Abstract
We previously showed that equilibrative nucleoside transporter 1 (ENT1) is a primary ribavirin transporter in human hepatocytes. However, because the role of this transporter in the antiviral mechanism of the drug remains unclear, the present study aimed to elucidate the role of ENT1 in ribavirin antiviral action. OR6 cells, a hepatitis C virus (HCV) replication system, were used to evaluate both ribavirin uptake and efficacy. The ribavirin transporter in OR6 cells was identified by mRNA expression analyses and transport assays. Nitrobenzylmercaptopurine riboside (NBMPR) and micro-RNA targeted to ENT1 mRNA (miR-ENT1) were used to reduce the ribavirin uptake level in OR6 cells. Our results showed that ribavirin antiviral activity was associated with its accumulation in OR6 cells, which was also closely associated with the uptake of the drug. It was found that the primary ribavirin transporter in OR6 cells was ENT1 and that inhibition of ENT1-mediated ribavirin uptake by NBMPR significantly attenuated the antiviral activity of the drug as well as its accumulation in OR6 cells. The results also showed that even a small reduction in the ENT1-mediated ribavirin uptake, achieved in this case using miR-ENT1, caused a significant decrease in its antiviral activity, thus indicating that the ENT1-mediated ribavirin uptake level determined its antiviral activity level in OR6 cells. In conclusion, our results show that by facilitating its uptake and accumulation in OR6 cells, ENT1 plays a pivotal role in the antiviral effectiveness of ribavirin and therefore provides an important insight into the efficacy of the drug in anti-HCV therapy.
INTRODUCTION
Chronic hepatitis C is a major cause of liver cirrhosis and hepatocellular carcinoma, and a combination of interferon-α (IFN-α) and ribavirin is a standard anti-hepatitis C virus (HCV) therapy. Since the addition of ribavirin to IFN-α significantly improves the rate of sustained virologic response (SVR) (40 to 60% in genotype 1 patients) (5), the drug plays a key role in current anti-HCV therapy.
Ribavirin, a purine nucleoside analog, is phosphorylated intracellularly to form mono-, di-, and tri-phosphates, which then accumulate within cells at high concentrations (4, 13). While the primary anti-HCV mechanisms of the drug are still under debate, it is considered likely that the important actions take place within the cells themselves, and several mechanisms have been proposed to explain what occurs there. These include inhibition of inosine monophosphate dehydrogenase (reviewed in references 4 and 7 and references therein). Additionally, a recent study revealed that ribavirin potentiates IFN-α action by augmenting IFN-stimulated induction of gene expression (16).
Taking into consideration the above-mentioned mechanisms, it is reasonable to assume that the uptake of ribavirin into hepatocytes is a prerequisite for its antiviral activity. Since ribavirin is a hydrophilic molecule, import of the drug into cells requires host nucleoside transporters, which are divided into two families: equilibrative nucleoside transporters (such as ENT1 to ENT4) and concentrative nucleoside transporters (such as CNT1 to CNT3) (9). ENTs are facilitated transporters, while CNTs are sodium-dependent active transporters. These transporters differ in tissue distribution, substrate preference, and inhibitor sensitivity. For example, sensitivities to inhibition by nitrobenzylmercaptopurine riboside (NBMPR) are different between ENT1 and ENT2 (20).
Our recent investigations into the ribavirin uptake system in human hepatocytes determined that ENT1 is a primary ribavirin uptake transporter (6). In addition, Morello et al. (12) reported the association of an intronic single nucleotide polymorphism (SNP) of the SLC29A1 (ENT1) gene with rapid virologic response (RVR; defined as an undetectable serum HCV RNA level at week 4) of treatment of genotype-1 Caucasian patients. More recently, Tsubota and colleagues revealed that another intronic SNP in the SLC29A1 gene is associated with SVR, as well as RVR, in genotype-1 Japanese patients (18). Based on these findings, it can be hypothesized that ENT1 plays an essential role in ribavirin anti-HCV activity.
In the present study, along with a detailed characterization of ribavirin uptake and its relationship to antiviral activity, we tested the above-mentioned hypothesis through the use of OR6 cells, which have been established as an efficient replication system for the HCV RNA genome. The HCV replication level was evaluated by monitoring the level of Renilla luciferase activity (8), which enabled us to simultaneously evaluate both ribavirin uptake and its antiviral activity.
MATERIALS AND METHODS
Cell culture.
OR6 cells were cloned from ORN/C-5B/KE cells (derived from Huh-7 cells) supporting genome-length HCV RNA (strain O of genotype 1b) containing the Renilla luciferase reporter gene, and the cells were cultured as described previously (8). Huh-7 cells were obtained from the Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). The Huh-7 cells were cultured at 37°C with 5% CO2–95% air in RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin.
