Abstract
Elevated levels of mucins present in bronchiectatic airways predispose patients to bacterial infections and reduce the effectiveness of antibiotic therapies by directly inactivating antibiotics. Consequently, new antibiotics that are not inhibited by mucins are needed to treat chronic respiratory infections caused by Pseudomonas aeruginosa and Staphylococcus aureus. In these studies, we demonstrate that fosfomycin synergistically enhances the activity of tobramycin in the presence of mucin. The bactericidal killing of a novel 4:1 (wt/wt) combination of fosfomycin-tobramycin (FTI) is superior (>9 log10 CFU/ml) relative to its individual components fosfomycin and tobramycin. Additionally, FTI has a mutation frequency resulting in an antibiotic resistance >3 log10 lower than for fosfomycin and 4 log10 lower than for tobramycin for P. aeruginosa. Mechanistic studies revealed that chemical adducts are not formed, suggesting that the beneficial effects of the combination are not due to molecular modification of the components. FTI displayed time-kill kinetics similar to tobramycin and killed in a concentration-dependent fashion. The bactericidal effect resulted from inhibition of protein biosynthesis rather than cell wall biosynthesis. Studies using radiolabeled antibiotics demonstrated that tobramycin uptake was energy dependent and that fosfomycin enhanced the uptake of tobramycin in P. aeruginosa in a dose-dependent manner. Lastly, mutants resistant to fosfomycin and tobramycin were auxotrophic for specific carbohydrates and amino acids, suggesting that the resistance arises from mutations in specific active transport mechanisms. Overall, these data demonstrate that fosfomycin enhances the uptake of tobramycin, resulting in increased inhibition of protein synthesis and ultimately bacterial killing.
INTRODUCTION
The airways of cystic fibrosis (CF) and non-CF bronchiectasis patients are restricted by sputum, a thick viscous secretion containing elevated levels of mucin, DNA from inflammatory cells, filamentous actin (F-actin), lipids, and peptides (16, 24, 47). The altered biophysical properties of sputum impair mucocilliary clearance, resulting in airway obstructions, and predispose the patients to bacterial infections (47). Whole sputum (18, 43) and individual components of sputum, including mucin, DNA, and F-actin bundles, also reduce the activity of cationic antibiotics and peptides (7, 42), thereby contributing to development of bacterial resistance (29, 32). Furthermore, microscopic analysis of sputum from CF patients demonstrates that Pseudomonas aeruginosa grows in aggregations or microcolonies and exhibits decreased susceptibility to tobramycin (TOB) due to production of biofilms (4, 26, 53). Development of antibiotic resistance is clearly a concern for patients with bronchiectasis. New antibiotic therapies with improved activity in the lung environment are needed to treat chronic airway infections.
Fosfomycin-tobramycin (FTI) is an antibiotic combination consisting of a 4:1 (wt/wt) ratio of fosfomycin (FOF) and tobramycin (28). It has broad-spectrum in vitro activity against both Gram-negative and Gram-positive bacterial pathogens commonly found in CF and non-CF bronchiectasis patients, including methicillin-resistant Staphylococcus aureus (MRSA). FTI is also highly active in vivo. In a chronic rat lung infection model with P. aeruginosa, twice-a-day (BID) aerosol treatment with FTI achieved complete eradication of the infection (>6 log10 CFU/lung) after 3 days. Furthermore, the in vivo studies demonstrated that FTI had >1 log10 killing relative to the component weights of fosfomycin and tobramycin and confirmed the enhanced antibacterial activities of FTI observed in the present in vitro study. Together, these data suggest that FTI may be an appropriate aerosol antibiotic therapy for CF and non-CF bronchiectasis patients.
Inhalation of nebulized solutions of antibiotics delivers high concentrations of drug directly to the site of infection. However, the physiology of normal and diseased lung inhibits antibiotic activity, thus reducing the effectiveness of the therapy. Standardized methods for evaluating antibiotics are often not predictive of drug activity in the lung environment. Mucin, which is present in large quantities in bronchiectatic airways, has been used as an in vitro tool to study the bioavailability of antibiotics and to evaluate the mucolytic activity of expectorants (10, 48, 62). We used mucin as an in vitro model of airway sputum to provide additional insights for evaluating activities of antibiotics in the lung environment. Purified porcine gastric mucin (PGM) was used as the source of mucin in this research, because it has a glycoprotein-carbohydrate composition similar to that of airway mucus, namely, high levels of MUC5AC and MUC5b (23, 41). Additionally, we have used this model to optimize various fosfomycin-aminoglycoside combinations, eventually leading to the selection of the 4:1 (wt/wt) ratio of fosfomycin-tobramycin currently in phase II clinical trials.
Our studies demonstrated that fosfomycin, the major component of FTI, enhanced the activity of tobramycin against P. aeruginosa in mucin and significantly reduced the development of antibiotic resistance. Mechanistic studies revealed that the enhancement arose from increased uptake of tobramycin, resulting in increased inhibition of protein synthesis and bactericidal killing. These data strongly suggest that the improved killing kinetics of FTI will minimize development of clinical antibiotic resistance. Lastly, our studies suggested that tobramycin is transported into P. aeruginosa by active transport systems designed to transport specific carbohydrates and amino acids (27).
(This work was presented in part at the American Thoracic Society Conference, Toronto, Ontario, Canada, 16 to 21 May 2008, and the 22nd Annual North American Cystic Fibrosis Conference, Orlando, FL, 23 to 25 October 2008.)
MATERIALS AND METHODS
Bacterial strains and antibiotics.
P. aeruginosa ATCC 27853 and S. aureus ATCC 29213 (American Type Culture Collection, Manassas, VA) were used in antibiotic susceptibility, single-step resistance, and mechanistic studies. P. aeruginosa PAO-1 parent strain and PAO-1 ISlacZ/hah or ISphoA/hah transposon mutants (PA0427, PA2018, PA2019, PA2020, PA2291, PA3186, and PA5235) were obtained from Colin Manoil, University of Washington, Seattle, WA (19). Fosfomycin disodium and tobramycin sulfate were obtained from Sigma-Aldrich (St. Louis, MO). FTI consisted of a 4:1 ratio (wt/wt basis) of fosfomycin and tobramycin. Glucose-6-phosphate (Sigma-Aldrich) was added to the bacteriological culture medium at a final concentration of 25 μg/ml for all evaluations of fosfomycin and FTI (22, 23) but not tobramycin. The enantiomer of fosfomycin, (+)-(1S,2R)-epoxypropylphosphonic acid disodium salt (Cy-230), was synthesized by a modification of Glamkowski incorporating a crystallization step with (−)-Ü-phenethylamine to isolate the desired isomer (15).
FTI susceptibility studies.
MICs were determined according to CLSI standards or by a modified agar plate dilution method (35, 36) that incorporated 20 g/liter of purified porcine gastric mucin (PGM; Sigma-Aldrich) in Mueller-Hinton agar (MHA; BBL, Sparks, MD). The MIC was defined as the lowest concentration of antibiotic that prevented visible growth after incubation at 35°C for 18 to 24 h.
Time-kill experiments were performed according to a modified CLSI method (37). Antibiotics were evaluated alone and in combination at multiples of the MIC in cation-adjusted Mueller-Hinton broth (CAMHB) (Remel; Lenexa, KS) containing 20 g/liter PGM. Bacterial cultures and antibiotic(s) were incubated at 37°C in a shaking water bath (200 rpm), and viability was assessed by the plate count method at 0, 1, 2, 4, 6, and 24 h. A no-drug control was run in each assay. Antibiotics that reduced the original inoculum by ≥3 log10 were considered bactericidal. Antibiotics that reduced the original inoculum by ≤2 log10 were considered bacteriostatic.
