Abstract
The yeast Saccharomyces cerevisiae senses glucose, its preferred carbon source, through multiple signal transduction pathways. In one pathway, glucose represses the expression of many genes through the Mig1 transcriptional repressor, which is regulated by the Snf1 protein kinase. In another pathway, glucose induces the expression of HXT genes encoding glucose transporters through two glucose sensors on the cell surface that generate an intracellular signal that affects function of the Rgt1 transcription factor. We profiled the yeast transcriptome to determine the range of genes targeted by this second pathway. Candidate target genes were verified by testing for Rgt1 binding to their promoters by chromatin immunoprecipitation and by measuring the regulation of the expression of promoter lacZ fusions. Relatively few genes could be validated as targets of this pathway, suggesting that this pathway is primarily dedicated to regulating the expression of HXT genes. Among the genes regulated by this glucose signaling pathway are several genes involved in the glucose induction and glucose repression pathways. The Snf3/Rgt2-Rgt1 glucose induction pathway contributes to glucose repression by inducing the transcription of MIG2, which encodes a repressor of glucose-repressed genes, and regulates itself by inducing the expression of STD1, which encodes a regulator of the Rgt1 transcription factor. The Snf1-Mig1 glucose repression pathway contributes to glucose induction by repressing the expression of SNF3 and MTH1, which encodes another regulator of Rgt1, and also regulates itself by repressing the transcription of MIG1. Thus, these two glucose signaling pathways are intertwined in a regulatory network that serves to integrate the different glucose signals operating in these two pathways.
The budding yeast Saccharomyces cerevisiae prefers glucose as a carbon source. Glucose elicits broad changes in the yeast cell that adapt it to use the sugar efficiently and exclusively of other available carbon sources. These changes include regulation of gene expression at the transcriptional (9, 20, 30), posttranscriptional (37, 60), translational (1, 37), and posttranslational (26, 27, 29) levels. For these adaptations to occur, the cell must sense glucose and transmit a signal to the appropriate effectors. Three glucose sensing systems are well documented for the budding yeast. Each system detects and transmits the glucose signal differently. One mechanism operates through the Snf1 protein kinase to cause a repression of gene expression when glucose levels are high. Another mechanism works through the Snf3 and Rgt2 glucose sensors to induce expression of genes encoding glucose transporters. A third glucose sensing mechanism employs the Gpr1 G-protein-coupled receptor and cyclic AMP as a second messenger (for a review, see reference 55). We have focused on the first two glucose sensing pathways, which exert their effects primarily by regulating gene expression.
The main player in the pathway through which glucose represses the expression of many genes involved in the utilization of alternative carbon sources and gluconeogenesis is the Snf1 protein kinase. When glucose is limiting, Snf1 is active and regulates transcription by catalyzing phosphorylation of the Mig1 transcriptional repressor and other repressors and activators (8, 34, 35, 68). The Snf1 protein kinase is activated under these conditions by its phosphorylation, catalyzed by one of three protein kinases (25, 46, 63). The addition of glucose inactivates the Snf1 kinase by stimulating its dephosphorylation, catalyzed by the Glc7-Reg1 protein phosphatase (41, 57). Transmission of the glucose signal to Snf1 involves Hxk2 (26), a hexokinase that catalyzes glucose phosphorylation, the first catalytic step of its metabolism (for a review, see reference 55). The Mig1 paralogue Mig2, which has essentially the same binding site as Mig1, also plays a role in glucose repression of the expression of some genes (an additional paralogue, Mig3, also binds to the same DNA sequence and contributes modestly to glucose repression [43]). The mechanism of glucose regulation of Mig2 function is unknown, but it is clear that the Snf1 protein kinase does not regulate Mig2 activity (43).
A separate glucose sensing pathway mediates glucose induction of the expression of HXT genes encoding glucose transporters (reviewed in references 30, 51, and 55). Glucose is sensed by two glucose sensors in the cell membrane, Snf3 and Rgt2, which generate an intracellular signal in the presence of glucose that induces HXT gene expression. The ultimate target of the pathway is Rgt1, a transcription factor that binds to and represses the expression of HXT genes in the absence of glucose. Two other proteins, the paralogues Mth1 and Std1, are required for Rgt1 to repress HXT gene expression. Mth1 and Std1 interact with the glucose sensors (38, 62) and with Rgt1 (V. Brachet, unpublished data). The glucose signal inhibits Rgt1-mediated repression by stimulating the degradation of Std1 and Mth1 (19, 39). Thus, activation of the Snf3 and Rgt2 glucose sensors by extracellular glucose generates an intracellular signal that derepresses the expression of HXT genes by inhibiting the function of the Rgt1 repressor.
The Snf1-Mig1 glucose repression pathway affects the expression of many genes (43, 70), but only six genes are known to be targets of the Snf3/Rgt2-Rgt1 glucose induction pathway, and all of them are HXT genes coding for glucose transporters (5, 16, 40, 52). While profiling the yeast genome to identify other genes regulated by the Snf3/Rgt2-Rgt1 pathway, we discovered that the glucose repression and glucose induction pathways are interlocked in an elaborate network of autoregulatory and cross-pathway-regulatory circuits.
MATERIALS AND METHODS
Media.
Yeast strains were grown on standard rich medium (2% Bacto Peptone [Difco], 1% yeast extract) or synthetic yeast nitrogen base media (0.17% yeast nitrogen base without ammonium sulfate [Difco] and with 0.5% ammonium sulfate), supplemented with appropriate amino acids, nitrogenous bases, and 2 or 4% glucose (high-glucose media), 2% galactose, or 2% raffinose, or as indicated in the tables.
Strains and plasmids.