Luciferase reporter assay.
OR6 cells were plated 1 day prior to the assay on 24-well plates at 1.5 × 104 to 2.5 × 104 cells/well, followed by treatment with ribavirin (Wako, Osaka, Japan) in the absence of G418 and at the indicated concentrations for 24, 48, and 72 h. The cells were then subjected to the luciferase assay using a dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's protocol. For data normalization, the protein contents were determined with a Pierce 660-nm protein assay reagent (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's protocol. The relative luciferase activity value of the untreated or vehicle treated cells (dimethyl sulfoxide [DMSO] for NBMPR and sterile water for others) was set to 100%. NBMPR (Sigma, St. Louis, MO), hypoxanthine (MP Biomedicals, Solon, OH), and formycin B (Berry & Associates, Ann Arbor, MI) were included in inhibition analyses at various concentrations.
Western blot analysis.
OR6 cells treated with ribavirin at various concentrations in the absence of G418 for 24, 48, and 72 h were harvested and homogenized. The homogenates (60 μg/well) were resolved in a sodium dodecyl sulfate (SDS)–15% polyacrylamide gel and then transferred onto a nitrocellulose membrane. The membrane was blocked with 5% skim milk and then incubated with either antibodies against the HCV core protein (2,000-fold dilution; Institute of immunology, Tokyo, Japan) or antibodies against β-actin (500-fold dilution; Sigma). Immunocomplexes were detected with enhanced chemiluminescence (ECL) Western blotting detection reagents (GE Healthcare, Giles, United Kingdom).
Accumulation assay.
OR6 cells were plated 1 day prior to the assay on 24-well plates, after which the cells were incubated with 0.5 μCi/ml [3H]ribavirin (Moravek Biochemicals, Brea, CA) and nonradiolabeled ribavirin at various concentrations. NBMPR was included in inhibition analyses at concentrations of 0.1, 1, 3, 10, 31, and 100 μM. After treatment for 9.6, 24, 48, or 72 h, the cells were washed twice with ice-cold Na+-free Krebs-Henseleit buffer (KHB) and lysed with 0.2% SDS. Radioactivity was measured using a liquid scintillation counter (LSC 5100; Aloka, Tokyo, Japan). The protein contents were determined as described above. To completely inhibit ENT-mediated ribavirin uptake, 30 μM dipyridamole (Wako) was used in the same experimental sets (20). The data were calculated by subtracting the accumulation values obtained with dipyridamole from those without dipyridamole at the same ribavirin concentrations. All assays were performed at 37°C.
Transport assays.
Transport assays were performed using the previously described centrifugal filtration method (6). OR6 cells were collected and resuspended in ice-cold Na+-containing KHB or Na+-free KHB at 1.4 × 106 cells/ml. NBMPR, troglitazone (Wako), hypoxanthine, and formycin B were included in the inhibition analyses. Since the rate of ribavirin uptake by OR6 cells was linear for at least 60 s in the preliminary assays, the incubation time was set to 30 s. The radioactivity and protein contents of the cells used in the assay were measured as described above. The same experiments were also performed at 4°C, and the data were obtained by subtracting the uptake levels at 4°C from those at 37°C at the same ribavirin concentrations.
Total RNA preparation, cDNA synthesis, reverse transcription-PCR (RT-PCR), and quantitative real-time PCR (qPCR).
Total RNA preparation, cDNA synthesis, RT-PCR, and qPCR were performed using previously described procedures (6). Among the nucleoside transporters, ENT1, ENT2, CNT2, and CNT3 mRNAs were examined by RT-PCR because they have been identified as ribavirin transporters (21). The primers for RT-PCR and qPCR are listed in Table S1 in the supplemental material. The UPL universal probes used were no. 9 (ENT1), no. 48 (ENT2), and no. 60 (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]).
Knockdown of ENT1 mRNA expression in OR6 cells.
The BLOCK-iT Pol II miR RNAi expression vector kit (Invitrogen) was used to suppress ENT1 mRNA expression in OR6 cells. The oligonucleotide containing micro-RNA targeted to ENT1 mRNA (miR-ENT1) was cloned into the pcDNA6.2-GW/EmGFP-miR vector. The control plasmid pcDNA6.2-GW/EmGFP-miR-neg, carrying an insert that is not known to target any identified vertebrate genes (miR-Neg), was used as a negative control. The sequences of inserts are shown in Table S1 in the supplemental material. The plasmids were transfected into OR6 cells using Lipofectamine LTX (Invitrogen). Two days after transfection, the culture medium was replaced with fresh medium containing 4 μg/ml blasticidin to obtain cells stably expressing miR-ENT1 (OR6/miR-ENT1) and cells stably expressing miR-Neg (OR6/miR-Ng).
Data analysis.