Single-step resistance to FTI.
Development of resistance after a single exposure to antibiotic was determined for P. aeruginosa ATCC 27853. Late-log-phase cultures containing 109 to 1010 CFU were spread onto MHA (BBL) plates containing 20g/liter PGM and 4×, 8×, or 16× the MIC of FTI, fosfomycin, or tobramycin, respectively. The culture plates were incubated at 35°C for 48 h, and the number of colonies on each plate was enumerated manually. The frequency of resistance was calculated by dividing the number of bacteria growing at the defined antibiotic concentration by the number of bacteria in the inoculum (29).
Fosfomycin-tobramycin chemical adducts.
FTI (512 μg/ml) was prepared in phosphate-buffered saline (PBS; pH 7.4) and incubated for 24 h at room temperature (RT), 37°C, or 89°C. An Agilent 1100 series high-performance liquid chromatography (HPLC) system equipped with a liquid chromatography/mass selective detector (LC/MSD) ion trap mass spectrometer and ChemStation data acquisition/data analysis software (Agilent Technologies, Santa Clara, CA) was used to detect potential chemical adducts of fosfomycin and tobramycin. Peak separation was effected using a SymmetryShield RP18 analytical column, 4.6-mm internal diameter (i.d.) by 150-mm length, with 3.5-μm packing (Waters Corporation, Milford, MA). The samples were eluted with 5% glacial acetic acid and 0.25% pentafluoropropionic acid (PFPA) in water as mobile phase A, and 5% glacial acetic acid and 0.25% PFPA in acetonitrile as mobile phase B. An elution gradient was applied from 0% to 34% mobile phase B over 25 min. Peaks were eluted directly from the column into the electrospray ionization source of the ion trap mass spectrometer. Ionization was in positive mode, using nitrogen as a drying gas at 10 liters/min and 350°C. Mass spectra were acquired over a range of 150 to 1,300 m/z. Antibacterial activities of FTI incubated at RT, 37°C, or 89°C were evaluated by MIC.
Inhibition of protein and cell wall biosynthesis.
Protein and cell wall peptidoglycan biosyntheses were determined by measuring the incorporation of a tritiated (3H) l-amino acid mixture (Ala, Arg, Asp, Glu, Gly, His, Ile, Leu, Lys, Phe, Pro, Ser, Thr, Tyr, Val; GE Healthcare Bio-Sciences Corp., Piscataway, NJ; catalog no. TRK440; 1.8 TBq/mmol) and 3H-labeled N-acetylglucosamine ([3H]NAG; GE Healthcare Bio-Sciences; catalog no. TRK376; 296 GBq/mmol), respectively (3, 50). An overnight culture of P. aeruginosa ATCC 27853 was diluted 1:1,000 in 50 ml CAMHB and 20 g/liter PGM in a 125-ml Erlenmeyer flask and incubated at 37°C, with shaking (200 rpm) for 1.5 h. Two milliliters of early-log-phase cultures (∼2 × 107 CFU/ml) were pulsed with 10 μCi of tritiated amino acids (3H-aa) (1.93 GBq/milliatom carbon) or 10 μCi of [3H]NAG (296 GBq/mmol) for 1 h at 37°C, 200 rpm. FTI, fosfomycin, or tobramycin were then added to cultures and incubated as described above for up to an additional 4 h. At various time points, 100-μl aliquots (triplicate) of culture were removed, and macromolecules were precipitated with 10% trichloroacetic acid (TCA) (VWR International, Radnor, PA). All samples were treated identically to reduce variation in quenching. Precipitates were harvested onto glass fiber filters (GFC) (PerkinElmer, Waltham, MA), washed two times with 35 ml of normal saline to remove unincorporated isotope, followed by one wash with 35 ml of 90% ethanol (VWR). Counts per minute (CPM) were determined using a Wallac MicroBeta TriLux (PerkinElmer).
Tobramycin uptake.
Antibiotic uptake was determined by measuring incorporation of [3H]tobramycin (540 mCi/mmol; Moravek Biochemicals, Brea, CA). An overnight culture of P. aeruginosa ATCC 27853 was diluted in nutrient broth (NB) (Difco & BBL, Sparks, MD) to an optical density at 625 nm (OD625) of 0.013 and incubated at 37°C with shaking (250 rpm) until it reached an OD625 of ∼0.5. Cells were harvested by centrifugation (6,000 × g, room temperature, 5 min), washed once in NB, and resuspended in prewarmed NB to an OD625 of 0.25. Unlabeled fosfomycin was added at 0, 0.05, 0.1, 1, 10, and 100 μg/ml, and the cultures were incubated for 3 min at 37°C with shaking (250 rpm). [3H]tobramycin (2.3 μg/ml) was added to each tube, and the cultures were incubated at 37°C with shaking (250 rpm) for an additional 2 min. Five-milliliter volumes were filtered through 0.45-μm-pore-size nitrocellulose membrane filters (Whatman Inc., Florham Park, NJ), presoaked with 410 mM MgCl2 (VWR), and CPM was determined with a MicroBeta scintillation counter. All samples were treated identically to reduce variation in quenching. The effect of electron transport uncouplers and divalent cations on [3H]-tobramycin uptake was determined by incorporating 1 mM sodium cyanide (Sigma-Aldrich) or 1 mM MgCl2 (Sigma-Aldrich) into the NB 3 min prior to the addition of [3H]tobramycin as described above.
Carbon and nitrogen auxotrophies of fosfomycin and tobramycin-resistant mutants.
Late-log-phase cultures (109 to 1010 CFU) of P. aeruginosa ATCC 27853 and PAO-1 were spread onto Mueller-Hinton agar (BBL, Sparks, MD) plates containing 4×, 8×, or 16× the MIC of FTI, fosfomycin, or tobramycin, respectively (1, 3, 5, 37). The culture plates were incubated at 37°C for 48 h, and individual colonies were purified by streaking to isolation. MIC values of representative mutants were compared to the parent strains to confirm increases in MIC. Spontaneous mutants having decreased susceptibility to fosfomycin and tobramycin were evaluated for their ability to utilize a variety of carbohydrates and amino acids as sole carbon and nitrogen sources (1, 3, 5, 25, 37). Single colonies were inoculated into normal saline and adjusted to 1 × 108 CFU/ml. Suspensions (1 × 104 CFU) were replica plated using a 48-pin replica plater (Sigma-Aldrich) onto M9 (Becton Dickinson and Company, Sparks, MD) agar plates supplemented with MgSO4 (2 mM), CaCl2 (0.1 mM), and 5 g/liter carbohydrate or 5 g/liter amino acid. Plates were incubated at 37°C for 48 h, and growth was assessed visually. Carbohydrates: Fru, d-fructose; Glc, d-glucose; G3P, sn-glycerol-3-phosphate; Gly, glycerol; Mtl, d-mannitol; NAG, N-acetylglucosamine; Rib, d-ribose; Suc, succinate; G6P, d-glucose-6-phosphate; F6P, d-fructose-6-phosphate; Spm, spermidine; Put, putrescine. l-Amino acids: Ala, alanine; Gly, glycine; Pro, proline; Trp, tryptophan, Asn, asparagine; Gln, glutamine; Asp, aspartic acid; Glu, glutamate; Lys, lysine; Arg, arginine; His, histidine.
RESULTS
FTI susceptibility studies.
