Abstract
The ciliated protozoan Tetrahymena thermophila undergoes extensive programmed DNA rearrangements during the development of a somatic macronucleus from the germ line micronucleus in its sexual cycle. To investigate the relationship between programmed DNA rearrangements and transposable elements, we identified several members of a family of non-long terminal repeat (LTR) retrotransposons (retroposons) in T. thermophila, the first characterized in the ciliated protozoa. This multiple-copy retrotransposon family is restricted to the micronucleus of T. thermophila. The REP (Tetrahymena non-LTR retroposon) elements encode an ORF2 typical of non-LTR elements that contains apurinic/apyrimidinic endonuclease (APE) and reverse transcriptase (RT) domains. Phylogenetic analysis of the RT and APE domains indicates that the element forms a deep-branching clade within the non-LTR retrotransposon family. Northern analysis with a probe to the conserved RT domain indicates that transcripts from the element are small and heterogeneous in length during early macronuclear development. The presence of a repeated transposable element in the genome is consistent with the model that programmed DNA deletion in T. thermophila evolved as a method of eliminating deleterious transposons from the somatic macronucleus.
Developmentally programmed DNA rearrangements occur in a wide variety of organisms (reviewed in reference 5). Functions such as altering gene dosage or directly regulating gene expression have been assigned to many but not all examples of programmed DNA rearrangements. A clinically important example of a programmed DNA rearrangement is V(D)J recombination (2). In addition, a variety of mammalian parasites use programmed DNA rearrangements to vary their surface antigens to avoid host immune response (4). The function of other programmed DNA rearrangements is not as clear. The extensive genome rearrangements that occur during nuclear development in the ciliated protozoa provide an example of programmed DNA rearrangements with poorly understood function.
Like all ciliated protozoa, the oligohymenopheran Tetrahymena thermophila displays nuclear dimorphism with a mostly transcriptionally silent diploid germ line nucleus (micronucleus) and a polyploid and transcriptionally active somatic nucleus (macronucleus) within the same cell. The macronucleus develops from a mitotic product of the micronucleus during conjugation. When two cells of different mating types conjugate, the micronucleus in each divides meiotically and mitotically to generate a haploid gametic nucleus that is reciprocally exchanged and fuses with that of its partner to form a zygotic nucleus. This zygotic nucleus divides and from one of the products develops a new macronucleus, while the old macronucleus is concurrently degraded. In T. thermophila, macronuclear development involves extensive programmed DNA rearrangements, including chromosome fragmentation, DNA amplification, and site-specific interstitial DNA deletion (12). Interstitial DNA deletion is responsible for the elimination of approximately 10 to 15% of the germ line genome representing more than 5,000 single and multicopy elements. The size of these internal eliminated sequences in T. thermophila ranges from 0.6 kb to over 22 kb. Internal eliminated sequences have not yet been found in the coding sequence in T. thermophila, but two are located within introns (11, 24).
One model describing the possible function of micronucleus-limited DNA and possible reasons for its absence from the macronucleus proposes that it has a specific function in the germ line micronucleus, serving as centromeres or matrix attachment regions, for example (12, 26). Another model (32) suggests that micronucleus-limited DNA elements represent the remains of an ancient transposon invasion and are eliminated from the streamlined, somatic nucleus to prevent the interruption of essential genes. This model suggests that internal eliminated sequences retain only the cis elements necessary for recognition by a trans-acting excision machinery provided by the host genome. These models are not necessarily mutually exclusive: repetitive DNA families including transposable elements are often found in centromeric or telomeric regions in higher organisms (35, 43).
The finding that small RNAs, the TWI1 gene product, and histone methylation have roles in DNA elimination in T. thermophila (47, 52) provides a possible molecular link between transposable elements and programmed DNA rearrangements. Twi1p belongs to the PPD family, a group of proteins containing conserved Piwi and PAZ domains, some members of which are involved in RNA interference as well as the silencing of transposable elements (51). These observations led to the development of the scan RNA model of DNA elimination (47) that suggests that small RNAs are used by the cell to “template” regions of DNA for elimination.
Transposable elements may be divided into two general classes, DNA transposons and RNA-based retrotransposons. Retrotransposons transpose via an RNA intermediate mediated by a reverse transcriptase (RT) (reviewed in reference 16). Retrotransposons are classified into two main groups. The first group of elements are flanked by long terminal repeats (LTRs) and transpose through a double-stranded DNA intermediate synthesized from the RNA template and then integrate with a mechanism similar to that used by DNA transposons (reviewed in reference 16). LTR retrotransposons contain gag and pol genes and are structurally similar to retroviruses, such as human immunodeficiency virus. Non-LTR elements (retroposons) lack terminal repeats and transpose by a mechanism termed target-primed reverse transcription, reverse transcribing a cDNA copy of their RNA directly into the nicked chromosomal target site (36). Non-LTR retrotransposons (also called long interspersed nuclear elements) are widespread in eukaryotes but absent from the model yeast Saccharomyces cerevisiae.
There are several examples of non-LTR retrotransposons with potential specific chromosomal function. The TART and HeT-A non-LTR retrotransposons transpose to the ends of the chromosomes and compensate for the absence of canonical telomeres in Drosophila melanogaster (35). In Giardia lamblia two families of non-LTR retrotransposons are found exclusively at the ends of chromosomes in head-to-tail arrays, where they may form a buffer between telomeric sequence and single-copy genes (1). In humans, the Lyon repeat hypothesis postulates that long interspersed nuclear elements play a role in the developmentally regulated silencing of the female X chromosome (37, 38).
DNA-based transposons have been characterized in the micronuclear genome of several spirotrichous ciliates (14, 28-30). In T. thermophila, several transposon-like families have been identified. The Tel-1 family is restricted to the micronucleus and structurally resemble transposons (10). The Tel-1 elements are found in the micronucleus, adjacent to internal blocks of tandemly repeated C4A2 telomeric repeat and do not contain open reading frames (ORFs). The Tlr family is also micronucleus limited (54), with the best-characterized family member, Tlr1, containing an 825-bp terminal inverted repeat separated by a long (22-kb) internal region (56). The internal conserved region contains numerous ORFs, some transposon-like, including a retrovirus-like integrase (22), and several that appear to be viral in origin (56). Sequences flanking most Tlr and Tel-1 elements are micronucleus limited, suggesting that they are eliminated as parts of larger tracts of micronucleus-limited sequence (56). Although there has been some speculation that the Tlr family could transpose through an RNA intermediate (22), no canonical retrotransposons have been found to date in the ciliated protozoa.