Statistical analysis was performed using Student's t test. The four-parameter logistic model was used to calculate the 50% effective concentration (EC50).
RESULTS
Concentration- and time-dependent anti-HCV activity and accumulation of ribavirin in OR6 cells.
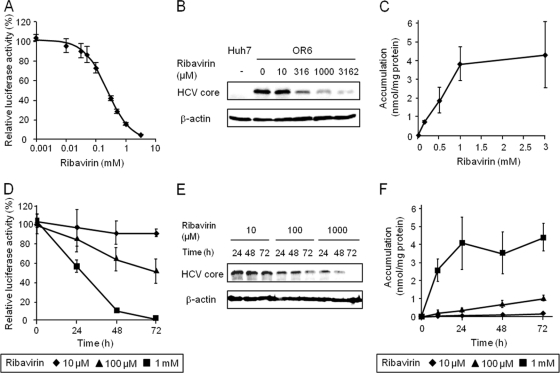
The inhibitory effects of ribavirin (1 to 3,162 μM) on HCV replication in OR6 cells were analyzed by monitoring the luciferase activity and HCV core protein expression levels. It was found that the HCV replication activity and core protein levels decreased in a ribavirin concentration-dependent manner (Fig. 1A and B), while the level of ribavirin accumulation increased in a saturable manner (Fig. 1C). Next, the time course of anti-HCV activity of ribavirin at concentrations of 10, 100, and 1,000 μM was examined. The results of our examination showed that, similar to the concentration-dependent profile, the HCV replication activity and core protein amounts decreased over time at each of the ribavirin concentrations tested (Fig. 1D and E) and that the levels of ribavirin accumulation increased linearly or saturably over time (Fig. 1F). These results suggest that ribavirin exerts concentration- and time-dependent antiviral activity that could be associated with the concentration- and time-dependent intracellular accumulation of the drug.
Fig 1.
Concentration- and time-dependent profiles of anti-HCV activity and accumulation of ribavirin in OR6 cells. (A) OR6 cells were treated with ribavirin at concentrations of 0, 1, 10, 31, 50, 100, 316, 500, 1,000 and 3,162 μM for 48 h. The value of relative luciferase activity in the absence of ribavirin was set to 100%. (B) Expression levels of HCV core protein in OR6 cells treated with ribavirin for 48 h were examined by Western blot analysis. β-Actin was used as a loading control. Huh-7 cells were used as a negative control. (C) OR6 cells were treated with ribavirin at concentrations of 0.1, 0.5, 1, and 3 mM for 48 h, after which the radioactivity within the cells was determined. (D) OR6 cells were treated with ribavirin. The value of relative luciferase activity in the absence of ribavirin at each time point was set to 100%. (E) Expression levels of HCV core protein in OR6 cells treated with ribavirin were examined by Western blot analysis. (F) OR6 cells were treated with ribavirin, after which the radioactivity within the cells was determined. Values are means and standard deviations (SD) of the relative luciferase activity or the accumulation for three independent experiments. Each experiment was performed in duplicate. For Western blotting, the representative result for three independent assays was shown.
Identification of the ribavirin uptake transporter in OR6 cells.
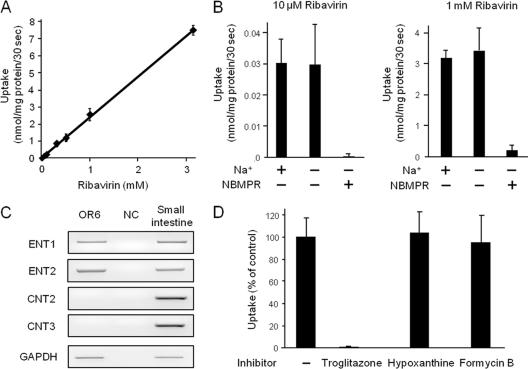
To identify the ribavirin uptake transporter in OR6 cells, we characterized the uptake profile of the drug and the nucleoside transporters mRNA expression in the cells. The ribavirin (1 to 3,162 μM) uptake level in Na+-plus KHB was found to increase linearly up to 3 mM (Fig. 2A), and the uptake activities of the drug (nmol/mg protein/30 s) at 10, 100 (data not shown), and 1,000 μM were recorded as 0.03 ± 0.01, 0.33 ± 0.02 and 3.2 ± 0.3, respectively (Fig. 2B). The removal of Na+ from the transport medium did not affect the uptake activities at any of the ribavirin concentrations tested, indicating that all the uptake activities of the drug were sodium independent. These activities were mostly abolished by the addition of 100 μM NBMPR, an inhibitor of ENT1 and ENT2. Consistently, the results of RT-PCR showed that ENT1 and ENT2 mRNAs were abundantly expressed in OR6 cells, while hardly any CNT2 and CNT3 mRNAs were expressed (Fig. 2C). During the above-described experiments, we found that a low concentration of NBMPR (100 nM) failed to inhibit ribavirin uptake by OR6 cells (M. Iikura, unpublished data). Considering that ENT1-mediated nucleoside uptake is generally sensitive to NBMPR inhibition at 100 nM (20), it was hypothesized that ENT2 should have contributed to ribavirin uptake in OR6 cells. However, our previous results indicated that ENT2 cannot transport ribavirin (6). Therefore, to clearly distinguish between ENT1- and ENT2-mediated ribavirin uptake, inhibition analysis was performed using troglitazone (60 μM), hypoxanthine (5 mM), and formycin B (50 μM). Troglitazone has been reported to specifically inhibit ENT1 activity (10). Hypoxanthine and formycin B, at the indicated concentrations, were previously reported to preferentially inhibit ENT2 activity (3, 22), and we confirmed the inhibitory effects of these compounds on ENT2 activity by using HeLa cells (see Fig. S1 in the supplemental material). The results of the inhibition analysis showed that troglitazone completely inhibited the ribavirin uptake activity, while neither hypoxanthine nor formycin B inhibited uptake of the drug in OR6 cells (Fig. 2D). Taken together, the results indicated that, even though the affinity of ENT1 of OR6 cells for NBMPR was somehow reduced, ENT1 was exclusively responsible for the ribavirin uptake in OR6 cells.