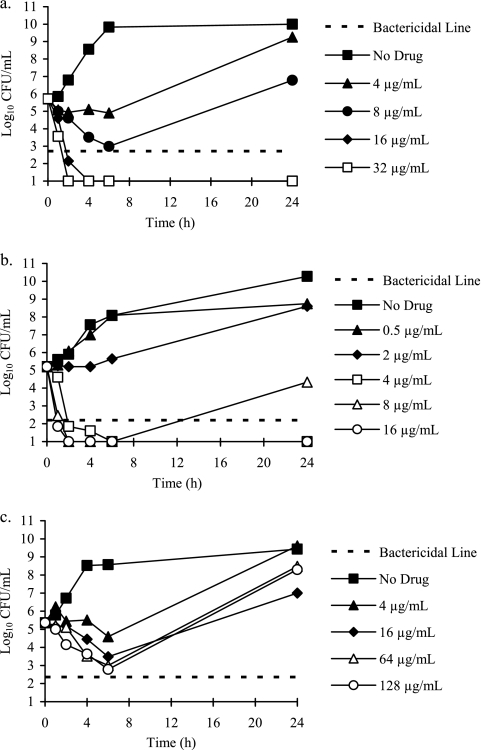
We studied the effects of mucin, a major component of normal airway mucus and sputum, on the antibacterial activities of a novel fosfomycin-tobramycin combination, because mucin is known to reduce the effectiveness of cationic antibiotics used to treat respiratory infections in CF and non-CF bronchiectasis patients (30, 43). Time-kill experiments conducted in the presence of mucin demonstrated that both FTI and tobramycin were rapidly bactericidal and exhibited concentration-dependent killing against P. aeruginosa. Increasing the concentrations of FTI and tobramycin increased both the rate and extent of bacterial killing (Fig. 1a and b). By comparison, fosfomycin was bacteriostatic for P. aeruginosa and demonstrated time-dependent killing (Fig. 1c).
Fig 1.
Time-kill curves for P. aeruginosa 27853 showing concentration-dependent killing by FTI (a) and tobramycin (b) and time-dependent killing by fosfomycin (c). Antibiotics were evaluated at multiples of the MIC in CAMHB supplemented with 2 g/liter of PGM.
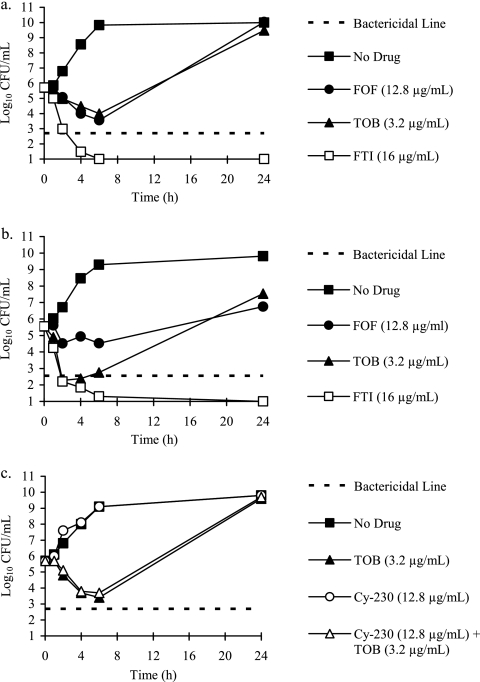
The activity of FTI relative to its component weights of fosfomycin and tobramycin against P. aeruginosa ATCC 27853 is shown in Fig. 2a. At 4× MIC (16 μg/ml), the killing activity of FTI was superior (>9 log10 CFU) relative to its components fosfomycin (12.8 μg/ml) and tobramycin (3.2 μg/ml). FTI rapidly reached bactericidal killing (1 to 2 h), while tobramycin and fosfomycin alone exhibited bacteriostatic killing. Additionally, FTI remained bactericidal at 24 h, while regrowth of the bacterial culture was observed with tobramycin and fosfomycin. A 4:1 (wt/wt) fosfomycin enantiomer (Cy-230)-tobramycin combination did not demonstrate enhanced killing relative to its components (Fig. 2c). FTI also demonstrated improved activity relative to fosfomycin and tobramycin against S. aureus ATCC 29213 (Fig. 2b).
Fig 2.
Fosfomycin enhanced the killing of tobramycin against P. aeruginosa ATCC 27853 (a) and S. aureus ATCC 29213 (b), relative to the component weights of fosfomycin and tobramycin. The fosfomycin enantiomer CY-230 and tobramycin combination did not demonstrate enhanced killing relative to its components against P. aeruginosa (c). Time-kill curves were conducted in CAMHB supplemented with 2 g/liter of PGM. Antibiotics were tested at the following concentrations: fosfomycin, 12.8 μg/ml, plus tobramycin, 3.2 μg/ml (open squares); fosfomycin, 12.8 μg/ml (closed circles); tobramycin, 3.2 μg/ml (closed triangles); fosfomycin enantiomer Cy-230, 12.8 μg/ml, plus tobramycin, 3.2 μg/ml (open triangles); and cy-230, 12.8 μg/ml (open circles). Closed squares, no drug; dotted line, bactericidal killing.
Table 1 shows the spontaneous mutation frequencies (SMF) of P. aeruginosa ATCC 27853 after a single exposure to antibiotic in the presence of mucin. At 16× the MIC, the mutation frequency in the presence of FTI was ≥3 log10 lower than for fosfomycin alone and 4 log10 lower than for tobramycin alone. FTI remained superior to fosfomycin at 8× the MIC but comparable to fosfomycin and tobramycin at 4× the MIC. Tobramycin at 4× and 8× the MIC resulted in confluent lawns.
Table 1.
Spontaneous mutation frequencies of P. aeruginosa ATCC 27853 resulting in development of antibiotic resistance
| Antibiotic | Mucin MIC (μg/ml) | Fold MICa |
||
|---|---|---|---|---|
| 4× | 8× | 16× | ||
| FTI | 4 | 5 × 10−6 | 6 × 10−7 | <5 × 10−9 |
| Fosfomycin | 4 | 7 × 10−6 | 3 × 10−6 | 3 × 10−6 |
| Tobramycin | 0.5 | TNTC | TNTC | 2 × 10−5 |
TNTC, too numerous to count.
Fosfomycin-tobramycin chemical adducts.
The chemical structures of fosfomycin, tobramycin, and five possible adducts are presented in Fig. S1 in the supplemental material. No chemical adducts were detected by HPLC/mass spectrometry (MS) after incubation of FTI at RT or 37°C for 24 h (see Fig. S2 in the supplemental material). Three adducts were formed after incubation at 89°C for 24 h, and the activity of the resulting mixture against P. aeruginosa ATCC 27853 had a 2-fold increase in MIC relative to FTI. Since the three adducts were positional isomers and isobaric, their exact identities were not determined. Incubation of FTI at −20°C, RT, or 37°C did not alter antimicrobial activity.
Inhibition of macromolecular biosynthesis.
