Abstract
Cryptococcus neoformans strains resistant to azoles due to mutations causing alterations in the ERG11 gene, encoding lanosterol 14α-demethylase, have rarely been reported. In this study, we have characterized a C. neoformans serotype A strain that is resistant to high concentrations of fluconazole (FLC). This strain, which was isolated from an FLC-treated patient, contained five missense mutations in the ERG11 gene compared to the sequence of reference strain H99. Molecular manipulations of the ERG11 gene coupled with susceptibility to triazole revealed that a single missense mutation resulting in the replacement of tyrosine by phenylalanine at amino acid 145 was sufficient to cause the high FLC resistance of the strain. Importantly, this newly identified point mutation in the ERG11 gene of C. neoformans afforded resistance to voriconazole (VRC) but increased susceptibility to itraconazole (ITC) and posaconazole (PSC), which are structurally similar to each other but distinct from FLC/VRC. The in vitro susceptibility/resistance of the strains with or without the missense mutation was reflected in the therapeutic efficacy of FLC versus ITC in the animals infected with the strains. This study shows the importance of the Y145F alteration of Erg11 in C. neoformans for manifestation of differential susceptibility toward different triazoles. It underscores the necessity of in vitro susceptibility testing for each FLC-resistant C. neoformans clinical isolate against different groups of azoles in order to assist patient management.
INTRODUCTION
Cryptococcus neoformans is the most common cause of life-threatening fungal meningoencephalitis in HIV-infected patients as well as the sporadic cause of the infection in immunocompetent patients worldwide. Prompt diagnosis and proper treatment are crucial, as cryptococcal meningitis is invariably fatal if untreated (6, 12). Fluconazole (FLC), a triazole, is the drug of choice for maintenance therapy of cryptococcosis in AIDS as well as in non-AIDS patients, due to its efficacy, excellent central nervous system penetration, and low toxicity (5, 22, 30). Long-term usage of FLC for the treatment of fungal infections in AIDS patients, however, led to the emergence of resistant strains (10, 18, 20, 28). There have been several reports on the isolation of FLC-resistant C. neoformans strains, mostly from AIDS patients undergoing FLC maintenance therapy (1–4, 18). These isolates were associated with clinical failure during the maintenance therapy, leading to a series of recurrences (15). Unlike the strains of Candida albicans isolated from recurrent cases of esophageal candidiasis in AIDS patients undergoing prolonged azole therapy, however, the cryptococcal strains isolated from most recurrent cases of central nervous system infection showed susceptibility to azoles similar to that of the primary strains (5). The cause of therapy failure without alterations in azole susceptibility remained unexplained until azole heteroresistance was characterized in cryptococcal pathogens (24, 25, 27). Characterization of more than 200 clinical and environmental isolates in our laboratory revealed that all the strains of C. neoformans and C. gattii are endowed with an innate ability to produce minor clonal subpopulations that can transiently adapt to high concentrations of FLC. In a recent study of heteroresistance in C. neoformans, we encountered a C. neoformans serotype A strain isolated from an AIDS patient undergoing FLC maintenance therapy that showed an exceptionally high level of heteroresistance (128 μg/ml) (24). Moreover, the resistance to triazoles in this strain was limited to FLC and to voriconazole (VRC); the strain remained susceptible to itraconazole (ITC) and posaconazole (PSC). While dissecting the cause of this high FLC resistance of the strain, we identified a missense mutation in the cryptococcal cytochrome P450 lanosterol 14α-demethylase (P450Dm, Erg11) that was found to be critical for resistance to FLC and VRC and yet enhanced sensitivity to ITC or PSC.
We further demonstrated that mice infected with the strains carrying this missense mutation benefited more from treatment with ITC, while mice infected with the wild-type strain lived longer with FLC treatment. This is the first study to identify a residue in the cryptococcal Erg11 that is critical for differential susceptibility to two different kinds of triazoles: FLC/VRC and ITC/PSC.
MATERIALS AND METHODS
Strains and media.
C. neoformans serotype A reference strain H99 and MRL862, another serotype A clinical isolate collected as part of the SENTRY antimicrobial surveillance program, were the parental strains used in the study. The ERG11 gene from both strains was cloned and sequenced. The strains designated TES12, TES9, and TES10 were constructed (in the H99 background) by molecular manipulation of the ERG11 gene cloned from H99 or from MRL862. Strains were stored in 25% glycerol at −80°C until use and were maintained on YEPD (1% yeast extract, 2% peptone, 2% glucose) agar plates at 30°C. RPMI 1640 medium (Sigma) buffered with 0.165 M MOPS (morpholinepropanesulfonic acid; pH 7) was used for antifungal microdilution susceptibility testing.