Fig 2.
Identification of the ribavirin uptake transporter in OR6 cells. (A) The concentration dependence of ribavirin uptake (concentrations are given in the legend to Fig. 1A) by OR6 cells was analyzed in Na+-containing KHB. (B) Ribavirin uptake by OR6 cells was analyzed in Na+-containing KHB and Na+-free KHB. In inhibition assays, the effect of 100 μM NBMPR on ribavirin uptake was analyzed in Na+-free KHB. (C) ENT1, ENT2, CNT2, CNT3 and GAPDH mRNA expression was examined by RT-PCR. Small intestine cDNA was used as a PCR control. NC, nontemplate control. Representative results from one of three independent analyses are shown. (D) To clearly distinguish between ENT1- and ENT2-mediated ribavirin uptake, inhibition analysis of ribavirin (100 μM) uptake by OR6 cells was performed in Na+-free KHB in the absence of inhibitor (−) or the presence of troglitazone (ENT1 inhibitor, 60 μM), hypoxanthine (ENT2 inhibitor, 5 mM), or formycin B (ENT2 inhibitor, 50 μM). The value of the transport activity of the control (no inhibitor) was set to 100%. In the above-described experiments, each value is the mean plus SD from three independent experiments, each performed in duplicate.
Effect of inhibition of ribavirin uptake on its anti-HCV activity.
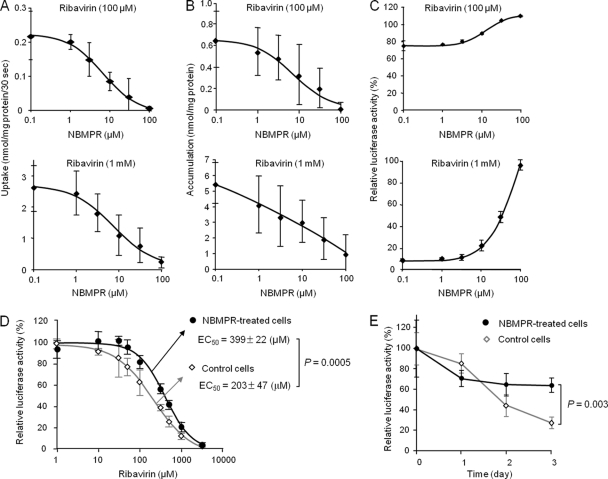
After it was determined that ENT1 was responsible for ribavirin uptake in OR6 cells, the role of ENT1 in the anti-HCV activity of the drug (100 μM and 1 mM) was examined by chemical inhibition of ENT1-mediated ribavirin uptake in OR6 cells. Since troglitazone itself somewhat repressed HCV replication in OR6 cells (Iikura, unpublished), NBMPR was used as an ENT1 inhibitor. As shown in Fig. 3A, NBMPR decreased the level of ribavirin uptake in a dose-dependent manner and, accordingly, decreased the accumulation level of the drug in a dose-dependent manner (Fig. 3B). In association with these decreases, it was determined that the ribavirin antiviral effect was weakened by NBMPR in a concentration-dependent manner (Fig. 3C). We confirmed that ENT1 protein expression was not changed in the cells treated with the highest ribavirin and NBMPR concentrations for 48 h (see Fig. S2 in the supplemental material). To further clarify the importance of ENT1-mediated ribavirin uptake in its antiviral effects, the concentration and time dependencies of the antiviral effects of the drug were examined in cells treated with NBMPR or its vehicle (0.1% DMSO). The concentration of NBMPR was set to 7 μM, which is near the EC50 against ENT1 activity calculated from the results of Fig. 3A, indicating that the ENT1 activity level of NBMPR-treated cells was approximately half that of the vehicle-treated cells. As shown in Fig. 3D, the EC50 of ribavirin in the NBMPR-treated cells was 399 ± 22 μM, which was significantly higher than that of the vehicle-treated cells (203 ± 47 μM, P = 0.0005) (The results of the individual experiments are shown in Fig. S3 in the supplemental material.) In addition, the response to ribavirin in the NBMPR-treated cells was significantly delayed in comparison to that in the vehicle-treated cells (Fig. 3E). We also examined the constraining effects of ENT2 inhibitors on ribavirin antiviral activity but found that hypoxanthine (5 mM) and formycin B (50 μM) had no effect (see Fig. S4 in the supplemental material). Furthermore, NBMPR, hypoxanthine and formycin B were found to have no effect on HCV replication activity in the above-described experiments (see Fig. S4 in the supplemental material), and NBMPR (7 μM) failed to affect telaprevir antiviral activity (see Fig. S5 in the supplemental material).