Yeast strains used in this study are listed in Table 1. The dominant RGT2-1 and SNF3-1 alleles were introduced into the chromosome by the transplacement method of Lundblad et al. (42). RGT2-1 (pBM3946) was inserted into pRS306 as a 3.3-kb EcoRI-BamHI fragment (from the pBM3270 plasmid [49]); SNF3-1 (pBM3948) was inserted into pRS306 as a 3.8-kb HindIII-SalI fragment (from the pBM3259 plasmid [50]). RGT2-1 was integrated into RGT2 by transforming yeast cells (FM391) to Ura+ with pBM3946 cut with SphI (cuts 712 bp upstream of RGT2); SNF3-1 was integrated into SNF3 by transforming cells to Ura+ with pBM3948 cut with ClaI (cuts 490 bp upstream of SNF3). The SNF3 and RGT2 duplications were resolved by selecting for Ura− segregants on 5-fluoroorotic acid plates, and segregants that had retained the dominant RGT2-1 and SNF3-1 mutations were identified based on their constitutive expression of HXT1 (recognized as blue colonies on galactose media containing X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] after transformation with an HXT1-lacZ reporter, pBM2636 [52]). Gene promoters were fused to lacZ in either the 2μm plasmids YEp357R and YEp367R (45) or the centromere plasmid pBM4088 (S.-W. Ho, unpublished data) by the gap repair technique (48). Promoters were amplified from genomic DNA extracted from YM4127 or FM391 according to the method described by Hoffman and Winston (23). The resulting plasmids are listed in Table 2. We constructed lexA-MIG2 expressed from the MIG2 promoter (pBM4258) by replacing by the gap repair technique the ADH1 promoter in pBM3091 (44) with the MIG2 promoter amplified with the primers OM2703 and OM2704 (cutting at the unique PacI site in the ADH1 promoter). The repressor activity of lexA-Mig2 was assayed by using two CYC1-lacZ reporters: lexO-less upstream activation sequence (UAS) (CYC1)-lacZ (pLG312s) (22) and 4xlexO-UAS (CYC1)-lacZ (JK1621) (31). The latter is identical to the former except for four lexA-binding operator sites inserted upstream of the CYC1 UAS. Yeast transformations were performed by using the method of Schiestl and Gietz (61), when high frequencies of transformation were required, or by using the one-step method of Chen et al. (11).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YM4127 | MATaura3-52 his3-200 ade2-101 lys2-801 leu2 trp1-903 tyr1-501 | 52 |
| YM4509 | MATaura3-52 his3-200 ade2-101 lys2-801 leu2 trp1-903 tyr1-501 rgtIΔ::hisG | 52 |
| BY4741 (FM391) | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | 6 |
| BY4742 (FM392) | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | 6 |
| BY4743 (FM393) | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2 | 6 |
| FM557 | BY4741 rgt1Δ::kanMX | 21 |
| YM6440 | BY4743 rgt1Δ::kanMX/rgt1Δ::kanMX | 21 |
| YM6545 | BY4741 RGT2-1 | This study |
| YM6546 | BY4741 RGT2-1 | This study |
| YM6548 | BY4741 SNF3-1 | This study |
| YM6554 | BY4743 RGT2-1/+ (cross: YM6546 × FM392) | This study |
| YM6557 | BY4743 SNF3-1/+ (cross: YM6548 × FM392) | This study |
| YM6247 | BY4742 rgt2::kanMX | 21 |
| YM6329 | BY4742 snf3::kanMX | 21 |
| YM6370 | BY4742 rgt2::kanMX snf3::kanMX | This study |
| FM558 | BY4741 trp1Δ::kanMX | 21 |
| YM6833 | BY4741 trp1Δ::kanMX MIG2-6HA-klTRP1 | This study |
| YM6835 | BY4741 trp1Δ::kanMX MIG3-6HA-klTRP1 | This study |
| YM6843 | BY4741 trp1Δ::kanMX MIG1-6HA-klTRP1 | This study |
| FM573 | BY4741 gal4Δ::kanMX | 21 |
| FM612 | BY4743 mig1Δ::kanMX/mig1Δ::kanMX | 21 |
| YM6682 | BY4743 mig1Δ::kanMX/mig1Δ::kanMX mig2Δ::kanMX/mig2Δ::kanMX | This study |
| YM6683 | BY4743 mig2Δ::kanMX/mig2Δ::kanMX mig3Δ::kanMX/mig3Δ::kanMX | This study |
| YM6684 | BY4743 mig1Δ::kanMX/mig1Δ::kanMX mig2Δ::kanMX/mig2Δ::kanMX mig3Δ::kanMX/mig3Δ::kanMX | This study |
TABLE 2.
Promoter lacZ fusions constructed for this studya
| Plasmid | lacZ vector | Promoter | Length of the cloned upstream region (bp) | Primer pair |
|---|---|---|---|---|
| pBM4270 | YEp357R | MIG2 | 1,459 | OM2392-OM2458 |
| pBM4270 | YEp357R | STD1 | 996 | OM2799-OM2800 |
| pBM4273 | YEp357R | YOR062C | 1,180 | OM2717-OM2718 |
| pBM4379 | YEp357R | MIG3 | 713 | OM2563-OM2564 |
| pBM4381 | YEp357R | YGL157W | 646 | OM3534-OM2566 |
| pBM4381 | YEp357R | YNL234W | 418 | OM3446-OM3447 |
| pBM4515 | YEp357R | YGL039W | 756 | OM3633-OM3634 |
| pBM4522 | YEp357R | HSF1 | 813 | OM3676-OM3677 |
| pBM4487 | YEp367R | SNF3 | 832 | OM3882-OM3883 |
| pBM4510 | YEp367R | PHM8 | 934 | OM3089-OM3090 |
| pBM4511 | YEp367R | AQR1 | 1,087 | OM4065-OM4066 |
| pBM4292 | pBM4088 | MTH1 | 1,476 | OM2812-OM3191 |
| pBM4296 | pBM4088 | MRK1 | 626 | OM2844-OM3190 |
| pBM4346 | pBM4088 | MIG2 | 1,459 | OM2875-OM2458 |
| pBM4500 | pBM4088 | VID24 | 686 | OM3448-OM3449 |
| pBM4501 | pBM4088 | HXT5 | 1,500 | OM3318-OM3441 |
| pBM4502 | pBM4088 | AHP1 | 967 | OM3088-OM3981 |
| pBM4512 | pBM4088 | CIT2 | 1,024 | OM4067-OM4068 |
| pBM4513 | pBM4088 | HOR2 | 1,092 | OM4069-OM4070 |
| pBM4514 | pBM4088 | PFK27 | 1,021 | OM3979-OM4071 |
| pBM4516 | pBM4088 | SKS1 | 1,340 | OM4072-OM4073 |
The gap repair procedure used for construction of the lacZ fusions is described in Materials and Methods. We also tested a YKR075C-lacZ fusion plasmid, pBM3469, and an MIG1-lacZ fusion plasmid, pBM3091, as described previously (43). Sequences of the primers are available upon request.
β-Galactosidase assay.
β-Galactosidase assays were performed according to the method of Rose et al. (56), with minor modifications. The reported lacZ activities (in nanomoles of hydrolyzed o-nitrophenyl-β-d-galactopyranoside [ONPG] per milligram of protein per minute) are averages of results from triplicate or duplicate assays of usually four different transformants. Standard deviations did not exceed the mean values by more than 10 to 25% for values higher than 100 U.
