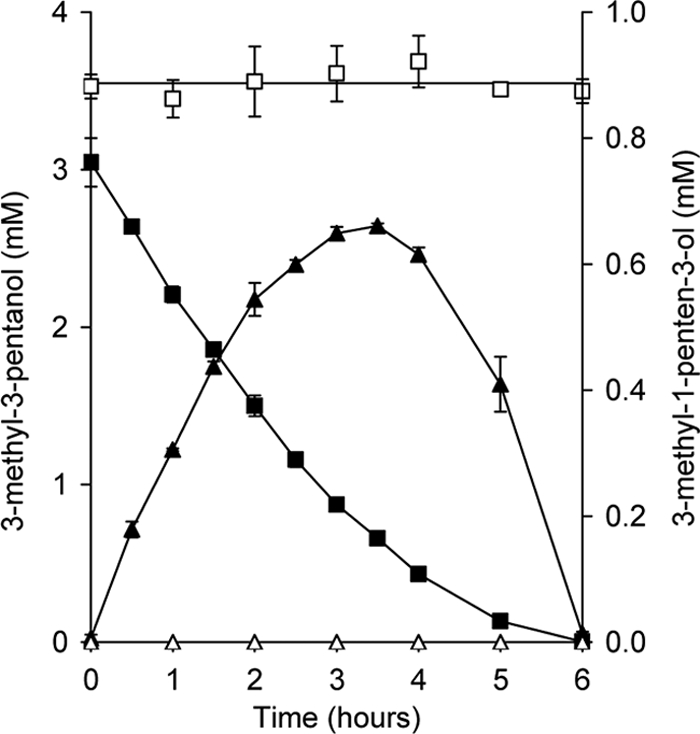
Accumulation of the desaturation product 3-methyl-1-penten-3-ol (closed triangles) by resting cells of wild-type A. tertiaricarbonis L108 from the tertiary alcohol substrate 3-methyl-3-pentanol (closed squares). Both L108(ΔmdpJ) K2 and L108(ΔmdpJ) K24 knockout mutant strains did not metabolize the tertiary alcohol (open squares). Accordingly, desaturation products were not detected (open triangles). Cells were pregrown on 2-methyl-3-buten-2-ol.
Vitamin dependence of tertiary alcohol degradation.
The vitamin requirements of strain L108 for the degradation of TAA, 3-methyl-3-pentanol, and TBA were studied. Biotin at >8 ng liter−1 was essential for growth on TAA and 3-methyl-3-pentanol, whereas TBA was still used as a growth substrate without biotin (Fig. 6). Interestingly, cobalamin was not essential for growth on TAA and 3-methyl-3-pentanol, whereas it was previously shown to be required for TBA metabolism in strain L108 as a component of a cobalamin-dependent CoA-carbonyl mutase for the conversion of the TBA metabolite 2-hydroxyisobutyric acid (35). Consequently, both TAA and 3-methyl-3-pentanol are obviously degraded via a totally different route not involving the mutase reaction.
Fig 6.