Fig 3.
Inhibitory effect of NBMPR on ribavirin uptake, accumulation, and anti-HCV activity. The ribavirin concentration used in these experiments (A to C) was 100 μM or 1 mM, while the NBMPR concentrations used were 0.1, 1, 3, 10, 31, and 100 μM. (A) The effect of NBMPR on ribavirin uptake by OR6 cells was analyzed in Na+-free KHB with NBMPR. Each value is the mean ± SD from five independent experiments, each performed in duplicate. (B) The effect of NBMPR on ribavirin accumulation in OR6 cells was analyzed by measuring the level of the drug within the cells, in the presence of NBMPR, for 48 h. Each value is the mean ± SD from three independent experiments, each performed in duplicate. (C) The effect of NBMPR on the anti-HCV activity of ribavirin in OR6 cells was analyzed by measuring the level of the luciferase activity, in the presence of NBMPR, for 48 h. The value of relative luciferase activity without ribavirin and NBMPR was set to 100%. Each value is the mean ± SD from three independent experiments, each performed in triplicate. (D) The concentration dependency of ribavirin antiviral action in the presence of NBMPR was examined. The ribavirin concentrations used are shown in the legend to Fig. 1A. The NBMPR concentration was set to 7 μM, which is near the EC50 of NBMPR calculated from the results in panel A. The value of relative luciferase activity in the absence of ribavirin was set to 100%. (E) The time dependency of ribavirin antiviral action in the presence of NBMPR was then examined. The ribavirin concentration was set to 150 μM, while the NBMPR concentration was still 7 μM. The value of relative luciferase activity in the absence of ribavirin at each time point was set to 100%.
These results clearly show that inhibition of ENT1-mediated ribavirin uptake significantly attenuates ribavirin antiviral effectiveness by reducing the accumulation level of the drug in the cells.
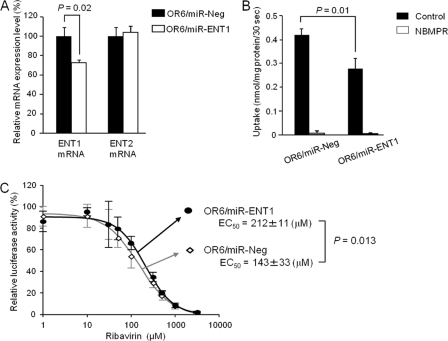
Effect of ENT1 mRNA knockdown on ribavirin anti-HCV activity.
The above-mentioned results prompted us to investigate whether a small change in ENT1 activity would similarly affect ribavirin antiviral effectiveness. miRNA targeted to ENT1 mRNA was used in this examination. We found that when stably expressed in OR6 cells (OR6/miR-ENT1), miR-ENT1 reduced the ENT1 mRNA expression level to 72.5 ± 3.4% of that of the control cells (OR6/miR-Ng) without affecting the ENT2 mRNA expression level (Fig. 4A). Accordingly, the ribavirin uptake level in OR6/miR-ENT1 cells was about 66.7 ± 14.0% of that in OR6/miR-Ng cells (Fig. 4B). To determine the degree to which this ENT1 mRNA knockdown affected ribavirin antiviral action, concentration dependencies of ribavirin action in OR6/miR-ENT1 and OR6/miR-Ng cells were characterized. We found that the EC50 of ribavirin in OR6/miR-ENT1 cells was 212 ± 11 μM, which was significantly higher than the EC50 in OR6/miR-Ng cells (143 ± 33 μM; P = 0.013) (The results of the individual experiments are shown in Fig. S3 in the supplemental material.) These results showed that even a small reduction in the ENT1 mRNA expression level could decrease the ribavirin uptake level, thus causing a reduction in the antiviral efficacy of the drug.
