Abstract
Candida albicans is an important human pathogen that displays a remarkable ability to detect changes in its environment and to respond appropriately by changing its cell morphology and physiology. Serum- and amino acid-based media are known to induce filamentous growth in this organism. However, the mechanism by which amino acids induce filamentation is not yet known. Here, we describe the identification and characterization of the primary amino acid sensor of C. albicans, Csy1. We show that Csy1p plays an important role in amino acid sensing and filamentation. Loss of Csy1p results in a lack of amino acid-mediated activation of amino acid transport and a lack of induction of transcription of specific amino acid permease genes. Furthermore, a csy1Δ/csy1Δ strain, lacking Csy1p, is defective in filamentation and displays altered colony morphology in serum- and amino acid-based media. These data provide the first evidence that C. albicans utilizes the amino acid sensor Csy1p to probe its environment, coordinate its nutritional requirements, and determine its morphological state.
Candida albicans is an opportunistic and dimorphic pathogenic fungus that is able to cause recalcitrant infections of skin, oral, gastrointestinal, and urogenital systems. Depending on host immunity, infection by this organism can be either superficial or hematogenously disseminated, resulting in life-threatening systemic candidiasis (19, 24, 27, 28, 38, 44, 45). C. albicans undergoes a reversible switch between yeast, pseudohyphal, and hyphal growth in response to various stimuli (36, 40). Serum, Lee's medium (rich in amino acids), high temperatures (37°C), and neutral pHs are among the conditions that positively influence hyphal morphogenesis (11, 40). Although serum is the most effective inducer of hyphae, the components of this medium responsible for this phenotypic switching have not yet been identified.
Studies with the yeast Saccharomyces cerevisiae have identified several plasma membrane nutritional sensors that are able to sense the levels of nutrients in the environment and induce specific signaling pathways to modulate the rate of uptake of such nutrients (15). Two sensors of glucose, Snf3p and Rgt2p, are involved in the regulation of glucose uptake, depending on the concentration of glucose in the environment (41). Although they are members of the sugar transport superfamily, Snf3p and Rgt2p are unable to transport glucose and do not restore the ability of a mutant lacking the glucose transporter genes, HXT-1 to -4, -6, and -7, to utilize glucose (33, 43). However, Snf3p and Rgt2p activation by low or high glucose concentrations results in the transcriptional regulation of HXT genes encoding low- and high-affinity glucose transporters (33, 41).
Similarly, S. cerevisiae possesses a sensor of amino acids, Ssy1p (10, 21, 25), which belongs to the amino acid permease (AAP) superfamily (14, 42, 50). Unlike other members of this family, Ssy1p does not transport amino acids, and it contains an N-terminal domain involved in sensing amino acids and activating downstream factors that regulate the expression of AAP genes (10, 21, 25). When amino acids are available in the environment, Ssy1p activates the expression of AAP genes, which then mediate the transport of amino acids (10, 21, 25). Thus, SSY1 encodes a regulator of transcription of AAP genes rather than an amino acid transporter (10). This idea is further supported by the finding that disruption of SSY1 results in the inhibition of uptake of several amino acids (13, 21). Furthermore, loss of the SSY1 gene is not compensated for by overexpression of AAP genes, and its overexpression does not rescue the loss of amino acid uptake in strains containing multiple deletions of amino acid transporter genes (10).
Expression of amino acid transporters in S. cerevisiae is also dependent on Ptr3p and Ssy5p, two peripheral membrane proteins that localize to the cytoplasmic face of the plasma membrane (14, 25). Ssy1p, Ptr3p, and Ssy5p are components of a sensor complex called SPS that resides in the plasma membrane and functions to transduce amino acid-derived signals (2, 14, 25). Interestingly, both ssy1Δ and ptr3Δ mutants exhibit enhanced haploid invasive growth compared to that of the wild type (25), suggesting an important role for the SPS complex in yeast filamentation. However, how Ssy1p regulates invasive growth is not well understood.