ERG11 gene manipulation.
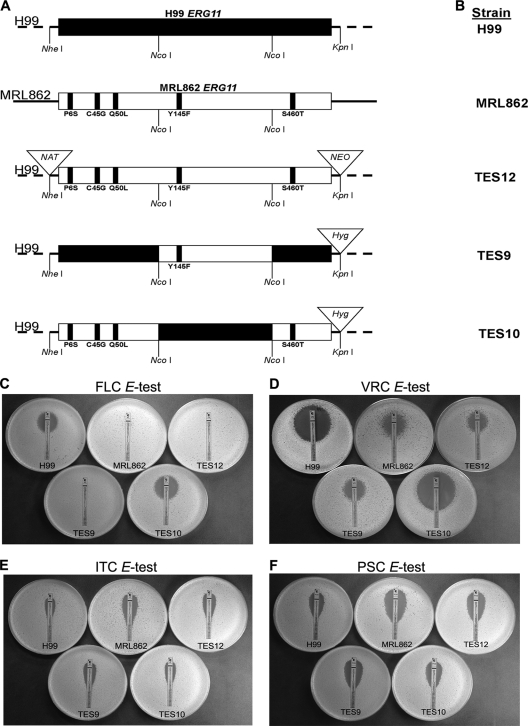
The ERG11 gene was cloned by PCR from genomic DNA of MRL862 as well as of H99. Three independent PCR clones were used to determine the EGR11 sequence. The sequencing was performed at the Laboratory of Molecular Technology (Frederick, MD). To replace the ERG11 gene in the H99 strain with that of strain MRL862, a plasmid (pES12) containing the entire EGR11 gene sequence of MRL862 along with 757 bp and 388 bp of the 5′ and 3′ flanking regions, respectively, was constructed. The plasmid pES12 also contained two drug resistance markers, those for resistance to nourseothricin (NAT) and neomycin (NEO), flanked by the upstream and downstream regions of H99 ERG11 to facilitate homologous integration. A linear ClaI-ApaI DNA fragment of pES12 was transformed into H99 using the biolistic transformation method. Transformants resistant to nourseothricin and neomycin were selected, and the clones whose ERG11 was replaced with that of pES12 were identified by PCR and confirmed by Southern blot analysis. This strain was designated TES12 (Fig. 1A). Plasmid pES9 was constructed by replacing the regions flanking the NcoI fragment of MRL862 ERG11 with the equivalent regions of H99 ERG11 and contained the mutation Y145F, which was suspected to be essential for the resistant phenotype. The plasmid pES10 contained MRL862 ERG11 in which the NcoI fragment was replaced with the NcoI fragment of H99 ERG11, leaving the Y145F mutation intact. Both pES9 and pES10 constructs contained the hygromycin drug resistance marker (HYG) in the 3′ flanking region between the junction of MRL862 ERG11 and H99 ERG11. Furthermore, linear ClaI-ApaI DNA fragments of pES9 and pES10 were separately introduced into the H99 strain in which ERG11 was replaced with that of MRL862 (TES12), and then transformants resistant to hygromycin but sensitive to neomycin and nourseothricin were selected (Fig. 1A). Putative transformants (TES9 and TES10) were identified by PCR and confirmed by Southern blot analysis.
Fig 1.
Importance of Y145F. (A) Outline of the strategy for ERG11 gene manipulation. Diagrams show a map of the ERG11 gene in H99 (no mutation), MRL862, and various constructs containing the amino acid substitutions. (B) Designations of strains with various ERG11 alleles. TES12 was created by replacing the H99 ERG11 gene with the MRL862 ERG11 gene, which contained all 5 point mutations; TES9 contained only the Y145F amino acid substitution, and TES10 contained the other four missense mutations (see Materials and Methods). (C to F) Effect of Y145F mutation on susceptibility to azoles. Etest plastic strips impregnated with a gradient of FLC (C), VRC (D), ITC (E), or PSC (F) were placed onto YEPD plates inoculated with the indicated strains. Growth inhibition was observed after 48 h at 30°C.