Fig 4.
Effect of ENT1 mRNA knockdown on anti-HCV activity of ribavirin. (A) The expression levels of ENT1 and ENT2 mRNA were determined by real-time PCR. Abundance is shown relative to the level of ENT1 or ENT2 mRNA in OR6/miR-Ng cells. Each value is the mean plus SD from three independent experiments, each performed in duplicate. (B) Ribavirin (100 μM) uptake by OR6/miR-ENT1 and OR6/miR-Ng cells was analyzed in Na+-free KHB in the absence (control) or presence of 100 μM NBMPR. Each value is the mean plus SD of transport activity from three independent experiments, each performed in duplicate. (C) The concentration dependency of ribavirin in OR6/miR-Ng and OR6/miR-ENT1 cells was then examined. The ribavirin concentrations used are shown in the legend to Fig. 1A. The relative luciferase activity value in the absence of ribavirin in each cell line was set to 100%. Each value is the mean ± SD of relative luciferase activity from four independent experiments, each performed in triplicate.
Toxicological analyses.
Concurrent with the above-described experiments, the cytotoxic effects of ribavirin and other reagents on OR6 cells were examined independently and/or simultaneously (see the supplemental methods in the supplemental material). As shown in Table S2 and Fig. S6 of the supplemental materials, the lactate dehydrogenase (LDH) release assay results showed that no severe cytotoxicity in OR6 cells occurred in any treatments (less than 10%). Microscopic observation also showed that the cells were viable upon treatment with ribavirin (3,162 μM) together with NBMPR (100 μM) for 48 h (see Fig. S2 in the supplemental material). We further performed the MTS assay, which can detect different types of toxicity, to confirm the results of the LDH assay. The results showed that even though marginal toxicity was observed at the highest ribavirin and NBMPR concentrations tested (at most 25%), most treatments did not display severe cytotoxicity for OR6 cells (less than 10%; see Table S2 in the supplemental material).
DISCUSSION
In this paper, we provide results supporting our hypothesis that ENT1 plays an essential role in the anti-HCV activity of ribavirin through detailed characterization of the antiviral activity of the drug and its association with ENT1-mediated uptake in OR6 cells.
Our results showed that the concentration and time dependency of ribavirin antiviral activity was closely associated with its accumulation in OR6 cells. This association is supported by several reports. For example, it has been reported that larger ribavirin accumulations were associated with significant decreases in the intracellular GTP pool (13) or with higher antiviral potency against the Hantaan virus (14). Therefore, it is considered likely that continuous ribavirin accumulation in hepatic cells at the higher levels, which are achieved by the sustained and higher ribavirin extracellular concentrations, is critical to the antiviral efficacy of the drug.
Due to its hydrophilicity, ribavirin requires a “gate” to penetrate the plasma membrane of cells prior to its accumulation. Our results clearly show that ENT1 provides this gate, thus facilitating the drug's import into and accumulation in OR6 cells. Since we recently showed that ENT1 is also exclusively involved in ribavirin uptake in human hepatocytes, which has a ribavirin uptake profile similar to that of OR6 cells (6), it is considered likely that this ENT1 role can probably be extended to human hepatocytes as well. The mode of ENT1-mediated ribavirin uptake in OR6 cells, as well as human hepatocytes, was represented by a linear increase in the uptake level along with an increase in extracellular ribavirin concentration (6; also this study). This uptake feature was the most probable reason why the higher extracellular ribavirin concentration resulted in a stronger antiviral effect in OR6 cells but might also explain why clinical findings show that a higher exposure to ribavirin leads to the better virologic response in HCV genotype-1 patients (11, 17). Therefore, our results, together with the other findings, indicate that ENT1 plays an indispensable role in ribavirin antiviral activity.
The importance of ENT1 in ribavirin antiviral activity was further underscored by the results of both the ENT1 knockdown and uptake inhibition experiments using NBMPR. It is noteworthy that even a small reduction of ENT1 activity significantly weakened ribavirin's antiviral potency. These results indicate that increasing or decreasing ENT1 activity level in the cells results in stronger or weaker ribavirin efficacy by increasing or reducing the uptake of the drug, even if extracellular ribavirin concentrations and exposure durations are constant. Therefore, it can be concluded that the ENT1-mediated ribavirin uptake level determines the level of ribavirin antiviral activity in OR6 cells and, presumably, in human hepatocytes.