Unlike other organisms, little is known about how C. albicans responds to changing nutritional conditions. Here, we provide evidence for the presence of a coordinated regulatory process for amino acid uptake and filamentation mediated by amino acids in C. albicans. We identified the primary amino acid sensor gene responsible for these activities, CSY1, and showed that C. albicans lacking Csy1p has altered amino acid uptake and filamentation.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains (Table 1) used in this study were constructed from strain BWP17, which was derived from strain RM1000 (39) as described previously (51). Strains were routinely cultured in rich medium (YPDU), which contains 2% Bacto Peptone, 1% yeast extract, 20% dextrose, and 80 μg of uridine/ml, or in synthetic medium (SD), which contains 1.7% yeast nitrogen base, 0.5% ammonium sulfate, and 2% dextrose. Supplements were added as required to maintain cell growth at standard concentrations (51). Growth inhibition assays on amino acid analogs were performed by plating 2 × 104 wild-type (CAEB-1), heterozygote (CSY1/csy1Δ) (CAEB-3), homozygote (csy1Δ/csy1Δ) (CAEB-5), or complemented (csy1Δ/csy1Δ+CSY1) (CAEB-6) cells on SD plates containing 25 mM l-glutamic acid γ-hydrazide. Filamentation assays and colony morphology studies were performed by plating 2 × 104 yeast cells of mid-log-phase cultures on either YPD containing 10% fetal bovine serum, Lee's medium (31), Spider medium (35), synthetic low-ammonium dextrose (SLAD) medium (17), or N-acetylglycosamine medium (5). Plates were incubated at 37°C. Cell morphology was determined by scraping the surfaces of the colonies and examining the cells by light microscopy.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BWP17 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 51 |
| DAY286 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG ARG4::URA3::arg4::hisG/arg4::hisG | Aaron Mitchell |
| CAEB-1 | ura3::imm434/ura3::imm434 HIS1::his1::hisG/his1::hisG ARG4::URA3::arg4::hisG/arg4::hisG | This study |
| CAEB-2 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG csy1::ARG4/CSY1 | This study |
| CAEB-3 | URA3::ura3::imm434/ura3::imm434 HIS1::his1::hisG/his1::hisG arg4::hisG/arg4::hisG csy1::ARG4/CSY1 | This study |
| CAEB-4 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG csy1::ARG4/csy1::URA3 | This study |
| CAEB-5 | ura3::imm434/ura3::imm434 HIS1::his1::hisG/his1::hisG arg4::hisG/arg4::hisG csy1::ARG4/csy1::URA3 | This study |
| CAEB-6 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG csy1::ARG4/csy1::URA3 CSY1::HIS1 | This study |
Molecular biology.
Plasmid pGEM-HIS1-CSY1 was constructed as follows. The CSY1 open reading frame was PCR amplified from genomic DNA (300 ng) obtained from strain BWP17 by using primers pro-CSY1-5′ (GGGGACGTCGGGCCCAGCCAGAGTGAGAATTCGA) and term-CSY1-3′ (ACATGCATGCAAATGAAATTAAGAAATAGAG). The resulting PCR product was digested with the ApaI and SphI restriction enzymes and cloned into the ApaI and SphI sites of pGEM-HIS1 (51), thus creating the pGEM-HIS1-CSY1 plasmid.
Genomic DNA from C. albicans was prepared by glass bead lysis as described by Hoffman and Winston (20). For RNA preparation, overnight cultures of C. albicans cells grown at 37°C on liquid SD medium in the presence or absence of 10 mM histidine were diluted in the same medium to 107 cells/ml. Cells were harvested at mid-log phase, and total RNAs were prepared using the heat/freeze RNA isolation method as previously described (46). Southern and Northern blot analyses were performed as described by Maniatis and colleagues (37). Probes were generated by random priming according to the manufacturer's recommendations (Roche Diagnostics Corporation). Substrates for the probes specific to CSY1, CAN1, orf 6.7739, orf 6.4609, and ACT1 were generated by PCR using C. albicans genomic DNA as the template and the following primer pairs: probe-CSY1-5′ (CCACTGCTGGATTTGTCACG) and probe-CSY1-3′ (CCCTGTAATGCCAACGAATAAG), CAN1-5′ (GCAGTAAAGGCAACGGCAAAG) and CAN1-3′ (CACACAATCTCTTGGTGAAATGGC), orf 6.7739-5′ (CACCAGTGTCTCTACCTTGAGC) and orf 6.7739-3′ (GGATGCCAAAACAAACCTG), orf 6.4609-5′ (AGACCAGAAAGAGCCAATAACC) and orf 6.4609-3′ (CACAAGATAAACTCACGGGTGTAG), and ACT1-5′ (AAGCCCAATCCAAAAGAGG) and ACT1-3′ (GATAGAACCACCAATCCAGACAG), respectively.
Strain construction.
Strain CAEB-4 (Table 1), in which both copies of the CSY1 gene were deleted, was obtained after two successive transformations of strain BWP17 with PCR-generated targeting cassettes containing either ARG4 or URA3 positive markers flanked by 60 bp of homology to CSY1 (51) using the transformation conditions of De Backer and colleagues (7). The targeting products for CSY1 disruption were generated by PCR using 200 ng of plasmid DNA (pGEM-URA3 or pRS-Arg4ΔSpeI) (51), 1 μM primers CSY1-5′ (AGCCGACAGCATGTGGTATCACCATTTTCAACCACAAGTATTAGCGATACTGTATCATTAGTTTTCCCAGTCACGACGTT) and CSY1-3′ (TGACAACATAATAACCCCAGCAACTACTTCTCCTGCCACCCCTGTAGAAAAACTCAAAAATGTGGAATTGTGAGCGGATA), 5 μl of 10× PCR buffer (Invitrogen), 0.4 mM deoxynucleoside triphosphates, 2 mM MgCl2, and 0.5 μl of Taq DNA polymerase (1 U/μl; Invitrogen). The mixture was incubated at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 3 min. After a final extension at 68°C for 5 min, the reaction mixture was stored at either 4 or −20°C before further use. Gene replacement was confirmed by PCR analysis using primers O1 (ATGATAGACCTAGAAGGGTCA) and O2 (AATATGTACCAAGTTGCTTCCC). The csy1Δ/csy1Δ prototrophic strain, CAEB-5, was generated by transforming the histidine auxotrophic strain CAEB-4 with the NruI-digested pGEM-HIS1 vector. A wild-type prototrophic strain (CAEB-1) was generated by transforming DAY286 (Table 1) with NruI-digested pGEM-HIS1. The prototrophic CSY1-heterozygous strain, CAEB-3, was obtained by integration of NruI-digested pGEM-HIS1 and BclI-digested pGEM-URA3 into the HIS1 and URA3 loci of strain CAEB-2, respectively. The csy1Δ/csy1Δ+CSY1 complemented strain (CAEB-6) was generated by transforming the csy1Δ/csy1Δ strain (CAEB-4) with NruI-digested pGEM-HIS1-CSY1. The genotypes of these strains were further confirmed by PCR and Southern blot analyses.