Repeated sequences that are retrotransposon-like in origin comprise large parts of the genomes of higher organisms (13) but have not yet been identified in the ciliated protozoa. To investigate possible links between retrotransposons and genome rearrangements in the ciliated protozoa, we identified a family of non-LTR elements in T. thermophila. The REP (Tetrahymena non-LTR retroposon) elements appear to be phylogenetically isolated, consistent with the idea that non-LTR retrotransposons are transmitted vertically (16). Consistent with its micronucleus-limited position and the scan RNA model for DNA elimination in T. thermophila (47), REP element transcripts during early conjugation, including meiosis, are heterogeneous in size. The presence of a repeated transposable element within the micronuclear genome is consistent with the model that internal eliminated sequence excision in T. thermophila evolved as a method of ridding the somatic macronucleus of transposable elements.
MATERIALS AND METHODS
Cell strains.
T. thermophila strains CU428 [Mpr/Mpr (VII, mp-s)] and B2086 [Mpr+/Mpr+ (II, mp-s)] of inbreeding line B were provided by J. Gaertig, University of Georgia, Athens. Cells were cultured as previously described (19).
Isolation of micro- and macronuclei.
Micro- and macronuclei were isolated from strain CU428, grown to mid-log phase in 1× SPP (19) by the method of Gorovsky et al. (23) as modified by Howard and Blackburn (27), without the filtration step.
Library screening.
The pMBR micronuclear genomic library (provided by Kathleen Karrer, Marquette University, Milwaukee, Wis.) consists of partially MboI-digested micronuclear DNA from T. thermophila cloned into the SacI site of pUC19 (49). An aliquot (100 ng) of library DNA was transformed into Escherichia coli DH5αF′ made competent with rubidium chloride (50). The library was screened with standard procedures (50). Probes were gel-purified and labeled by random priming (50) with [α-32P]dATP (Amersham).
Southern blotting and DNA manipulations.
Standard molecular biology techniques were performed as described by Sambrook et al. (50) or by following supplier's instructions. DNA-modifying enzymes were obtained from New England Biolabs. Southern blots were hybridized in 1% sodium dodecyl sulfate (SDS)-5× SSPE-0.5% dry milk powder and washed three times at room temperature for 20 min each in 1× SSPE-0.1% SDS, once in 0.1× SSPE-0.1% SDS at 50°C, and imaged with either X-ray film or a Canberra Packard Instant Imager.
RNA isolation and Northern analysis.
Total cell RNA from T. thermophila was isolated with the Trizol reagent (BRL); 6 × 106 cells were concentrated into 100 μl, to which 1 ml of Trizol was added. Whole-cell RNA was purified according to instructions provided by the supplier. The RNA pellet was air-dried before being dissolved in RNase-free double-distilled H2O at ≈4 μg/μl. Northern analysis was performed by separating 20 μg of total RNA on a 1% agarose-0.66 M formaldehyde gel. RNA was transferred to a nylon filter after three 20-min washes in 10× SSC (50). Prehybridization and hybridization were performed at 42°C in 50% formamide-5× Denhardt's solution-1% SDS-5× SSC with sheared, denatured salmon sperm DNA at 100 μg/ml. Filters were washed three times for 15 min in 1× SSC-0.1% SDS at room temperature, and 1 time in 0.25× SSC-0.1% SDS at 42°C for 15 min after overnight incubation with the indicated probes. Filters were stripped of hybridized probe by boiling in 1× TE-1% SDS for 5 min, cooling to room temperature over 10 min, and then washing in 5× SSC for 5 min. The stripped filters were immediately covered with Saran Wrap before being imaged for verification and then stored at 4°C.
Oligonucleotides.
See Table 1 for the oligonucleotides used during this study.
TABLE 1.
Sequences of oligonucleotides used for PCR amplification in this studylegend
| Oligonucleotide | Sequencea |
|---|---|
| TTRTF | 5′-GTAATTCTGCTGGAAATGCTA |
| TTRTR | 5′-GATTGCTGAAGCGTACTATAG |
| RTFII | 5′-GAACAAACATTAAGCGATATT |
| TTPKF | 5′-GARATHCCHGGHYARTAY |
| TTPKR | 5′-GYDGTNARHGYRAADGG |
| RTR | 5′-GATTGCTGAAGCGTAC |
| REV1 | 5′-GAATTCGGATGGGATAGTGA |
| FOR3 | 5′-TGTCAGTTCTCTTGTATAAG |
aR = A + G; H = A + T + C; N = A + T + C + G; Y = T + C; D = A + T + G.
PCR conditions.
PCR was performed as described previously (19). Long-range PCR was performed with the Expand Long Template PCR system (Boehringer Mannheim) with conditions as specified by the supplier. To obtain 5′ sequence from REP6, we first used inverse PCR with primers targeted within the macronucleus-destined HEH2 gene (Fig. 1) to amplify and then sequence the macronuclear locus of the region. We then amplified the 5′ end of REP6 with a primer in the 5′ macronuclear-destined sequence (FOR3, Table 1) and one in REP6 (REV1, Table 1) with whole-cell DNA as a template.
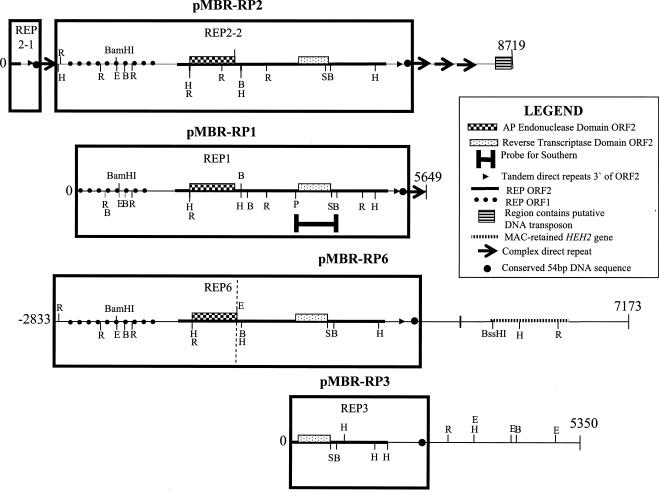
FIG. 1.
Genomic organization and partial restriction maps of pMBR-RP1 to pMBR-RP3 and pMBR-RP6 with additional genomic sequence for pMBR-RP6 (see Materials and Methods). The plasmids are numbered from 0 to the end of sequenced regions, with an additional 2,833 bp of micronuclear sequence added to pMBR-RP6 (−2833 to the vertical dotted line where cloned pMBR-RP6 sequence begins). The putative ORFs are transcribed from left to right. The DNA sequence corresponding to each REP element is boxed (see text for details). Abbreviations: H, HindIII; E, EcoRI; R, EcoRV; B, BglII; S, SacI; P, PstI.
DNA sequencing.
Sequencing was performed with automated cycle sequencing with dye-labeled dideoxy terminators and a PE/ABI 373a or 377 sequencer at the Core Molecular Biology Facility, York University, Toronto, Ontario, Canada.