The above-mentioned findings and suppositions prompt us to propose the following two possibilities (see Fig. S7 in the supplemental material). One is that patients with higher ENT1 activity levels in hepatocytes could more likely attain RVR (defined as a faster and stronger ribavirin antiviral effect in the early stage of the treatment) than those with lower ENT1 activity levels, when other factors affecting the treatment outcome are similar. The mechanisms underlying the interindividual difference in the hepatic ENT1 activity level remain unclear, but SNPs are promising candidates for the causal factors that result in the difference. Since two intronic SNPs have been revealed to be associated with RVR (and SVR) (12, 18), investigations should be conducted to determine whether these SNPs have a positive effect on the hepatic ENT1 expression level.
The other possibility is that the hepatic uptake of ribavirin by ENT1 could be hindered by coadministered chemicals, thus resulting in attenuation of the treatment response in some patients, as shown in Fig. 3. Although there have been no clinical reports supporting this possibility, preceding studies have been performed to determine whether hepatic uptake inhibition of pravastatin and metformin, which are hepatocyte-targeting drugs, reduces their effectiveness (1). These drugs are known substrates for hepatic organic ion transporters, and it has been shown that aberrations in these transporters significantly impair their in vivo functions (2, 15). Since, due to attendant complications or other chronic diseases, several drugs are often coprescribed along with ribavirin during treatment regimens, it may be worth considering whether interactions between ribavirin and other drugs at the point of ENT1-mediated uptake can affect the treatment response.
Exploration of these possibilities must await further studies aimed at clarification of the factors affecting the hepatic ENT1 activity level, including the above-described SNP studies and ribavirin-drug interaction studies. The results obtained from such studies could contribute not only to a better understanding of the mode of action of ENT1 on ribavirin antiviral activity but also to identification of the associated markers for RVR or null responses in clinical settings.
It should be noted that, unexpectedly, ENT1 activity was found to be insensitive to inhibition by NBMPR in the nanomolar range in OR6 cells. This was not due to nucleotide alterations in ENT1 cDNA of OR6 cells (Iikura, unpublished). Since OR6 cells were derived from Huh-7 cells, we examined the sensitivity of ENT1 to inhibition of NBMPR using Huh-7 cells and obtained results similar to those obtained with OR6 cells (Iikura, unpublished). Therefore, the lower sensitivity of ENT1 to NBMPR in OR6 cells was thought to have originated from the Huh-7 cells. Although the reason for the altered sensitivity of ENT1 to NBMPR remains unknown at this time, it is believed that the cell-specific posttranslational modification might be involved. It has been reported that defective glycosylation of ENT1 leads to decreased affinity for NBMPR (19). Therefore, it can be speculated that the type or structure of glycochain and/or other modifications could be responsible for decreased affinity of ENT1 of OR6/Huh-7 cells for NBMPR. Further studies aimed at ascertaining the reason might provide novel insights into the biology of ENT1.
Finally, we briefly discuss the static cytotoxic effects of ribavirin and NBMPR on OR6 cells. According to the results of toxicological analyses, these reagents (at most concentrations tested) did not cause severe toxicity in OR6 cells (less than 10%), and only marginal toxicity was found in treatment of the reagents at the highest concentrations tested in an MTS assay. In contrast, blasticidin S treatment (20 ng/ml) significantly damaged the cells (>50% in the MTS assay [Iikura, unpublished]). Therefore, it is assumed that OR6 cells possess inherent resistance to ribavirin and NBMPR, and this factor might be related to the relatively high EC50 of ribavirin. Although we do not know the reason for the behavior of the cells, it is unlikely that the limited toxicity would give rise to a question regarding the present results. In actuality, 100 μM NBMPR treatment, which caused marginal toxicity, did not affect HCV replication activity (see Fig. S4 in the supplemental material).
In conclusion, we have clearly demonstrated that ENT1 plays an indispensable role in ribavirin antiviral activity by facilitating the uptake and accumulation of the drug in OR6 cells, thereby indicating that ENT1 provides a gate that is essential to the success of ribavirin's mission. Our study limitations include an in vitro HCV model system using hepatoma cells and no in vivo evidence of association between hepatic ENT1 activity and ribavirin efficacy. Nevertheless, our results, together with the literature, strongly suggest that ENT1 also plays the determinant role in the antiviral efficacy of ribavirin in the human liver during the course of anti-HCV therapy. Accordingly, it is believed that our results, as well as the ideas described in this paper, will encourage further studies aimed at the clarification of the clinical importance of ENT1 in anti-HCV therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant (20790128) from the Ministry of Education, Sciences, Sports and Culture of Japan and partially supported by a Special Funds for Education and Research (Development of SPECT Probes for Pharmaceutical Innovation) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a research grant from the Nakatomi Foundation (Tokyo, Japan).