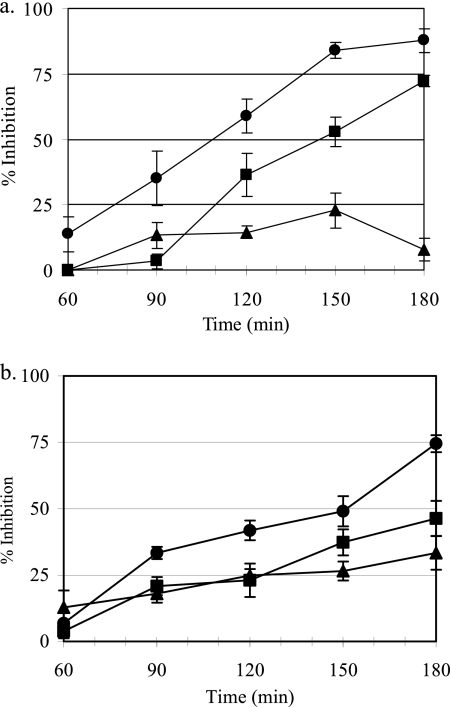
The effects of antibiotic concentration and time of bacterial exposure to antibiotic on protein and cell wall biosynthesis were determined by measuring the uptake of 3H-amino acids and [3H]NAG, respectively. FTI inhibited protein and cell wall biosynthesis to a greater degree than either fosfomycin or tobramycin. Inhibition of protein synthesis by FTI was dose dependent, like that of tobramycin, and occurred at concentrations close to the MIC. Increasing the concentration of FTI also resulted in increased inhibition of cell wall biosynthesis; however, protein biosynthesis was inhibited to a greater degree than cell wall biosynthesis (Table 2). By comparison, increasing the concentration of fosfomycin did not result in increased inhibition of either protein or cell wall biosynthesis. Time-response studies also demonstrated that FTI was acting primarily through inhibition of protein synthesis (Fig. 3a) and correlated well with the onset of bactericidal activity in time-kill studies (Fig. 1a). The inhibition of protein synthesis by FTI at 8 μg/ml was rapid (time to 50% inhibition [T50] = 108 min) compared to 6.4 μg/ml fosfomycin (T50 = 145 min) and 1.6 μg/ml tobramycin (T50 = >180 min). Tobramycin had less activity against its target, protein synthesis, than fosfomycin. This is likely due to the inhibitory action of mucin as well as the concentration of tobramycin used in this experiment. In contrast, FTI (8 μg/ml) caused a more gradual inhibition of cell wall biosynthesis (T50 = 152 min), while neither fosfomycin (6.4 μg/ml) nor tobramycin (1.6 μg/ml) reached 50% inhibition within 180 min (Fig. 3b).
Table 2.
Effects of antibiotic concentration on protein and cell wall synthesis in P. aeruginosa ATCC 27853
| Antibiotic (concn [μg/ml]) | % inhibitiona |
|
|---|---|---|
| Protein | Cell wall | |
| FTI (16) | 92 | 59 |
| FTI (8) | 50 | 26 |
| FTI (4) | 28 | 11 |
| Fosfomycin (12.8) | 9 | 3 |
| Fosfomycin (6.4) | 21 | 14 |
| Fosfomycin (3.2) | 0 | 8 |
| Tobramycin (3.2) | 42 | 36 |
| Tobramycin (1.6) | 6 | 10 |
| Tobramycin (0.8) | 2 | 4 |
Percentage of inhibition at 2 h relative to the no-drug control.
Fig 3.
Time-response studies demonstrating that FTI acts primarily through inhibition of protein synthesis. The inhibitory effects of FTI and the component weights of fosfomycin and tobramycin on protein (a) and cell wall biosynthesis (b) in P. aeruginosa ATCC 27853 grown in CAMHB supplemented with 2 g/liter of PGM. FTI, 8 μg/ml (closed circles); tobramycin, 1.6 μg/ml (closed triangles); and fosfomycin, 6.4 μg/ml (closed squares). Values represent the means ± standard deviations (SD) from four independent experiments.
Tobramycin uptake.
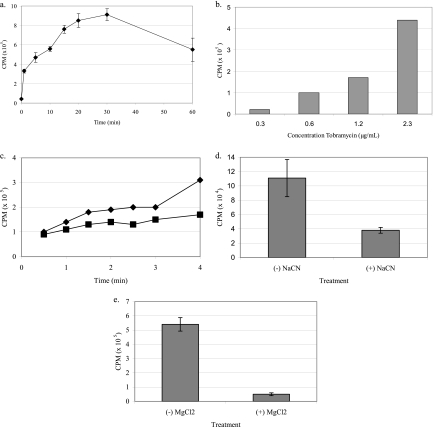
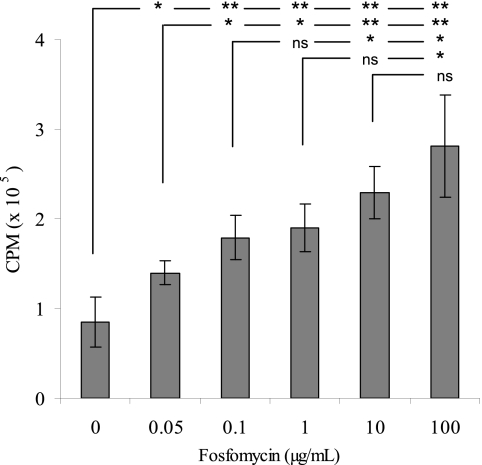
[3H]tobramycin uptake in P. aeruginosa was characterized by determining the effects of (i) time of incubation of antibiotic and bacteria, (ii) antibiotic concentration, (iii) incubation temperature, (iv) electron transport uncouplers, and (v) divalent cations. Uptake of [3H]tobramycin occurred rapidly and in a time (Fig. 4a)- and concentration-dependent fashion (Fig. 4b). The rate and degree of tobramycin uptake decreased with incubation temperature (Fig. 4c). When P. aeruginosa was incubated with 1 mM NaCN, [3H]tobramycin transport was inhibited, demonstrating an energy-dependent transport mechanism (Fig. 4d). Furthermore, [3H]tobramycin uptake was inhibited by the divalent cation Mg2+ (Fig. 4e). Fosfomycin increased the uptake of [3H]tobramycin in P. aeruginosa in a dose-dependent manner (Fig. 5). The addition of 10 μg/ml fosfomycin resulted in a 170% increase in [3H]tobramycin uptake relative to the no-fosfomycin control.
Fig 4.
Active transport characteristics of [3H]tobramycin (2.3 μg/ml) in P. aeruginosa PAO-1. (a) Accumulation occurred in a rapid, time-dependent fashion; (b) uptake at 5 min was concentration dependent; (c) accumulation over 5 min was temperature dependent (cultures grown at 37°C [closed triangle] versus 25°C [closed square]); (d) the electron transport inhibitor NaCN (1 mM) blocked uptake, demonstrating energy dependence; (e) uptake was inhibited by the divalent cation MgCl2 (1 mM).
Fig 5.
Fosfomycin induces uptake of [3H]tobramycin (2.3 μg/ml) in P. aeruginosa ATCC 27853 in a dose-dependent fashion. Incorporation of [3H]tobramycin into bacterial cells was determined 5 min after addition of unlabeled fosfomycin (0, 0.05, 0.1, 1, 10, or 100 μg/ml). Values represent the means ± SD from four independent experiments. Statistical differences were evaluated by Student's t test (*, P < 0.05; **, P < 0.005).
Carbon and nitrogen auxotrophies of fosfomycin- and tobramycin-resistant mutants.
The P. aeruginosa PAO-1 parent strain was able to utilize a wide variety of carbohydrates (Table 3) and l-amino acids (Table 4) as sole carbon and nitrogen sources. Two different classes of tobramycin-resistant mutants were identified in this study. C400 was able to utilize glucose, succinate, and putrescine but was auxotrophic to fructose, glycerol-3-phosphate, glycerol, mannitol, N-acetyl-d-glucosamine, ribose, glucose-6-phosphate, fructose-6-phosphate, and spermidine. C401 was auxotrophic to fructose, glycerol-3-phosphate, N-acetyl-d-glucosamine, and spermidine. Two classes of fosfomycin isolates were also identified and referred to as C398 and C399. C398 was broadly auxotrophic, while C399 was auxotrophic to glycerol-3-phosphate, maltose, and ribose. Auxotrophies for amino acids are also shown in Table 4. Both C400 and C401 were unable to utilize tryptophan, while C400 was also auxotrophic for glycine, lysine, arginine, and histidine. C398 was broadly auxotrophic, while C399 displayed utilization patterns identical to those of the PAO-1 parent strain.
