Abstract
In Saccharomyces cerevisiae, Ras proteins connect nutrient availability to cell growth through regulation of protein kinase A (PKA) activity. Ras proteins also have PKA-independent functions in mitosis and actin repolarization. We have found that mutations in MOB2 or CBK1 confer a slow-growth phenotype in a ras2Δ background. The slow-growth phenotype of mob2Δ ras2Δ cells results from a G1 delay that is accompanied by an increase in size, suggesting a G1/S role for Ras not previously described. In addition, mob2Δ strains have imprecise bud site selection, a defect exacerbated by deletion of RAS2. Mob2 and Cbk1 act to properly localize Ace2, a transcription factor that directs daughter cell-specific transcription of several genes. The growth and budding phenotypes of the double-deletion strains are Ace2 independent but are suppressed by overexpression of the PKA catalytic subunit, Tpk1. From these observations, we conclude that the PKA pathway and Mob2/Cbk1 act in parallel to determine bud site selection and promote cell cycle progression.
Ras proteins are highly conserved, small monomeric GTPases that function as molecular switches in signal transduction pathways to regulate cell growth and differentiation in response to various environmental cues. In Saccharomyces cerevisiae, Ras proteins are encoded by two genes, RAS1 and RAS2, and are particularly responsive to changes in nutrient concentration (4, 5, 55, 57). Deletion of either RAS gene alone does not affect growth on rich media. However, deletion of both results in decreased mass accumulation and an attendant terminal G1 cell cycle arrest. Detailed analysis of this phenotype has demonstrated that Ras proteins redundantly fulfill an essential function in coupling cell growth to nutrient availability (29, 35, 37, 56, 60).
Ras proteins maintain an essential basal level of cyclic AMP (cAMP) through their activation of adenylate cyclase, the only Ras effector protein identified in S. cerevisiae (41). Restoration of fermentable sugar to starved cells elicits Ras stimulation of adenylate cyclase, resulting in a transient increase in intracellular cAMP (10, 15, 60). This cAMP spike is not essential for growth but does accelerate both resumption of growth of stationary-phase cells and germination of spores upon nutrient addition (25). cAMP binds to the inhibitory regulatory subunit of protein kinase A (PKA), thereby releasing the catalytic subunit to phosphorylate numerous substrates affecting metabolism and transcription (reviewed in references 5 and 58). The catalytic subunits are encoded by three genes, TPK1, TPK2, and TPK3, which have both unique and redundant functions (43, 50, 59). Overexpression of any one of the catalytic subunits of PKA restores growth to ras1Δ ras2Δ strains (59).
In addition to its role in PKA activation, Ras participates in mitosis through a PKA-independent mechanism (38). Deletion of the Ras-related GTPase RSR1 in a ras1Δ ras2Δ strain (but not in a RAS+ strain) results in a late mitotic arrest that cannot be suppressed by TPK1 overexpression but can be suppressed by overexpression of any of a number of genes involved in mitotic exit, including DBF2, CDC5, CDC15, and SPO12 (38). These genes function to control the localization and the activity of the protein phosphatase Cdc14 (reviewed in reference 18). Cdc14 plays an integral role in controlling the cell's decision to exit from mitosis and enter G1. Although this genetic evidence supports a function of Ras in exit from mitosis, the exact role of Ras in this process has not been resolved.
Ras proteins also function, independent of PKA, in regulating actin cytoskeleton polarity. The actin cytoskeleton normally polarizes to regions of cell growth. Upon shifting wild-type cells to 37°C, the cytoskeleton temporarily depolarizes, but re-polarizes upon continued incubation at the elevated temperature (22). In contrast, the cytoskeleton in ras2Δ cells subjected to the same treatment remains depolarized. Overexpression of a PKA catalytic subunit does not suppress the actin polarity defect.
To further understand the role of Ras in S. cerevisiae, we performed a genetic screen to identify mutations that cause lethality or slow growth in a ras2Δ background. The product encoded by one such mutation, MOB2, exhibits homology with the mitotic exit network component encoded by MOB1, although Mob2 does not appear to be a member of the mitotic exit network (34). Instead, Mob2 is a component of the RAM signaling network, which localizes and activates the Swi5-like transcription factor Ace2 in the daughter cell nucleus to direct daughter cell-specific transcription of several genes involved in cell separation (8, 39, 64). In addition to Mob2, the RAM signaling network consists of an Ndr/Warts family kinase, Cbk1 (cell wall biosynthesis kinase 1), and the conserved gene products Hym1, Kic1, Pag1, and Sog2. Accordingly, deletion of MOB2, CBK1, HYM1, PAG1, KIC1, SOG2, or ACE2 results in cells with a cell separation defect (2, 8, 13, 39, 48, 64).
Cbk1 and Mob2 also have Ace2-independent functions. First, cbk1Δ and mob2Δ cells are round rather than ovoid due at least partially to an inability of daughter cells to grow apically (2, 64). Second, CBK1Δ cells exhibit altered bud site selection. Deletion of CBK1 in cells that normally exhibit a bipolar budding pattern results in cells that are able to choose properly the initial bud site but lose the ability in subsequent generations to form buds solely at the poles (2, 48). Finally, deletion of CBK1 affects mating projection formation, yet another type of polarized growth. After prolonged exposure of cbk1Δ cells to pheromone, most cells have one or more small protrusions on the surface instead of the normal-size mating projection (2). Mob2, Pag1, Sog2, and Kic1 interact with Cbk1 and affect its localization and activity (13, 39). Hym1 interacts with Kic1 and Sog2. Deletion of genes encoding RAM members confers cell separation, bipolar budding, apical growth, and shmoo formation defects similar to those in cbk1Δ strains (2, 8, 13, 39, 64).
In the current study, we show that deletion of MOB2, CBK1, HYM1, KIC1, or PAG1 confers a synthetic slow-growth phenotype with a ras2 deletion. This slow-growth phenotype is due to decreased PKA activation, because it is suppressed by overexpression of TPK1 and is independent of the daughter cell-specific transcription program driven by the Cbk1/Mob2 complex. Flow cytometry and microscopic analysis of synchronized cells indicate that ras2Δ mob2Δ strains have a cell cycle defect in G1/S progression. Since this delay is accompanied by an increase in size, the phenotype differs from that previously described for Ras pathway mutants, which exhibit a G1 arrest without an accompanying size increase (26, 61). In addition to the synthetic slow-growth defect, ras2Δ mob2Δ mutations cause synthetic budding defects that are suppressed by overexpression of TPK1, demonstrating a novel role for the Ras/PKA signal pathway in bud site selection. Thus, the Ras/PKA pathway and the Mob2/Cbk1 pathway share a function important for cell cycle progression and bud site selection.
MATERIALS AND METHODS
Yeast strains and plasmids.
Standard yeast media were used (6). All yeast strains used are listed in Table 1 and were derived from the S288C background. mob2Δ (Y3012), cbk1Δ (Y3016), hym1Δ (Y3027), ace2Δ (Y3026), pag1Δ (Y3088), swe1Δ (Y3154), and bub2Δ (Y3017) deletion strains were constructed by PCR-based gene disruption (6) or PCR amplification of deleted loci from either the Saccharomyces Genome Deletion Project (65) or YHQ2h5 (from Haiyan Qi). RAS2 and KIC1 deletion strains were constructed as described previously (54, 60). Y3084, Y3085, Y3087, and Y3086 were created by digesting B2521 with HincII (Y3084, Y3085, Y3087) or BsaBI (Y3086) and subsequent transformation into Y3014, Y3025, Y3015, and Y3024. Y3155 was created by digesting B2575 with Tth111I and subsequent two-step gene replacement in Y2813. The presence of the mutations was confirmed by DNA sequencing of a PCR product containing the ACE2 or CDC28 region of interest from the aforementioned strains. Remaining strains listed in Table 1 produced in this study were derived by standard genetic techniques utilizing strains listed above and their parents.
TABLE 1.