Triazoles and MIC determination.
FLC was purchased from Hospira, Inc. (Lake Forest, IL), and ITC (Sporanox) was purchased from Janssen Pharmaceutica N.V. (Beerse, Belgium) for the treatment of infected animals. MICs for FLC, ITC, PSC, and VRC were determined using Etest strips according to the Etest technical guide (AB Biodisk, Solna, Sweden), with slight modification. Overnight YEPD cultures were diluted in sterile 0.9% NaCl to an optical density at 600 nm (OD600) of 0.05 and plated on YEPD agar plates. Etest strips were overlaid, plates were incubated at 30°C for 48 h, and the susceptibility endpoint was read at the first growth inhibition ellipse. The susceptibilities of the isolates to azole antifungals were also performed using the standardized CLSI M27-A3 broth dilution method (8).
Sterol analysis.
Sterol profiles of the strains were analyzed as described previously (24). Briefly, the cells were grown overnight in YEPD broth at 30°C. Each culture was adjusted to an OD600 of 0.3 and divided into three samples: one supplemented with 64 μg/ml FLC, the second supplemented with 4 μg/ml ITC, and the third not supplemented with any drug. All samples were grown at 30°C for 3 h, and the cells were harvested by centrifugation and washed once with sterile distilled water. The pellets were resuspended in 9 ml methanol; 4.5 ml 60% (wt/vol) KOH was added together with 5 μg cholesterol (used as an internal recovery standard). Cell suspensions were heated to 75°C in a water bath for 2 h to complete the saponification; the sterols were then extracted with hexane and analyzed by gas chromatography-mass spectrometry (GC-MS) with an Agilent 6890 gas chromatograph (using an HP-1 fused silica column and a temperature program from 200°C [for 1 min] to 300°C at a rate of 10°C/min) coupled to a Waters GCT time of flight high-resolution mass spectrometer using MassLynx (version 4.0) software. The relative amount of each sterol was obtained by comparing the area under the curve for each sterol with that for the internal standard of cholesterol in the chromatogram.
Animal experimental study.
All animal studies were approved by the institutional animal care and use committee at NIAID, NIH. For the in vivo susceptibility to azoles, the isolates were tested in female BALB/c mice (weight, 20 g) by using the experimental model of systemic cryptococcosis. The mice were challenged intravenously with an inoculum of 5 × 104 CFU/mouse. FLC and/or ITC was administered intraperitoneally at a concentration of 10 mg/kg of body weight/day. Treatment was initiated 24 h after infection and was continued for 10 days for mice infected with H99-derived strains and 20 days for mice infected with the MRL862 strain. Ten animals were used for each strain. The mice were observed through day 30, and mortality was recorded daily.
Statistics.
Survival data from the animal experiments were analyzed using a two-group Wilcoxon test. An unpaired t test was used to compare differences in sterol content between isolates containing point mutations in ERG11 and the H99 wild-type strain. A P value of less than 0.05 was considered to be significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the MRL862 and H99 ERG11 genes are JQ044789 and JQ044790, respectively.
RESULTS AND DISCUSSION
Detection of mutations in the ERG11 gene in an FLC-resistant clinical isolate of C. neoformans (MRL862).