Sequence assembly and analysis.
The pMBR clones were separately subjected to transposon-based random insertion with the GPS-1 genome priming system (New England Biolabs, Mississauga, Ontario). Restriction enzyme mapping was used to identify a series of subclones that when individually sequenced with universal primers provided for the GPS-1 kit for pMBR-RP1, pMBR-RP2, pMBR-RP4, and pMBR-RP6 were assembled by pairwise sequence alignments with BlastN (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). Gaps were filled by DNA sequencing with specifically designed oligonucleotides. When the entire sequence across each plasmid insert was obtained, it was scanned for open reading frames (ORFs) with homology to known proteins by searching the nonredundant protein databases with the BlastX algorithm. DNA sequences identified as containing a possible match to a known protein were translated with the ciliate genetic code and used to query the nonredundant protein databases with the BlastP algorithm. Reverse position-specific Blast was used to search these regions for potential conserved domains with the conserved domain database of NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Simple Modular Architecture Research Tool (SMART: http://smart.embl-heidelberg.de/) was also utilized for this purpose.
Multiple sequence alignments of the reverse transcriptase (RT) and apurinic/apyrimidinic endonuclease (APE) domains were performed by sequentially adding the sequence of the APE and RT domains of ORF2 from the T. thermophila REP elements from the pMBR-RP clones to the APE and RT profile alignments of Malik et al. (40) with the profile alignment option of ClustalW (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html#profile), where profile 1 was either accession number DS36736 [APE domain (40)] or ds36752 [RT domain (40)] (ftp://ftp.ebi.ac.uk/pub/databases/embl/align/) and profile 2 was the T. thermophila REP element sequences. Alignments were adjusted by hand and smaller versions were shaded by importing the ALN file into the Boxshade server (http://www.ch.embnet.org/software/BOX_form.html) and then editing the postscript file in Adobe Illustrator 9.0.
Phylogenetic analysis of the RT and APE domains.
The resulting profile alignments were used to construct a neighbor-joining tree (N = 1,000 replicates) with ClustalW (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html#trees) which was visualized with the Phylodendron Phylogenetic tree printer (http://www.es.embnet.org/Doc/phylodendron/treeprint-form.html) and edited as a postscript file in Adobe Illustrator 9.0.
Nucleotide sequence accession numbers.
GenBank accession numbers for the DNA sequences of pMBR-RP1 to pMBR-RP3 and pMBR-RP6 are AY371728 to AY371731, respectively.
RESULTS
T. thermophila contains a non-LTR retrotransposon family.
We used degenerate primers (TTPKF and TTPKR, Table 1) to amplify from T. thermophila whole-cell DNA a sequence of ≈700 bp that, when compared against the nonredundant NCBI database, yielded similarity to the RT domain of non-LTR retrotransposons. To amplify DNA sequence containing the RT domain, we used inverse PCR (48) of HindIII-digested T. thermophila whole-cell DNA with primers RTF and RTR (Table 1) to obtain 2.2 kb of sequence, a portion of which was used as a DNA probe to screen a T. thermophila micronuclear DNA library. Hybridizing clones were abundant, suggesting that the sequence is repetitive in the T. thermophila micronuclear genome. Four independent clones were sequenced in full. Pairwise and multiple sequence alignments of the four clones revealed regions of extensive DNA sequence similarity. Further sequence analysis revealed that the similarity arises due to the presence in each plasmid of either a full or partial non-LTR retrotransposon.
Partial restriction maps showing the micronuclear genomic structure of the four pMBR-clones are presented in Fig. 1. Plasmids pMBR-RP1 and -RP3 each contain partial sequence of one REP element (named REP1 and REP3, respectively; see boxed regions, Fig. 1). Plasmid pMBR-RP2 contains one full REP element (REP2-2) as well as partial sequence of a second REP element (REP2-1; see boxed region, Fig. 1). In addition, plasmid pMBR-RP6 contains partial sequence of an additional REP element (REP6; Fig. 1). We amplified additional DNA sequence from T. thermophila micronuclear DNA to complete the entire REP6 sequence (Fig. 1).
5′UTR of the REP elements.
The REP2-2 element is the only REP element identified within the four plasmids potentially to contain an entire REP element with an intact 5′ untranslated region (5′UTR). We verified the 5′ limit of REP element sequence with the additional amplified REP6 sequence (see Fig. 1 and Materials and Methods). We aligned REP2-2 and the complete REP6 sequence and identified a 243-bp region immediately upstream of ORF1 (Fig. 1) that is highly conserved in both REP6 and REP2-2 and is therefore predicted to comprise the entire 5′UTR region of the REP element. The upstream boundary of REP2-2 is therefore the predicted 3′UTR and/or flanking genomic sequence of REP2-1 (Fig. 1).
ORF1.
A potential ORF1 in REP elements was detected by translating the nucleotide sequence upstream of the previously identified ORF2 of REP1 and REP2-2 (Fig. 1) and aligning the predicted protein sequences. The 412-amino-acid protein conceptually encoded by ORF1 of REP2-2 does not contain any significant similarity to other proteins. REP1 contains the DNA encoding the C-terminal 368 amino acids of another ORF1. The overlapping regions of the two ORF1s are 95% identical at the amino acid level (see Fig. A in supplemental material, http://www.yorku.ca/ronp/). Analysis of the predicted amino acid sequence with SMART indicates the possible presence of a zinc finger related to the CCCH class (Fig. B in supplemental material, http://www.yorku.ca/ronp/) as well as a coiled-coil region. The CCCH class of the zinc finger superfamily is found in many RNA-binding proteins and, to our knowledge, has not been found in any non-LTR retrotransposons described to date.
ORF2.
The main conserved feature that the REP elements share with other non-LTR retrotransposons is the 1,147-amino-acid ORF2. Conserved domain analysis of the ORF2 amino acid sequence identified two protein domains characteristic of non-LTR retrotransposons: a reverse transcriptase (RT) domain, and an apurinic/apyrimidinic endonuclease (APE) domain. Many non-LTR retrotransposons encode an APE domain which is related to the AP endonucleases that function in DNA repair (40, 44). The APE domain of at least one non-LTR retrotransposon has been shown to nick DNA with sequence specificity related to its target site (18), although not all non-LTR retrotransposons are associated with specific genomic target sites (40). A multiple sequence alignment of the REP APE domain with that of several other non-LTR retrotransposons is shown in Fig. 2. The REP APE is likely to encode a functional endonuclease, as there is excellent conservation of key residues that have been identified in the APE active site (45).
FIG. 2.
Multiple sequence alignment of the APE domain of REP1 and REP2-2 with APE domains of several non-LTR retrotransposons (see Materials and Methods for details). Key residues identified for APE activity (45) are indicated by an asterisk. Black and grey shading indicates identical and similar amino acid residues, respectively.