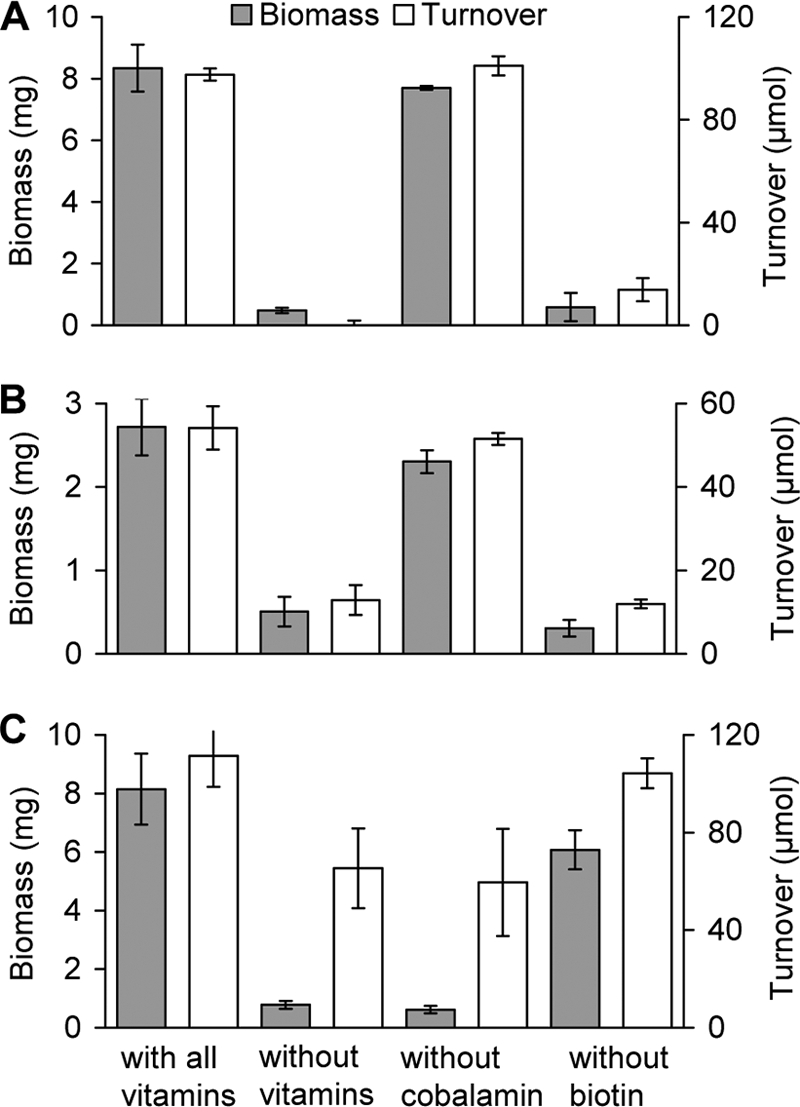
Comparison of vitamin dependences of wild-type A. tertiaricarbonis L108 cells grown on TAA (A), 3-methyl-3-pentanol (B), and TBA (C) as the sole source of energy. In all cases, biomass increase and substrate turnover refer to incubation periods required for the complete consumption of the respective tertiary alcohol substrates in the reference cultures supplemented with all vitamins of complete MSM. Incubation without vitamins was achieved by diluting cultures pregrown in complete MSM with cobalt- and vitamin-free MSM, resulting in a 2,500-fold dilution of cobalt and all vitamins. The same dilution was applied when only single vitamins, i.e., cobalamin or biotin, were omitted.
Leucine degradation pathway in strains PM1 and L108.
The occurrence of 3-methylcrotonic acid in the course of TAA conversion and the strict dependence of strain L108 on biotin for growth on this tertiary alcohol let us assume that 3-methylcrotonyl-CoA carboxylase is involved in the degradation pathway. Normally, this biotin-dependent enzyme is part of the leucine catabolism, catalyzing the carboxylation of 3-methycrotonyl-CoA produced from the branched-chain amino acid via 2-oxoisocaproic acid and isovaleryl-CoA (21). The genes encoding the relevant enzymes LiuABCDE are organized in the liu (leucine-isovalerate-utilizing) operon. Accordingly, a complete set of liu genes can be found on the chromosome of strain PM1. The sequencing of the corresponding DNA fragment in strain L108 gave a similar result (Fig. 7). Interestingly, a gene encoding a putative acyl-CoA synthetase is present only in the liu operon found in strain L108, while two extra genes, encoding a putative transmembrane protein (Mpe_A3357) and a hypothetical protein (Mpe_A3355), were found only in strain PM1. Another difference might be the regulation of gene expression, as genes encoding proteins belonging to the different regulator families TetR and AraC are found upstream from the liu genes in strains PM1 and L108, respectively.
Fig 7.
liu operons found in M. petroleiphilum PM1 and A. tertiaricarbonis L108. For comparison, the corresponding region present in the genome of Thauera sp. strain MZ1T is also shown (Tmz1t_0747 to Tmz1t_0753). Genes not yet related to leucine and isovaleric acid catabolism are highlighted in black.