Expression profiling using DNA microarrays.
Detailed protocols for the isolation of mRNA, the generation of Cy dye-labeled cDNA, and their hybridization to a microarray, have been described previously (28). Briefly, cells were grown in synthetic complete (SC) medium to an optical density at 600 nm (OD600) of 1.2, harvested by centrifugation, and broken with a Mini Beadbeater (BioSpec Products, Inc., Bartlesville, Okla.). Total RNA and poly(A)+ RNA were isolated with an RNeasy midi kit and an Oligotex kit from QIAGEN according to protocols provided by the manufacturer. cDNA samples labeled with Cy3 or Cy5 dye (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) were generated with Superscript reverse transcriptase (Gibco BRL, Rockville, Md.). Corning CMT S288C yeast gene arrays (Corning Incorporated Life Sciences, Acton, Mass.) were used for hybridization, according to the manufacturer's instructions. For each experiment, four arrays were used. On two of the arrays, the control sample labeled with Cy3 was mixed with the test sample labeled with Cy5 and the hybridization mixture. On the other two arrays, the control sample labeled with Cy5 was mixed with the test sample labeled with Cy3. A Molecular Dynamics GenIII laser scanner was used to acquire hybridization signals, according to the manufacturer's procedures. Array images were analyzed with ArrayVision software (versions 4 and 5; Imaging Research, Toronto, Canada) to obtain fluorescence signal intensities corresponding to each spotted open reading frame (ORF). The absolute intensity of each spot was then normalized by using the default parameters of the software.
For hybridizations with microarrays, RNAs were isolated from (i) diploid strains, namely, heterozygous RGT2-1/+ (YM6554) and SNF3-1/+ (YM6557) strains, a homozygous rgt1Δ strain (YM6440), and the wild-type BY4743 (FM393) grown in 2% galactose-SC medium to an OD600 of 1.2; and (ii) haploid strains, namely, snf3Δ (YM6329), rgt2Δ (YM6247), and snf3Δ rgt2Δ (YM6370) strains and the reference haploid BY4742 (FM392), all shifted to 2% glucose-SC medium for 4 h after growth in 2% galactose-SC medium to an OD600 of 1.2.
Expression profiling with high-density oligonucleotide arrays.
RNA was isolated from three separate cultures of YM4509 (rgt1Δ) and YM4127 (wild-type haploid strain) grown in rich 3% glycerol plus 3% lactate medium to an OD600 of 0.8 by the acidic hot phenol method described previously (24) (a detailed protocol is provided at R. Young's laboratory website [http://web.wi.mit.edu/young/expression/]) and pooled after enrichment fractionation with an Oligotex kit (Qiagen) to obtain a final concentration between 1 and 2 μg/μl. RNA was labeled (target synthesis) and hybridized at the GeneChip Core Facility in the Siteman Cancer Center, Washington University, St. Louis, Mo. (detailed protocols can be found at the institution website [http://pathbox.wustl.edu/∼mgacore/genechip.htm]). Data were analyzed by using expression analysis software from Affymetrix. Target RNA was hybridized to the yeast S98 Affymetrix oligoarrays.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays of Rgt1 binding in vivo to the promoters of candidate target genes were carried out as described previously (32).
Western analysis of tagged Mig1, Mig2, and Mig3.
Mig1, Mig2, and Mig3 were tagged at their C termini with six copies of the influenza virus hemagglutinin (HA) epitope tag in the genome of strain BY4741 trp1Δ (FM558; background, FM391 [Table 1]) as described by Knop et al. (33). Correct tagging of the genes was confirmed by the PCR verification assay (using corresponding primers C from the Saccharomyces Genome Deletion Project [SGD] website [http://www-sequence.stanford.edu/group/yeast_deletion_project/ ] and the primer KAN & HIS [33]). Strains YM6843 (MIG1-6HA), YM6833 (MIG2-6HA), and YM6835 (MIG3-6HA) (Table 1) were precultured in SC medium with 5% glycerol and 0.5% galactose and inoculated (at an OD600 of 0.1 to 0.2) into the same medium and into media containing 5% glycerol and 0.05% glucose (low-glucose medium) or 4% glucose (high-glucose medium). The cultures were grown until they reached an OD600 of approximately 1.0, and proteins were extracted as described by Knop et al. (33). Protein extracts were resolved through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (polyacrylamide gradient of 4 to 15%), blotted to a polyvinylidene difluoride membrane (Immobilon; Millipore), and detected with the use of a primary polyclonal antibody against hemagglutinin (Sigma), a secondary antibody against rabbit immunoglobulin G (IgG) conjugated to hydroxyperoxidase, and an enhanced chemiluminescence detection system (Pierce).
RESULTS
Identification of target genes of the glucose induction pathway.
To search for targets of the Snf3/Rgt2 glucose sensing pathway, we performed gene expression profiling of the yeast genome using mutants with different defects in the signal transduction pathway. RGT2-1 and SNF3-1 are dominant mutations that cause the glucose sensors to always generate a signal, leading to constitutive expression of the HXT genes (49). The HXT genes are also constitutively expressed in a mutant lacking the Rgt1 repressor (52). Thus, we looked for genes whose expression is increased in the absence of glucose in RGT2-1, SNF3-1, and rgt1Δ mutants relative to that of the wild-type strain. Conversely, expression of the HXT genes cannot be induced by glucose in an snf3 rgt2 mutant, which lacks glucose sensors (52). We therefore looked for genes whose level of expression is decreased in snf3 rgt2 cells growing on glucose relative to that in wild-type cells.
The expression of 43 genes was increased at least 1.8-fold relative to that in the wild type in an rgt1Δ mutant growing on galactose (Table 3). Twenty-nine of these genes (Table 3) are particularly good candidates for Rgt1 targets because they have at least one of three additional features: (i) their levels of expression in one or both dominant mutants (RGT2-1 and SNF3-1) growing on galactose are increased at least 1.7-fold relative to that in the wild type, (ii) their expression is decreased at least twofold relative to that of the wild type in an snf3 rgt2 null mutant grown on glucose, or (iii) the increase in their transcript level can be reproduced by a different detection method (oligonucleotide arrays). The promoters of most of these genes have apparent Rgt1 binding sites (32) that are conserved in the orthologous promoters of other Saccharomyces species, suggesting that they are functional (13). It is reassuring that the well-known Rgt1 targets, HXT1 to HXT4, have all three features (except for HXT2, which for unknown reasons did not show induction in oligonucleotide array experiments) and are among the genes with the largest increases in expression of the rgt1Δ mutant growing on galactose. Two other HXT genes coding for glucose transporters, HXT5 and HXT8, seem to be targets of this glucose induction pathway (Table 3).