Structurally, RT enzymes adopt a hand structure that has a palm containing the active site and fingers and a thumb that loosely hold the RNA template while the RT moves laterally. Xiong and Eickbush (58) found that there are seven blocks of amino acid identity common to the RT of all retroelements. The REP family contains the seven classic domains 1 to 7, as is expected for a non-LTR retrotransposon (Fig. 3). Burke et al. (6) extended non-LTR RT homology to 11 blocks of identity (including domains 0, 2a, 8, and 9) by comparing the three-dimensional structure of the RT domain of the retrovirus human immunodeficiency virus (33) to the RT domain of the R2 non-LTR retrotransposon. The highly conserved PGPD motif in domain 0 (Fig. 3) is absent in the REP family. Burke et al. (6) suggest that this domain is believed, in combination with domain 2a, which is present in the REP elements (Fig. 3), to extend the active site of the non-LTR RT by giving additional or longer fingers compared with that of human immunodeficiency virus RT.
FIG. 3.
Multiple sequence alignment of the RT domain of REP1, REP2-2, REP3, and REP6 with that of several non-LTR retrotransposons (see Materials and Methods for details). RT subdomains are indicated by a corresponding number above the conserved block of amino acid sequence (see text for details). With respect to domain 9, non-LTR retrotransposons are divided into two subgroups based upon the presence or absence of a glycine-rich tetramer (domain 9 versus 9*: see boxed region). Shading of amino acid residues is as in Fig. 2.
Domain 8 appears to be relatively conserved between the REP elements and non-LTR retrotransposons (Fig. 3). Domains 9 and 9* have been used to divide non-LTR transposons into two subgroups (Fig. 3) based upon the presence or absence of a glycine-rich tetramer (40). The multiple sequence alignment (Fig. 3) places the REP elements into the subgroup without the glycine-rich tetramer (domain 9, Fig. 3). It is believed that the function of domain 8 and 9 is to extend the thumb. Domain 7 was identified as the thumb region of human immunodeficiency virus RT (33). It has recently been suggested that the presence of domains 0 and 2a (more/longer fingers) and 8 and 9 (extended thumb) allows the RT to more completely wrap around its RNA template. The biological relevance of this would be higher processivity of the non-LTR RT compared to those of retroviruses and LTR retrotransposons (3). The biological relevance of the absent domain 0 in the REP element RT is not known. The ORF2 of the T. thermophila REP element does not appear to encode a C-terminal DNA-binding domain or an RNase H domain similar to human long interspersed nuclear elements.
3′UTR of REP elements.
Pairwise or multiple sequence alignments of the sequence immediately downstream of the stop codon of each of the five identified full or partial REP elements do not show sequence identity except that four out of the five immediately follow the stop codon with the sequence 5′-AAT/GAA-3′. Four of the five REP elements contain two to eight copies of different tandemly repeated DNA sequence (Fig. C in supplemental material, http://www.biol.yorku.ca/ronp/). The REP element of pMBR-RP3 does not appear to have this sequence organization (Fig. 1). The only conserved DNA sequence common to all five REP elements is a highly conserved 54-bp sequence that is found a distance of 404 bp to 932 bp from the respective REP element stop codon (Fig. 4).
FIG. 4.
Multiple sequence alignment of the conserved 54-bp sequence found 3′ of ORF2 of the REP elements. An arrow represents the point within the conserved sequence where there is a small amount of overlap with the repetitive sequence described in Fig. 5. The numbering begins at the stop codon of ORF2 of the respective REP element. The black shading indicates nucleotides that are identical in all five sequences.
REP elements 1, 2-1, and 2-2 have a ≈350-bp imperfect repeat (Fig. 5) that immediately follows the conserved 54-bp sequence. One of these repeats is downstream of the conserved sequence of both REP1 and REP2-1 while three follow the conserved sequence of REP2-2 (Fig. 5). No 350-bp elements are found downstream of REP3 and REP6.
FIG. 5.
Multiple sequence alignment of the ≈350-bp imperfectly repeated sequence found immediately downstream of the conserved 54-mer (Fig. 4; included here as a part of REP1, REP2-1, and REP2-2-1). The sequence similarity between the repeats of REP2-1, REP2-2-1, and REP2-2-2 extends further than that of REP1 and REP2-2-3. Nucleotide residues that are identical in all five sequences are shaded black.
Genomic organization of each REP element.
A summary of the important features of each of the pMBR plasmids is shown in Fig. 1. The 5′ boundary of pMBR-RP1 exists within the putative coding sequence of ORF1 of REP1 (Fig. 1) while the 3′ pMBR-RP1 boundary almost immediately follows the ≈350-bp repeated sequence element (Fig. 5).
Plasmid pMBR-RP2 contains an entire REP element (REP2-2) as well as the 3′ terminal sequence of another (REP2-1, Fig. 1). The 5′boundary of the plasmid pMBR-RP2 exists within the C-terminal 19 amino acids of ORF2 of REP2-1 (Fig. 1). Therefore, pMBR-RP2 should also contain the 3′UTR of REP2-1. The putative 3′-flanking sequence of REP2-1 contains one copy of the ≈350-bp repeat while three copies follow REP2-2 (Fig. 1). From the analysis of REP2-2 and REP6, we estimate that the size of an intact REP element is approximately 6.1 kb.
Downstream of the three imperfect repeats that flank the REP2-2 element is a region that contains a sequence that appears to be the C terminus of an ORF encoded by a DNA transposon of the Tc-1 family. Only partial sequence of the putative Tc-like ORF is available as the 3′ boundary of pMBR-RP2 interrupts the putative coding region of this DNA transposon (Fig. 1). The putative T. thermophila Tc-like element is organized in opposite orientation to the two REP elements (Fig. 1).
The 5′ end of pMBR-RP3 contains the C-terminal 508 amino acids of ORF2 of the pMBR-RP3 REP element. The ≈350-bp sequence that flanks the REP1 and both REP2 elements does not similarly flank REP3. No significant matches were found when the pMBR-RP3 sequence downstream of the conserved 54-bp sequence was used in a BlastX search against the public protein database.
The 5′ end of pMBR-RP6 contains the C-terminal 820 amino acids of the REP6 ORF2. The pMBR-RP6 sequence downstream of the conserved 54-bp sequence (Fig. 1) contains high similarity to human KIAA1279 (31), a gene of unknown function expressed in the human brain. This gene (HEH2) is expressed during vegetative growth of T. thermophila and its protein product has been localized to basal bodies (21). We used Southern blotting and PCR to determine that the HEH2 gene is macronuclearly retained (data not shown). The REP6 locus has been mapped to the right arm of micronuclear chromosome 2 (Hamilton and Orias, personal communication).
REP elements are restricted to the germ line micronucleus.