Strains used in this study
| Strain | Mating type | Genotype | Plasmid | Source |
|---|---|---|---|---|
| Y797 | MATa | his3Δ1 leu2-3,12 ura3-52 trp1-289 ras2Δ gal | B844 | Lab collection |
| Y2465 | MATα | his3Δ1 leu2-3,112 ura3-52 trp1-289 ras2Δ | B844 | Lab collection |
| Y2811 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 | Lab collection | |
| Y2812 | MATα | ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | Lab collection | |
| Y2813 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 ras2::LEU2 | This study | |
| Y3011 | MATα | his3Δ1 leu2-3,112 ura3-52 trp1-289 ras2Δ mob2-1 | B844 | This study |
| Y3012 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 mob2::HIS3 | This study | |
| Y3013 | MATα | ura3-52 leu2-3,112 trp1Δ1 his3Δ200 ras2::URA3 | This study | |
| Y3014 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 | This study | |
| Y3015 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ras2::URA3 mob2:HIS3 | This study | |
| Y3016 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 cbk1::HIS3 | This study | |
| Y3017 | MATa | ura3-52 leu2-3,112 trp1Δ1 his3Δ200 mob2::HIS3 bub2::kanMX | This study | |
| Y3018 | MATa | ura3-52 leu2-3,112 his3 mob2::HIS3 ras2Δ | B844 | This study |
| Y3019 | MATα | ura3-52 leu2-3,112 his3 ade2-101 cbk1::HIS3 ras2Δ | B844 | This study |
| Y3020 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 | This study | |
| Y3021 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 ras2::URA3/ras2::URA3 | This study | |
| Y3022 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 mob2::HIS3/mob2::HIS3 | This study | |
| Y3023 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 ras2::URA3/ras2::URA3 mob2::HIS3/mob2::HIS3 | This study | |
| Y3024 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ras2::URA3 | This study | |
| Y3025 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 mob2::HIS3 | This study | |
| Y3026 | MATα | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ace2::kanMX | This study | |
| Y3027 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 hym1::HIS3 | This study | |
| Y3028 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 ras2::URA3 | This study | |
| Y3082 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 cbk1::HIS3/cbk1::HIS3 | This study | |
| Y3083 | MATa/MATα | ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 ADE2/ade2-101 TRP1/trp1Δ1 cbk1::HIS3/cbk1::HIS3 ras2::URA3/ras2::URA3 | This study | |
| Y3084 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ACE2G128E | This study | |
| Y3085 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 mob2::HIS3 ACE2G128E | This study | |
| Y3086 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ras2::URA3 ACE2G128E | This study | |
| Y3087 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 ras2::URA3 mob2:HIS3 ACE2G128E | This study | |
| Y3088 | MATa | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 pag1::HIS3 | This study | |
| Y3089 | MATα | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 kic1::URA3 | This study | |
| Y3090 | MATα | ura3-52 leu2-3,112 his3Δ200 ade2-101 ras2::URA3 | This study | |
| Y3154 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 ras2::LEU2 swe1::KAN | This study | |
| Y3155 | MATa | ura3-52 leu2-3,112 ade2-101 his3Δ200 ras2::LEU2 cdc28Y19F | This study | |
| Y3157 | MATα | ura3-52 leu2-3,112 his3Δ200 trp1Δ1 mob2::HIS3 ras2::URA3 | This study |
Plasmids used in this study are listed in Table 2. MOB2 and ACE2 were amplified from genomic DNA by PCR using genomic DNA from strain Y2812 and primers hybridizing 500 bp upstream and downstream from the initial codon and stop codon, respectively. The MOB2 PCR product was first subcloned into pGEM-T (Promega), and then the MOB2 EagI fragment was subcloned into the EagI site of pRS413 (B2467). The primers used to amplify ACE2 contained SpeI and XhoI restriction sites that were used to clone the PCR product into the SpeI and XhoI sites of pRS415 (B2461). The ACE2G128E mutation was introduced into this plasmid by using the Quickchange mutagenesis kit (B2462; Stratagene). The presence of the mutation was confirmed by DNA sequencing. ACE2G128E was subcloned into pRS404, using the SpeI and XhoI sites to create B2521. CDC28Y19F was subcloned from pRS414-CDC28Y19F (from Yu Jiang) into the SpeI and XhoI sites of pRS406 to create B2575.
TABLE 2.
Plasmids used in this study
| Name | Vector | Description | Source or reference |
|---|---|---|---|
| B560 | pUC8 | ras2::LEU2 | 29 |
| B561 | pUC8 | ras2::URA3 | 29 |
| B844 | pGS20 | CEN4 ARS1 URA3 RAS2 | Lab collection |
| B1436 | YCp-LEU | CEN4 ARS1 LEU2 RAS2 | 17 |
| B1373 | YEp13 | LEU2 REP3 2μm-ori TPK1 | 59 |
| B2461 | pRS415 | CEN6 ARS4 LEU2 ACE2 | This study |
| B2462 | pRS415 | CEN6 ARS4 LEU2 ACE2G128E | This study |
| B2467 | pRS413 | CEN6 ARS4 HIS3 MOB2 | This study |
| B2521 | pRS404 | TRP1 ACE2G128E | This study |
| B2575 | PRS406 | URA3 CDC28Y19F | This study |
Genetic screen.
ras2Δ synthetic mutations were generated by treating Y2465 with ethyl methanesulfonate such that 89% of the cells treated were killed (6). Cells were plated on yeast extract-peptone-dextrose medium (YEPD) at an appropriate density, incubated for 2 days at 30°C, and replica plated to synthetic complete medium (SC) and SC plus 5-fluoroorotic acid (5-FOA). Colonies that grew only on SC were transformed with a CEN LEU2 RAS2 plasmid and tested for 5-FOA resistance to show dependence upon RAS2. Strains exhibiting plasmid-dependent 5-FOA sensitivity were crossed to strain Y797, sporulated, and subjected to tetrad analysis. Tetrads exhibiting 2:2 segregation of 5-FOA sensitivity were further analyzed. From 13,000 mutagenized colonies, 44 FOA-sensitive colonies whose phenotype was due to a single mutation as determined by segregation analysis and dependent upon RAS2 were isolated. Of these, 28 were recessive and 16 were dominant. A YCp50-LEU-based genomic library was used to clone the recessive mutation in Y3011 by complementation. Genomic inserts on plasmids conferring 5-FOA resistance were subcloned into pRS405 and integrated (circular integration) into Y797 such that the locus was marked with LEU2. The resulting strains were crossed to Y3011 to determine linkage. The genes present on the appropriate complementing genomic library plasmid were individually subcloned, transformed into Y3011, and tested for their ability to confer 5-FOA resistance.
FUN 1 staining.
Exponentially growing cells were harvested and stained with FUN 1 (6.3 μM; Molecular Probes) according to the manufacturer's instructions. Cells were mounted onto cover slips using low-melt-point agarose (1% wt/vol) in GH buffer (10 mM HEPES-KOH [pH 7.2], 2% [wt/vol] glucose) and visualized by using an integrated DeltaVision system (Applied Precision). This included a Nikon Eclipse TE200 inverted microscope, with a 100× (numerical aperture, 1.4) objective. A Princeton Instruments charge-coupled device camera was used to capture images.
LY staining.
Exponentially growing cells (5 × 106) were pelleted, resuspended in YEPD (90 μl) containing lucifer yellow (LY) (10 μl; Molecular Probes), and grown for 1 h at 30°C. Cells were then washed with ice-cold buffer (50 mM succinate, NaOH [pH 5], 100 mM NaCl, 10 mM MgCl2, 20 mM NaN3) and mounted as described previously (49). LY staining was then visualized by using an integrated DeltaVision system as described above.
Bud scar and actin staining.
To examine budding patterns, exponentially growing cells (2 ml) were formaldehyde fixed (3.7% final concentration) and stained with calcofluor white (Fluorescent Brightener 28; Sigma; final concentration, 0.17 mg/ml) as described previously (6). Cells were placed on a glass slide, and bud scars were visualized with a fluorescent Zeiss Photomicroscope IIIRS with a 63× (numerical aperture, 1.25) objective and a UV filter set. Cells that had undergone one or two budding events were analyzed. Buds were scored as distal (positioned in the third of the cell opposite the birth scar), proximal (positioned in the third of the cell next to the birth scar), or medial (positioned in the middle third of the cell). In the case of strains containing a MOB2 deletion, chains or clusters of cells were analyzed based upon the location of a cell with a birth scar (2). Axial budding was examined by calcofluor white staining and time-lapse microscopy. To examine actin polarization in mitotic cells, exponentially growing cells were fixed and stained with rhodamine phalloidin (2.2 μM; Molecular Probes) followed by staining with calcofluor white as described previously (6). After being stained, cells were washed with phosphate-buffered saline (PBS) (20) and resuspended in mounting medium (100 μl; 6). An aliquot of stained cells (5 μl) was placed on a 1% (wt/vol) low-melt-point agarose-PBS patch which had been allowed to set on a concavity slide (PGC Scientifics) and covered with a cover slip (22 by 22 mm). The cells were then visualized by using the integrated DeltaVision system described above. Thirty-five optical sections were taken at 0.2-μm intervals, deconvolved, and flattened using softWoRx v. 2.50 (Applied Precision).