Amino acid transport assays.
Overnight cultures of cells grown at 30 or 37°C on SD medium in the presence or absence of 10 mM amino acids (except for tyrosine, which was added at 2.5 mM) were diluted in 20 ml of the same medium to an optical density at 600 nm (OD600) of 0.2. Cells were harvested at an OD600 of 0.65 by centrifugation at 2,000 × g for 10 min at 4°C, washed twice with cold phosphate-buffered saline (PBS), and resuspended in 700 μl of SD medium. A 100-μl volume of this suspension was added to 150 μl of SD medium containing 0.2 μCi of [3H]-labeled amino acids (24 to 120 Ci/mmol; Amersham Pharmacia). After incubation of this mixture at 30 or 37°C, amino acid uptake was stopped by addition of 5 ml of ice-cold PBS. Cells were collected on a Whatman GF/C glass microfiber filter, washed three times with cold PBS, air dried, and counted in a scintillation counter (18). All uptake studies were performed in duplicate, and average values were determined.
RESULTS
Substrate specificity of sensing and activation of amino acid transport in C. albicans.
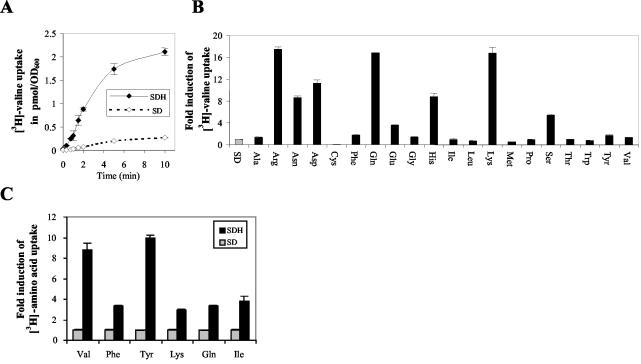
The ability of C. albicans to switch between yeast, pseudohyphal, and hyphal forms is an important component of its pathogenesis. Serum- and amino acid-based media (e.g., Lee's medium) are known to induce these morphological transitions. However, the molecular bases for amino acid-mediated morphogenesis are unknown. To understand how C. albicans responds to external amino acids, we examined the effects of these nutrients in the wild-type strain on the transport properties of AAPs in ammonium-based minimal medium at 37°C. The transport of radiolabeled valine indicated that the initial rate and overall uptake of this branched amino acid were increased at least ninefold in wild-type C. albicans cells grown in the presence of histidine over those in cells grown in the absence of this amino acid (Fig. 1A). The transport of radiolabeled valine was also examined in the wild-type strain in the presence or absence of the nonpolar amino acids leucine, methionine, phenylalanine, and tryptophan and the polar uncharged amino acids threonine and tyrosine, all known to mediate Ssy1p-dependent activation of amino acid uptake and transcription of AAP genes in S. cerevisiae (9, 14, 15, 21). None of the substrates tested had any effect on the uptake of valine in C. albicans (Fig. 1B). These results suggest that the activation of amino acid uptake in C. albicans differs from that of S. cerevisiae and is triggered by different substrates.
FIG. 1.
Induction of amino acid transport in wild-type C. albicans. (A) Time-dependent [3H]valine transport in the wild-type strain (CAEB-1) in ammonium-based medium lacking (SD) or containing (SDH) 10 mM histidine. (B) [3H]valine transport of wild-type cells in minimal medium without (shaded bar) or with (solid bars) 10 mM l-α-amino acids. (C) Uptake of [3H]-labeled valine, phenylalanine, lysine, glutamine, and isoleucine in minimal medium lacking (shaded bars) or containing (solid bars) histidine.
To define the substrates responsible for activation of amino acid uptake in C. albicans, we examined the uptake of radiolabeled valine in the wild-type strain for the remaining 12 amino acids. Among all the substrates tested, arginine, asparagine, aspartic acid, glutamine, glutamic acid, histidine, lysine, and serine were found to induce valine transport (Fig. 1B). Arginine, glutamine, and lysine resulted in the highest levels of induction, with ∼16 to 18-fold increases in valine uptake. Histidine, aspartic acid, and asparagine caused ∼10-fold, serine caused ∼7-fold, and glutamic acid caused ∼4-fold induction of valine uptake. To assess whether histidine-mediated activation was specific for valine, we examined the effects of histidine on the transport of radiolabeled phenylalanine, tyrosine, lysine, glutamine, and isoleucine. As with valine, transport of these substrates was also increased 3- to 10-fold when histidine was added (Fig. 1C).