TABLE 3.
Known and potential targets of Rgt1a
| ORF | Gene name | Protein function or characteristic | Ratio of spot intensity of the mutant to that of the wild typeb
|
No. of Rgt1 sites/ no. of conserved sitesc | |||
|---|---|---|---|---|---|---|---|
| rgt1Δ Gal | RGT2-1 Gal | SNF3-1 Gal | snf3Δ rgt2Δ Glu | ||||
| YHR092C | HXT4 | Glucose transporter | ↑23.1 (↑10.5) | ↑2.4 | ↑10.6 | ↓0.2 | 5/2 |
| YDR345C | HXT3 | Glucose transporter | ↑13.3 (↑16.9) | ↑2.8 | ↑5.3 | ↓0.2 | 11/7 |
| YMR011W | HXT2 | Glucose transporter | ↑13.0 (NC) | ↑1.9 | ↑5.2 | ↓0.3 | 3/2 |
| YKR075C | Similarity to N terminus of Reg1 | ↑12.6 (↑2.1) | ↑2.0 | ↑8.1 | ↓0.2 | 7/5 | |
| YGL157W | Similarity to dihydroflavonol 4-reductase | ↑7.9 (↑6.6) | ↑2.5 | ↑4.6 | ↓0.1 | 2/1 | |
| YHR094C | HXT1 | Glucose transporter | ↑7.2 (↑51.4) | ↑2.4 | ↑3.8 | ↓0.1 | 11/4 |
| YOR047C | STD1 | Regulator of Rgt1 | ↑4.5 (↑3.5) | NC | ↑2.7 | ↓0.4 | 2/2 |
| YHR096C | HXT5 | Glucose transporter | ↑4.0 (NC) | NC | ↑2.1 | ↓0.3 | 4/0 |
| YGL209W | MIG2 | Glucose-dependent repressor | ↑3.4 (↑12.5) | ↑2.0 | ↑6.8 | NC | 9/4 |
| YNL234W | Heme-binding globin-like protein | ↑3.1 (NC) | NC | ↑1.8 | ↓0.5 | 2/1 | |
| YOR062C | Similarity to N terminus of Reg1 | ↑3.0 (↑5) | NC | ↑1.7 | ↓0.2 | 4/1 | |
| YNL065W | AQR1 | MFSg transporter; resistance to monocarboxylic acids | ↑2.6 (NC) | NC | ↑1.7 | NC | 5/4 |
| YLR109W | AHP1 | Alkyl hydroperoxide reductase; redox homeostasis | ↑2.5 (NC) | NC | ↑1.7 | ↓0.1 | 2/2 |
| YER037W | PHM8 | Involved in phosphate metabolism? | ↑2.4 (NC) | ↑2.2 | ↑2.3 | ↓0.2 | 1/1 |
| YOR338W | Uncharacterized ORF (SGD) | ↑2.2 (NC) | NC | NC | NC | 0 | |
| YKL036C | Dubious ORF (SGD) | ↑2.1 (A) | NC | NC | NC | 2 | |
| YCR005C | CIT2 | Peroxisomal citrate synthase | ↑2.1 (↑4) | ↑1.7 | NC | ↓0.5 | 3/2d |
| YER028C | MIG3 | Possible glucose-dependent repressor | ↑2.0 (↑4.6) | NC | NC | NC | 4/2 |
| YGL039W | Similarity to dihydroflavonol 4-reductase | ↑2.0 (NC) | NC | NC | ↓0.5 | 2/0 | |
| YKL035W | UGP1 | UDP-glucose pyrophosphorylase | ↑2.0 (NC) | NC | NC | NC | 1/0 |
| YOL016C | CMK2 | Calmodulin-dependent protein kinase | ↑2.0 (↑3.4) | ↑1.7 | NC | NC | 1/0 |
| YHR087W | Uncharacterized ORF (SGD) | ↑2.0 (NC) | NC | NC | ↓0.2 | 1/0 | |
| YMR316W | DIA1 | Regulation of invasive growth? | ↑1.9 (NC) | ↑1.7 | NC | NC | 3/0 |
| YER062C | HOR2 | Glycerol-1-phosphatase | ↑1.9 (NC) | NC | NC | ↓0.3 | 2/1 |
| YJL214W | HXT8 | Glucose transporter | ↑1.9 (↑1.8) | NC | ↑2.9 | NC | 5/0 |
| YBR105C | VID24 | Vacuolar protein targeting | ↑1.9 (↑2.6) | NC | NC | NC | 4/3 |
| YDR423C | CAD1 | Jun family of transcription factors | ↑1.9 (NC) | NC | NC | NC | 0 |
| YHR097C | Uncharacterized ORF (SGD) | ↑1.9 (NC) | NC | NC | ↓0.5 | 0 | |
| YOL046C | Dubious ORF (SGD) | ↑1.9 (A) | NC | NC | NC | 2 | |
| YBR067C | TIP1 | Cell wall mannoprotein | ↑1.9 (NC) | NC | NC | ↑3.8 | 5/1 |
| YDR277C | MTH1 | Regulator of Rgt1 | ↑1.8 (↑2.1) | NC | ↑2.0 | NC | 4/2 |
| YOL136C | PFK2 | 6-Phosphofructo-2-kinase; regulation of glycolysis | ↑1.8 (NC) | NC | ↑1.8 | NC | 2/2 |
| YFL054C | Glycerol transporter | ↑1.8 (↑3.5) | NC | ↑2.0 | NC | 3/2 | |
| YDR001C | NTH1 | Neutral trehalase; stress response | ↑1.8 (NC) | NC | NC | NC | 2/2 |
| YPL026C | SKS1 | Protein kinase; multicopy suppressor of snf3 | ↑1.8 (NC) | NC | NC | NC | 6/3e |
| YLR413W | Uncharacterized ORF (SGD) | ↑1.8 (NC) | NC | NC | NC | 1/0 | |
| YDL062W | Dubious ORF (SGD) | ↑1.8 (A) | NC | NC | NC | 1 | |
| YMR136W | GAT2 | GATA zinc finger toxin factor | ↑1.8 (NC) | NC | NC | NC | 2/1 |
| YFR016C | Uncharacterized ORF (SGD) | ↑1.8 (NC) | NC | NC | NC | 0 | |
| YKR098C | UBP11 | Ubiquitin-specific protease | ↑1.8 (NC) | NC | NC | NC | 4/3 |
| YKR076W | ECM4 | Cell wall organization | ↑1.8 (NC) | NC | NC | NC | 7/5f |
| YLR194C | Uncharacterized ORF (SGD) | ↑1.8 (↑1.8) | NC | NC | ↓0.3 | 1/0 | |
| YAL061W | Putative polyol dehydrogenase | ↑1.8 (↑1.7) | NC | NC | NC | 1/0 | |
Genes whose transcript levels were induced to increase at least 1.8-fold on galactose in a homozygous rgt1Δ/rgt1Δ strain (YM6440) in microarray hybridizations (averages of ratios of mutant spot intensities to wild-type intensities are shown). Boldface type indicates that the ORF or gene was a good candidate for an Rgt1 target (see the text).