To examine actin polarization in mating projections, cells were grown overnight at 30°C in SC-LEU, pH 3.5 (5 ml). Overnight cultures were diluted into fresh SC-LEU, pH 3.5 (2 ml), to a density of 3 × 106 cells/ml and allowed to grow for 2 h at 30°C. Cells were then incubated with α-factor (10 μg/ml) for 3 h, fixed with formaldehyde, stained with rhodamine phalloidin and calcofluor white, and visualized as described for mitotic cells.
Cell cycle experiments.
Cells were grown in YEPD or SC-LEU and synchronized with α-factor (5 μg/ml) as described previously (3). Aliquots were removed before α-factor addition, just after release, and every 15 or 20 min after release. Cells were fixed as described previously (6), treated with RNase A (0.25 mg/ml in sodium citrate buffer [pH 7.2] overnight at 37°C) and proteinase K (2 mg/ml; 2 h at 50°C), and stained with Sytox green (1 μM in sodium citrate [pH 7.2]; Molecular Probes). Fluorescence and forward scatter was analyzed using a Becton Dickinson FACScan flow cytometer and Cell Quest software (Becton Dickinson). At least 30,000 events were analyzed for each sample. Percentages of cells in G1, S, and G2 were determined by using the Dean-Jett-Fox algorithm, which is part of the FlowJo flow cytometry analysis package (version 4; Tree Star, Inc.). Synchronized fixed cells were also stained with 50 ng of 4′,6-diamidino-2′phenylindole (DAPI) per ml in PBS (6), sonicated briefly, and examined microscopically. Alternatively, cells were synchronized with α-factor, washed, and mounted on a concavity slide containing a YEPD-agarose (1% [wt/vol]) pad. Time-lapse microscopy was performed at 5-min intervals.
Trehalose assay.
Cells were grown in triplicate for 4 days in YEPD (5 ml) at 30°C. Trehalose levels were measured as described previously (30). Purified porcine kidney trehalase and a glucose oxidase/peroxidase assay kit were purchased from Sigma. Trehalose levels were determined by subtracting the amount of glucose present in mock-treated extracts from the amount of glucose in trehalase-treated extracts and comparing to a standard curve. Protein concentration was determined using the Bio-Rad protein assay according to the manufacturer's instructions.
RNA isolation and Northern analysis.
Total RNA was isolated by extraction with hot acidic phenol (1). Fifteen micrograms of total RNA was fractionated on a 1% agarose gel (0.1 M sodium borate buffer [pH 8.3], 7.5% formaldehyde), transferred to a Nytran SuperCharge membrane (Schleicher & Schüll), baked, and probed with 32P-labeled (Random Primers Labeling System; Invitrogen) PCR-generated DNA according to the manufacturer's directions. Signal intensity was quantified using a STORM 860 imaging system (Molecular Dynamics).
RESULTS
mob2Δ confers a synthetic slow-growth phenotype with ras2Δ.
To further our understanding of the cellular processes affected by Ras proteins in yeast, we conducted a screen to identify mutations that confer lethality or slow growth in a ras2Δ strain. One mutant isolated was suppressed by introduction of either MOB2 or RAS2 on a low-copy-number vector into the mutant background (Fig. 1A). Linkage analysis verified that the synthetic growth phenotype was due to a mutation at or near the MOB2 locus (data not shown). Deletion of MOB2 confers a synthetic growth phenotype with ras2Δ (Fig. 1B). Together, these results demonstrate that the mutation conferring a synthetic slow-growth phenotype with ras2Δ in the original strain isolated is in MOB2. Deletion of MOB2 alone does not affect growth in the S288C background used at 30, 14, or 37°C (data not shown). Deletion of MOB2 does not affect the growth of strains containing deletions of RAS1 or GPA2, a Gα homologue that affects cAMP levels in a pathway parallel to Ras (data not shown). Mutations that downregulate Ras signaling, such as a ras2C318S allele or the srv2-1 allele (14, 25), do not confer a slow-growth phenotype to a mob2Δ strain (data not shown).
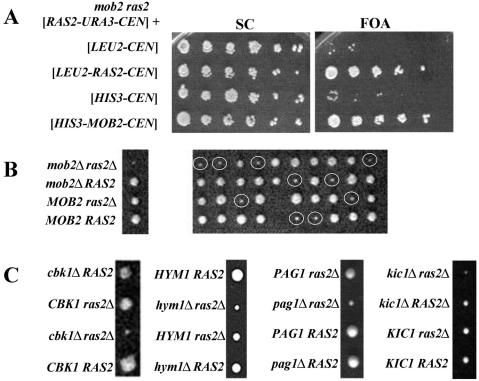
FIG. 1.
mob2Δ confers a synthetic slow-growth phenotype to a ras2Δ strain. (A) Strain Y3011 (ras2 mob2 [CEN-RAS2-URA3]) harboring, from top to bottom, [CEN-LEU2], [CEN-LEU2-RAS2], [CEN-HIS3], or [CEN-HIS3-MOB2] were grown in SC liquid medium, spotted onto SC and SC plus FOA plates in fivefold serial dilutions, and incubated at 30°C. (B) Tetrad analysis of Y3012 (mob2Δ) × Y3013 (ras2Δ). A representative tetratype tetrad is labeled and shown at the left of the tetrads. At least 10 tetrads were analyzed with consistent results (mob2Δ ras2Δ spore clones are circled). (C) cbk1Δ, hym1Δ, pag1Δ, and kic1Δ confer a synthetic slow-growth phenotype with ras2Δ. In all subpanels, representative tetratype tetrads are labeled and shown. For each strain, at least 10 tetrads were analyzed with results fully consistent with those shown. (Left to right) Tetrad analysis of Y3013 (ras2Δ) × Y3016 (cbk1Δ), Y3013 (ras2Δ) × Y3027 (hym1Δ), Y3090 (ras2Δ) × Y3088 (pag1Δ), and Y2813 (ras2Δ) × Y3089 (kic1Δ).
Mob2 physically associates with the Cbk1 kinase, which localizes to regions of cell growth as well as the nucleus (8, 64). Mob2 is required for daughter cell specific-nuclear localization of Cbk1 and Ace2. Other members of the Cbk1 signaling pathway include Hym1, Pag1, and Kic1 (2, 39). Tetrad analysis shows that deletion of CBK1, HYM1, PAG1, or KIC1 confers a synthetic slow-growth phenotype in a ras2Δ strain (Fig. 1C). Deletion of SGT2, ECM10, or SEC28, which encode gene products that have been found in a complex with Cbk1, does not affect growth of either a mob2Δ or ras2Δ strain (23; data not shown). Together, these data suggest that RAS2 and MOB2/CBK1/HYM1/PAG1/KIC1 function in parallel pathways, either of which is necessary for efficient cell growth.
Deletion of CBK1 affects the yeast's ability to form mating projections in response to pheromone and, in the case of cells exhibiting a bipolar budding pattern, to correctly position the next bud. The polarisome, consisting of Spa2, Bud6, Pea2, and Bni1, also plays a role in these pathways (reviewed in references 45 and 46). To determine if Mob2 or Ras2 was acting in a pathway parallel to the polarisome, strains containing single deletions in each component (spa2Δ, bud6Δ, pea2Δ, or bni1Δ) were crossed to strains containing either a MOB2 or a RAS2 deletion. Tetrad analysis demonstrated that none of the progeny from this cross had a slow-growth phenotype (data not shown). Since no synthetic effects were observed, this experiment suggests that neither RAS2 nor MOB2 acts in a pathway essential for growth that is parallel with the polarisome.
Overexpression of TPK1 suppresses the mob2Δ ras2Δ synthetic growth defect.
To test whether Tpk mediated the Ras function that acts in parallel with Mob2, we determined whether Tpk overexpression could suppress the slow-growth phenotype of a ras2Δ mob2Δ strain. As evident from Fig. 2A, overexpression of TPK1 in a ras2Δ mob2Δ strain or a ras2Δ cbk1Δ strain rescues their slow-growth phenotype.
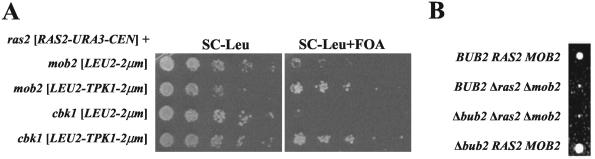
FIG. 2.