Table 3.
Growth properties of tobramycin- or fosfomycin-resistant mutants of P. aeruginosa PAO-1 on carbohydrates and polyamines
| Strain | MIC (μg/ml)a |
Growth on carbohydrateb |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOB | FOF | Fru | Glc | G3P | Gly | Mtl | NAG | Rib | Suc | G6P | F6P | Spm | Put | |
| PAO-1 | 0.5 | 128 | + | + | + | + | + | + | + | + | + | + | + | + |
| C400 | 8 | 128 | − | + | − | − | − | − | − | + | − | − | − | + |
| C401 | 8 | 128 | − | + | − | + | + | − | + | + | + | + | − | + |
| C398 | 0.5 | 4096 | − | − | − | − | − | − | − | − | − | − | − | − |
| C399 | 0.5 | 512 | + | + | − | + | − | + | − | + | + | + | + | + |
FOF, fosfomycin; TOB, tobramycin.
+, growth; −, no growth.
Table 4.
Growth properties of tobramycin- and fosfomycin-resistant mutants of P. aeruginosa PAO-1 on l-amino acids
| Strain | MIC (μg/ml)a |
Growth on amino acidsb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOB | FOF | Ala | Gly | Pro | Trp | Asn | Gln | Asp | Glu | Lys | Arg | His | |
| PAO-1 | 0.5 | 128 | + | + | + | + | + | + | + | + | + | + | + |
| C400 | 8 | 128 | + | − | + | − | + | + | + | + | − | − | − |
| C401 | 8 | 128 | + | + | + | − | + | + | + | + | + | + | − |
| C398 | 0.5 | 4096 | − | − | − | − | − | − | − | − | − | − | − |
| C399 | 0.5 | 512 | + | + | + | + | + | + | + | + | + | + | + |
FOF, fosfomycin; TOB, tobramycin.
+, growth; −, no growth.
Characterization of transport mutants.
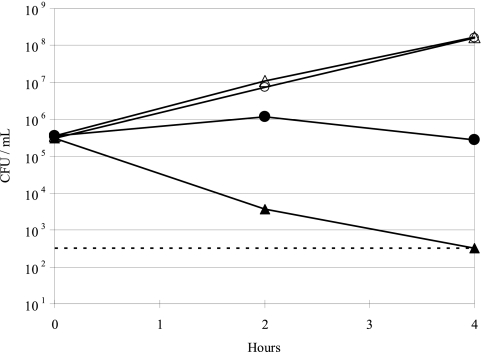
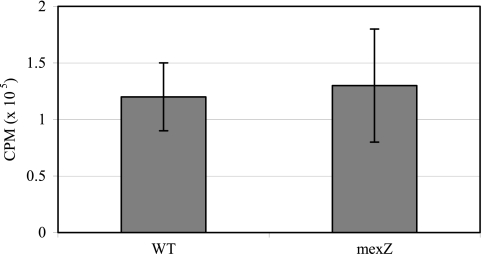
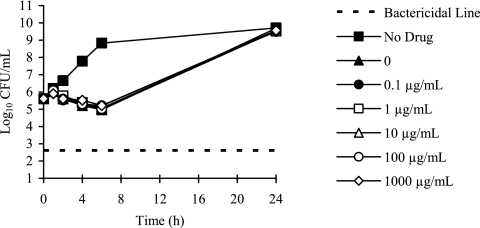
To gain further insights into potential transport systems identified from the carbohydrate and amino acid auxotrophy studies, we characterized select PAO-1 mutants with transposon insertions in genes encoding glucose porins, efflux transporters, and a glycerol-3-phosphate transporter (Table 5). Both the oprB and the glucose-sensitive porin mutants showed a 2-fold increase in tobramycin MIC relative to the parent PAO-1 strain. In time-kill experiments, tobramycin (1 μg/ml) prevented growth of the oprB mutant but resulted in 3 log10 killing of PAO-1 at 4 h (Fig. 6.) The oprB mutant was auxotrophic for glycerol (Table 5). Mutations in efflux transporter genes mexX, mexY, and oprM resulted in a 2- to 4-fold decrease in tobramycin MIC, while the mexZ mutant had a 2-fold increase in MIC. Uptake of [3H]tobramycin in the mexZ mutant was not significantly different from the PAO-1 parent strain (Fig. 7), and none of the efflux transporter mutants had carbohydrate or amino acid auxotrophies. Mutation of the glpT gene resulted in high levels of fosfomycin resistance and a glycerol-3-phosphate auxotrophy, but no change in tobramycin susceptibility (Table 5). Time-kill studies demonstrated that killing of P. aeruginosa with fosfomycin (4 μg/ml) is not enhanced by the addition of glucose-6-phosphate to the CAMHB (Fig. 8.).
Table 5.
Characteristics of P. aeruginosa PAO-1 transposon mutantsa
| Mutant | Gene | Description | MIC (μg/ml) |
Carbohydrate and amino acid auxotrophies | |
|---|---|---|---|---|---|
| TOB | FOF | ||||
| PAO-1 | Parent strain, burn wound | 0.5 | 128 | None | |
| PA2291 | Glucose-sensitive porin | 1.0 | 128 | ND | |
| PA3186 | oprB | Outer membrane porin OprB precursor | 1.0 | 128 | Glycerol |
| PA0427 | oprM | Outer membrane protein OprM precursor | 0.125 | 128 | None |
| PA2018 | mexY | RND efflux membrane fusion protein | 0.25 | 128 | None |
| PA2019 | mexX | RND multidrug efflux transporter | 0.25 | 128 | ND |
| PA2020 | mexZ | Transcriptional regulator | 1.0 | 128 | None |
| PA5235 | glpT | Glycerol-3-phosphate transporter | 0.5 | 4096 | G3P |
ND, not determined; FOF, fosfomycin; TOB, tobramycin.
Fig 6.
Time-kill curves demonstrating that a P. aeruginosa PAO-1 oprB mutant (closed circles) was less susceptible to 1 μg/ml tobramycin than the PAO-1 parent strain (closed triangles). No-drug controls were included for both the PAO-1 parent (open triangles) and PAO-1 oprB mutant (open circles).
Fig 7.
Inactivation of mexZ, the transcriptional regulator of mexXY efflux transport system, does not alter [3H]tobramycin uptake. P. aeruginosa PAO-1 parent strain and the mexZ mutant PA2020 were incubated with 2.3 μg/ml [3H]tobramycin for 30 min. Values represent the means ± SD from seven independent experiments. Statistical differences were evaluated by Student's t test (P = 0.689).
Fig 8.
Time-kill curves of P. aeruginosa 27853 demonstrating that the addition of 0.1 to 1,000 μg/ml of glucose-6-phospahte (G6P) to CAMHB does not improve killing by fosfomycin (4 μg/ml).
DISCUSSION
We demonstrate here that fosfomycin enhances the activity of tobramycin in the presence of mucin by increasing active uptake of tobramycin, resulting in greater inhibition of protein synthesis. In contrast, tobramycin alone had greatly reduced antibacterial activity in the presence of mucin. This finding was in agreement with that of Mendelman et al., who reported that 16× the tobramycin MIC was required for bactericidal killing of P. aeruginosa in CF sputum (30). Antibiotic activity in the lung microenvironment is of particular concern in the management of CF and non-CF bronchiectasis patients (44). Current inhaled therapies deliver high concentrations of antibiotic to the airways but rarely eradicate the infections (14, 61). Repeated exposure to antibiotics combined with suboptimal reductions of bacterial CFU led to selection of antibiotic-resistant bacterial populations (32, 33). P. aeruginosa and S. aureus isolates, particularly those from CF patients, are resistant to many of the currently approved antibiotics (11, 31, 49). Consequently, existing antibiotic monotherapies are becoming less effective for treating bacterial infections because of increasing resistance.