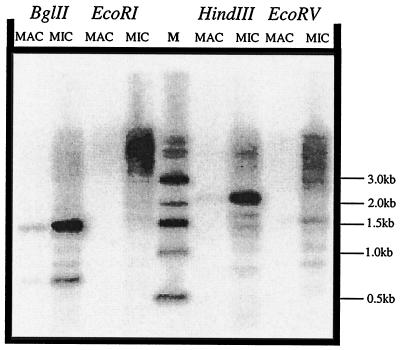
The possible relationship between transposable elements and programmed DNA elimination in the ciliated protozoa led us to predict that REP elements would not be found in the somatic macronucleus. Southern analysis of DNA from macronuclei and micronuclei probed with the conserved RT domain showed strong hybridization to micronuclear but not to macronuclear DNA (Fig. 6).
FIG. 6.
Southern analysis of restriction-digested T. thermophila macronuclear and micronuclear DNA probed with a DNA fragment complementary to the REP1 RT domain (PstI/BglII fragment of pMBR-RP1). The blot was subsequently stripped and reprobed to verify that equal amounts of macronuclear and micronuclear DNA preparations were used (data not shown). Abbreviations: M, 1-kb ladder (New England Biolabs); MAC, macronuclear DNA; MIC, micronuclear DNA.
Nucleotide substitutions in ORF2 are nonrandom with respect to codon position.
Nucleotide substitutions in several of the open reading frames of the transposon-like Tlr elements are nonrandom with respect to their codon position, with the majority occurring in the third position (22, 56), implying that the ORFs are under selective pressure to preserve their function. We analyzed two regions of ORF2 of the identified REP elements for codon position of nucleotide changes: the APE domain of REP1 and REP2-2, and the RT domain of REP1, REP2-2, REP3, and REP6. If the REP elements were not under selective pressure to maintain a functional protein, nucleotide changes should occur at random with an equal frequency at each position. According to χ2 analysis, nucleotide changes are nonrandom with respect to codon position (P < 0.001), with the majority occurring in the third position (Table 2). In agreement with this result, considering only the RT domain, 93% of the identified 215-nucleotide polymorphisms (Table 2) lead to either identical or similar amino acids, as defined by the Structure-Genetic matrix scoring system (17). Although the sample size of the APE domain is smaller, the result is striking, with 100% of nucleotide polymorphisms leading to identical or similar amino acids (Table 2).
TABLE 2.
Nucleotide changesa
| Amino acid | No. of changes at nucleotide position:
|
Total no. of nucleotides (%)
|
||||||
|---|---|---|---|---|---|---|---|---|
| First
|
Second
|
Third
|
||||||
| I | II | I | II | I | II | I | II | |
| Identical | 4 | 16 | 0 | 0 | 26 | 153 | 30 (77) | 169 (79) |
| Similar | 6 | 16 | 2 | 8 | 1 | 6 | 9 (23) | 30 (14) |
| Dissimilar | 0 | 4 | 0 | 10 | 0 | 2 | 0 (0) | 16 (7) |
| Total | 10 | 36 | 2 | 18 | 27 | 161 | 39 | 215 |
A comparison of nucleotide changes in the first, second, or third position within the codons of either of the indicated domains of the REP element ORF2. Amino acid changes were scored with respect to whether they were identical, similar, or dissimilar. I, APE domain of REP1 and REP2-2. II, RT domain of REP1, REP2-2, REP3, and REP6.
The noncanonical genetic code of T. thermophila (25) dictates that the “universal” stop codons TAA and TAG encode glutamine (Q), ensuring that T → C transitions in the first nucleotide of TAA or TAG codons are silent. For the RT domain, 11 out of 16, or 69%, nucleotide changes resulting in the same amino acid were one of these two transitions. A similar result, 11 out of 18, or 60%, was obtained when the entire ORF2 of REP1 and REP2-2 was considered.
Phylogeny of the T. thermophila REP elements.
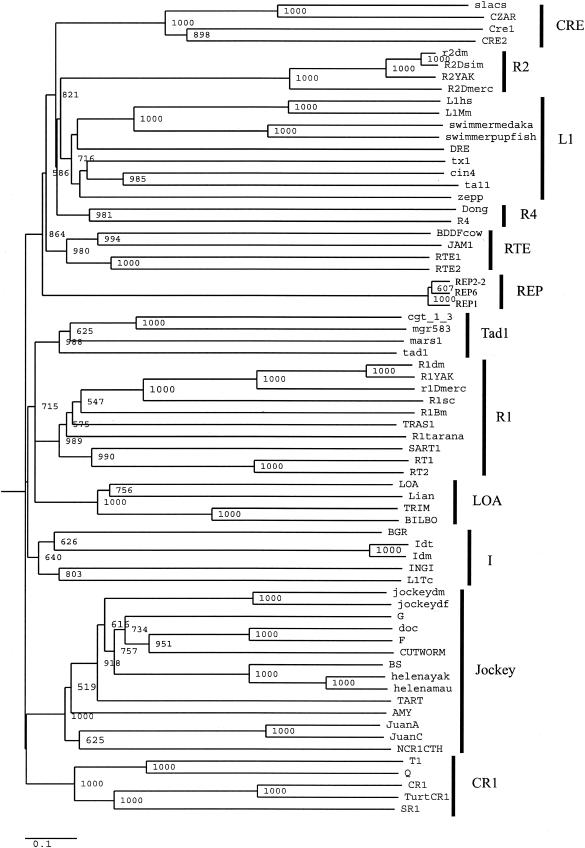
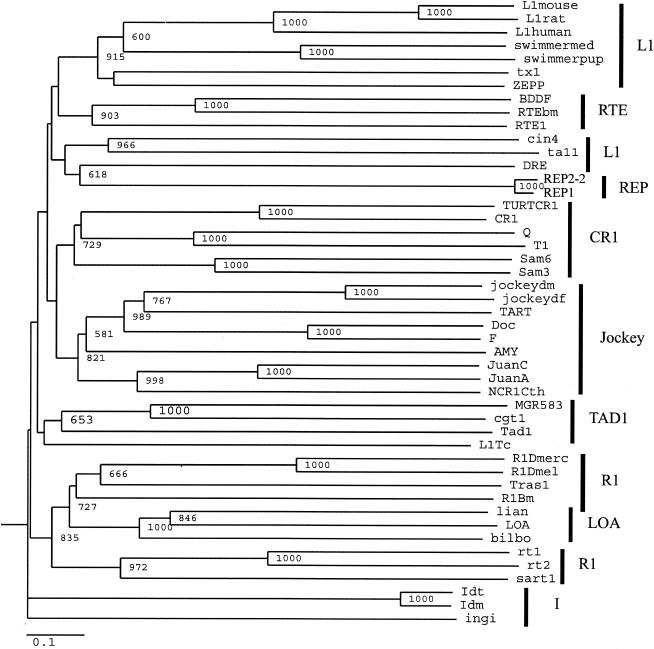
The above analysis indicates that the REP elements have evolved within the context of the T. thermophila genome. Since non-LTR retrotransposons are primarily transmitted vertically (40), and to date no non-LTR retrotransposons have been identified in the ciliated protozoa, we would predict that REP elements should be grouped apart in molecular phylogenies of the non-LTR retrotransposon family. We aligned separately the RT domains (REP1, REP2-2, and REP3) and the APE domains (REP1 and REP2-2) with the respective domains from non-LTR retrotransposons (40) deposited in the EMBL database (Fig. 2 and 3 and Materials and Methods). Possible phylogenetic relationships inferred from neighbor-joining trees constructed from the alignments of the RT and the APE domains are presented in Fig. 7 (RT) and Fig. 8 (APE).