Overexpression of TPK1 suppresses the mob2Δ ras2Δ and cbk1Δ ras2Δ slow-growth phenotypes. (A) Y3018 (ras2Δ mob2Δ [CEN-URA3-RAS2]) and Y3019 (ras2Δ cbk1Δ [CEN-URA3-RAS2]) were transformed with YEp13 (LEU2 2μm) and B1373 (LEU2 TPK1 2μm) and analyzed for growth on SC-Leu and SC-Leu plus FOA as described in the legend to Fig. 1A. (B) Representative tetratype tetrad from tetrad analysis of a diploid resulting from crossing Y3013 and Y3017. More than 10 tetrads were analyzed with consistent results.
Since the role of Ras in mitotic exit is Tpk independent, the above result suggests that this particular function of Ras in mitotic exit does not cause the synthetic slow-growth phenotype of ras2Δ mob2Δ strains (38). To further analyze this, we examined the effect of BUB2 deletion on the mob2Δ ras2Δ slow-growth phenotype. In the absence of Bub2, the mitotic exit network is active and cells do not arrest in the presence of the microtubule-destabilizing agent nocadazole (reviewed in reference 16). If Mob2 and Ras2 function upstream of BUB2 in the mitotic exit network, then deletion of BUB2 should eliminate the growth delay of the mob2Δ ras2Δ mutant; however, deletion of BUB2 has no effect on the growth rate of the mob2Δ ras2Δ double mutant (Fig. 2B). Furthermore, overexpression of Cdc14, the downstream target of the mitotic exit network, does not suppress the growth defect of the double mutant (data not shown). Thus, the growth defect observed in the mob2Δ ras2Δ strain is independent of the role of Ras in mitosis.
Deletion of MOB2 does not increase trehalose levels of a ras2Δ strain.
Strains with reduced PKA activity exhibit phenotypes characteristic of increased stress resistance, including high trehalose levels and heat shock resistance. Inhibition of Cbk1 activity results in similar phenotypes that are independent of cAMP and PKA (63). To test whether deletion of both RAS2 and MOB2 results in increased trehalose levels, we measured trehalose levels in ras2, mob2, cbk1, ras2 mob2, and ras2 cbk1 cells. As evident from the data in Table 3, deletion of RAS2 promoted increased trehalose levels, as expected, but this level was not further increased by deletion of either MOB2 or CBK1.
TABLE 3.
Deletion of MOB2 does not increase trehalose levels of a ras2Δ straina
| Strain genotype | Trehalose (mg/mg of protein) |
|---|---|
| Wild type | 3.6 ± 0.7 |
| mob2Δ | 1.6 ± 0.9 |
| cbk1Δ | 7.4 ± 0.3 |
| ras2Δ | 47 ± 5 |
| mob2Δ ras2Δ | 30 ± 4 |
| cbk1Δ ras2Δ | 35 ± 1 |
Strains Y3020 (wild type), Y3021 (ras2Δ), Y3022 (mob2Δ), Y3023 (mob2Δ ras2Δ), Y3082 (cbk1Δ), and Y3083 (cbk1Δ ras2Δ) were grown in YEPD, and trehalose levels were determined as described in Materials and Methods. Numbers represent the averages for three independent cultures and the standard deviations of the means.
The slow-growth phenotype of the mob2Δ ras2Δ strain does not involve ACE2.
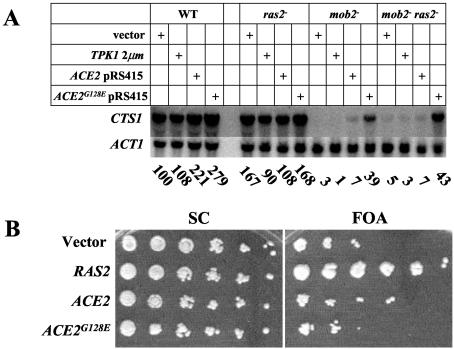
Previous studies have shown that one function of the Mob2/Cbk1 pathway is to activate and direct localization of the Swi5-like transcription factor Ace2 to daughter cell nuclei (8, 64). Deletion of CBK1 results in a cell separation defect that is at least partly due to decreased ACE2-dependent transcription of genes, such as chitinase, that are involved in degrading the cell wall after cytokinesis (8, 48). The ACE2 mutation ACE2G128E partially suppresses the cbk1Δ cell separation defect (48). Ace2G128E localizes to both the mother and daughter cell nuclei even in the absence of Cbk1 or Mob2 and thereby restores expression of genes impaired in cbk1Δ or mob2Δ strains (8, 48). Thus, as expected, the level of chitinase (encoded by CTS1) RNA is drastically reduced in both the mob2Δ and the mob2Δ ras2Δ strain (Fig. 3A). The ACE2G128E allele partially rescues steady-state levels of CTS1 in both these strains (Fig. 3A). Also, the ACE2G128E allele partially restores the mob2Δ and mob2Δ ras2Δ cell separation defect (data not shown). Expression of the ACE2G128E allele in the mob2Δ ras2Δ strain does not rescue the slow-growth phenotype (Fig. 3B), and deletion of ACE2 does not confer a synthetic slow-growth phenotype in a ras2Δ strain (data not shown). Thus, the cell cycle delay observed in the mob2Δ ras2Δ strain cannot be explained by impaired Ace2 activity.
FIG. 3.
The slow growth of a mob2Δ ras2Δ strain is not due to loss of Ace2 localization or activation. (A) Northern blot of CTS1 and ACT1 RNA from strains Y3020 (wild type [WT]), Y3021 (ras2/ras2), Y3022 (mob2/mob2), and Y3023 (ras2/ras2 mob2/mob2) transformed with plasmids carrying the indicated markers (YEp13, B1373, B2461, or B2462). Normalized values for the levels of CTS1 mRNA are indicated below the Northern blot. (B) Y3018 (ras2Δ mob2Δ [CEN-URA3-RAS2]) containing a plasmid carrying the indicated marker was spotted onto SC and SC plus FOA as for Fig. 1A.
Actin polarization in mitotic cells is not affected in the mob2Δ ras2Δ strain.
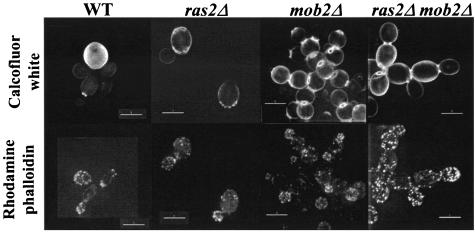
The cortical patch structures formed by yeast actin polymers exhibit marked distribution changes during the cell cycle that correlate with polarized cellular growth. In the strain background used by Ho and Bretscher (22), growth of a ras2Δ strain is reduced and the actin cytoskeleton is depolarized at 37°C. Since the role of Ras in actin repolarization is PKA independent and overexpression of TPK1 suppresses the growth defect of a mob2Δ ras2Δ strain, defects in actin polarization likely do not contribute to the mob2Δ ras2Δ synthetic growth defect. To confirm that actin polarization was not affected by the mutations, the mob2Δ ras2Δ strain was stained with rhodamine-conjugated phalloidin. No significant difference was observed in the polarization of cortical patches in small to medium size buds between the mob2Δ ras2Δ strain and the mob2Δ, ras2Δ, or wild-type strain (Fig. 4). Additionally, the mob2Δ ras2Δ strain does not exhibit slower growth at 37°C than at 30°C, as would be expected if actin repolarization was affected (data not shown). Mutants having a defect in actin polarization often exhibit an endocytosis defect that can be monitored by uptake of fluorescent vacuolar markers such as LY and FM464 (reviewed in reference 45). The mob2Δ ras2Δ strain does not have a synthetic defect in LY endocytosis at 30°C (wild type [Y3014], 90% vacuolar staining; mob2Δ ras2Δ [Y3015], 92% vacuolar staining; n ≥ 150).
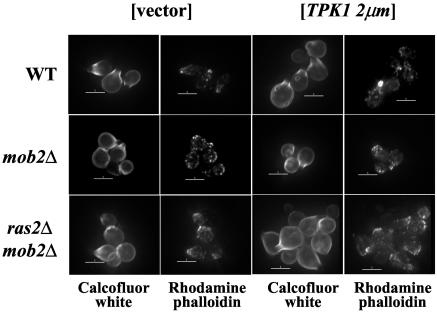
FIG. 4.
Filamentous actin is polarized in a mob2Δ ras2Δ strain. Calcofluor white (upper panels) and rhodamine phalloidin (lower panels) staining of strains Y3020 (wild type [WT]), Y3021 (ras2Δ/ras2Δ), Y3022 (mob2Δ/mob2Δ), and Y3023 (ras2Δ/ras2Δ mob2Δ/mob2Δ) is shown. Bars = 5 μm.