The MRL862 strain exhibited unusually high resistance to FLC, with high MICs (according to the CLSI method) and a heteroresistance level of 128 μg/ml (Table 1). The heteroresistant clones grown at 128 μg/ml FLC were able to adapt to FLC concentrations of up to 400 μg/ml (24). In the present study, these highly resistant clones that could tolerate 400 μg/ml FLC reverted back to the original level of heteroresistance at 128 μg/ml after 63 daily transfers in drug-free YEPD broth. Clinical isolates of C. neoformans with such a high FLC resistance level have rarely been reported, and we suspected that MRL862 may contain a mutation(s) in ERG11 that encodes the lanosterol 14α-demethylase, the target of azoles. The ERG11 gene of MRL862 was obtained by PCR, and its sequence was compared to that of ERG11 of H99, the genome-sequenced serotype A reference strain. The sequence of the ERG11 gene from MRL862 revealed a total of 17 nucleotide changes within its exons compared to the sequence of ERG11 from H99. Among these, 5 were considered the candidate mutations responsible for FLC resistance, while the remaining 12 changes were silent mutations (see Fig. S1 in the supplemental material). One of the five missense mutations was the replacement of tyrosine with phenylalanine at amino acid 145 (Y145F) in the catalytic domain. This amino acid is highly conserved in cytochrome P450 ERG11/Cyp51 in the B′ helix/BC loop in most fungi and animals but not in plants or bacteria (see Fig. S1 in the supplemental material) (13). Azole resistance associated with alterations in the target gene ERG11 has been extensively studied in C. albicans and Aspergillus fumigatus but not in C. neoformans (9, 14, 16, 21). The C. neoformans ERG11 gene sequence is conserved in other basidiomycetes (20). However, which residues of Erg11 are associated with azole resistance is poorly understood in basidiomycetous fungi in general and C. neoformans in particular. Although the functional importance of the tyrosine replacement by phenylalanine has not been analyzed, several studies have reported strains of C. albicans and Histoplasma capsulatum with the corresponding mutation, Y132F and Y136F, respectively, in the ERG11 gene that were isolated from patients treated with triazoles (7, 11, 16, 19, 29). To characterize the functional importance of the Y145F mutation in the ERG11 gene of strain MRL862, we first replaced the ERG11 gene in H99 with the genomic copy of MRL862 ERG11 by biolistic transformation (Fig. 1A; TES12). Further molecular manipulations were carried out in TES12 by swapping various forms of mutations into the ERG11 gene to determine the role of Y145F in FLC resistance of C. neoformans (see Materials and Methods).
Table 1.
MICs of azole compounds for isolates used in the study
| Strain [amino acid substitution(s)] | MIC (μg/ml)a |
||||
|---|---|---|---|---|---|
| FLCb | FLCc | VRCc | ITCc | PSCc | |
| H99 | 8 | 16 | 0.25 | 1 | 0.5 |
| MRL862 (P6S, C45G, Q50L, Y145F, S460T) | 128 | >256 | 2 | 0.094 | 0.125 |
| TES12 (P6S, C45G, Q50L, Y145F, S460T) | 128 | >256 | 3 | 0.38 | 0.25 |
| TES9 (Y145F) | 128 | >256 | 2 | 0.38 | 0.25 |
| TES10 (P6S, C45G, Q50L, S460T) | 8 | 12 | 0.19 | 0.75 | 0.5 |
Triazole MICs of each strain were consistent between three independent tests.
MICs determined by microdilution method.
MICs determined by Etest.
Antifungal susceptibility.
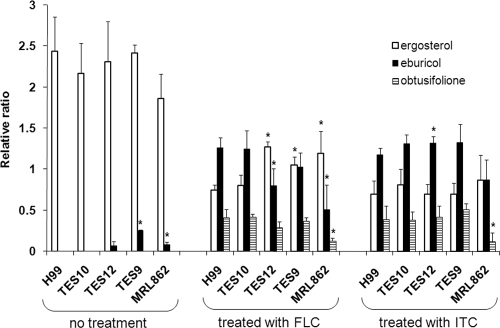
Table 1 and Fig. 1 show the results of triazole susceptibility testing of strains carrying different ERG11 alleles. The MIC of FLC for the H99 strain was between 8 and 16 μg/ml, while the MRL862 strain exhibited MICs of 128 to 256 μg/ml. The FLC MIC of TES12, a derivative of the H99 strain carrying the ERG11 gene isolated from MRL862 that contained all five missense mutations, was 128 to 256 μg/ml. This result strongly suggested that the resistance of MRL862 to FLC is linked to the five missense mutations in its ERG11 gene. Moreover, the TES9 strain, which contained only one missense mutation, Y145F, in the ERG11 gene showed the same FLC MIC as the MRL862 and TES12 strains. This demonstrated that a single Y145F substitution in Erg11 can confer high resistance to FLC. This conclusion was supported by the relatively low FLC MIC of the TES10 strain (8 to 16 μg/ml), which contained the other four missense mutations in Erg11 (P6S, C45G, Q50L, and S460T). The substitutions of the four amino acids in Erg11 of MRL862, therefore, were unrelated to the FLC resistance in vitro.