The ratio of gene transcript levels, determined from hybridization of cellular RNA probes to DNA microarrays, of heterozygous RGT2-1 (YM6554) and SNF3-1 (YM6557) strains grown on galactose and a haploid snf3Δ rgt2Δ strain (YM6370) grown on glucose (Glu) to the gene transcript levels of wild-type (diploid and haploid) strains. The same data from the hybridization of RNA (from an rgt1Δ haploid strain [YM4509] compared to that in the wild type [YM4127] grown in rich 3% glycerol plus 3% lactate medium) to an oligonucleotide array (Affymetrix) are in parentheses. A, Affymetrix hybridization analysis software designated the transcript as being absent; NC, transcript levels that were not changed significantly in the mutant relative to those in the wild type. Symbols: ↑, increased transcript level (≥1.7-fold) in the mutant compared to that in the wild type; ↓, decreased transcript level (≤0.5-fold) in the mutant compared to that in the wild type.
Ratio of the number of potential Rgt1 binding sites (CGGANNA) (33) in the promoter to the number of these sites that are conserved in the orthologous promoters of other Saccharomyces species, as described in reference 14.
No Rgt1 binding sites lie in the 201 bp between CIT2 and YCR006C (classified as dubious in the SGD), but three potential Rgt1 binding sites lie within 1,200 bp upstream of the CIT2 ATG.
Only one Rgt1 binding site is in the intergenic region, but five additional sites lie in the upstream 558-bp ORF (YPL025C, a likely gene since a two-hybrid interaction has been reported between YPL025C and MIG1 [65]).
ECM4 and YKR075C are divergently transcribed and so share upstream sequences. These conserved potential Rgt1 binding sites are rather remote from the ECM4 ATG, and the closest three sites are not conserved, so it is doubtful that Rgt1 regulates ECM4 expression.
MFS, major facilitator superfamily.
We attempted to verify the most promising candidate Rgt1 target genes using two experimental approaches. First, we characterized the transcriptional regulation of several of these genes by fusing their promoters to lacZ and measuring the amount of β-galactosidase activity produced (Tables 4 to 6). For analysis, we mostly selected promoters containing at least two potential Rgt1 binding sites, at least one of which is evolutionarily conserved. Second, we assessed the ability of Rgt1 to bind to the same set of promoters in vivo by using chromatin immunoprecipitation (ChIP) assays (Fig. 1). Fifteen of the genes that we tested bind Rgt1 in cells grown on galactose but not glucose (Fig. 1), suggesting that Rgt1 directly regulates their expression.
TABLE 4.
Newly identified targets of Rgt1 with unknown functionsa
| Promoter fusion | Activity of lacZ reporter
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
rgt1Δ strain
|
RGT2-1 strain
|
SNF3-1 strain
|
||||||
| Gal | Raf | Glu | Gal | Glu | Gal | Glu | Gal | Glu | |
| YKR075c-lacZ | 612.8 | 2,985.6 | 183.5 | 10,324.8 | 1,925.5 | 1,434.3 | 532.0 | 4,877.2 | 1,215.5 |
| YOR062c-lacZ | 44.7 | 88.8 | 305.0 | 540.2 | 200.8 | 139.3 | 258.4 | 307.1 | 208.2 |
| YGL157w-lacZ | 98.4 | 181.9 | 704.9 | 1,094.0 | 456.6 | 200.8 | 569.7 | 589.6 | 592.7 |
| YNL234w-lacZ | 2.5 | 5.3 | 12.9 | 19.4 | 12.3 | 6.4 | NT | 14.7 | NT |
Strains transformed with plasmids carrying the indicated lacZ fusions (Table 1) were pregrown in galactose medium selective for the plasmid, inoculated into media with the indicated carbon source at an OD600 of approximately 0.1, and grown until mid-log phase (usually 5 to 6 h). Activities of lacZ reporters (in nanomoles of ONPG per milligram of protein per minute) were measured in cellular extracts prepared from the following cultures: BY4741 (FM391) (wild type), FM557 (rgt1Δ), YM6546 (RGT2-1), and YM6549 (SNF3-1). Plasmids used were pBM3469 (43) (pYKR075C-lacZ), pBM4273 (pYOR062C-lacZ), pBM4381 (pYGL157W-lacZ), and pBM4458 (pYNL234W-lacZ). All plasmids carry the 2-micron origin of replication. Raf, raffinose; Glu, glucose; NT, not tested.
TABLE 6.
Newly identified targets of Rgt1: transcriptional regulatorsa
| Promoter fusion | Activity of lacZ reporter
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type
|
rgt1Δ strain
|
RGT2-1 strain
|
SNF3-1 strain
|
|||||
| Gal | Glu | Gal | Glu | Gal | Glu | Gal | Glu | |
| STD1-lacZ 2μm | 113.1 | 2,302.0 | 2,216.0 | 1,825.4 | 380.8 | 2,007.3 | 1,184.8 | 2,100.3 |
| MTH1-lacZ CEN | 635.7 | 117.3 | 1,396.0 | 449.3 | 1,118.0 | 286.2 | 1,780.8 | 450.5 |
| MIG2-lacZ 2μm | 150.8 | 3,702.7 | 3,803.9 | 2,446.3 | 1,724.1 | 2,689.9 | 3,422.7 | NT |
| MIG3-lacZ 2μm | <0.2 | 152.4 | 51.2 | 42.3 | 3.2 | 155.6 | 14.6 | 134.9 |
Expression of the Rgt1 target genes in the indicated mutants. Assays were done as described in Table 4, footnote a. Strains used were BY4741 (FM391) (wild type), FM557 (rgt1Δ), YM6546 (RGT2-1), and YM6549 (SNF3-1). Plasmids used were pBM4270 (pSTD1-lacZ), pBM4292 (pMTH1-lacZ), pBM4268 (pMIG2-lacZ), and pBM4379 (pMI63-lacZ). Glu, glucose; NT, not tested; CEN, centromere-containing plasmid.