Footnotes
Published ahead of print 9 January 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Bachmakov I, Glaeser H, Fromm MF, König J. 2008. Interaction of oral antidiabetic drugs with hepatic uptake transporters: Focus on organic anion transporting polypeptides and organic cation transporter 1. Diabetes 57:1463–1469 [DOI] [PubMed] [Google Scholar]
- 2. Becker ML, et al. 2009. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 9:242–247 [DOI] [PubMed] [Google Scholar]
- 3. Burke T, Lee S, Ferguson PJ, Hammond JR. 1998. Interaction of 2′,2′-difluorodeoxycytidine (gemcitabine) and formycin B with the Na+-dependent and -independent nucleoside transporters of Ehrlich ascites tumor cells. J. Pharmacol. Exp. Ther. 286:1333–1340 [PubMed] [Google Scholar]
- 4. Dixit NM, Perelson AS. 2006. The metabolism, pharmacokinetics and mechanisms of antiviral activity of ribavirin against hepatitis C virus. Cell. Mol. Life Sci. 63:832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 6. Fukuchi Y, Furihata T, Hashizume M, Iikura M, Chiba K. 2010. Characterization of ribavirin uptake systems in human hepatocytes. J. Hepatol. 52:486–492 [DOI] [PubMed] [Google Scholar]
- 7. Hofmann WP, Herrmann E, Sarrazin C, Zeuzem S. 2008. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int. 28:1332–1343 [DOI] [PubMed] [Google Scholar]
- 8. Ikeda M, et al. 2005. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem. Biophys. Res. Commun. 329:1350–1359 [DOI] [PubMed] [Google Scholar]
- 9. Kong W, Engel K, Wang J. 2004. Mammalian nucleoside transporters. Curr. Drug Metab. 5:63–84 [DOI] [PubMed] [Google Scholar]
- 10. Leung GP, Man RY, Tse CM. 2005. Effect of thiazolidinediones on equilibrative nucleoside transporter-1 in human aortic smooth muscle cells. Biochem. Pharmacol. 70:355–362 [DOI] [PubMed] [Google Scholar]
- 11. Lindahl K, Stahle L, Bruchfeld A, Schvarcz R. 2005. High-dose ribavirin in combination with standard dose peginterferon for treatment of patients with chronic hepatitis C. Hepatology 41:275–279 [DOI] [PubMed] [Google Scholar]
- 12. Morello J, et al. 2010. Influence of a single nucleotide polymorphism at the main ribavirin transporter gene on the rapid virological response to pegylated interferon-ribavirin therapy in patients with chronic hepatitis C virus infection. J. Infect. Dis. 202:1185–1191 [DOI] [PubMed] [Google Scholar]
- 13. Smee DF, Matthews TR. 1986. Metabolism of ribavirin in respiratory syncytial virus-infected and uninfected cells. Antimicrob. Agents Chemother. 30:117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y, Chung DH, Chu YK, Jonsson CB, Parker WB. 2007. Activity of ribavirin against Hantaan virus correlates with production of ribavirin-5′-triphosphate, not with inhibition of IMP dehydrogenase. Antimicrob. Agents Chemother. 51:84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tachibana-Iimori R, et al. 2004. Effect of genetic polymorphism of OATP-C (SLCO1B1) on lipid-lowering response to HMG-CoA reductase inhibitors. Drug Metab. Pharmacokinet. 19:375–380 [DOI] [PubMed] [Google Scholar]
- 16. Thomas E, et al. 2011. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology 53:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsubota A, Hirose Y, Izumi N, Kumada H. 2003. Pharmacokinetics of ribavirin in combined interferon-alpha 2b and ribavirin therapy for chronic hepatitis C virus infection. Br. J. Clin. Pharmacol. 55:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsubota A, et al. 2011. Contribution of ribavirin transporter gene polymorphism to treatment response in peginterferon plus ribavirin therapy for HCV genotype 1b patients. Liver Int. [Epub ahead of print.10.1111/j.1478–3231.2011.02727.x [DOI] [PubMed] [Google Scholar]
- 19. Vickers MF, et al. 1999. Functional production and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cerevisiae. Interaction of inhibitors of nucleoside transport with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q). Biochem. J. 339:21–32 [PMC free article] [PubMed] [Google Scholar]
- 20. Ward JL, Sherali A, Mo ZP, Tse CM. 2000. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. J. Biol. Chem. 275:8375–8381 [DOI] [PubMed] [Google Scholar]
- 21. Yamamoto T, et al. 2007. Ribavirin uptake by cultured human choriocarcinoma (BeWo) cells and Xenopus laevis oocytes expressing recombinant plasma membrane human nucleoside transporters. Eur. J. Pharmacol. 557:1–8 [DOI] [PubMed] [Google Scholar]
- 22. Yao SY, et al. 2002. Functional and molecular characterization of nucleobase transport by recombinant human and rat equilibrative nucleoside transporters 1 and 2. J. Biol. Chem. 277:24938–24948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.