DISCUSSION
While the metabolic sequences responsible for tert-butyl alkyl ether and TBA degradation in bacteria have been largely elucidated, practically nothing is known about the metabolism of tert-amyl alkyl ethers and TAA. The work presented here revealed that the putative Rieske nonheme mononuclear iron oxygenase MdpJ functions as a desaturase enabling the degradation of the TAME and TAEE metabolite TAA via hemiterpenes to intermediates of the leucine catabolism.
Comparison of the wild type and the two knockout mutants of A. tertiaricarbonis L108 clearly demonstrated that MdpJ is essential for TAA degradation. However, the latter compound is not converted to diols by hydroxylation reactions, but instead, the hemiterpene 2-methyl-3-buten-2-ol is formed. This implies that MdpJ shows desaturase activity when attacking tertiary alcohols, thereby causing the formation of a double bond by the removal of two hydrogen radicals from vicinal carbon atoms. A similar multifunctionality was found previously for the MdpJ-related Rieske nonheme mononuclear iron naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 (11). Besides showing mono- and dihydroxylations of aromatic compounds, naphthalene dioxygenase is also catalyzing double-bond formations (26). On the other hand, diiron center-containing desaturases not only are responsible for double-bond formation in fatty acids but also can catalyze hydroxylations (41). The specificity of catalysis depends mainly on the substrate structure and its mobility at the catalytic site of the enzyme (5, 49). TBA does not possess vicinal carbons allowing hydrogen removal and double-bond formation. Consequently, only hydroxylation can occur, resulting in the diolic reaction product 2-methylpropan-1,2-diol. In contrast, the structure of TAA would allow both hydroxylation and desaturation reactions. Interestingly, MdpJ obviously catalyzes mainly double-bond formation in this alcohol, as TAA was predominantly converted to hemiterpenes by resting cells of strain L108 when pregrown on TBA and incubated in the presence of a translation inhibitor.
Sequence comparisons revealed that MdpJ is quite unique. Thus far, all mdpJ genes and the corresponding proteins detected in pure cultures and enrichments associated with MTBE or TBA degradation were highly similar (3, 38). The amino acid sequences of MdpJ found for strains PM1 and L108, for example, showed 97% identity. A BLAST search with the complete MdpJ sequence of strain PM1 as a query against the NCBI nonredundant database (November 2011) resulted in only a couple of MdpJ-like proteins as closest matches, with about 60% identity to the PM1 sequence. These predicted proteins all belong to strains of Bordetella parapertussis, Bordetella bronchiseptica, and Achromobacter xylosoxidans, which are known to cause respiratory diseases and other infections but have not often been associated with the degradation of fuel oxygenate ethers or tertiary alcohols. Hence, the origin of MdpJ remains quite enigmatic. However, at least A. xylosoxidans strains 6A and MCM1/1 were recently demonstrated to degrade MTBE (3a, 46).