We also examined the drug sensitivity of these strains to various other triazoles. VRC is a triazole structurally similar to FLC, with the major difference being the substitution of a fluoropyrimidine group in place of one of the triazole moieties in FLC. All strains exhibited similar patterns of resistance toward VRC as toward FLC: H99 and TES10 were more susceptible to VRC (MIC ranges, 0.19 to 0.25 μg/ml) than the MRL862, TES12, and TES9 strains (MIC ranges, 2 to 3 μg/ml) (Fig. 1D). ITC and PSC are related azoles, and though they target the same enzyme in ergosterol biosynthesis, they are structurally very different from FLC and VRC by having long side chains. Interestingly, MRL862 was highly susceptible to ITC (MIC, 0.094 μg/ml; Fig. 1E) and PSC (MIC, 0.125 μg/ml; Fig. 1F). In comparison with MRL862, H99 displayed ITC and PSC MICs of 1 μg/ml and 0.5 μg/ml, respectively. The strains TES12 and TES9, which contained Y145F in the H99 background, were slightly more susceptible to ITC (MIC, 0.38 μg/ml) and PSC (MIC, 0.25 μg/ml), while TES10 had susceptibility similar to that of H99 to both azoles (ITC MIC, 0.75; PSC MIC, 0.5 μg/ml).
Other missense mutations in Erg11/Cyp51 that result in differential susceptibility to different azoles have been reported in C. albicans (16) as well as in A. fumigatus (9, 14). Replacement of tyrosine with a histidine in amino acid 447 (Y447H) in the C. albicans Erg11 resulted in resistance to FLC and ITC but maintenance of a high susceptibility to VRC. Similarly, mutations in the amino acid 54 of the A. fumigatus Cyp51A, replacing glycine with arginine, glutamate, or tryptophan, resulted in resistance to ITC/PSC but not to VRC (14). These findings indicate that there are multiple amino acid residues of Erg11/Cyp51 in each species that can differentially affect the interaction of the enzyme with triazoles.
Molecular model of C. neoformans ERG11.
The molecular model of cryptococcal Erg11 has been published (23). However, the location of Y145 and its position relative to the substrate (eburicol) or azoles have not been clearly described. We performed homology modeling and molecular dynamics to understand the structural importance of Y145 in the Erg11 molecule (see Fig. S2 in the supplemental material). We hypothesize that such approaches might explain the different effects observed with FLC and VRC as well as ITC and PSC. The specific domains in the Erg11 mutant which enable stable docking of these triazoles may be structurally different from those in the wild type. However, we failed to use methods of molecular dynamics with our models to corroborate our experimental observations with in silico analysis (data not shown). How Y145F is affecting the interaction of Erg11-substrate or Erg11-azoles may require X-ray crystallography or multiple-dimensional nuclear magnetic resonance. Nevertheless, these techniques are difficult to apply to large membrane proteins such as Erg11.
Sterol profiles of the isolates.
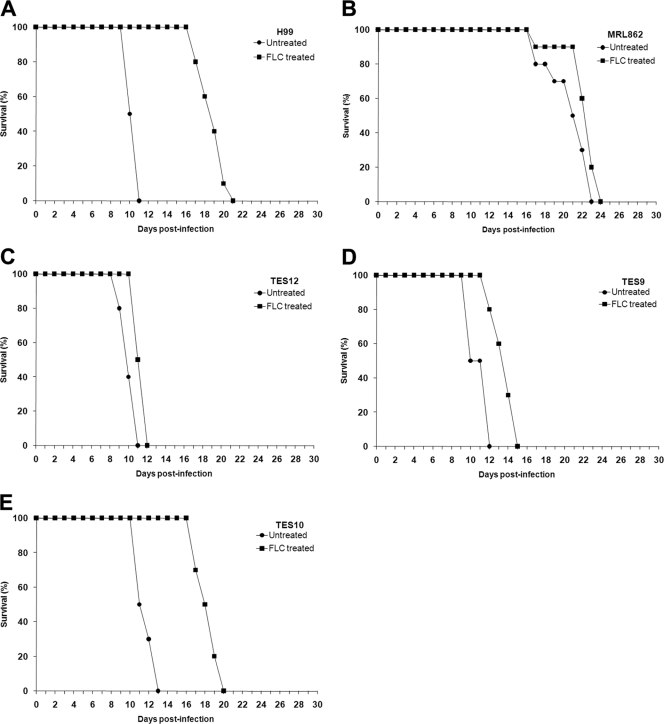
We have previously shown that FLC treatment of the H99 strain resulted in a decrease of ergosterol content accompanied by an increase of eburicol and obtusifolione, which was designated an unidentified sterol intermediate (24). To understand the specific impact of Erg11 dysfunction, the sterol profiles in the mutants were analyzed. Without azole treatment, ergosterol was the major fraction of the total sterol content and a minor peak of ergost-7-enol was detected in all strains (see Fig. S3A in the supplemental material). These results are consistent with our previous observations (24). However, the strains containing the Y145F mutation, TES9, TES12, and MRL862, exhibited a low level of eburicol without any drug treatment (Fig. 2; see Fig. S3A in the supplemental material). Following FLC treatment, the ergosterol content was dramatically reduced in strains containing Y145 (H99 and TES10) and eburicol became the predominant sterol (Fig. 2; see Fig. S3B in the supplemental material). This result corroborates the classic pattern of cryptococcal sterol 14α-demethylase activity inhibited by azoles (17, 24). In strains containing the Y145F mutation, the ergosterol content was also reduced, while the eburicol amount increased under FLC treatment. However, the relative levels of ergosterol were significantly higher and those of eburicol were significantly lower in strains containing the Y145F mutation than in strains without the mutation (Fig. 2; P < 0.05). Similarly, upon FLC treatment the relative amounts of obtusifolione were lower in strains containing the Y145F mutation than in strains that did not, although the differences were not statistically significant, except for those for the MRL862 strain (P < 0.05). These results suggest that the differences in the sterol levels in the presence or absence of FLC most likely were the result of the Y145F mutation.
Fig 2.
Histogram of relative amounts of major sterol fractions from isolates treated with FLC and ITC. The sterol contents of each strain from untreated, FLC-treated, or ITC-treated cultures were analyzed by gas chromatography. Data are presented as the relative ratio between the sterol contents to the internal cholesterol standard. Bars represent standard deviations of three biological repeats. *, P < 0.05 compared to H99 in each set of treatments.
Accumulation of obtusifolione, a 14-methylated 3-ketosteroid, in the sterol profile of the strains treated with either FLC or ITC was noteworthy and is in agreement with the published reports (17, 26). Sterol analysis revealed eburicol to be the predominant intermediate sterol present in the total sterol fraction of the strains treated with FLC and ITC. This is consistent with blockage of the ergosterol biosynthetic pathway at the C-14 demethylation step. Similarly, in strains treated with ITC, the eburicol and obtusifolione contents increased and ergosterol content decreased (Fig. 2; see Fig. S3C in the supplemental material). However, the relative differences in the contents of these sterols were not significant in most of the strains compared to H99, except for eburicol in TES12.
Therapeutic effect in mice.
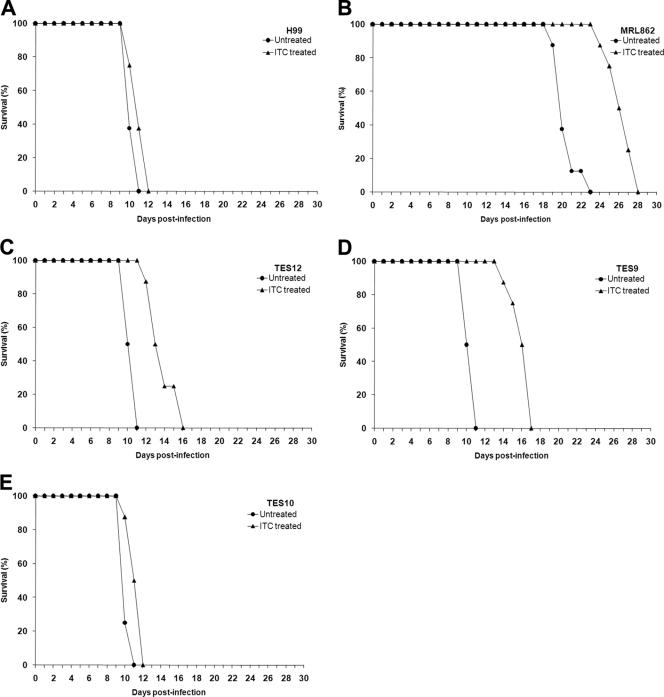
Since MRL862 was isolated from a case of cryptococcosis refractory to FLC treatment, we attempted to determine the therapeutic effect of azoles in mice infected with MRL862 and the H99-derived strains (with or without the Y145F mutation). BALB/c mice were inoculated intravenously with the aforementioned isolates. The infected mice were divided into two groups; one group was treated with azole drugs daily, and the other group received vehicle only (see Materials and Methods). Figure 3 demonstrates the survival rate of infected mice with or without FLC treatment. MRL862, H99, and the three ERG11-modified H99 strains caused 100% mortality in both the control (no treatment) and the FLC-treated groups. However, in the groups infected with H99 or TES10 (Y145), the mice survived longer with FLC treatment than the untreated controls (Fig. 3A and E; P < 0.0001). FLC therapy had no effect in mice infected with the MRL862 strain (Fig. 3B; P = 0.0702) or the TES12 strain (containing all five