In this study, experiments comparing the rate and degree of killing of FTI to the component weights of fosfomycin and tobramycin show that fosfomycin enhances the activity of tobramycin by 9 log10 CFU/ml against P. aeruginosa and S. aureus. Identical experiments incorporating the enantiomer of fosfomycin in combination with tobramycin did not show a similar enhancement, suggesting that the absolute stereochemistry of fosfomycin and/or biological activity are required to reduce the inhibitory effects of mucin on tobramycin. Previous studies have shown that fosfomycin enhances the time-kill kinetics of gentamicin (46) and amikacin (45) against methicillin-resistant S. aureus and P. aeruginosa 59957 RyC, respectively. Here, we demonstrate the utility of the phenomenon for the enhancement of tobramycin activity by fosfomycin in the presence of mucin. The high antibacterial activity of FTI in mucin was also reflected in resistance studies. The spontaneous mutations resulting in resistance after a single exposure to FTI was 3 to 4 log10 lower than those of fosfomycin and tobramycin alone and decreased with increasing drug concentrations. By comparison, tobramycin had a very high SMF even at 16× MIC. Previous reports have shown that P. aeruginosa biofilms grown on mucin-coated surfaces develop large cellular aggregates and have increased tolerance to tobramycin (26). Our susceptibility data demonstrate that fosfomycin enhances the antibacterial activities of tobramycin in mucin, resulting in bactericidal killing and thus reducing the development of resistance.
The molecular mechanism by which fosfomycin enhances the activity of tobramycin is unknown. It is possible that adducts formed between tobramycin and fosfomycin could possess improved antibacterial activity. Aminoglycosides and some β-lactams chemically interact when combined to form new chemical entities via nucleophilic cleavage of the β-lactam ring by one of the amino groups of tobramycin (17). Based on the number of primary amines on tobramycin, formation of at least five adducts with fosfomycin is possible. However, no chemical adducts of fosfomycin and tobramycin were detected by HPLC/MS after incubation of FTI at RT or 37°C for 24 h. This suggests the enhanced antibacterial activities were not due to new chemical entities. Furthermore, the three adducts formed at 89°C had 2-fold-lower activity than FTI incubated at RT or 37°C.
It is also plausible that when fosfomycin and tobramycin are combined at a 4:1 (wt/wt) ratio, uptake of one or both of the antibiotics into the bacterial cell is enhanced, resulting in improved activity. The major component of FTI, fosfomycin, is a phosphonic acid derivative that inhibits the first step of peptidoglycan biosynthesis in the bacterial cell wall by irreversibly binding to the enzyme phosphoenolpyruvate (UDP-N-acetylglucosamine enolpyruval-transferase) (21). The minor component, tobramycin, prevents protein biosynthesis by causing translational errors and by inhibiting translocation (55, 60). Based on these well-characterized mechanisms of action, we provide several lines of evidence that strongly suggest that the enhanced activities of FTI are due to increased uptake of tobramycin rather than fosfomycin: (i) time-kill curves demonstrated that both FTI and tobramycin were bactericidal and killed in a concentration-dependent fashion, while fosfomycin was bacteriostatic and killed in a time-dependent fashion; (ii) macromolecular biosynthesis studies showed that FTI and tobramycin inhibited protein synthesis in a concentration-dependent fashion; (iii) FTI inhibited protein synthesis faster and to a greater degree than cell wall biosynthesis at a concentration (8 μg/ml) and in a time frame (2 to 4 h) that corresponded to bactericidal killing; (iv) the addition of 10 μg/ml fosfomycin resulted in a 170% increase in [3H]tobramycin uptake relative to that of the no-fosfomycin control.
The higher levels of cell-associated tobramycin could be explained by passive uptake due to a compromised cell wall, increased facilitated diffusion across the outer membrane via a porin, or increased active uptake across the cytoplasmic membrane through a carrier-mediated transport system. Alternatively, increased accumulation of tobramycin could be due to impaired efflux. The concentration of fosfomycin used in our studies had little effect on cell wall biosynthesis, rendering the first of these possibilities unlikely. To probe for explanations associated with transport phenomena, fosfomycin- and tobramycin-resistant mutants were generated, and their ability to grow on single carbon sources was examined. Two classes of tobramycin-resistant mutants each with distinct carbohydrate utilization patterns were identified in this study. Mutant C400 was unable to utilize fructose, glycerol, and mannitol as sole carbon sources. The P. aeruginosa porin OprB is known to facilitate the diffusion of glucose, glycerol, fructose, and mannitol (63, 64), and uptake of these sugars is inducible (40). Consistent with our findings, Bryan and Kwan demonstrated that a streptomycin-resistant P. aeruginosa mutant generated by transposon mutagenesis was unable to transport glucose and glycerol and suggested that this was due to functional loss of OprB (6). In our investigations, we demonstrated that oprB (PA3186) and a glucose-sensitive porin (PA2291) mutant had 2-fold increases in tobramycin MIC, suggesting a possible role in transporting tobramycin across the outer membrane of P. aeruginosa. Decreased susceptibility to tobramycin was confirmed for the oprB mutant in time-kill experiments. Auxotrophies of these mutants were not fully characterized, but the oprB mutant was auxotrophic for glycerol. These data are consistent with previous reports showing that glycerol is transported into P. aeruginosa by OprB (63). Since glycerol uptake is inducible (63) and since fosfomycin is also a substrate for glycerol transporters (8, 21), fosfomycin may be inducing uptake of tobramycin via OprB. However, C400 had a 16-fold increase in tobramycin MIC compared to a 2-fold increase for the oprB mutant. This suggests that the high level of tobramycin resistance in C400 is due to multiple independent mutations.
Since both fosfomycin and tobramycin have cytoplasmic targets, it is also possible that tobramycin enters the cytoplasm of P. aeruginosa through active transport systems normally used for transporting carbohydrates or amino acids (1, 39) and that fosfomycin enhances this uptake. Interestingly, C400 was also auxotrophic for hexose-6-phosphate sugars (G6P and F6P) and G3P, which are transported into Escherichia coli and S. aureus by two phosphoenopyruvate:sugar phosphotransferase systems (PTS), the uptake of hexose-6-phosphate (UhpT) and glycerol-3-phosphate (GlpT) transport, respectively. Fosfomycin is also transported into E. coli and S. aureus via GlpT and UhpT, and mutation of these transporters results in fosfomycin resistance and auxotrophies for their respective substrates. P. aeruginosa contains the GlpT transport system (51, 52), but the UhpT transport system appears to be absent (8). As a result, uptake and susceptibility to fosfomycin is not increased by the addition of G6P to bacteriological culture medium, as is the case with E. coli (21). Interestingly, the closest homolog to UhpT in P. aeruginosa is GlpT (8). We also demonstrated in this study that a glpT transposon mutant (PA5235) had a 16-fold increase in fosfomycin MIC, severely impaired growth on G3P, but no auxotrophies for either G6P or F6P. The tobramycin MIC of the glpT mutant was unchanged, demonstrating that the G6P and F6P auxotrophies and tobramycin resistance of C400 were not the result of a mutation in glpT.
The tobramycin-resistant mutant C401 was auxotrophic for G3P, NAG, and Fru, suggesting a distinct mutation(s) from C400. NAG and Fru are actively transported into P. aeruginosa by the phosphotransferase system (20). Amino sugars like NAG are used for synthesis of the peptidoglycan component of the bacterial cell wall, the target of fosfomycin (21, 25). It is possible that fosfomycin causes stimulation of cell wall biosynthesis genes and induces uptake of tobramycin due to its structural similarity to NAG. Tobramycin consists of two aminoglucose molecules covalently linked to a 2-deoxystreptamine nucleus (60). The aminoglucose moieties have structural similarity to NAG and d-glucose (40, 54).