When yeast cells are exposed to pheromone, they undergo a cell cycle arrest and form elongated projections. ras2Δ strains form mating projections similar to those of wild-type cells, while cbk1Δ and mob2Δ cells do not efficiently form mating projections, suggesting an inability to maintain polarized growth (2, 64). mob2Δ ras2Δ strains also have a defect in mating projection formation; the cells do not form long projections, and the actin patches are not consistently concentrated at the tip of the projections (Fig. 5; data not shown). However, the phenotype of the mob2Δ mutant is too severe to determine if the double mutant has a synthetic effect. Overexpression of TPK1 does not significantly alter the positions of the actin patches (Fig. 5).
FIG. 5.
Morphological changes and formation of mating projections in response to pheromone. Calcofluor white (first and third columns) and rhodamine phalloidin (second and fourth column) staining of strains Y3014 (wild type [WT]), Y3025 (mob2Δ), and Y3015 (mob2Δ ras2Δ) containing Yep13 (vector) or B1373 (TPK1 2μm) and treated with α-factor as described in Materials and Methods is shown.
Deletion of MOB2 and RAS2 has a synthetic effect on bud site selection.
Diploid cells normally bud at a site in the distal or proximal pole with respect to the birth scar, a process called bipolar budding. Since deletion of MOB2 affects bipolar budding, we analyzed budding patterns in mob2 and ras2 mob2 strains. Exponentially growing diploid strains containing homozygous deletions in RAS2, MOB2, or both RAS2 and MOB2 were stained with calcofluor white, and the positions of the first and second budding events were analyzed. As shown in Table 4, deletion of RAS2 does not significantly affect the budding pattern at either the first or second bud stage. In contrast, deletion of MOB2 does not affect the position of the initial bud but has an effect on bud site selection at the second bud stage (Table 4). In approximately 20% of mob2Δ/mob2Δ cells, the second bud does not originate from either pole, whereas in wild-type cells about 5% exhibit nonbipolar budding (P < 0.001). This defect is similar to that of a homozygous cbk1Δ/cbk1Δ strain (data not shown; 2, 48). The mob2Δ ras2Δ/mob2Δ ras2Δ strain exhibited not only an increase in mislocalization of the second bud but also increased mislocalization of the first bud (Table 4; P < 0.0005). Thus, Ras2 and Mob2 both contribute to the precision of bipolar budding.
TABLE 4.
ras2Δ and mob2Δ exhibit synthetic bud site selection defects: diploid budding patterna
| Strain genotype | First bud
|
Second bud
|
||||
|---|---|---|---|---|---|---|
| Distal | Proximal | Medial | Distal | Proximal | Medial | |
| Wild type | 99 ± 2 | 1.1 ± 1.2 | 0.3 ± 0.3 | 92 ± 4 | 4.7 ± 1.7 | 3.3 ± 2.4 |
| ras2Δ | 98 ± 1 | 1.9 ± 0.9 | 0.9 ± 0.7 | 93 ± 4 | 3.0 ± 1.4 | 6.0 ± 2.9 |
| mob2Δ | 97 ± 2 | 2.4 ± 1.7 | 1.6 ± 0.8 | 81 ± 4 | 1.3 ± 1.3 | 18 ± 3b |
| ras2Δ mob2Δ | 82 ± 8 | 1.6 ± 0.8 | 17 ± 8c | 68 ± 4 | 2.9 ± 0.8 | 29 ± 3d |
| Wild type (vector) | 97 ± 2 | 2.3 ± 1.2 | 0.3 ± 0.5 | 76 ± 7 | 17 ± 2.4 | 6.3 ± 5.0 |
| mob2Δ (vector) | 95 ± 4 | 0 ± 0 | 4.9 ± 3.5 | 84 ± 2 | 0.3 ± 0.5 | 16 ± 2 |
| mob2Δ (TPK1) | 93 ± 5 | 0.3 ± 0.5 | 6.6 ± 4.9 | 85 ± 8 | 0.3 ± 0.5 | 14 ± 7 |
| ras2Δ mob2Δ (vector) | 77 ± 10 | 1.9 ± 1.6 | 21 ± 8e | 68 ± 6 | 3.3 ± 2.7 | 29 ± 3f |
| ras2Δ mob2Δ (TPK1) | 94 ± 3 | 0.3 ± 0.5 | 5.2 ± 3.7 | 73 ± 22 | 7.3 ± 9.6 | 19 ± 13f |
Diploid strains Y3020 (wild type), Y3021 (ras2Δ/ras2Δ), Y3022 (mob2Δ/mob2Δ), and Y3023 (ras2Δ/ras2Δ mob2Δ/mob2Δ), harboring no plasmid or carrying YEp13 (vector) or B1373 (TPK1), were grown to logarithmic phase and scored for positions of the first and second bud by calcofluor white staining as described in Materials and Methods. Numbers represent the average percentages (±standard deviations) of three independent experiments in which at least 100 of each budding event (first buds, second buds) were counted. In all cases, the least-significant P value of each of the three independent experiments is reported.
P < 0.001 for percent medial buds = percent wild-type medial buds.
P < 0.0005 for percent medial buds = percent wild-type medial buds, and P < 0.005 for percent medial buds = percent mob2Δ medial buds
P < 0.000005 for percent medial buds = percent wild-type medial buds
P < 0.0001 for percent medial buds = percent wild-type medial buds, and P < 0.002 for percent medial buds = percent ras2Δ mob2Δ (TPK1) medial buds.
P < 0.005 for percent medial buds = percent wild-type medial buds.
To determine if Ace2 functions downstream from Mob2 with respect to bud site selection, the dominant ACE2G128E allele was expressed in the mob2Δ ras2Δ/mob2Δ ras2Δ strain on a low-copy-number vector and the positions of the first and second buds were analyzed as before. Although the presence of this allele partially rescues the CTS1 transcriptional defect of a mob2Δ ras2Δ/mob2Δ ras2Δ strain, it does not affect the bipolar budding defect (data not shown). This is consistent with data demonstrating that the ACE2G128E allele does not suppress the bipolar budding defect of a cbk1Δ strain (48). In contrast, overexpression of TPK1 did restore the bipolar budding pattern to approximately that observed in the mob2Δ/mob2Δ strain at both the first and second bud stages in the mob2Δ/mob2Δ ras2Δ/ras2Δ strain (Table 4). Thus, the PKA pathway, but not Ace2, affects bud site selection in bipolar budding cells.
Haploid cells bud adjacent to the site at which the previous bud emerged, a process called axial budding. Deletion of RAS2 and MOB2 caused a defect in axial budding not seen by deletion of either gene alone. Exponentially growing wild-type, ras2Δ, mob2Δ, and ras2Δ mob2Δ cells were stained with calcofluor white, and their budding patterns were analyzed (Table 5). Deletion of neither MOB2 nor RAS2 alone significantly affected the fidelity of axial budding, while deletion of both significantly increased bipolar budding at the expense of axial budding (Table 5, P < 2 × 10−7). Time-lapse experiments of α-factor-synchronized cells confirmed a decrease in axial budding in a mob2Δ ras2Δ strain relative to either the wild type or a mob2Δ strain (more than 50 budding events analyzed; P < 0.002; data not shown). TPK1 overexpression in a mob2 ras2 strain suppressed the axial budding defect (Table 5).
TABLE 5.
ras2Δ and mob2Δ exhibit synthetic bud site selection defects: haploid budding patterna
| Strain genotype | Axial | Bipolar | Random |
|---|---|---|---|
| Wild type | 97 ± 2 | 2.5 ± 2.1 | 0 ± 0 |
| ras2Δ | 89 ± 1 | 11 ± 1 | 0 ± 0 |
| mob2Δ | 94 ± 5 | 5.7 ± 5.2 | 0.5 ± 0.4 |
| ras2Δ mob2Δ | 65 ± 10b | 34 ± 10 | 0.8 ± 0.6 |
| Wild type [vector] | 98 ± 1 | 1.7 ± 1.7 | 0.3 ± 0.5 |
| Wild type (TPK1) | 97 ± 2 | 2.7 ± 2.5 | 0.3 ± 0.5 |
| ras2Δ mob2Δ (vector) | 56 ± 13c | 39 ± 12 | 4.7 ± 1.2 |
| ras2Δ mob2Δ (TPK1) | 92 ± 3 | 8.0 ± 1.4 | 0 ± 1.2 |
Haploid strains Y3084 (wild type), Y3085 (ras2Δ), Y3086 (mob2Δ), and Y3087 (ras2Δ mob2Δ), harboring no plasmid or carrying YEp13 (vector) or B1373 (TPK1), were grown to logarithmic phase and scored for position of the first bud by calcofluor-white staining as described in Materials and Methods. Numbers represent the average percentages (±standard deviations) of three independent experiments in which at least 100 cells were counted.