missense mutations) (Fig. 3C; P = 0.0606). Mice infected with the TES9 (with Y145F) strain responded more poorly to FLC than those infected with H99 and/or TES10 (Fig. 3D; P = 0.002). In contrast, ITC treatment did not prolong survival of the mice infected with H99 or TES10 (no Y145F) (Fig. 4A and E; P > 0.05). However, a higher efficacy of ITC treatment was discernible in the animals infected with strains containing the Y145F mutation (MRL862, TES12, or TES9) (Fig. 4B to D; P < 0.001), although all mice eventually succumbed to infection. The inocula in these experiments were rather high (5 × 104 cells/mouse via the intravenous route), and the differences in the efficacy of FLC and ITC treatment could have been more pronounced with a lower inoculum, depending on the status of the infecting strains' Erg11 145 residue. These animal data corroborated the in vitro susceptibility of the strains to the different triazoles and confirm the importance of the Y145F missense mutation for differential susceptibility toward FLC versus ITC.
Fig 3.
Effects of FLC treatment on the survival of mice. The mice were challenged intravenously with the designated strains with an inoculum of 5 × 104 CFU/mouse. FLC was administered intraperitoneally at a concentration of 10 mg/kg/day. Survival rates (percentages) for FLC-treated animals and untreated controls are plotted against the number of days after inoculation.
Fig 4.
Effects of ITC treatment on the survival of mice. The mice were challenged intravenously with the designated strains with an inoculum of 5 × 104 CFU/mouse. ITC was administered intraperitoneally at a concentration of 10 mg/kg/day. Survival rates (percentages) for ITC-treated animals and untreated controls are plotted against the number of days after inoculation.
The present study on the characterization of the Erg11 mutation in MRL862 has several significant aspects. First, the MRL862 strain showed the highest MIC as well as the highest level of heteroresistance to FLC (128 μg/ml) observed in more than 100 environmental and clinical isolates screened (24). Second, the MRL862 strain was highly resistant to FLC and VRC but relatively sensitive to ITC and PSC. Third, differences in susceptibility to the two groups of triazoles were reflected in the treatment efficacy in mice infected with this strain.
Recently, we have reported that C. neoformans strains acquire resistance to high concentrations of FLC by duplications of multiple chromosomes in response to drug pressure (25). Indeed, the MRL862 strain produced heteroresistant clones that could tolerate FLC concentrations up to 400 μg/ml, and the heteroresistant clones contained two copies of chromosome 1 (data not shown). Importantly, it has been shown that the duplication of chromosome 1 is closely associated with two of its resident genes: ERG11 and AFR1, the major transporter of azoles in C. neoformans (25). Upon repeated transfer in drug-free medium, the MRL862 heteroresistant clones that could tolerate 400 μg/ml FLC returned to the original level of heteroresistance (128 μg/ml) by losing the extra copy of chromosome 1. These observations indicate that the mutations in Erg11 do not interfere with the mechanism of heteroresistance, an innate mechanism of azole resistance in C. neoformans. Our findings also suggest that it is important to test susceptibilities to different triazole drugs before choosing the appropriate one to treat patients infected by FLC-resistant C. neoformans strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ashok Varma for critical discussions and reading of the manuscript.