Transcriptional regulation of AAP genes by amino acids.
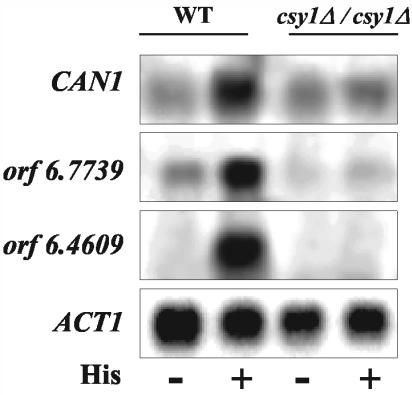
To assess whether amino acid-mediated activation of amino acid transport in C. albicans occurs via induction of transcription of specific AAP genes, we examined the transcription levels of the arginine permease gene CAN1 and two putative AAP genes, orf 6.7739 and orf 6.4609, in the presence and absence of histidine. orf 6.7739 and orf 6.4609 encode proteins which share high homology with the specific permeases Hip1p, Gnp1p, Tat1p, Tat2p, and Agp1p of S. cerevisiae. Although CAN1, orf 6.7739, and orf 6.4609 transcripts could be detected in the absence of histidine, addition of this amino acid resulted in a dramatic increase in their transcript levels (Fig. 2). As a control, the transcript levels of the actin gene, ACT1, remained the same under both conditions (Fig. 2). These data suggest that amino acid-mediated activation of amino acid uptake is dependent on induction of the transcription of specific AAP genes.
FIG. 2.
Transcription levels of AAP genes in wild-type and csy1Δ/csy1Δ strains in the presence of histidine. Northern blot analysis was performed on RNA isolated from wild-type (WT) (CAEB-1) and csy1Δ/csy1Δ (CAEB-5) strains grown to mid-log phase in minimal medium in the absence (−) or presence (+) of 10 mM histidine. The probes used are derived from the CAN1, orf 6.7739, orf 6.4609, and ACT1 genes as described in Materials and Methods.
The amino acid sensor Csy1p is important in the amino acid-mediated activation of amino acid uptake and transcription of AAP genes.
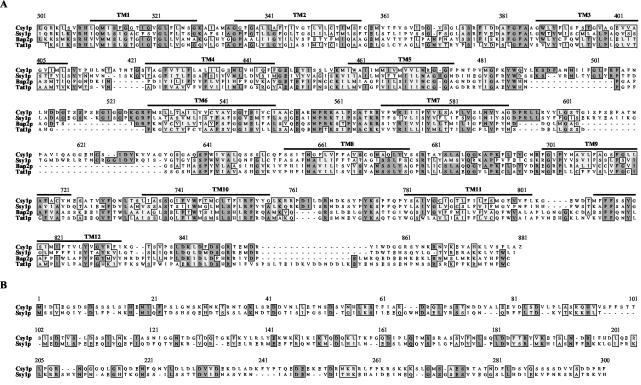
To examine whether the amino acid-dependent increase in amino acid uptake and transcription of specific AAP genes requires a membrane sensor, we cloned the putative amino acid sensor gene of C. albicans, CSY1, by using three specific bioinformatic criteria: (i) homology of its primary sequence to members of the AAP family, including S. cerevisiae Ssy1p, (ii) presence of an additional N- or C-terminal extension that might play a role in nutrient sensing, and (iii) predicted topology similar to that of Ssy1p. CSY1 encodes a protein of 881 amino acids that displays 43% identity and 64% similarity to the amino acid sensor of S. cerevisiae, Ssy1p. The homology between Csy1p, Ssy1p, and two other members of the AAP family, Bap2p and Tat1p, begins with amino acid 307 of Csy1p and continues throughout the remainder of the sequence (Fig. 3A). The N-terminal domain of Ssy1p has been shown to play a critical role in amino acid sensing (14, 25). Interestingly, pairwise alignment revealed only a low degree of homology (17% identity and 16% similarity) between the N-terminal domains of Ssy1p and Csy1p (Fig. 3B). Like Ssy1p, topology prediction indicated the presence of 12 putative hydrophobic transmembrane domains and two large extracellular loops that connect transmembrane domains V and VI and transmembrane domains VII and VIII in Csy1p.
FIG. 3.
Csy1p sequence alignment. (A) Alignment of C. albicans Csy1p with the S. cerevisiae amino acid sensor Ssy1p, the branched-chain AAP Bap2p, and the tyrosine transporter Tat1p. (B) Sequence alignment of the N-terminal extensions of Ssy1p and Csy1p. Dark shading, identical residues; light shading, similar residues. The predicted membrane-spanning domains (TM1 to TM12) for Csy1p are indicated by solid lines above the aligned proteins and are numbered sequentially.