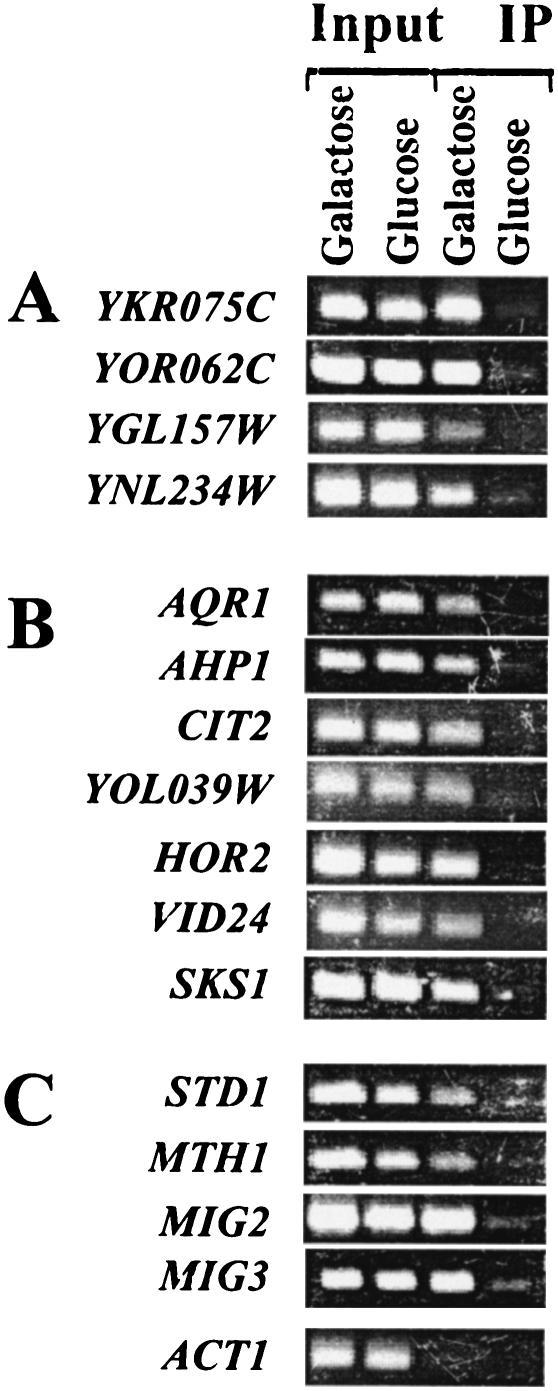
FIG. 1.
ChIP assay using an antibody against native Rgt1. PCR products amplified with primers specific for sequences upstream of the indicated genes are shown. Panels A, B, and C show results of ChIP experiments for genes presented in Tables 4, 5, and 6, respectively. Rgt1 levels have been shown to be similar in cells growing on galactose and glucose (33). IP, immunoprecipitate.
The newly identified targets of Rgt1 include several genes without assigned functions (Table 4). Two of the genes, YKR075C and YOR062C, are paralogues that have modest similarity to the N-terminal part of Reg1, the regulatory subunit of the Glc7 protein phosphatase that inactivates Snf1 protein kinase in response to high levels of glucose (41). The expression of YOR062C is regulated by Rgt1 and induced by glucose (Table 4); the expression of YKR075C is induced by raffinose (equivalent to low levels of glucose) due to regulation by Rgt1 and repressed by high levels of glucose through the action of the Mig1 and Mig2 repressors (43). The regulation of these two genes by glucose and their similarity to Reg1, which acts in glucose signaling pathways, suggest that they may be involved in glucose metabolism or regulation. It is not obvious how the proteins encoded by the two other newly identified Rgt1 targets—Ygl157, which is similar to oxidoreductases with dihydroflavanol 4-reductase activity (Gene Ontology Consortium, SGD; http://www.geneontology.org/), and Ynl234, which is similar to the globins and has a functional heme-binding domain (59)—might be involved in glucose signaling or metabolism. It is likely that Rgt1 directly regulates these genes because it binds to their promoters in vivo (Fig. 1A).
Several of the remaining genes that are candidates for regulation by Rgt1 have been reported to be induced by glucose: AQR1 (7, 67), HOR2 (7), VID24 (12), and PFK27 (2, 4, 7). We observed relatively modest induction of expression of most of these genes due to the deletion of RGT1 (Tables 3 and 5). Rgt1 clearly binds in vivo to the promoters of these genes (Fig. 1B), so they are likely its direct targets. Independent evidence that SKS1 is a direct target of Rgt1 comes from the observation that its promoter derepresses HXT gene expression when it is present in high copy, probably by titrating Rgt1 (69).
TABLE 5.
Potential Rgt1 target genesa
| Promoter fused to lacZ | β-Galactosidase activity of cells grown on galactose
|
||
|---|---|---|---|
| Wild type | rgt1Δ strain | Repression (fold) | |
| AQR1 | 362.8 | 1,605.7 | 4.4 |
| AHP1 | 278.0 | 1,329.0 | 4.8 |
| PHM8 | 401.3 | 574.5 | 1.4 |
| CIT2 | 79.1 | 134.7 | 1.7 |
| YGL039W | 216.3 | 244.4 | 1.1 |
| HOR2 | 162.5 | 591.5 | 3.6 |
| VID24 | 54.2 | 119.9 | 2.2 |
| PFK27 | 84.8 | 239.6 | 2.8 |
| SKS1 | 109.1 | 1,069.1 | 9.8 |
Analysis was done as described for Table 4 (footnote a). Strains used were BY4741 (FM391) (wild type) and FM557 (rgt1Δ). Plasmids used were pBM4511 (pAQR1-lacZ), pBM4502 (pAHP1-lacZ), pBM4510 (pPHM8-lacZ), pBM4512 (pCIT2-lacZ), pBM4515 (pYGL039W-lacZ), pBM4513 (pHOR2-lacZ), pBM4514(pPFK27-lacZ), and pBM4516 (pSKS1-lacZ).