P < 2 × 10−7 for percent nonaxial buds = percent wild-type nonaxial buds and for percent nonaxial buds = percent mob2Δ nonaxial buds.
P < 6 × 10−8 for percent nonaxial buds = percent wild-type nonaxial buds.
Deletion of both mob2 and ras2 affects growth rate but not viability.
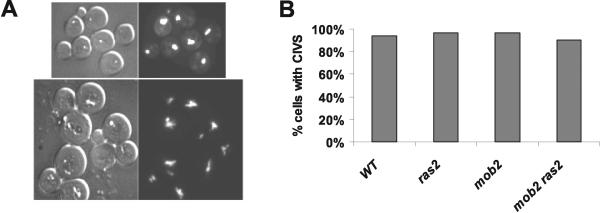
In order to determine cell viability, we used FUN 1 staining as an assay of live cells. Live cells metabolize the fluorescent reagent FUN 1, resulting in the formation of cylindrical intravacuolar structures (CIVS) (Fig. 6A). About 90% of mob2Δ ras2Δ cells exhibited positive staining with FUN 1 compared with 94% of wild-type cells (Fig. 6B). Similarly, we detected no loss in viability, as measured by plating efficiency, with any of the mutant strains compared to the wild type (data not shown). Thus, mob2 ras2 strains do not exhibit reduced viability.
FIG. 6.
Slow-growth phenotype of mob2Δ ras2Δ strains is caused by a growth delay, not loss in viability. (A) Differential interference contrast (left panels) and FUN 1 (right panels) staining of wild-type strain Y3014 (upper panels) and mob2Δ ras2Δ strain Y3015 (lower panels). (B) Percent cells (n ≥ 200) exhibiting positive staining with FUN 1 as measured by the appearance of cylindrical intracellular vacuolar structures (CIVS).
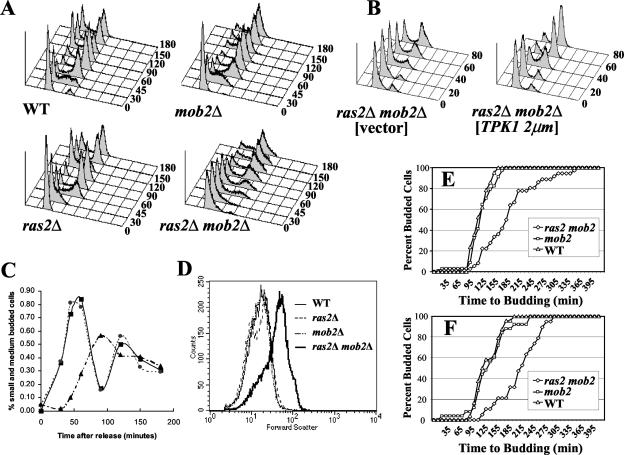
To characterize the slow-growth defect of the mob2Δ ras2Δ strain, we examined wild-type, ras2Δ, mob2Δ, and mob2Δ ras2Δ strains by flow cytometry following release of cells from an α-factor-induced G1 arrest (Fig. 7A). As anticipated, deletion of MOB2 resulted in a G2/M defect. Time-lapse microscopy demonstrated that this was due to inefficient cell separation of the mob2Δ strain (data not shown; 8, 64). Most (>72%) of the wild-type, ras2Δ, and mob2Δ cells exited G1 by 45 min after release from the α-factor arrest. In contrast, <50% of the mob2Δ ras2Δ cells exited G1 phase at this time. This suggests that the mob2Δ ras2Δ cells have a G1 delay. Since overexpression of TPK1 suppressed the growth defect of a mob2Δ ras2Δ strain, it was likely that TPK1 overexpression would overcome the observed G1/S delay. Flow cytometry analysis following release of cells synchronized in G1 with α-factor was used to determine if overexpression of TPK suppressed the G1/S cell cycle delay observed in mob2Δ ras2Δ cells. As observed previously, mob2Δ ras2Δ cells exhibited a G1/S delay; however, this was suppressed by overexpression of TPK1 (Fig. 7B).
FIG. 7.
The slow growth of a mob2Δ ras2Δ strain is due to a G1 delay. (A) Strains Y3084 (wild type [WT]), Y3085 (mob2Δ), Y3086 (ras2Δ), and Y3087 (mob2Δ ras2Δ) were synchronized in G1 as described in Materials and Methods. Cells were collected at the indicated times (minutes), stained with Sytox green, and analyzed by fluorescence-activated cell sorting (FACS) as described in Materials and Methods. (B) Strain Y3087 (mob2Δ ras2Δ) was transformed with YEp13 (vector) or B1373 (TPK1 2μm) and synchronized in G1 as described in Materials and Methods. Cells were collected at the indicated times, stained with Sytox green, and analyzed by FACS as described in Materials and Methods. (C) Percentages of budded cells as a function of time following release from α-factor block. ▪, wild type (Y3084); •, mob2Δ (Y3085); ▴, mob2Δ ras2Δ (Y3087). (D) Forward scatter of asynchronous Y3084, Y3085, Y3086, and Y3087 fixed cells. (E) Cells of the indicated strains were synchronized with α-factor and then plated on YEPD and examined by time-lapse microscopy over two generations. Shown is the cumulative percent mother cells with the indicated time interval between the emergence of the first and second buds. (F) Shown is the cumulative percent daughter cells from the experiment for which results are shown in panel E with the indicated interval between birth and emergence of the first bud.
Microscopic examination of DAPI-stained cells synchronized as in the flow cytometry experiment supports the G1/S delay in mob2Δ ras2Δ cells. Wild-type, ras2Δ, and mob2Δ cells begin budding about 30 min after release from α-factor arrest, while mob2Δ ras2Δ cells begin budding 60 min after release (Fig. 7C; data not shown). Forward-angle light scattering in a flow cytometer provides a measure of the cell size distribution. Disruption of either RAS2 or MOB2 alone did not significantly affect the overall cell size distribution; however deletion of both MOB2 and RAS2 resulted in an increased average cell size (Fig. 7D).
The observed increase in the length of time mob2Δ ras2Δ cells spend in G1 following α-factor arrest could be due to an increase in α-factor sensitivity. To address this, wild-type, mob2Δ, and mob2Δ ras2Δ cells were synchronized with α-factor and washed and division time was analyzed using time-lapse microscopy. Cells were followed for two division cycles, and the time between the formation of the first and second buds from the same mother cell was measured (Fig. 7E). On average, the second bud from mob2Δ ras2Δ mother cells appeared approximately 200 min after the emergence of the first bud following release from the arrest, whereas wild-type cells and mob2Δ cells produced a second bud within an average of 118 and 123 min, respectively (Fig. 7E; more than 30 cells analyzed; P < 10−17). mob2Δ ras2Δ daughter cells also took longer to bud (n = 19, approximately 250 min, P < 0.002) than wild-type (n = 24, average about 135 min) or mob2Δ (n = 25, approximately 140 min (Fig. 7F) cells. Even though the cells were initially synchronized with α-factor, the second- and third-generation mob2Δ ras2Δ cells analyzed still exhibited a significant cell cycle delay even though they had not been exposed to this treatment. This indicates that deletion of MOB2 and RAS2 results in a true delay in cell cycle progression rather than exacerbated α-factor sensitivity.
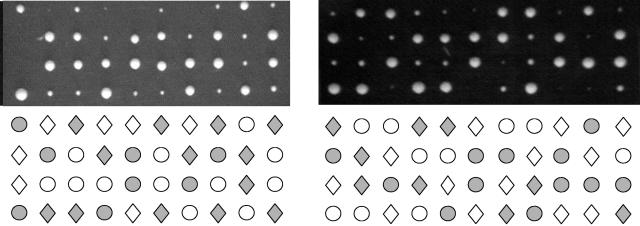
In Saccharomyces, a morphogenesis checkpoint delays cell cycle progression in response to insults that impair actin organization and/or bud formation (reviewed in reference 32). Various environmental and experimental insults that impair bud formation stabilize the Swe1 kinase, which phosphorylates Cdc28 at Y19 to inhibit kinase activity and cell cycle progression (32, 52, 53). To determine whether the cell cycle delay in ras2 mob2 strains results from an engagement of the morphogenesis checkpoint, we examined the effect of deleting SWE1 in a mob2Δ ras2Δ strain (Fig. 8). We found that either of these mutations exacerbated the slow-growth defect, such that the triple mutant was extremely sick or, in most cases, dead (Fig. 8). Similarly, the presence of a CDC28Y19F allele as the sole copy of CDC28 exacerbated the slow-growth defect of mob2Δ ras2Δ cells (Fig. 8). Due to the cell separation defect of a mob2Δ strain, we are unable to determine if G2 is longer in the mob2Δ ras2Δ strain than in the mob2Δ strain. The synthetic effect on bud site selection observed in the mob2Δ ras2Δ strain indicates a cell polarity defect, which may require a Swe1-imposed pause to give the cell sufficient time to complete its required tasks during division.