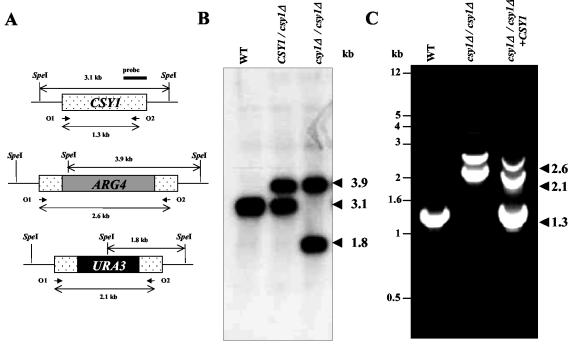
To determine the physiologic role of Csy1p and its importance in C. albicans survival and amino acid sensing, the two alleles of the CSY1 gene were disrupted by homologous recombination via two successive transformations to produce a csy1Δ/csy1Δ strain. Southern blot (Fig. 4A and B) and PCR (Fig. 4A and C) analyses, using a probe specific to the CSY1 gene and specific primers in the CSY1 upstream and downstream regions, confirmed the replacement of the two CSY1 chromosomal loci by ARG4 and URA3 cassettes. The csy1Δ/csy1Δ strain is viable and grows as well as the wild-type strain on rich or minimal media (data not shown). To assess whether Csy1p is important for the amino acid-mediated activation of transcription of AAP genes, we compared the transcription of CAN1, orf 6.7739, and orf 6.4609 in the csy1Δ/csy1Δ strain to that in the wild type in the presence and absence of histidine. Unlike those in the wild type, the transcript levels of CAN1, orf 6.7739, and orf 6.4609 in the csy1Δ/csy1Δ strain did not change in response to histidine (Fig. 2). As a control, the transcript levels of the actin gene, ACT1, in the csy1Δ/csy1Δ strain were similar to those of the wild type and were not affected by histidine (Fig. 2). These data suggest that Csy1p plays a critical role in amino acid-mediated induction of transcription of the C. albicans specific AAP genes.
FIG. 4.
Generation and molecular characterization of the csy1Δ/csy1Δ knockout. A CSY1 gene knockout was generated using the strategy described in Materials and Methods. (A) Schematic representation of the CSY1 genomic locus before and after replacement with ARG4 and URA3 cassettes. O1 and O2 are the two oligonucleotides used to confirm CSY1 gene disruption by PCR analysis. (B) Southern blot analysis of wild-type (WT), heterozygote (CSY1/csy1Δ), and homozygote (csy1Δ/csy1Δ) strains. Genomic DNAs were digested with SpeI. The probe used is a 0.7-kb fragment of CSY1. (C) Agarose gel electrophoresis showing the PCR products obtained with oligonucleotides O1 and O2 (see panel A) and genomic DNAs isolated from the wild-type, csy1Δ/csy1Δ, and csy1Δ/csy1Δ+CSY1 strains as templates.
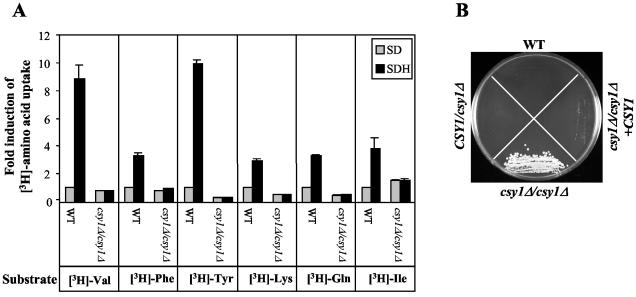
To further analyze the importance of Csy1p as a sensor of amino acids and regulator of amino acid uptake, we compared the transport of radiolabeled valine, phenylalanine, tyrosine, lysine, glutamine, and isoleucine in wild-type and csy1Δ/csy1Δ strains in the absence or presence of histidine. No differences in the uptake of those amino acids between the two strains could be detected in the absence of histidine (Fig. 5A). In the presence of histidine, no induction of uptake of those amino acids could be detected in the csy1Δ/csy1Δ strain, whereas a major induction of amino acid transport could be measured in the wild-type strain (Fig. 5A). As a control, histidine activation was also examined in the heterozygote (CSY1/csy1Δ) and complemented (csy1Δ/csy1Δ +CSY1) strains, where it exhibited the same regulatory profile as in the wild-type strain (data not shown). To further examine the defect in amino acid uptake upon loss of Csy1p, we analyzed the growth of wild-type, csy1Δ/csy1Δ, CSY1/csy1Δ, and csy1Δ/csy1Δ+CSY1 strains on media containing the amino acid analog l-glutamic acid γ-hydrazide. The csy1Δ/csy1Δ strain was more resistant to this compound than the wild-type, heterozygote, and complemented strains (Fig. 5B). Together, these data indicate that amino acid transport in the csy1Δ/csy1Δ mutant is altered and that amino acid-mediated activation of amino acid uptake requires a functional Csy1p protein.
FIG. 5.
Comparison of amino acid transport and sensitivity to the amino acid analog l-glutamic acid γ-hydrazide in the wild-type and csy1Δ/csy1Δ strains. (A) Uptake of amino acids in the wild-type (WT) (CAEB-1) and csy1Δ/csy1Δ (CAEB-5) strains. Cells were grown in SD medium lacking or supplemented with 10 mM histidine and were assayed as described in Materials and Methods. Standard deviations are shown. Uptake values were normalized to those of the wild-type strain in SD medium. (B) Growth of wild-type (CAEB-1), CSY1/csy1Δ (CAEB-3), csy1Δ/csy1Δ (CAEB-5), and csy1Δ/csy1Δ+CSY1 (CAEB-6) strains on SD plates containing 25 mM l-glutamic acid γ-hydrazide and incubated at 37°C for 1 week.
csy1Δ/csy1Δ cells show altered filamentation and colony morphology.