FIG.7.
Phylogenetic relationships of the T. thermophila REP element with other non-LTR retrotransposons based upon the RT domain as defined in Malik et al. (40). The tree is neighbor joining with bootstrap values indicated as number out of 1,000. Only bootstrap values of >50% are indicated. The amino acid divergence scale is indicated. The alignment and amino acid sequences were derived as described in Materials and Methods. The non-LTR elements are divided into clades based upon phylogenetic relationships. Clades are indicated by the black bars and are defined in Malik et al. (40).
FIG. 8.
Phylogenetic relationships of the T. thermophila REP element with other non-LTR retrotransposons based upon the APE domain as defined in Malik et al. (40). The tree is neighbor joining with bootstrap values indicated as number out of 1,000. Only bootstrap values of >50% are indicated. The amino acid divergence scale is indicated. The alignment and amino acid sequences were derived as described in Materials and Methods. The non-LTR elements are divided into clades based upon phylogenetic relationships. Clades are indicated by the black bars and as defined in Malik et al. (40).
Malik et al. (40) proposed the term clade to represent those non-LTR retrotransposons that are grouped together with strong phylogenetic support. By this criterion, from the RT phylogeny (Fig. 7), it appears that the T. thermophila REP elements form their own clade and are restricted to their own long branch in the phylogeny. This finding is supported by the APE phylogeny (Fig. 8): the Tetraymena REP elements form a long branch, although they are grouped together with the DRE element, a non-LTR retrotransposon of Dictyostelium discoideum (42). From the RT phylogeny (Fig. 7), it is seen that DRE is a long-branching member of the L1 clade, in agreement with the results of Malik et al. (40). There are several other small differences in this phylogeny, such as the RTE clade placed within the L1 clade (Fig. 8) and LOA within the R1 clade [(Fig. 8; also seen in Malik et al. (40)], although the bootstrap values at the nodes of these long-branching divergences are not high. This may be due to the fact that the APE phylogeny has less resolution than the RT phylogeny (40).
We conclude that the T. thermophila REP elements represent a unique clade of non-LTR retrotransposons. The grouping of the APE domain of the REP elements with that of the DRE element may signify that the APE domain of the REP elements is evolving under tight functional constraints. Consistent with this hypothesis, we did not detect nucleotide changes that resulted in dissimilar amino acids in the analysis of the APE domains of REP1 and REP2-2 (Table 2).
REP elements are transcribed during nuclear development in T. thermophila.
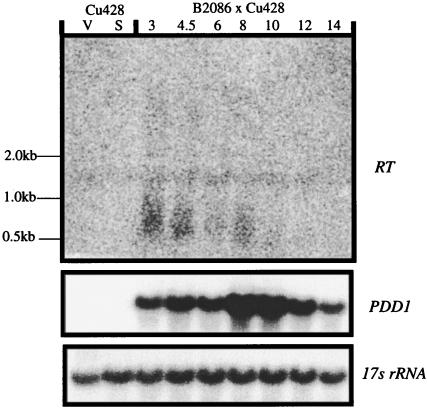
To analyze transcription of the REP elements during growth or nuclear development, we isolated whole cell RNA from growing, starved or conjugating T. thermophila and analyzed it by Northern blotting, probing with the RT of the REP element (Fig. 9). During early conjugation, we observed heterogeneously sized transcripts that ranged in size from ≈0.5 kb to 1.0 kb. They were visible as early as 3 h after the initiation of conjugation, a time corresponding to micronuclear meiosis (46).
FIG. 9.
Northern analysis of REP element transcription. Whole-cell RNA was separated on a 1% agarose-formaldehyde denaturing gel, transferred to a nylon filter, and probed with a DNA fragment corresponding to the RT domain of the REP element (corresponding to the EcoRV-BglII fragment of REP2-2 that encompasses the RT domain). The blot was also probed for the 17s rRNA as a loading control to test for RNA integrity as well as PDD1, a development-specific gene (39).
DISCUSSION
The REP elements of T. thermophila represent a new clade of non-LTR retrotransposons. We have characterized the first no n-LTR retrotransposon from the ciliated protozoa. Similar to all transposable elements identified to date in the ciliates, the REP family appears to be restricted to the micronucleus. Phylogenetic analysis suggests that the REP family is relatively divergent compared to identified non-LTR retrotransposon clades (40). This is consistent with the results of our analysis of the nature of nucleotide changes in the ORF2 that suggest that the REP elements have evolved in the context of the genome of T. thermophila (Table 2). These data are also consistent with the fact that maintenance of non-LTR retrotransposons in the genome of a particular organism is almost exclusively by vertical and not horizontal transmission (16).
Structure of the T. thermophila REP element.
The REP element shares many features with the non-LTR retrotransposon family. Many non-LTR retrotransposons encode an N-terminal ORF1 protein, most of which contain CCHC zinc finger motifs that are possibly involved in binding the element RNA after translation to import it back into the nucleus for transposition. The REP elements encode a putative ORF1 that contains an unusual CCCH zinc finger motif. The fact that some CCCH zinc finger-containing proteins have been shown to specifically bind 3′UTR mRNA (34) raises the possibility that ORF1 of the T. thermophila REP element has a similar function.
The REP ORF2 contains both an APE and an RT domain. All non-LTR retrotransposons encode an RT domain and an endonuclease which is either an N-terminal APE like the REP elements or one C-terminal of the RT similar to various prokaryotic restriction endonucleases (59). The APE domain of the REP element is predicted to be functional since key residues in the active site are conserved. Except for domain 0, key RT residues in the RT domain are also conserved. The function of this small domain is unknown at this point, but it is extremely well conserved within the non-LTR group of RTs (40). In combination with domain 2a, which is present in the REP elements (Fig. 4), it is believed to extend the active site of the non-LTR RT by giving additional or longer fingers compared with those of retroviral RTs. The conservation of key residues in both the APE and RT domains suggests that ORF2 remains catalytically active despite its phylogenetic age.