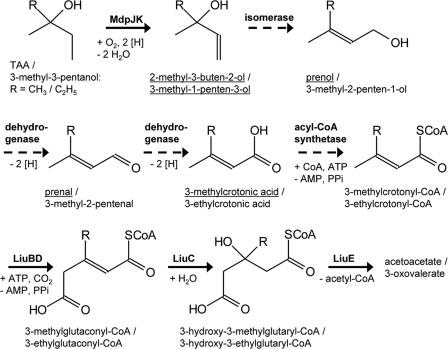
The initial attack of tertiary alcohols seems to be the result of recent evolutionary processes referred to as the significant release of fuel oxygenates into the environment in the last decades. However, all other enzymatic steps downstream from MdpJ catalysis required for TAA mineralization are related to metabolic sequences already established for a long time in nature for the degradation of nonxenobiotic compounds. As proposed previously by Malone and coworkers (28), the product of TAA desaturation, the unsaturated tertiary alcohol 2-methyl-3-buten-2-ol, is likely catabolized via the primary alcohol prenol and its corresponding aldehyde prenal (Fig. 8). The latter is then oxidized to 3-methylcrotonic acid. This metabolic sequence would require an allylic rearrangement previously shown for the interconversion of the monoterpenes linalool and geraniol (13). Recently, the enzyme responsible for the rearrangement in the bacterium Castellaniella defragrans strain 65Phen was identified (6). Besides isomerization, this so-called linalool dehydratase-isomerase also catalyzes the dehydration of linalool to myrcene, very similar to the allylic rearrangement and dehydration of unsaturated tertiary alcohols catalyzed by plant monoterpene cyclases (48). As the relevant structures in both hemi- and monoterpenes are identical, it could be that in all these cases, the same catalytic mechanism is employed. As already described for monoterpene cyclases, a uniform cationic intermediate which is formed after the removal of the hydroxyl group could be postulated (see Fig. S8 in the supplemental material). In the case of unsaturated alcohols, stabilization by mesomerism can be observed. Either deprotonation or capture by water then resulted in elimination or isomerization, respectively. With saturated alcohols, on the other hand, only the elimination reaction is possible. This dehydratase function of the postulated 2-methyl-3-buten-2-ol isomerase would explain the alkene formation from TBA and TAA observed previously for strains L108 and PM1 (38) and also the isoamylene and isoprene formation found in this study. However, conversion of saturated tertiary alcohols and hemiterpenes has not yet been tested with the enzyme from strain 65Phen or with terpene cyclases. In addition, a BLAST search based on the complete genome sequence of strain PM1 clearly confirms that a sequence homologous to the linalool dehydratase-isomerase is not present.
Fig 8.
New proposal for a bacterial degradation pathway for the tertiary alcohols TAA and 3-methyl-3-pentanol involving an initial desaturation step catalyzed by the Rieske nonheme mononuclear iron oxygenase MdpJ and its reductase, MdpK. Underlined compounds indicate metabolites detected by GC mass spectrometry analysis. Broken lines indicate postulated but not-yet-characterized enzymatic reactions (28).
The second step in hemiterpene catabolism (Fig. 8), the prenol-oxidizing activity, was described previously for NAD-dependent benzyl alcohol dehydrogenase-like enzymes found in Pseudomonas putida strains MB-1 and mt-2 as well as in Acinetobacter calcoaceticus NCIB 8250 (28). All these enzymes show rather broad substrate specificities for both allylic and aromatic alcohols, indicating that a hemiterpene-specific dehydrogenase is not required for oxidizing prenol to prenal. Nevertheless, prenol dehydrogenase activity was highly induced in strain MB-1 when cells were grown on 2-methyl-3-buten-2-ol (28). Likewise, prenol did not accumulate when TAA-degrading cells of strain L108 were pregrown on TAA or 2-methyl-3-buten-2-ol, indicating that either TAA itself or the resulting hemiterpene alcohols can act as an inducer. The final oxidation to 3-methylcrotonic acid has not been characterized so far. However, at least prenal dehydrogenase activity was detected in strain MB-1 (28). In summary, although the metabolic sequence from 2-methyl-3-buten-2-ol via prenol and prenal to 3-methylcrotonic acid is consistent with the observed accumulation of metabolites, the identity of the enzymes involved in bacterial hemiterpene catabolism remains unclear. On the basis of our findings and considering the substantial bioinformatic data already available for strains L108 and PM1, it now seems to be worthwhile to elucidate the biochemistry of hemiterpene degradation in these fuel oxygenate-metabolizing bacteria.
2-Methyl-3-buten-2-ol is part of the complex mixture of volatile organic compounds released by biota, mainly by plants (12), and involved in the tropospheric organic aerosol formation influencing the world climate (7, 22). Moreover, it has been found that this hemiterpene can be the dominating nonmethane volatile organic compound emitted, e.g., by a North American pine forest (17, 23). Currently, P. putida MB-1 and strains L108 and PM1 are the only bacteria known to be capable of using this hemiterpene as the sole source of carbon and energy (28). However, considering the presence of 2-methyl-3-buten-2-ol and other hemiterpenes in the environment for millions of years, hemiterpene degradation is likely widespread, and an efficient bacterial degradation pathway could have evolved. Consequently, linking the conversion of the truly xenobiotic TAA with an already well-established hemiterpene catabolism by the desaturase activity of MdpJ is straightforward, whereas the degradation of hydroxylation products would require the invention of several novel enzymatic steps (Fig. 1).