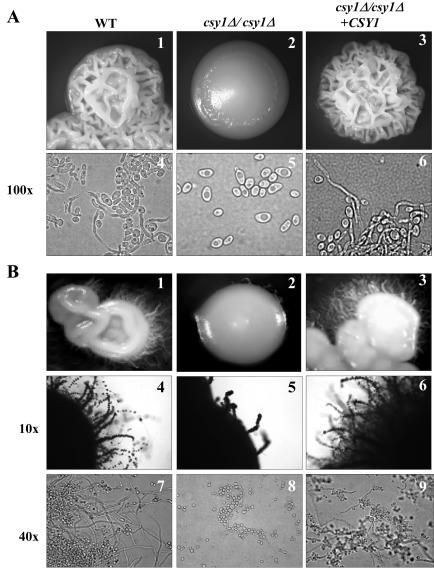
The importance of Csy1p in amino acid sensing led us to investigate the possible role of this sensor in nutrient-induced yeast-hypha differentiation in C. albicans. Thus, we compared the colony and cell morphologies of wild-type, csy1Δ/csy1Δ, and csy1Δ/csy1Δ+CSY1 strains on serum-based (Fig. 6A) and Lee's (Fig. 6B) media. Whereas wild-type and csy1Δ/csy1Δ+CSY1 strains formed wrinkled colonies at 37°C in serum-based and Lee's media after 48 h or 1 week of incubation, respectively, csy1Δ/csy1Δ mutant cells produced colonies with a smooth morphology (Fig. 6A and B). Microscopic analysis of the cells within the colonies revealed the formation of hyphae in wild-type and csy1Δ/csy1Δ+CSY1 strains, whereas csy1Δ/csy1Δ cells grew predominantly in the yeast form (Fig. 6). No significant morphological differences between wild-type, csy1Δ/csy1Δ, and csy1Δ/csy1Δ+CSY1 strains could be detected on N-acetylglucosamine, Spider, or SLAD solid media or serum-based and Lee's liquid media (data not shown).
FIG. 6.
csy1Δ/csy1Δ mutants are defective in filamentation and colony morphology. Wild-type (WT) (CAEB-1), csy1Δ/csy1Δ (CAEB-5), and csy1Δ/csy1Δ+CSY1 (CAEB-6) strains were grown at 37°C on solid plates containing either 10% serum (A) or Lee's medium (B). Colony morphologies (A1 to 3; B1 to 6) and cell morphologies (A4 to 6; B7 to 9) were assessed by light microscopy.
DISCUSSION
The AAP family consists of a large number of membrane proteins that mediate the transport of amino acids across the plasma membrane (50). These permeases share significant sequence homology and predicted topology. In S. cerevisiae at least two members of this family, Hnm1p and Ssy1p, are not involved in amino acid transport sensu stricto. Hnm1p is involved in the transport of choline, which is the first step in the CDP-choline pathway for the synthesis of phosphatidylcholine from choline (4). Ssy1p is an amino acid sensor and a critical component of the trimeric SPS complex, which responds to amino acids and transduces signals to modulate the transcription of amino acid-metabolizing genes via activation of the transcriptional factors Stp1p and Stp2p (1, 8, 10, 21, 23, 25). Amino acid transporters are further divided into specific and nonspecific permeases. General AAPs, such as Gap1p, are nonspecific permeases and transport all common l-amino acids, various d-amino acids, and several related compounds in S. cerevisiae (22). Most other members of the AAP family in S. cerevisiae encode transporters with narrow substrate specificities (15).
We demonstrated that amino acid transport in C. albicans is regulated by specific substrates. Eight amino acids—arginine, asparagine, aspartic acid, glutamine, glutamic acid, histidine, lysine, and serine—were found to increase amino acid uptake. The amino acid-mediated activation of amino acid transport in ammonium-based medium was also examined at 30°C and yielded similar results (data not shown). We further confirmed that this activation of amino acid transport correlates with induction of expression of CAN1 and two putative AAP genes, orf 6.7739 and orf 6.4609, identified by genome sequencing, in the presence of histidine. Recent studies have referred to orf 6.7739 and orf 6.4609 as general AAPs (3, 34). However, the expression pattern of those two genes in ammonium-based (Fig. 2) and proline-based (unpublished data) media in the presence and absence of histidine and their dependence on Csy1p are reminiscent of S. cerevisiae AGP1 and GNP1 and of S. cerevisiae BAP2 and BAP3, respectively, but not of GAP1. Future genetic and biochemical studies of orf 6.7739, orf 6.4609, and other putative AAPs of C. albicans to determine their transport properties and substrate specificities are warranted.