FIG. 8.
Impairment of the morphogenesis checkpoint exacerbates the slow-growth phenotype of a mob2Δ ras2Δ strain. (Left) Tetrad analysis of Y3154 (swe1Δ ras2Δ) × Y3157 (mob2Δ ras2Δ). Genotypes of spore clones are shown below the panel (MOB2, circle; mob2, diamond; SWE1, filled symbol; swe1, open symbol). (Right) Tetrad analysis of Y3157 (mob2Δ ras2Δ) × Y3155 (cdc28 Y19F ras2Δ). Individual segregants were analyzed by PCR amplification and KpnI digestion of the CDC28 locus to determine the presence of the cdc28Y19F mutation. Genotypes of spore clones are shown below the panel (MOB2, circle; mob2 diamond; CDC28, filled symbol; cdc28Y19F, open symbol).
DISCUSSION
The MOB2/CBK pathway is required for efficient cell cycle progression in a ras2Δ strain.
In a genetic screen to further our understanding of Ras functions in S. cerevisiae, we found that mutation of MOB2 results in a slow-growth phenotype in the absence of RAS2. The viability of mob2Δ ras2Δ strains is similar to that of the wild type, but the cells grow more slowly due to a G1/S delay that differs from the G0/G1 arrest formerly described for PKA mutants (26). The mob2Δ ras2Δ cells exhibit a G1/S delay accompanied by an increase in cell size, indicative of a cell cycle defect. In contrast, cAMP/PKA pathway mutants have a growth defect that causes a G0/G1 arrest with no size increase. Mob2 is part of a signaling network, including Cbk1, Hym1, Pag1, and Kic1, which functions to activate the Ace2 transcription factor and to control polarized morphogenesis (2, 8, 11, 13, 27, 39, 48, 64), and deletion of any of these genes confers slow growth in a ras2Δ background. Thus, the Cbk1 signaling pathway shares a function with Ras2 in cell cycle progression.
Deletion of MOB2 alone in the S288C background used in this study did not affect cell viability. In contrast, deletion of CBK1, MOB2, HYM1, PAG1, or KIC1 is lethal in the S288C background used by the Saccharomyces Deletion Project Consortium (SDPC). However, deletion of SSD1 can suppress the lethality of deleting CBK1 in the SDPC background (13, 27, 39). Since deletion of SSD1 in the strain background used in this study does not affect the growth rate of either a mob2Δ or a mob2Δ ras2Δ strain, this strain probably does not contain the SSD1-v allele (data not shown).
mob2Δ ras2Δ strains are defective in bud site selection.
Mob2 has a role in bipolar bud site selection in diploid cells, similar to that previously described for Cbk1. In the absence of MOB2, diploid cells show increased random budding, a defect that is exacerbated by concurrently eliminating RAS2. Furthermore, ras2 mob2 strains, but neither single mutant alone, exhibit significant bipolar budding as haploids, which under normal circumstances exclusively bud axially. Both budding defects are suppressed by overexpression of the catalytic subunit of PKA, implicating a role of the Ras/PKA pathway in bud site selection. Previous studies have demonstrated that loss of PKA activity affects the switch from bipolar to unipolar budding in nitrogen-starved cells, but the identity of the kinase target and the molecular mechanism driving the switch in bud site selection are unknown (43). The axial budding defect observed in ras2 mob2 cells is likely the result of the cell cycle delay causing loss of transient spatial cues; however, the bipolar defect should not be affected in the same way (7). Many genes affect the process of bud site selection, which involves first marking the positions of the two poles and then selecting the appropriate pole for forming the new bud. Mutations in either BEM4 or SUR4 give phenotypes similar to those observed for mob2 ras2 strains (40). Bem4 interacts with Rho1 and Cdc42, proteins involved in bud emergence but not pole marking. Thus, the synthetic effects of Ras2 and Mob2 on budding may well involve efficiency of recognizing the existing cell polarity rather than establishing it. In addition, these components may suggest potential targets for the convergence of the Ras/PKA and Mob2/Cbk1 pathways.
What are the downstream targets affected in the mob2Δ ras2Δ strain?
Previous studies have identified PKA-dependent and PKA-independent roles of the RAS genes. Ras proteins have PKA-independent functions in mitotic exit and in actin repolarization following heat shock. Since overexpression of TPK1, encoding a catalytic subunit of PKA, rescues growth of mob2Δ ras2Δ strains, defects in neither of these cellular roles account for slow growth of the double mutant. Furthermore, neither deletion of BUB2 nor overexpression of CDC14 affects growth of a mob2Δ ras2Δ strain, although either of these conditions would be expected to affect growth if a defect in the mitotic exit network was involved. In addition, the mob2Δ ras2Δ strain grows similarly at 30 and 37°C; if actin polarity was affected, a decrease in growth would be expected at the higher temperature. Therefore, Ras signaling through PKA is important for cellular proliferation of a mob2Δ strain.
An important role of Mob2, together with Cbk1, is to direct localization of the transcription factor Ace2 into the daughter cell nucleus, resulting in the activation of several genes involved in cell separation (8, 64). Currently, Ace2 is the only known downstream target of Cbk1. We have shown that a mutant allele of ACE2, ACE2G128E, does not suppress the slow-growth phenotype of a mob2Δ ras2Δ strain, although it partially rescues transcriptional defects of mob2 and mob2Δ ras2Δ strains. Importantly, deletion of ACE2 does not confer a synthetic slow-growth phenotype to a ras2Δ strain. Consequently, the Mob2/Cbk1 complex is likely signaling to a downstream target other then Ace2, which is important for cellular proliferation in the absence of RAS2.
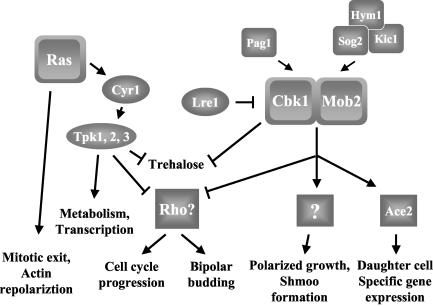
A likely common target of Ras/PKA and Mob2/Cbk1 is Rho1 (Fig. 9). Rho1 plays a dual role in the regulation of cell morphology: as a regulatory subunit for 1,3-β-glucan synthase and an activator of Pkc1 (12, 21, 28, 36, 42, 44, 47). Activation of Pkc1 by Rho1 results in actin cytoskeleton polarization and cellular integrity preservation via activation of the mitogen-activated protein kinase cascade. Overexpression of LRE1, a negative regulator of CBK1, rescues the 1,3-β-glucan synthesis of a strain containing mutations in the catalytic subunits of 1,3-β-glucan synthase, suggesting that Cbk1 negatively regulates 1,3-β-glucan synthase (48, 51, 63). Epistasis analysis suggests that LRE1 functions upstream from GTP association with RHO1 (51). Consistent with the Cbk1 pathway negatively regulating Rho1, deletions in genes encoding Rho1 GTPase-activating proteins, SAC7 and BEM2, confer a synthetic slow-growth defect in cbk1Δ and hym1Δ strains (27). These results suggest that loss of Cbk1 signaling may cause constitutive activation of Rho1 signaling. Consistent with a role of RHO1 in bipolar bud site selection, a mutant allele of BEM2, bem2-101, or deletion of ROM2, which encodes the RHO1 exchange factor, confers a bipolar budding defect (31, 40). Data suggesting that the Ras/cAMP pathway modulates cell wall biosynthesis also exists. The WSC genes function to activate RHO1 (9, 19, 24, 33, 62). Heat shock sensitivity of a wsc1Δ wsc2Δ wsc3Δ strain is suppressed by deletion of RAS2, and overexpression of WSC1 suppresses the heat shock sensitivity of a ras1Δ ras2Δ strain overexpressing CYR1 (62). Finally, we found that a dominant activated allele of RHO1 confers a slow-growth defect to a ras2Δ strain, consistent with the hypothesis that the synthetic slow-growth phenotype of the mob2Δ ras2Δ strain is due to an inability to properly regulate RHO1 (L. Schneper and J. R. Broach, unpublished results). Thus, the observed growth and budding defects of mob2Δ ras2Δ strains could be explained by their upregulation of Rho1.