The product of prenal oxidation, 3-methylcrotonic acid, could be metabolized via the leucine degradation route. This would require activation to the corresponding CoA ester, putatively by an AMP-forming acyl-CoA synthetase (Fig. 8). 3-Methylcrotonyl-CoA is then carboxylated (LiuBD) to 3-methylglutaconyl-CoA. After the addition of water (LiuC), the resulting 3-hydroxy-3-methylglutaryl-CoA is split (LiuE) to acetoacetate and acetyl-CoA. In addition, the involvement of the biotin-dependent carboxylase LiuBD in the TAA degradation pathway is indicated by the biotin dependence observed for strain L108. In line with a rising demand for this vitamin when employing a biotin-dependent enzymatic step in a dissimilatory route, biotin-auxotrophic strain L108 cannot grow on TAA in MSM containing only about 8 ng liter−1 biotin, while TBA is still used as a growth substrate under these conditions.
It is likely that MdpJ is also employed for tertiary alcohol degradation in strain PM1 (18). However, it is quite surprising that this strain shows a temporary accumulation of TAA when grown on TAME. The nearly identical mdpJ gene environments in strains PM1 and L108, likely due to a recent horizontal gene transfer event, imply similar enzymatic activities for TAA conversion. Consequently, differences in the degradation pathways more likely exist downstream from the MdpJ activity. Accordingly, a comparison of the liu operons in strains PM1 and L108 revealed low levels of similarity regarding the sequence, length, and number of genes, indicating only a distant phylogenetic relationship. The leucine degradation pathway belongs to the branched-chain amino acid catabolism present in many bacteria (21). In the case of betaproteobacteria, the activation of the liu genes is normally regulated by LiuR and/or LiuQ, belonging to the MerR and TetR families of transcriptional regulators, respectively. As was reported previously (21), in strain PM1, the operon is under the control of LiuQ and consists of the degradation genes liuABCDE. In strain L108, on the other hand, two additional genes not present in strain PM1 are found directly upstream from liuA, encoding a transcription factor of the AraC family and a putative acyl-CoA synthetase (Fig. 7). A similar genetic organization is also present in the betaproteobacterium Thauera sp. strain MZ1T. Possibly, the AraC-like transcriptional factor is better suited to activate the genes of the liu operon when not leucine but only its catabolites are supplied as the sole carbon source. In addition, in a pathway running via the free 3-methylcrotonic acid (Fig. 8), a specific 3-methylcrotonyl-CoA synthetase is required, which is not necessary when the catabolic route starts from leucine. The putative acyl-CoA synthetase encoded by the additional gene found in the liu operon of strain L108 may play this role, likely resulting in a more efficient degradation of leucine catabolites.
Tertiary alcohols, such as TBA and TAA, are rarely found in nature. However, anthropogenic sources do exist and tend to pollute the environment. Besides fuel oxygenate ether degradation, the hydroxylation of branched-chain alkanes, such as isobutane, 2-methylbutane, and higher homologues, at the tertiary carbon position by various monooxygenases may lead to tertiary alcohol formation (9, 19, 33). In this context, it can also be speculated whether MdpJ or similar enzymes might be suitable for the desaturation of larger aliphatic compounds bearing a tertiary alcohol group, e.g., the C9 alcohols 2,3,5-trimethyl-2-hexanol, 3,6-dimethyl-3-heptanol, and 2-methyl-2-octanol, formed in the course of the bacterial degradation of nonylphenols, which are widely used as surfactants in cleaning products (15, 16). At least the C6 alcohol 3-methyl-3-pentanol can be attacked by MdpJ and is converted to the expected unsaturated tertiary alcohol. Moreover, strain L108 is also able to use it as the sole source of carbon and energy, suggesting that not only MdpJ but also the enzymes involved in hemiterpene and leucine catabolism are able to process molecules larger than C5.