Our finding that C. albicans regulates its amino acid uptake in a manner similar to that of S. cerevisiae suggested that this regulation might involve an upstream regulatory sensor that monitors the availability of amino acids internally and/or externally and transduces signals to regulate the transcription of AAP genes. Accordingly, we identified and characterized Csy1p, the C. albicans homolog of the S. cerevisiae amino acid sensor. Our results provided several lines of evidence indicating that Csy1p is the primary amino acid sensor of C. albicans involved in the regulation of AAPs. Loss of Csy1p, although mutants remained viable, resulted in (i) lack of induction of specific AAPs, (ii) lack of activation of amino acid uptake in response to external amino acids, and (iii) resistance to the amino acid analog l-glutamic acid γ-hydrazide. The expression levels of the CSY1 gene in the wild-type strain remained the same in the absence and presence of histidine (data not shown), suggesting that Csy1p-dependent amino acid-mediated transcriptional activation of AAP genes does not require an increase in Csy1p expression but reflects an activation of its sensor activity. Furthermore, similar levels of amino acid transport were measured in both the wild-type and csy1Δ/csy1Δ strains in the absence of external amino acids. These results suggest that C. albicans utilizes Csy1p to sense amino acids in its environment, and they confirm recent data for S. cerevisiae indicating that sensing by Ssy1p occurs via its direct interaction with external amino acids (16).
Although Csy1p and Ssy1p share a high degree of homology in their C-terminal domains, their N-terminal extension domains, which are absent from other AAPs and are known to be critical for the sensing function in S. cerevisiae, are highly divergent. Our data suggest that this high degree of divergence in this domain might account for the differences in ligand specificity and sensing between C. albicans and S. cerevisiae. We showed that with the exception of serine, the seven other inducers of amino acid uptake in C. albicans found in this study (arginine, asparagine, aspartic acid, glutamine, glutamic acid, histidine, and lysine) have previously been shown to have little or no effect on the Ssy1p-dependent expression of AGP1, which encodes a broad-specificity AAP in S. cerevisiae (15, 21). Tyrosine, phenylalanine, leucine, tryptophan, methionine, threonine, alanine, and cysteine, which have high or intermediate effects on AGP1 expression, were found to have no effect on valine uptake in C. albicans. Our studies strongly support a role for Csy1p in amino acid sensing; however, at this stage we cannot exclude the possibility that this protein might also have an intrinsic amino acid transport activity.
This study was prompted by previous observations that Lee's medium, which is rich in amino acids, induces the C. albicans yeast-hypha morphological transition, demonstrating that amino acids can play a role in this process and suggesting a possible involvement of Csy1p in the regulatory mechanism controlling hyphal morphogenesis. Our study demonstrated that C. albicans cells lacking Csy1p show altered colony morphology and hyphal formation in serum- and amino acid-based solid media, but not in N-acetylglucosamine, Spider, and SLAD media, which do not contain amino acids. These results support the idea that CSY1 is an important component of the signaling pathway controlling serum- and amino acid-induced hyphal morphogenesis in C. albicans. However, it is not yet clear why no differences between wild-type and csy1Δ/csy1Δ strains could be detected on serum-based and Lee's liquid media.
In C. albicans, two signaling pathways, the mitogen-activated protein kinase (MAPK) and cyclic AMP (cAMP)-dependent protein kinase (PKA) pathways, have thus far been implicated in yeast-hypha morphogenesis (32). The MAPK cascade consists of the kinases Cst20p, Ste11p, Hst7p, and Cek1p, which are responsible for the activation of the transcription factor Cph1p (6, 26, 29, 30, 35). The cAMP-PKA pathway involves a protein kinase A, encoded by the TPK2 gene, which acts upstream of the transcriptional factor Efg1p (47, 48). Both the MAPK and cAMP-PKA pathways are activated by a common upstream factor, Ras1p (30); however, how these pathways are activated by external stimuli is not known.
Tripathi and colleagues have shown that amino acid starvation promotes pseudohyphal, but not hyphal, growth in C. albicans and that this response is dependent on Cagcn4p (49). However, this morphogenetic switch differs from that induced by serum, amino acids (proline, arginine, alanine, histidine, isoleucine, lysine), and serum filtrates of <1 kDa (12), all of which are known to induce hyphal forms. Together, these data suggest that serum- or amino acid-mediated hyphal formation and starvation-mediated pseudohyphal formation are two separate processes.
In summary, our studies provide the first evidence that the amino acid sensor Csy1p is an important regulatory membrane protein playing a critical role in amino acid transport and filamentation in C. albicans. Further studies will determine how Csy1p senses amino acids, activates the transcription of AAP genes, and regulates the C. albicans yeast-hypha morphogenetic transition.
Acknowledgments
This work was supported by the University of Connecticut Health Center Fund, the U.S. Army Medical Research and Material Command (to C.B.M.) and The Robert Leet and Clara Guthrie Patterson Trust (to C.B.M. and R.Z.). E.B. is supported by a fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) of Brazil.
We thank Aaron Mitchell and Vincent Bruno, Columbia University, for providing strains and plasmids. We are grateful to the Stanford DNA Sequencing and Technology Center for the CSY1 sequence. We thank Bruno Andre, Universite Libre de Bruxelles, and Stephen K. Wikel, Gabriella Pessi, Guillermo Kociubinski, Kamal ElBissati, Teresa C. Santiago, and Kenneth Bourell, University of Connecticut Health Center, for critical reading and help with manuscript preparation.