FIG. 9.
Model for the roles of Ras and Mob2 in S. cerevisiae. Ras proteins have PKA-independent functions in mitotic exit and actin repolarization and PKA-dependent functions affecting metabolism, transcription, and cell growth. Cbk1, together with Mob2, regulates the Ace2 transcription factor, effecting daughter cell-specific gene expression. Cbk1 and Mob2 have additional Ace2-independent functions in regulating apical growth and mating projection formation. Hym1 interacts with Kic1 and Sog2, and this complex along with Pag1 interacts with Cbk1 to promote its localization and function. Data presented in this report demonstrate that the Ras/PKA and Cbk1/Mob2 pathways play parallel roles in cell growth and bud site selection, likely through regulation of Rho1 (see Discussion for details). Cbk1 and the Ras/PKA pathway independently repress cellular trehalose levels through a different mechanism.
Acknowledgments
We thank Mark Rose, Virginia Zakian, Waheeda Khalfan, Elbert Chiang, Haiyan Qi, Yu Jiang, and Angelika Amon for generous gifts of plasmids and strains; Mark Rose, Peter Houston, and Sean Clark for training and assistance on yeast microscopy; David Drubin and Eric Weiss for their thoughtful comments, advice on actin staining, and sharing of information prior to publication; Katrin Düvel, Patricia Melloy, Cemile Güldal, and Peter Houston for critical suggestions regarding the manuscript; Christina deCoste and Andrew Taggart for advice and assistance with flow cytometry and cell synchronization; the Princeton University SynSeq Facility for DNA sequencing and oligonucleotide synthesis; and Fang Li for preparation of media.
This work was supported by NIH grant CA41086 to J.R.B. and by NRSA grant 5F32 GM19308 to L.S.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology, vol. 2. John Wiley and Sons, Inc., New York, N.Y.
- 2.Bidlingmaier, S., E. L. Weiss, C. Seidel, D. G. Drubin, and M. Snyder. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breeden, L. L. 1997. Alpha-factor synchronization of budding yeast. Methods Enzymol. 283:332-341. [DOI] [PubMed] [Google Scholar]
- 4.Broach, J. R. 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7:28-33. [DOI] [PubMed] [Google Scholar]
- 5.Broach, J. R., and R. J. Deschenes. 1990. The function of ras genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54:79-139. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Chant, J., and J. R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 9.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vendittis, E., A. Vitelli, R. Zahn, and O. Fasano. 1986. Suppression of defective RAS1 and RAS2 functions in yeast by an adenylate cyclase activated by a single amino acid change. EMBO J. 5:3657-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorland, S., M. L. Deegenaars, and D. J. Stillman. 2000. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154:573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drgonova, J., T. Drgon, K. Tanaka, R. Kollar, G. C. Chen, R. A. Ford, C. S. Chan, Y. Takai, and E. Cabib. 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272:277-279. [DOI] [PubMed] [Google Scholar]
- 13.Du, L. L., and P. Novick. 2002. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedor-Chaiken, M., R. J. Deschenes, and J. R. Broach. 1990. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61:329-340. [DOI] [PubMed] [Google Scholar]
- 15.Field, J., J. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner, R. D., and D. J. Burke. 2000. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 10:154-158. [DOI] [PubMed] [Google Scholar]
- 17.Garrett, S., and J. Broach. 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3:1336-1348. [DOI] [PubMed] [Google Scholar]
- 18.Geymonat, M., S. Jensen, and L. H. Johnston. 2002. Mitotic exit: the Cdc14 double cross. Curr. Biol. 12:R482-R484. [DOI] [PubMed] [Google Scholar]
- 19.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow, E., and D. Lane. 1988. Antibodies, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8:1211-1214. [DOI] [PubMed] [Google Scholar]
- 22.Ho, J., and A. Bretscher. 2001. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell 12:1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 24.Jacoby, J. J., S. M. Nilius, and J. J. Heinisch. 1998. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 258:148-155. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y., C. Davis, and J. R. Broach. 1998. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17:6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston, G. C., J. R. Pringle, and L. H. Hartwell. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79-98. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, P., B. Nelson, M. D. Robinson, Y. Chen, B. Andrews, M. Tyers, and C. Boone. 2002. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162:1091-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 271:9193-9196. [DOI] [PubMed] [Google Scholar]
- 29.Kataoka, T., S. Powers, C. McGill, O. Fasano, J. Strathern, J. Broach, and M. Wigler. 1984. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 37:437-445. [DOI] [PubMed] [Google Scholar]
- 30.Kienle, I., M. Burgert, and H. Holzer. 1993. Assay of trehalose with acid trehalase purified from Saccharomyces cerevisiae. Yeast 9:607-611. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y. J., L. Francisco, G. C. Chen, E. Marcotte, and C. S. Chan. 1994. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J. Cell Biol. 127:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lew, D. J. 2000. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 10:47-53. [DOI] [PubMed] [Google Scholar]
- 33.Lodder, A. L., T. K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luca, F. C., and M. Winey. 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto, K., I. Uno, and T. Ishikawa. 1985. Genetic analysis of the role of cAMP in yeast. Yeast 1:15-24. [DOI] [PubMed] [Google Scholar]
- 36.Mazur, P., and W. Baginsky. 1996. In vitro activity of 1, 3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271:14604-14609. [DOI] [PubMed] [Google Scholar]
- 37.Mbonyi, K., M. Beullens, K. Detremerie, L. Geerts, and J. M. Thevelein. 1988. Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 8:3051-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morishita, T., H. Mitsuzawa, M. Nakafuku, S. Nakamura, S. Hattori, and Y. Anraku. 1995. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science 270:1213-1215. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, L. Pratt, A. Zougman, L. D. B. McBroom, T. R. Hughes, C. Boone, and F. Luca. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed]
- 40.Ni, L., and M. Snyder. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikawa, J., S. Cameron, T. Toda, K. M. Ferguson, and M. Wigler. 1987. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1:931-937. [DOI] [PubMed] [Google Scholar]
- 42.Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai. 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5931-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J. L. Carpentier, L. S. Klig, and M. A. Payton. 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12:4896-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 46.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 47.Qadota, H., C. P. Python, S. B. Inoue, M. Arisawa, Y. Anraku, Y. Zheng, T. Watanabe, D. E. Levin, and Y. Ohya. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1, 3-beta-glucan synthase. Science 272:279-281. [DOI] [PubMed] [Google Scholar]
- 48.Racki, W. J., A. M. Becam, F. Nasr, and C. J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riezman, H. 1985. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell 40:1001-1009. [DOI] [PubMed] [Google Scholar]
- 50.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono, M. Minemura-Asakawa, S. Ishihara, T. Watanabe, and Y. Ohya. 2002. Dissection of upstream regulatory components of the Rho1p effector, 1, 3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics 162:663-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sia, R. A., E. S. Bardes, and D. J. Lew. 1998. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 17:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sia, R. A., H. A. Herald, and D. J. Lew. 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7:1657-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan, D. S., S. Biggins, and M. D. Rose. 1998. The yeast centrin, Cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatchell, K. 1993. RAS genes in the budding yeast Saccharomyces cerevisiae, p. 147-188. In J. Kurjan, and B. J. Taylor (ed.), Signal transduction. Prokaryotic and simple eukaryotic systems. Academic Press, San Diego, Calif.
- 56.Tatchell, K., L. C. Robinson, and M. Breitenbach. 1985. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc. Natl. Acad. Sci. USA 82:3785-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thevelein, J. M. 1994. Signal transduction in yeast. Yeast 10:1753-1790. [DOI] [PubMed] [Google Scholar]
- 58.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 59.Toda, T., S. Cameron, P. Sass, M. Zoller, and M. Wigler. 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277-287. [DOI] [PubMed] [Google Scholar]
- 60.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 61.Tokiwa, G., M. Tyers, T. Volpe, and B. Futcher. 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371:342-345. [DOI] [PubMed] [Google Scholar]
- 62.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Versele, M., and J. M. Thevelein. 2001. Lre1 affects chitinase expression, trehalose accumulation and heat resistance through inhibition of the Cbk1 protein kinase in Saccharomyces cerevisiae. Mol. Microbiol. 41:1311-1326. [DOI] [PubMed] [Google Scholar]
- 64.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al., D. A. Lashkari, D. Morris, and M. Mittmann. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]