The majority of non-LTR retrotransposons contain poly(A) tails in their 3′UTR, the direct result of reverse transcription and transposition of a polyadenylated transcript. Several contain tandem repeats in their 3′UTR (8, 41). These repeats suggest that an abortive cDNA synthesis step by the RT may exist in these non-LTR retrotransposons and this may be mechanistically related to telomere addition (8). The 3′UTR of REP elements is unusual in that there is no evidence for poly(A) tails and four of the five that we have analyzed contain tandem repeats of very different lengths.
Genomic organization of T. thermophila REP elements.
One common motif at the 3′ end of the REP elements is a conserved 54-bp sequence. Due to the absence of sequence conservation in the 3′UTR, it is unclear at this point whether this sequence represents a conserved sequence in the REP element itself or forms part of a REP target site for transposition into genomic DNA. Three of the five REP elements are associated with a ≈350-bp sequence that appears to be repeated in the micronuclear genome. The fact that two of the REP elements are not associated with this repeat implies that the repeat may be a preferred but not exclusive target site for REP retrotransposition. There are other examples of retrotransposons associated with repetitive sequence. For example, Genie 1 is an ancient lineage of site-specific non-LTR retrotransposons in G. lamblia that is found within a telomeric repeat (7) and centromeric sequence in Arabidopsis thaliana is composed primarily of a repetitive sequence interspersed with retroelements (43).
The REP elements are restricted to the T. thermophila micronucleus. Our preliminary data from Southern blotting experiments have shown that each of the REP elements is flanked by a micronucleus-limited sequence (T. A. Thing, N. Vythilingum, J. S. Fillingham, and R. E. Pearlman, unpublished data). Thus, it is likely that REP elements are eliminated as parts of larger tracts of micronucleus-limited sequence, similar to the Tlr and Tel-1 elements.
T. thermophila REP elements: implication for genome stability.
There are several reports of selection acting to limit missense mutations from accumulating in the open reading frames of transposons in various ciliated protozoa (15, 22, 55, 56). We have determined that ORF2 of the T. thermophila REP elements is subject to the same phenomenon. The L1 element, a human non-LTR retrotransposon, has the property of cis preference in that its two ORFs predominantly mobilize only the RNA that encodes it (53). As a non-LTR retrotransposon, the REP elements may also be subject to cis preference, which would dictate that, to remain mobile, a particular transposon must retain its own functional proteins and not rely on them to be provided in trans.
A micronucleus-specific source of RT may have implications for T. thermophila micronucleus genome stability that extend beyond REP element transposition. It has been speculated that the Trl element could transpose through an RNA intermediate but it does not encode an RT (56). It is possible that the RT of the REP elements could function in trans and catalyze the transposition of various sequences in the micronuclear genome.
The restriction of REP elements to the transcriptionally silent micronucleus is consistent with all transposons described to date in the ciliated protozoa. One model to describe the evolution of programmed DNA deletion in the ciliates suggests that it originated to rid the somatic macronucleus of potentially deleterious transposons. This model suggests that nontransposon internal eliminated sequences represent degenerate transposons that have yielded their hypothetical “excisase” in trans, retaining only the necessary cis-acting signals for programmed elimination (32). The recent discovery of a piwi-related gene with a function in developmentally programmed genome rearrangement in T. thermophila (47) supports this model.
The molecular characterization of the piwi gene family has provided a molecular link between the RNA interference phenomenon and genome surveillance (51). The scan RNA model of programmed DNA rearrangements presented by Mochizuki et al. (47) proposes that germ line-specific sequence is transcribed and processed to small “scan RNAs” that are involved in targeting regions of DNA for programmed deletion in T. thermophila. Further evidence in support of this model has been provided by Yao et al. (60), who found that injection of double-stranded RNA into the cell during early conjugation triggers deletion of the targeted genomic regions during macronuclear development. In addition, Wuitschick and Karrer have shown that the multiple-copy Tlr elements contain multiple, redundant cis-acting sequences within their micronucleus-limited sequences that are able to promote programmed DNA deletion, suggesting that Tlr elements are subject to an RNA interference-like system that detects the presence of multiple-copy sequence within the micronucleus (57). The fact that the REP elements are transcribed during early macronuclear development in a manner similar to other micronuclear-limited sequences (9) is consistent with the hypothesis that programmed DNA deletion in T. thermophila is a method of eliminating transposable elements from the macronucleus.
Preliminary dot-blotting analysis indicates that there are between 40 and 175 REP elements per micronucleus (N. Vythilingum, J. S. Fillingham, and R. E. Pearlman, unpublished). The fact that there is selection on them to conserve the encoded proteins suggests to us that the T. thermophila micronucleus is under threat of transposon mobilization. This raises the question of whether developmentally programmed DNA deletion is the only method of transposon inactivation in T. thermophila, or whether there is an additional growth-specific mechanism that functions in the transcriptionally silent micronucleus. It will be of interest to determine whether REP elements are capable of transposition during vegetative growth. We have previously identified a growth-associated piwi-related gene in an expressed sequence tag sequencing project (20). Caenorhabditis elegans mutants carrying mutations in the piwi-related gene rde-1 exhibit transposon mobilization (51). The analysis of micronuclear stability of the REP elements in mutants defective in this gene may yield some insight into molecular mechanisms that ensure genome stability in somatic nuclei in eukaryotes.
Acknowledgments
We thank Anita Samardzic for expert technical assistance. We also thank Nora Tsao, Emina David, and Noah Fine for helpful discussions throughout the course of this work. We thank Thomas H. Eickbush (University of Rochester, N.Y.) for help with phylogenetic analysis. DNA sequencing was done by Lee Wong (Core Molecular Biology Facility, York University).
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to R.E.P. J.S.F. was supported by a CIHR studentship and the York University President's Dissertation Scholarship.