In P. aeruginosa, arginine is actively transported by multiple systems, including the basic-amino-acid-specific porin OprD (20, 56, 57). Furthermore, arginine has been shown to increase tobramycin susceptibility in P. aeruginosa grown under anaerobic growth conditions (5). C400 was also auxotrophic for arginine, but we have not further evaluated known arginine transport mutants. Tamber et al. demonstrated that mutation of oprD and 18 homologues in P. aeruginosa did not alter tobramycin susceptibilities (57). However, MICs were determined under aerobic conditions, and their role in tobramycin resistance may have been underestimated. It is plausible that fosfomycin is inducing a broad substrate transport system(s) normally expressed only in the presence of arginine and under microaerophilic or anaerobic conditions and that tobramycin is actively transported across the cytoplasmic membrane.
The fosfomycin-resistant mutant C398 was pan-auxotrophic for carbohydrates and amino acids. In E. coli, transport of carbohydrates requires the cyclic AMP receptor protein-cyclic AMP complex (CRP-cAMP) (2, 59). Mutation of genes in this complex leads to fosfomycin resistance and broad carbohydrate auxotrophies (1, 2, 9, 59). It is possible that C398 may have mutations in the CRP. The second fosfomycin-resistant mutant C399 was auxotrophic for G3P, Mtl, and ribose but did not have any amino acid auxotrophies.
The MexXY-OprM efflux system is a major aminoglycoside resistance mechanism in P. aeruginosa (13). Since active uptake and efflux are both energy dependent, it is also plausible that fosfomycin inhibits tobramycin efflux, resulting in increased accumulation of tobramycin. In this study, we developed several lines of evidence that strongly demonstrate that fosfomycin is not inhibiting tobramycin efflux. (i) Time course studies demonstrated that fosfomycin induced a rapid accumulation of [3H]tobramycin within 5 min, while [3H]tobramycin efflux was not evident until 30 to 60 min. (ii) mexX, mexY, oprM, and mexZ mutants had 2-fold changes in tobramycin MICs relative to the wild-type parent PAO-1 strain, but there were no changes in fosfomycin MICs. If fosfomycin was inhibiting tobramycin efflux via MexXY-OprM, we would expect that mutation of these genes would also cause changes in fosfomycin MIC. (iii) Despite >20 years of clinical usage of fosfomycin in treating urinary tract infections caused by Gram-negative bacteria, fosfomycin-resistant efflux mutants have not been reported. The major fosfomycin resistance mechanisms in clinical isolates are due to mutations of cya, ptsI, glpT, and uhpT genes or to the presence of FosA (38). (iv) MexZ, a negative regulator of MexXY-OprM, is the most frequent clinically derived efflux mutation resulting in aminoglycoside resistance in P. aeruginosa isolates from CF patients (13, 34). In this study, an mexZ mutant had a 2-fold increase in tobramycin MIC, but it did not show a decrease in [3H]tobramycin uptake as would be expected with activation of MexXY-OprM efflux. These data also strongly demonstrate that tobramycin efflux is not the underlying mechanism. (v) mexY and mexZ mutants did not have auxotrophies for either carbohydrates or amino acids, demonstrating a different resistance mechanism than the spontaneous mutants isolated in this study. (vi) The characteristics of [3H]tobramycin uptake were similar to those of active transport of carbohydrates and amino acids in P. aeruginosa (12, 22, 58). Specifically, we observed an approximate 5-fold change in tobramycin uptake in 5 to 10 min and that tobramycin uptake was inducible with fosfomycin and concentration dependent over 0.3 to 2.3 μg/ml. Saturation was not achieved, because concentrations of tobramycin of >4 μg/ml are rapidly bactericidal for P. aeruginosa. Tobramycin uptake was also temperature dependent, and rates decreased from 37°C to 25°C. Lastly, tobramycin uptake was energy dependent and could be inhibited with sodium azide. Taken together, these data strongly demonstrate that fosfomycin is inducing the active uptake of tobramycin rather than inhibiting tobramycin efflux.
The objective of this study was to gain insights into the synergistic activity of a novel 4:1 fosfomycin-tobramycin combination in the presence of mucin. We have demonstrated for the first time that fosfomycin enhances the active transport of tobramycin in P. aeruginosa. In our model, increased levels of tobramycin, reaching the target, result in greater inhibition of protein synthesis, greater bacterial killing, and ultimately a lower frequency of resistance. Furthermore, we provide good insights into the systems that transport tobramycin across the outer and cytoplasmic membranes. It is unclear from the present studies whether fosfomycin and tobramycin share the same transport system, or if fosfomycin is inducing tobramycin uptake via coordinate regulation of a different transport system. Additional studies utilizing a variety of molecular approaches are warranted to conclusively identify the genes involved in the activation and transport of tobramycin. Lastly, while the present study focused primarily on P. aeruginosa, we have also shown that FTI has enhanced activity against S. aureus. Both P. aeruginosa and S. aureus are known to transport and metabolize many of the same carbohydrates and amino acids and also share transport systems for fosfomycin. Therefore, it is plausible that fosfomycin is also inducing tobramycin uptake in S. aureus via the same mechanism.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Gilead Sciences, Inc.
We thank Robert Mourey and Will Watkins for critical review of the manuscript.
D.L.M, W.R.B., T.F.K., J.H.T., and J.V. own stock and options in Gilead Sciences, Inc. L.M.B. and J.L.S. have no conflicts of interest to declare.