REFERENCES
- 1.Andreasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, F., and B. Andre. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41:489-502. [DOI] [PubMed] [Google Scholar]
- 3.Biswas, S., M. Roy, and A. Datta. 2003. N-Acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 4.Carman, G. M., and S. A. Henry. 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38:361-399. [DOI] [PubMed] [Google Scholar]
- 5.Cassone, A., P. A. Sullivan, and M. G. Shepherd. 1985. N-Acetyl-d-glucosamine-induced morphogenesis in Candida albicans. Microbiologica 8:85-99. [PubMed] [Google Scholar]
- 6.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Backer, M. D., D. Maes, S. Vandoninck, M. Logghe, R. Contreras, and W. H. Luyten. 1999. Transformation of Candida albicans by electroporation. Yeast 15:1609-1618. [DOI] [PubMed] [Google Scholar]
- 8.de Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen, and R. J. Planta. 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28:974-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Didion, T., M. Grausland, C. Kielland-Brandt, and H. A. Andersen. 1996. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178:2025-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didion, T., B. Regenberg, M. U. Jorgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, J. F. 2000. Regulation of dimorphism in Candida albicans. Contrib. Microbiol. 5:98-111. [DOI] [PubMed] [Google Scholar]
- 12.Feng, Q., E. Summers, B. Guo, and G. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg, H., C. F. Gilstring, A. Zargari, P. Martinez, and P. O. Ljungdahl. 2001. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42:215-228. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 16.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 18.Grauslund, M., T. Didion, M. C. Kielland-Brandt, and H. A. Andersen. 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269:275-280. [DOI] [PubMed] [Google Scholar]
- 19.Hermann, P., Z. Berek, G. Nagy, K. Kamotsay, and F. Rozgonyi. 2001. Pathogenesis, microbiological and clinical aspects of oral candidiasis (candidosis) (a review). Acta Microbiol. Immunol. Hung. 48:479-495. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles, A. Urrestarazu, and B. Andre. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jauniaux, J. C., and M. Grenson. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39-44. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 24.Jradeh, E. F., S. A. Al-Kharashi, and K. F. Tabbar. 2001. Candida keratitis in a patient with candidiasis of the fingernails. Eur. J. Ophthalmol. 11:380-382. [DOI] [PubMed] [Google Scholar]
- 25.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohler, J. R., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kontoyiannis, D. P., B. T. Reddy, H. A. Torres, M. Luna, R. E. Lewis, J. Tarrand, G. P. Bodey, and I. I. Raad. 2002. Pulmonary candidiasis in patients with cancer: an autopsy study. Clin. Infect. Dis. 34:400-403. [DOI] [PubMed] [Google Scholar]
- 28.Laine, P. O., J. C. Lindqvist, S. O. Pyrhonen, L. M. Teerenhovi, S. M. Syrjanen, and J. H. Meurman. 1993. Lesions of the oral mucosa in lymphoma patients receiving cytostatic drugs. Eur. J. Cancer B Oral Oncol. 29B:291-294. [DOI] [PubMed] [Google Scholar]
- 29.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. Gow, A. J. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 32.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, H., and R. F. Gaber. 1996. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 7:1953-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Limjindaporn, T., R. A. Khalaf, and W. A. Fonzi. 2003. Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50:993-1004. [DOI] [PubMed] [Google Scholar]
- 35.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 36.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Mootha, V. V., M. L. Schluter, and A. Das. 2002. Intraocular hemorrhages due to warfarin fluconazole drug interaction in a patient with presumed Candida endophthalmitis. Arch. Ophthalmol. 120:94-95. [PubMed] [Google Scholar]
- 39.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 40.Odds, F. C. 1985. Morphogenesis in Candida albicans. Crit. Rev. Microbiol. 12:45-93. [DOI] [PubMed] [Google Scholar]
- 41.Ozcan, S., J. Dover, A. G. Rosenwald, S. Wolfl, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulsen, I. T., M. K. Sliwinski, B. Nelissen, A. Goffeau, and M. H. Saier, Jr. 1998. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 430:116-125. [DOI] [PubMed] [Google Scholar]
- 43.Reifenberger, E., K. Freidel, and M. Ciriacy. 1995. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16:157-167. [DOI] [PubMed] [Google Scholar]
- 44.Saiman, L., E. Ludington, J. D. Dawson, J. E. Patterson, S. Rangel-Frausto, R. T. Wiblin, H. M. Blumberg, M. Pfaller, M. Rinaldi, J. E. Edwards, R. P. Wenzel, and W. Jarvis. 2001. Risk factors for Candida species colonization of neonatal intensive care unit patients. Pediatr. Infect. Dis. J. 20:1119-1124. [DOI] [PubMed] [Google Scholar]
- 45.Sallah, S., J. Y. Wan, N. P. Nguyen, P. Vos, and G. Sigounas. 2001. Analysis of factors related to the occurrence of chronic disseminated candidiasis in patients with acute leukemia in a non-bone marrow transplant setting: a follow-up study. Cancer 92:1349-1353. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonneborn, A., D. P. Bockmuhl, M. Gerads, K. Kurpanek, D. Sanglard, and J. F. Ernst. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386-396. [DOI] [PubMed] [Google Scholar]
- 48.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Belle, D., and B. Andre. 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13:389-398. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]