Many genes exhibit an increased level of expression in the snf3Δ rgt2Δ double mutant compared to that in the wild type (see Table S1 in the supplemental material). Most of these genes are known to be repressed by glucose and/or are induced in the diauxic phase of growth (i.e., after glucose depletion) (15). An increased expression of glucose-repressed genes in the snf3Δ rgt2Δ mutant growing on glucose was expected (51) and is almost certainly an indirect effect of reduced glucose metabolism as a result of the severely reduced glucose transport capacity of this mutant, which weakens or abolishes the glucose signal that regulates the Snf1 protein kinase (18). This increased expression is also a direct effect of the reduction of MIG2 (and possibly MIG3) expression (see below).
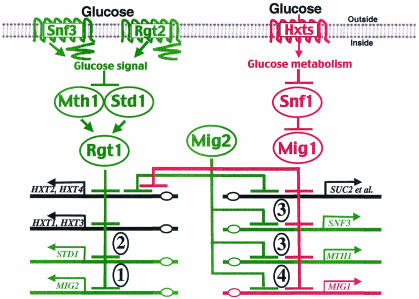
The glucose repression and glucose induction pathways are intertwined in a regulatory network.
Some of the most notable targets of the Snf3/Rgt2-Rgt1 glucose induction pathway encode components of the glucose repression and glucose induction pathways. These results, together with findings from previous studies (43, 47, 52, 71), reveal that these two glucose signal transduction pathways are intertwined in a regulatory network (Fig. 2). We can recognize four inter- and intrapathway controls (Fig. 2, circled numbers 1 to 4): (i) the regulation of glucose repression by the glucose induction pathway through Snf3/Rgt2-mediated induction of MIG2 expression, (ii) the autoregulation of the glucose induction pathway through Snf3/Rgt2-mediated glucose induction of STD1 expression, (iii) the regulation of glucose induction by the glucose repression pathway through Mig1- and Mig2-mediated repression of MTH1 and SNF3 expression, and (iv) the autoregulation of the glucose repression pathway through Mig1-mediated repression of MIG1 expression. Each of these branches of the regulatory network is described below.
FIG. 2.
Interwoven regulatory network of glucose sensing pathways. The components shown in green respond to the glucose signal generated by the Rgt2 and Snf3 glucose sensors, and the components shown in red respond to the glucose signal that affects the function of the Snf1 kinase. The genes shown with black lines are the ultimate targets of these two glucose signaling pathways. Lines ending in arrows denote activation, and lines ending in bars denote inhibition. Circled numbers refer to the four types of control described in the text.
Regulation of the Mig2 (and Mig3) glucose repressors by the glucose induction pathway.
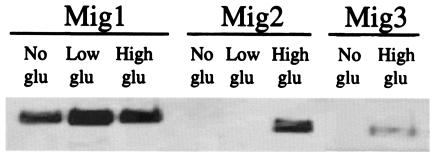
The Snf3/Rgt2 glucose induction pathway contributes to glucose repression by regulating expression of MIG2 and MIG3 (Tables 6 and 7), which encode glucose-activated repressors that collaborate with Mig1 to repress expression of many glucose-repressed genes (Table 8) (43). Rgt1 is likely a direct regulator of these genes because it binds to their promoters in vivo (Fig. 1C). MIG2 (and MIG3) expression is induced by high levels of glucose and is constitutively induced in SNF3-1 and RGT2-1 mutants (Table 6). This regulation is also apparent in the levels of these two proteins: Mig2 (and Mig3) is detectable only in extracts of cultures grown on glucose (with Mig3 being less abundant than Mig2) (Fig. 3). We conclude that one of the outputs of the glucose signal that is generated by the Rgt2 and Snf3 glucose sensors is glucose repression of expression of genes that are targets of Mig2 (and Mig3).
TABLE 7.
MIG2 expression is regulated by the Snf3/Rgt2-Rgt1 pathwaya
| Relevant genotype | β-Galactosidase levels in MIG2-lacZ cells
|
|
|---|---|---|
| Gal | Glu | |
| Wild type | 7.0 | 468.6 |
| rgt1Δ | 1,201.8 | 536.7 |
| rgt2Δ | 3.8 | 301.1 |
| snf3Δ | 0.2 | 328.1 |
| rgt2Δ snf3Δ | 0.2 | 4.2 |
Assays were done as described in Table 4, footnote a. Strains used were BY4741 (FM391) wild type, FM557 (rgt1Δ), YM6247 (rgt2Δ), YM6329 (snf3Δ), YM6370 (rgt2Δ snf3Δ). The plasmid used was pBM4346 (pMIG2-lacZ [CEN]). Glu, glucose.
TABLE 8.
MTH1, MIG1, SNF3, HXT2, and MRK1 expression is glucose repressed by Mig1, Mig2, and Mig3a
| Relevant genotypeb | β-Galactosidase activity in glucose-grown (and raffinose-grown) cells
|
||||
|---|---|---|---|---|---|
| MTH1-lacZ | MIG1-lacZ | SNF3-lacZ | HXT2-lacZ | MRK1-lacZ | |
| Wild type | 24.1 (216.0) | 238 (311.4) | 8.2 (143.0) | 22.1 (176.4) | 1.1 (18.8) |
| mig1Δ | 85.1 | 287.2 | 11.0 | 28.5 | 1.8 |
| mig1Δ mig2Δ | 248.0 | 771.4 | 60.1 | 298.7 | 4.6 |
| mig2Δ mig3Δ | 40.6 | 302.8 | 11.6 | 51.9 | 1.6 |
| mig1Δ mig2Δ mig3Δ | 381.9 | 1,088.5 | 104.0 | 447.0 | 12.5 |
Analysis was done as described for Table 4 (footnote a). Strains used were BY4743 (wild type), YM6430 (mig1Δ/mig1Δ), YM6682 (mig1Δ mig2Δ/mig1Δ mig2Δ), YM6683 (mig2Δ mig3Δ/mig2Δ mig3Δ), and YM668 (mig1Δ mig2Δ mig3Δ/mig1Δmig2Δmig3Δ). Plasmids used were pBM4292 (pMTH1-lacZ [CEN]), pBM3190 (pMIG1-lacZ [2μm]), pBM4487 (pSNF3-lacZ [2μm]), pBM4440 (pHXT2-lacZ [CEN]), and pBM4296 (pMRK1-lacZ [CEN]).
All strains are homozygous diploids.
FIG. 3.