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Published ahead of print 12 December 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Armengou A, Porcar C, Mascaro J, Garcia-Bragado F. 1996. Possible development of resistance to fluconazole during suppressive therapy for AIDS-associated cryptococcal meningitis. Clin. Infect. Dis. 23:1337–1338 [DOI] [PubMed] [Google Scholar]
- 2. Assing K, Birgens H, Arendrup M. 2003. Cryptococcus neoformans var neoformans resistant to fluconazole in an HIV-negative patient with chronic lymphocytic leukemia. Clin. Microbiol. Infect. 9:441–444 [DOI] [PubMed] [Google Scholar]
- 3. Berg J, Clancy CJ, Nguyen MH. 1998. The hidden danger of primary fluconazole prophylaxis for patients with AIDS. Clin. Infect. Dis. 26:186–187 [DOI] [PubMed] [Google Scholar]
- 4. Bicanic T, Harrison T, Niepieklo A, Dyakopu N, Meintjes G. 2006. Symptomatic relapse of HIV-associated cryptococcal meningitis after initial fluconazole monotherapy: the role of fluconazole resistance and immune reconstitution. Clin. Infect. Dis. 43:1069–1073 [DOI] [PubMed] [Google Scholar]
- 5. Brandt ME, et al. 2001. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates in the United States: 1992 to 1994 and 1996 to 1998. Antimicrob. Agents Chemother. 45:3065–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casadevall A, Perfect JR. 1998Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 7. Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 48:2124–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2008Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd edCLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Diaz-Guerra TM, Mellado E, Cuenca-Estrella M, Rodriguez-Tudela JL. 2003. A point mutation in the 14alpha-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friese G, Discher T, Fussle R, Schmalreck A, Lohmeyer J. 2001. Development of azole resistance during fluconazole maintenance therapy for AIDS-associated cryptococcal disease. AIDS 15:2344–2345 [DOI] [PubMed] [Google Scholar]
- 11. Goldman GH, et al. 2004. Evaluation of fluconazole resistance mechanisms in Candida albicans clinical isolates from HIV-infected patients in Brazil. Diagn. Microbiol. Infect. Dis. 50:25–32 [DOI] [PubMed] [Google Scholar]
- 12. Kwon-Chung KJ, Bennett JE. 1992Medical mycology.Lea & Febiger, Philadelphia, PA [Google Scholar]
- 13. Lepesheva GI, Virus C, Waterman MR. 2003. Conservation in the CYP51 family. Role of the B′ helix/BC loop and helices F and G in enzymatic function. Biochemistry 42:9091–9101 [DOI] [PubMed] [Google Scholar]
- 14. Mann PA, et al. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob. Agents Chemother. 47:577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mondon P, et al. 1999. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob. Agents Chemother. 43:1856–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morio F, Loge C, Besse B, Hennequin C, Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66:373–384 [DOI] [PubMed] [Google Scholar]
- 17. Nes WD, et al. 2009. Sterol 24-C-methyltransferase: an enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch. Biochem. Biophys. 481:210–218 [DOI] [PubMed] [Google Scholar]
- 18. Paugam A, et al. 1994. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin. Infect. Dis. 19:975–976 [DOI] [PubMed] [Google Scholar]
- 19. Perea S, et al. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Revankar SG, et al. 2004. Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 324:719–728 [DOI] [PubMed] [Google Scholar]
- 21. Rodero L, et al. 2003. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47:3653–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saag MS, et al. 1999. A comparison of itraconazole versus fluconazole as maintenance therapy for AIDS-associated cryptococcal meningitis. National Institute of Allergy and Infectious Diseases Mycoses Study Group. Clin. Infect. Dis. 28:291–296 [DOI] [PubMed] [Google Scholar]
- 23. Sheng C, et al. 2009. Three-dimensional model of lanosterol 14 alpha-demethylase from Cryptococcus neoformans: active-site characterization and insights into azole binding. Antimicrob. Agents Chemother. 53:3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 53:2804–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6:e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanden Bossche H, et al. 1993. Effects of itraconazole on cytochrome P-450-dependent sterol 14 alpha-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob. Agents Chemother. 37:2101–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varma A, Kwon-Chung KJ. 2010. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob. Agents Chemother. 54:2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Venkateswarlu K, Taylor M, Manning NJ, Rinaldi MG, Kelly SL. 1997. Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob. Agents Chemother. 41:748–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wheat LJ, et al. 2006. Activity of newer triazoles against Histoplasma capsulatum from patients with AIDS who failed fluconazole. J. Antimicrob. Chemother. 57:1235–1239 [DOI] [PubMed] [Google Scholar]
- 30. Zonios DI, Bennett JE. 2008. Update on azole antifungals. Semin. Respir. Crit. Care Med. 29:198–210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.