Footnotes
Published ahead of print 9 January 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alper MD, Ames BN. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic-AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J. Bacteriol. 133:149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Artman M, Werthamer S. 1974. Use of streptomycin and cyclic adenosine 5“-monophosphate in the isolation of mutants deficient in CAP protein. J. Bacteriol. 120:542–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baum EZ, et al. 2001. Identification and characterization of new inhibitors of the Escherichia coli murA enzyme. Antimicrob. Agents Chemother. 45:3182–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjarnsholt T, Jensen PO, Fiandaca MJ. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558 [DOI] [PubMed] [Google Scholar]
- 5. Borriello G, et al. 2006. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50:382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryan LE, Kwan S. 1981. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrate reductase, and aerobic transport. Antimicrob. Agents Chemother. 19:958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bucki R, Sostarecz AG, Byfield FJ. 2007. Resistance of the antibacterial agent ceragenin CSA-13 to inactivation by DNA or F-actin and its activity in cystic fibrosis sputum. J. Antimicrob. Chemother. 60:535–545 [DOI] [PubMed] [Google Scholar]
- 8. Castaneda-Garcia A, et al. 2009. The glycerol-3-phosphate permease glpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191:6968–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cordaro JC, et al. 1976. Fosfomycin resistance: selection method for internal and extended deletions of the phosphoenolpyruvate:sugar phosphotransferase genes of Salmonella typhimurium. J. Bacteriol. 128:785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis SS, et al. 1985. A new mucotropic agent—in vitro and in vivo evaluation of 2-alpha-thenoylthiopropionylglycine (bronchoplus). Eur. J. Respir. Dis. 67:94–102 [PubMed] [Google Scholar]
- 11. Driffield K, et al. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61:1053–1056 [DOI] [PubMed] [Google Scholar]
- 12. Eagon RG, Phibbs PV., Jr 1971. Kinetics of transport of glucose, fructose, and mannitol by Pseudomonas aeruginosa. Can. J. Biochem. 49:1031–1041 [DOI] [PubMed] [Google Scholar]
- 13. Feliziani S, et al. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geller DE, Pitlick WH, Nardella PA. 2002. Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest 122:219–226 [DOI] [PubMed] [Google Scholar]
- 15. Glamkowski EJ, et al. 1970. A new synthesis of the antibiotic phosphonomycin. J. Org. Chem. 35:3510–3512 [DOI] [PubMed] [Google Scholar]
- 16. Henke MO, et al. 2007. MUC5AC and MUC5B mucins increase in cystic fibrosis airway secretions during pulmonary exacerbation. Am. J. Respir. Crit. Care. Med. 175:816–821 [DOI] [PubMed] [Google Scholar]
- 17. Huh K, et al. 1994. Structures and biological activities of tobramycin-ticarcillin adducts. J. Pharm. Sci. 83:763–767 [DOI] [PubMed] [Google Scholar]
- 18. Hunt BE, et al. 1995. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob. Agents Chemother. 39:34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobs MA, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson DA, et al. 2008. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 4:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kahan FM, et al. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364–386 [DOI] [PubMed] [Google Scholar]
- 22. Kay WW, Gronlund AF. 1969. Amino acid transport in Pseudomonas aeruginosa. J. Bacteriol. 97:273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirkham S, et al. 2002. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem. J. 361:537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirkham S, et al. 2008. MUC5B is the major mucin in the gel phase of sputum in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care. Med. 178:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komatsuzawa H, et al. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221–1231 [DOI] [PubMed] [Google Scholar]
- 26. Landry RM, An D, Hupp JT. 2006. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol. Microbiol. 59:142–151 [DOI] [PubMed] [Google Scholar]
- 27. Li W, Lu CD. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189:5413–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacLeod DL, et al. 2009. Antibacterial activities of a fosfomycin/tobramycin combination: a novel inhaled antibiotic for bronchiectasis. J. Antimicrob. Chemother. 64:829–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mendelman PM, et al. 1985. Aminoglycoside penetration, inactivation, and efficacy in cystic fibrosis sputum. Am. Rev. Respir. Dis. 132:761–765 [DOI] [PubMed] [Google Scholar]
- 31. Mirakhur A, et al. 2003. Fosfomycin therapy for multiresistant Pseudomonas aeruginosa in cystic fibrosis. J. Cyst. Fibros. 2:19–24 [DOI] [PubMed] [Google Scholar]
- 32. Mueller M, de la Pena A, Derendorf H. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: kill curves versus MIC. Antimicrob. Agents Chemother. 48:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mueller M, de la Pena A, Derendorf H. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: distribution in tissue. Antimicrob. Agents Chemother. 48:1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulcahy LR, et al. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192:6191–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 6th ed NCCLS document M7-A6. NCCLS, Wayne, PA [Google Scholar]
- 36. National Committee for Clinical Laboratory Standards 2003. Performance standards for antimicrobial susceptibility testing; approved standard, 14th ed NCCLS document M100-S13. NCCLS, Wayne, PA [Google Scholar]
- 37. National Committee for Clinical Laboratory Standards 1999. Methods for determining bactericidal activity of antimicrobial agents; approved standard. NCCLS document M26-A. NCCLS, Wayne, PA [Google Scholar]
- 38. Nilsson AI, et al. 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 47:2850–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nishijyo T, Haas D, Itoh Y. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 40:917–931 [DOI] [PubMed] [Google Scholar]
- 40. Phibbs PV, Jr, Eagon RG. 1970. Transport and phosphorylation of glucose, fructose, and mannitol by Pseudomonas aeruginosa. Arch. Biochem. Biophys. 138:470–482 [DOI] [PubMed] [Google Scholar]
- 41. Phillips JE, et al. 2006. An enzyme-linked immunosorbent assay (ELISA) for the determination of mucin levels in bronchoalveolar lavage fluid. J. Pharmacol. Toxicol. Methods 53:160–167 [DOI] [PubMed] [Google Scholar]
- 42. Purdy Drew KR, et al. 2009. Cationic amphiphiles increase activity of aminoglycoside antibiotic tobramycin in the presence of airway polyelectrolytes. J. Am. Chem. Soc. 131:486–493 [DOI] [PubMed] [Google Scholar]
- 43. Ramphal R, et al. 1988. The binding of anti-pseudomonal antibiotics to macromolecules from cystic fibrosis sputum. J. Antimicrob. Chemother. 22:483–490 [DOI] [PubMed] [Google Scholar]
- 44. Ramsey BW, et al. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 340:23–30 [DOI] [PubMed] [Google Scholar]
- 45. Rodriguez A, Vicente MV, Olay T. 1985. Experimental endocarditis and fosfomycin. Drugs Exp. Clin. Res. 11:55–62 [PubMed] [Google Scholar]
- 46. Rodriguez A, Vicente MV, Olay T. 1987. Single- and combination-antibiotic therapy for experimental endocarditis caused by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1444–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rose MC, Voynow JA. 2006. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 86:245–278 [DOI] [PubMed] [Google Scholar]
- 48. Saggers BA, Lawson D. 1966. Some observations on the penetration of antibiotics through mucus in vitro. J. Clin. Pathol. 19:313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saiman L, et al. 2002. Synergistic activities of macrolide antibiotics against Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. Antimicrob. Agents Chemother. 46:1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarubbi E, et al. 2004. Mode of action of the microbial metabolite GE23077, a novel potent and selective inhibitor of bacterial RNA polymerase. Eur. J. Biochem. 271:3146–3154 [DOI] [PubMed] [Google Scholar]
- 51. Schweizer HP, Po C. 1996. Regulation of glycerol metabolism in Pseudomonas aeruginosa: characterization of the glpR repressor gene. J. Bacteriol. 178:5215–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Silo-Suh L, et al. 2005. Adaptations of Pseudomonas aeruginosa to the cystic fibrosis lung environment can include deregulation of zwf, encoding glucose-6-phosphate dehydrogenase. Antimicrob. Agents Chemother. 187:7561–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sriramulu DD, et al. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667–676 [DOI] [PubMed] [Google Scholar]
- 54. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 55. Taber HW, Mueller JP, Miller PF. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamber S, Hancock RE. 2006. Involvement of two related porins, OprD and OpdP, in the uptake of arginine by Pseudomonas aeruginosa. FEMS Microbiol. Lett. 260:23–29 [DOI] [PubMed] [Google Scholar]
- 57. Tamber S, Ochs MM, Hancock REW. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 188:45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsay SS, Brown KK, Gaudy ET. 1971. Transport of glycerol by Pseudomonas aeruginosa. J. Bacteriol. 108:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsuruoka T, Miyata A, Yamada Y. 1978. Two kinds of mutants defective in multiple carbohydrate utilization isolated from in vitro fosfomycin-resistant strains of Escherichia coli K-12. J. Antibiot. 31:192–201 [DOI] [PubMed] [Google Scholar]
- 60. Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Valcke Y, Pauwels R, Van der Straeten M. 1990. Pharmacokinetics of antibiotics in the lungs. Eur. Respir. J. 3:715–722 [PubMed] [Google Scholar]
- 62. Viswanathan H, et al. 2006. MUC5B secretion is up-regulated in sinusitis compared with controls. Am. J. Rhinol. 20:554–557 [DOI] [PubMed] [Google Scholar]
- 63. Williams SG, Greenwood JA, Jones CW. 1994. The effect of nutrient limitation on glycerol uptake and metabolism in continuous cultures of Pseudomonas aeruginosa. Microbiology 140:2961–2969 [DOI] [PubMed] [Google Scholar]
- 64. Wylie JL, Worobec EA. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177:3021–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.