Our finding that TAA and the higher homologue 3-methyl-3-pentanol are not hydroxylated to the corresponding diols but that unsaturated alcohols are formed might be surprising at first sight. However, by linking the desaturase reaction with hemiterpene and branched-chain amino acid catabolism, an efficient linear degradation pathway has evolved. The alternative hydroxylation can lead to a significant number of metabolites, including stereoisomers (Fig. 1). Consequently, a large number of enzymatic steps would be required for the complete mineralization of the tertiary alcohols. This branching of the degradation pathway can be prevented only by employing highly specific enzymatic catalysts. However, especially at the beginning of pathway evolution, when only enzymes not well adapted to a new substrate can be recruited, it is unlikely that the catalysis of a highly selective monooxygenase could be employed, resulting in only one hydroxylation product. In a theoretic study based on the YATP concept, we have already shown that fuel oxygenates, such as MTBE and ETBE, are formally good growth substrates, allowing high theoretical biomass yields (30). However, when including an extended Monod equation, it became obvious that low growth rates would result in maintenance requirements too high for supporting productive degradation. In this connection, it is quite consistent that not a complicated pathway with several branched metabolic sequences has evolved for degrading TAME and TAEE metabolites but only a superior linear route via hemiterpenes is employed in A. tertiaricarbonis L108 and likely also in M. petroleiphilum PM1.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the UFZ within the CITE program. We are grateful to the DBU (Deutsche Bundesstiftung Umwelt) for financial support of F.S. (AZ: 20008/994).
We thank C. Schumann (UFZ) and M. Neytschev (UFZ) for technical assistance and B. Würz (UFZ) for excellent analytical advice.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amberg A, Rosner E, Dekant W. 2000. Biotransformation and kinetics of excretion of tert-amyl-methyl ether in humans and rats after inhalation exposure. Toxicol. Sci. 55:274–283 [DOI] [PubMed] [Google Scholar]
- 3. Aslett D, Haas J, Hyman M. 2011. Identification of tertiary butyl alcohol (TBA)-utilizing organisms in BioGAC reactors using 13C-DNA stable isotope probing. Biodegradation 22:961–972 [DOI] [PubMed] [Google Scholar]
- 3a. Basbera MJ, Mateo E, Monkaityte R, Constanti M. 2011. Biodegradation of methyl tert-butyl ether by newly identified soil microorganisms in a simple mineral solution. World J. Microbiol. Biotechnol. 27:813–821 [Google Scholar]
- 4. Bastida F, et al. 2010. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol. Ecol. 73:370–384 [DOI] [PubMed] [Google Scholar]
- 5. Behrouzian B, Buist PH. 2002. Fatty acid desaturation: variations on an oxidative theme. Curr. Opin. Chem. Biol. 6:577–582 [DOI] [PubMed] [Google Scholar]
- 6. Brodkorb D, Gottschall M, Marmulla R, Lüddeke F, Harder J. 2010. Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J. Biol. Chem. 285:30436–30442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan AW, et al. 2009. Photooxidation of 2-methyl-3-buten-2-ol (MBO) as a potential source of secondary organic aerosol. Environ. Sci. Technol. 43:4647–4652 [DOI] [PubMed] [Google Scholar]
- 8. Chauvaux S, et al. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubbels BL, Sayavedra-Soto L, Arp DJ. 2007. Butane monooxygenase of ‘Pseudomonas butanovora’: purification and biochemical characterization of a terminal-alkane hydroxylating diiron monooxygenase. Microbiology 153:1808–1816 [DOI] [PubMed] [Google Scholar]
- 10. Fayolle F, Vandecasteele J-P, Monot F. 2001. Microbial degradation and fate in the environment of methyl tert-butyl ether and related fuel oxygenates. Appl. Microbiol. Biotechnol. 56:339–349 [DOI] [PubMed] [Google Scholar]