REFERENCES
- 1.Arkhipova, I. R., and H. G. Morrison. 2001. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 3.Bibillo, A., and T. H. Eickbush. 2002. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 277:34836-34845. [DOI] [PubMed] [Google Scholar]
- 4.Borst, P. 2002. Antigenic variation and allelic exclusion. Cell 109:5-8. [DOI] [PubMed] [Google Scholar]
- 5.Borst, P., and D. R. Greaves. 1987. Programmed gene rearrangements altering gene expression. Science 235:658-667. [DOI] [PubMed] [Google Scholar]
- 6.Burke, W. D., H. S. Malik, J. P. Jones, and T. H. Eickbush. 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16:502-511. [DOI] [PubMed] [Google Scholar]
- 7.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 8.Chaboissier, M. C., D. Finnegan, and A. Bucheton. 2000. Retrotransposition of the I factor, a non-long terminal repeat retrotransposon of Drosophila, generates tandem repeats at the 3′ end. Nucleic Acids Res. 28:2467-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry, J. M., and E. H. Blackburn. 1985. The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell 43:747-758. [DOI] [PubMed] [Google Scholar]
- 11.Chilcoat, N. D., and A. P. Turkewitz. 1997. In vivo analysis of the major exocytosis-sensitive phosphoprotein in Tetrahymena. J. Cell Biol. 139:1197-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne, R. S., D. L. Chalker, and M. C. Yao. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 30:557-578. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, C. 2002. Mouse genome: a forage in the junkyard. Nature 420:458-459. [DOI] [PubMed] [Google Scholar]
- 14.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doak, T. G., D. J. Witherspoon, C. L. Jahn, and G. Herrick. 2003. Selection on the genes of Euplotes crassus Tec1 and Tec2 transposons: evolutionary appearance of a programmed frameshift in a Tec2 gene encoding a tyrosine family site-specific recombinase. Eukaryot. Cell. 2:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eickbush, T. H., and H. S. Malik. 2002. Origins and evolution of retrotransposons, p. 1111-1144. In N. L. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 17.Feng, D. F., M. S. Johnson, and R. F. Doolittle. 1984. Aligning amino acid sequences: comparison of commonly used methods. J. Mol. Evol. 21:112-125. [DOI] [PubMed] [Google Scholar]
- 18.Feng, Q., G. Schumann, and J. D. Boeke. 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fillingham, J. S., D. Bruno, and R. E. Pearlman. 2001. Cis-acting requirements in flanking DNA for the programmed elimination of mse2.9: a common mechanism for deletion of internal eliminated sequences from the developing macronucleus of Tetrahymena thermophila. Nucleic Acids Res. 29:488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fillingham, J. S., N. D. Chilcoat, A. P. Turkewitz, E. Orias, M. Reith, and R. E. Pearlman. 2002. Analysis of expressed sequence tags (ESTs) in the ciliated protozoan Tetrahymena thermophila. J. Eukaryot. Microbiol. 49:99-107. [DOI] [PubMed] [Google Scholar]
- 21.Fine, N. A. 2003. M.Sc. thesis. York University, Toronto, Ontario, Canada.
- 22.Gershan, J. A., and K. M. Karrer. 2000. A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein. Nucleic Acids Res. 28:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorovsky, M. A., M. C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 24.Heinonen, T. Y., and R. E. Pearlman. 1994. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J. Biol. Chem. 269:17428-17433. [PubMed] [Google Scholar]
- 25.Horowitz, S., and M. A. Gorovsky. 1985. An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. USA 82:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horsfall, W. H., and R. E. Pearlman. 1988. Micronuclear DNA sequence from Tetrahymena do not confer mitotic stability on ARS plasmids in Saccharomyces. Genome 30:690-696. [Google Scholar]
- 27.Howard, E. A., and E. H. Blackburn. 1985. Reproducible and variable genomic rearrangements occur in the developing somatic nucleus of the ciliate Tetrahymena thermophila. Mol. Cell. Biol. 5:2039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter, D. J., K. Williams, S. Cartinhour, and G. Herrick. 1989. Precise excision of telomere-bearing transposons during Oxytricha fallax macronuclear development. Genes Dev. 3:2101-2112. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs, M. E., and L. A. Klobutcher. 1996. The long and the short of developmental DNA deletion in Euplotes crassus. J. Eukaryot. Microbiol. 43:442-452. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, M. E., A. Sanchez-Blanco, L. A. Katz, and L. A. Klobutcher. 2003. Tec3, a new developmentally eliminated DNA element in Euplotes crassus. Eukaryot. Cell 2:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuno, R., T. Nagase, K. Ishikawa, M. Hirosawa, N. Miyajima, A. Tanaka, H. Kotani, N. Nomura, and O. Ohara. 1999. Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 6:197-205. [DOI] [PubMed] [Google Scholar]
- 32.Klobutcher, L. A., and G. Herrick. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 56:1-62. [DOI] [PubMed] [Google Scholar]
- 33.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of human immunodeficiency virus-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 34.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 35.Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar, and F. M. Sheen. 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75:1083-1093. [DOI] [PubMed] [Google Scholar]
- 36.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 37.Lyon, M. F. 2000. LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc. Natl. Acad. Sci. USA 97:6248-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyon, M. F. 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80:133-137. [DOI] [PubMed] [Google Scholar]
- 39.Madireddi, M. T., R. S. Coyne, J. F. Smothers, K. M. Mickey, M. C. Yao, and C. D. Allis. 1996. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87:75-84. [DOI] [PubMed] [Google Scholar]
- 40.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 41.Malik, H. S., and T. H. Eickbush. 1998. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 15:1123-1134. [DOI] [PubMed] [Google Scholar]
- 42.Marschalek, R., J. Hofmann, G. Schumann, R. Gosseringer, and T. Dingermann. 1992. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol. Cell. Biol. 12:229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martienssen, R., and W. R. McCombie. 2001. The first plant genome. Cell 105:571-574. [DOI] [PubMed] [Google Scholar]
- 44.Martin, F., C. Maranon, M. Olivares, C. Alonso, and M. C. Lopez. 1995. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 247:49-59. [DOI] [PubMed] [Google Scholar]
- 45.Martin, F., M. Olivares, M. C. Lopez, and C. Alonso. 1996. Do non-long terminal repeat retrotransposons have nuclease activity? Trends Biochem. Sci. 21:283-285. [PubMed] [Google Scholar]
- 46.Martindale, D. W., C. D. Allis, and P. J. Bruns. 1982. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp. Cell Res. 140:227-236. [DOI] [PubMed] [Google Scholar]
- 47.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 48.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers, M. B., and K. M. Karrer. 1989. Cloning of Tetrahymena genomic sequences whose message abundance is increased during conjugation. Dev. Biol. 131:261-268. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. T. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 52.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 53.Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells, J. M., J. L. Ellingson, D. M. Catt, P. J. Berger, and K. M. Karrer. 1994. A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol. Cell. Biol. 14:5939-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witherspoon, D. J., T. G. Doak, K. R. Williams, A. Seegmiller, J. Seger, and G. Herrick. 1997. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol. Biol. Evol. 14:696-706. [DOI] [PubMed] [Google Scholar]
- 56.Wuitschick, J. D., J. A. Gershan, A. J. Lochowicz, S. Li, and K. M. Karrer. 2002. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 30:2524-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wuitschick, J. D., and K. M. Karrer. 2003. Diverse sequences within Tlr elements target programmed DNA elimination in Tetrahymena thermophila. Eukaryot. Cell. 2:678-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao, M. C., P. Fuller, and X. Xi. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300:1581-1584. [DOI] [PubMed] [Google Scholar]