- 11. Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175–190 [DOI] [PubMed] [Google Scholar]
- 12. Fisher AJ, Baker BM, Greenberg JP, Fall R. 2000. Enzymatic synthesis of methylbutenol from dimethylallyl diphosphate in needles of Pinus sabiniana. Arch. Biochem. Biophys. 383:128–134 [DOI] [PubMed] [Google Scholar]
- 13. Foss S, Harder J. 1997. Microbial transformation of a tertiary allylalcohol: regioselective isomerization of linalool to geraniol without nerol formation. FEMS Microbiol. Lett. 149:71–75 [Google Scholar]
- 14. Fraile J, et al. 2002. Monitoring of the gasoline oxygenate MTBE and BTEX compounds in groundwater in Catalonia (Northeast Spain). Sci. World J. 2:1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gabriel FLP, et al. 2005. A novel metabolic pathway for degradation of 4-nonylphenol environmental contaminants by Sphingomonas xenophaga Bayram. J. Biol. Chem. 280:15526–15533 [DOI] [PubMed] [Google Scholar]
- 16. Gabriel FLP, Giger W, Guenther K, Kohler H-PE. 2005. Differential degradation of nonylphenol isomers by Sphingomonas xenophaga Bayram. Appl. Environ. Microbiol. 71:1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldan PD, Kuster WC, Fehsenfeld FC, Montzka SA. 1993. The observation of a C5 alcohol in a North American pine forest. Geophys. Res. Lett. 20:1039–1042 [Google Scholar]
- 18. Hristova KR, et al. 2007. Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol. Appl. Environ. Microbiol. 73:7347–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imai T, et al. 1986. Microbial oxidation of hydrocarbons and related compounds by whole-cell suspensions of the methane-oxidizing bacterium H-2. Appl. Environ. Microbiol. 52:1403–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reference deleted.
- 21. Kazakov AE, et al. 2009. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J. Bacteriol. 191:52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiendler-Scharr A, et al. 2009. New particle formation in forests inhibited by isoprene emissions. Nature 461:381–384 [DOI] [PubMed] [Google Scholar]
- 23. Kim S, et al. 2010. Emissions and ambient distributions of biogenic volatile organic compounds (BVOC) in a ponderosa pine ecosystem: interpretation of PTR-MS mass spectra. Atmos. Chem. Phys. 10:1759–1771 [Google Scholar]
- 24. Kolb A, Püttmann W. 2006. Comparison of MTBE concentrations in groundwater of urban and nonurban areas in Germany. Water Res. 40:3551–3558 [DOI] [PubMed] [Google Scholar]
- 25. Lechner U, et al. 2007. Aquincola tertiaricarbonis gen. nov., sp. nov., a tertiary butyl moiety-degrading bacterium. Int. J. Syst. Evol. Microbiol. 57:1295–1303 [DOI] [PubMed] [Google Scholar]
- 26. Lee K, Gibson DT. 1996. Toluene and ethylbenzene oxidation by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62:3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopes Ferreira N, Malandain C, Fayolle-Guichard F. 2006. Enzymes and genes involved in the aerobic biodegradation of methyl tert-butyl ether (MTBE). Appl. Microbiol. Biotechnol. 72:252–262 [DOI] [PubMed] [Google Scholar]
- 28. Malone VF, et al. 1999. Characterization of a Pseudomonas putida allylic alcohol dehydrogenase induced by growth on 2-methyl-3-buten-2-ol. Appl. Environ. Microbiol. 65:2622–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran MJ, Zogorski JS, Squillace PJ. 2005. MTBE and gasoline hydrocarbons in ground water of the United States. Ground Water 43:615–627 [DOI] [PubMed] [Google Scholar]
- 30. Müller RH, Rohwerder T, Harms H. 2007. Carbon conversion efficiency and limits of productive bacterial degradation of methyl tert-butyl ether and related compounds. Appl. Environ. Microbiol. 73:1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Müller RH, Rohwerder T, Harms H. 2008. Degradation of fuel oxygenates and their main intermediates by Aquincola tertiaricarbonis L108. Microbiology 154:1414–1421 [DOI] [PubMed] [Google Scholar]
- 32. Nakatsu CH, et al. 2006. Methylibium petroleiphilum PM1T gen. nov., sp. nov., a new methyl tert-butyl ether (MTBE) degrading methylotroph of the beta-Proteobacteria. Int. J. Syst. Evol. Microbiol. 56:983–989 [DOI] [PubMed] [Google Scholar]
- 33. Patel RN, Hou CT, Laskin AI, Felix A. 1982. Microbial oxidation of hydrocarbons: properties of a soluble methane monooxygenase from a facultative methane-utilizing organism, Methylobacterium sp. strain CRL-26. Appl. Environ. Microbiol. 44:1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poitrat E. 1999. The potential of liquid biofuels in France. Renew. Energy 16:1084–1089 [Google Scholar]
- 35. Rohwerder T, Breuer U, Benndorf D, Lechner U, Müller RH. 2006. The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72:4128–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salanitro JP. 1995. Understanding the limitations of microbial metabolism of ethers used as fuel octane enhancers. Curr. Opin. Biotechnol. 6:337–340 [Google Scholar]
- 37. Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schäfer F, et al. 2011. Alkene formation from tertiary alkyl ether and alcohol degradation by Aquincola tertiaricarbonis L108 and Methylibium spp. Appl. Environ. Microbiol. 77:5981–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schäfer F, et al. 2007. Growth of Aquincola tertiaricarbonis L108 on tert-butyl alcohol leads to the induction of a phthalate dioxygenase-related protein and its associated oxidoreductase subunit. Eng. Life Sci. 7:512–519 [Google Scholar]
- 40. Schmidt R, Battaglia V, Scow K, Kane S, Hristova KR. 2008. Involvement of a novel enzyme, MdpA, in methyl tert-butyl ether degradation in Methylibium petroleiphilum PM1. Appl. Environ. Microbiol. 74:6631–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shanklin J, Guy JE, Mishra G, Lindqvist Y. 2009. Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J. Biol. Chem. 284:18559–18563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steffan RJ, McClay K, Vainberg S, Condee CW, Zhang D. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sumner SCJ, et al. 2003. Characterization of metabolites and disposition of tertiary amyl methyl ether in male F344 rats following inhalation exposure. J. Appl. Toxicol. 23:411–417 [DOI] [PubMed] [Google Scholar]
- 44. US Environmental Protection Agency 1997. Drinking water advisory: consumer acceptability advice and health effects analysis on methyl tertiary butyl ether (MTBE). EPA-822-F-97-008 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 45. van Wezel A, Puijker L, Vink C, Versteegh A, de Voogt P. 2009. Odour and flavour thresholds of gasoline additives (MTBE, ETBE and TAME) and their occurrence in Dutch drinking water collection areas. Chemosphere 76:672–676 [DOI] [PubMed] [Google Scholar]
- 46. Vosahlikova-Kolarova M, Krejcik Z, Cajthaml T, Demnerova K, Pazlarova J. 2008. Biodegradation of methyl tert-butyl ether using bacterial strains. Folia Microbiol. (Praha) 53:411–416 [DOI] [PubMed] [Google Scholar]
- 47. Weaver JW, Exum LR, Prieto LM. 2010. Gasoline composition regulations affecting LUST sites. EPA 600/R-10/001 Office of Research and Development, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 48. Wheeler CJ, Croteau R. 1986. Monoterpene cyclases: use of the noncyclizable substrate analog 6,7-dihydrogeranyl pyrophosphate to uncouple the isomerization step of the coupled isomerization-cyclization reaction. Arch. Biochem. Biophys. 246:733–742 [DOI] [PubMed] [Google Scholar]
- 49. Whittle EJ, Tremblay AE, Buist PH, Shanklin J. 2008. Revealing the catalytic potential of an acyl-ACP desaturase: tandem selective oxidation of saturated fatty acids. Proc. Natl. Acad. Sci. U. S. A. 105:14738–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.