Western blot analysis of tagged Mig1, Mig2, and Mig3. Strains YM6843 (MIG1-6HA), YM6833 (MIG2-6HA), and YM6835 (MIG3-6HA) were pregrown in 5% glycerol plus 0.5% galactose (no glu) and inoculated into the same medium and into a medium containing 5% glycerol plus 0.05% glucose (low glu) or 4% glucose (high glu) at an OD600 of 0.1 to 0.2. Protein extracts were prepared after the cultures reached an OD600 of approximately 1.0. For the MIG1-6HA strain, each lane was loaded with extract in a volume equivalent to a cell OD600 of 0.25. For MIG2-6HA, cells with an OD600 of 0.5 were loaded; for MIG3-6HA, cells with an OD600 of 1.0 were loaded. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 15% polyacrylamide gradient) and detected by immunoblotting. Loading of the lanes with expected amounts of protein was subsequently confirmed by staining the membrane with Ponceau Red (data not shown).
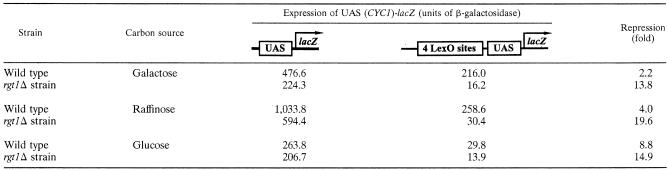
Glucose induction of MIG2 expression seems to account for all of the glucose regulation of Mig2 function, because when MIG2 expression is rendered constitutive by the deletion of RGT1, Mig2 repressor activity in cells growing on galactose or raffinose is virtually equal to its activity in glucose-grown wild-type cells (Table 9). Regulation of MIG2 expression by the Snf3/Rgt2-Rgt1 pathway explains why the Snf1 protein kinase, which regulates the function of Mig1, plays no role in regulating Mig2 (43).
TABLE 9.
Gene expressed from the MIG2-lexA-MIG2 promotera
The function of lexA-MIG2 expressed from the MIG2 promoter is regulated by Rgt1. Cells were pregrown in a 2% raffinose-0.05% glucose medium selective for the plasmids and inoculated at an OD600 of approximately 0.1 into fresh medium with the indicated carbon sources. After 5 to 6 h of growth, cellular extracts were prepared from the cultures, and β-galactosidase activity (in nanomoles of ONPG per milligram of protein per minute) was measured in the extracts. Strains used were BY4743 (FM393) (wild type [diploid]) and YM6440 (rgt1Δ/rgt1Δ). Plasmids used were pBM4258 (pMIG2-lexA-MIG2), pLG312s (lexO-less UAS-lacZ reporter) (22), and JK1621 (4x lexO-UAS-lacZ reporter) (31). The amount of repression caused by LexA (not attached to Mig2) in the wild-type strain was as follows: with raffinose, 2.9-fold; with galactose, 1.7-fold; and with glucose, 2.2-fold.
If glucose induction of MIG2 (and MIG3) expression is largely responsible for glucose activation of Mig2 (and Mig3) repressor function, then glucose-repressed genes that are Mig2 (and Mig3) targets should exhibit reduced expression in an rgt1 mutant (due to increased levels of Mig2). Indeed, the expression of some glucose-repressed genes is reduced in an rgt1Δ mutant growing on galactose (see Table S2 in the supplemental material). One of these genes, JEN1, is known to be regulated by Mig1 and Mig2 (3). The gene whose expression was most strongly and reproducibly reduced in our gene expression profiling experiments of an rgt1 mutant, MRK1, is regulated by all three Mig repressors (Table 8).
Feedback regulation of the glucose induction pathway.
Expression of STD1, which encodes a regulator of Rgt1, is induced by high levels of glucose due to regulation by Rgt1 (Table 6). It is likely that Rgt1 directly represses STD1 expression, because it binds to the STD1 promoter in vivo (Fig. 1C). Since Std1 promotes transcriptional repression by Rgt1 (38, 62), this process constitutes a feedback loop whose effect should be to dampen the glucose induction of HXT gene expression, since it should act to counteract the glucose-induced degradation of Std1.
MTH1 expression is not induced by glucose but is nevertheless modestly repressed by Rgt1 (Table 6). In addition, MTH1 expression is modestly induced by galactose due to the regulation by the Gal4 transcriptional activator (54). Thus, Gal4 sustains and Rgt1 attenuates MTH1 expression in galactose-grown cells. Activation of MTH1 expression by galactose would reinforce Rgt1-mediated repression of HXT genes encoding glucose transporters. However, the level of regulation of MTH1 expression by Gal4 and Rgt1 is modest, so this regulation probably serves only to fine-tune the glucose induction pathway.
Mig1 and Mig2 (and Mig3) mediate glucose repression of MTH1 and MIG1.
MTH1 expression is repressed by glucose (62) due to the combined action of Mig1 and Mig2 (and Mig3 to a lesser extent) (Table 8). Thus, Mig2 (and Mig3) regulates expression of a protein (Mth1) that collaborates with Rgt1 to repress their expression in the absence of glucose. This is a feedforward regulatory loop in the glucose induction pathway, the effect of which is to reinforce the effect of glucose on Mth1 function; the addition of glucose inhibits Mth1 function by inducing its degradation (19) and reduces MTH1 transcription through Mig2 (and Mig3)-mediated glucose repression. Finally, Mig1 and Mig2 (and Mig3) repress expression of MIG1 (43, 71). This process constitutes a feedback loop of the glucose repression pathway, the effect of which is to dampen Mig1-mediated glucose repression.
DISCUSSION
Our results suggest that a surprisingly small number of genes are targets of the glucose signaling pathway that operates through the Snf3 and Rgt2 glucose sensors and the Rgt1 repressor. In addition to the several HXT genes that are well-established targets of this signal transduction pathway (5, 16, 52), we could validate only seven other genes (MIG2, MIG3, STD1, YGL157W, YKR075C, YOR062C, and YNL234W) as significant targets of Rgt1. These results are consistent with those from other recent global profilings of the yeast transcriptome (2, 7). While the expression of several other genes changes when RGT1 is inactivated (Tables 3 and 5), the role of Rgt1 in regulating the expression of these genes is modest at best (Tables 3 and 5). We conclude that the Rgt2/Snf3-Rgt1 glucose signal transduction pathway is primarily dedicated to regulating the expression of HXT genes.
We can only speculate about the possible functions of the proteins encoded by the few newly discovered Rgt1 targets (YKR075C, YOR062C, YGL157W, and YNL234W). Because Ykr075 and Yor062 have similarity to Reg1, a regulatory subunit of the Glc7 PP1 protein phosphatase known to be involved in the Snf1 glucose signaling pathway, we suspect that they are involved in glucose sensing or metabolism. However, we have not been able to detect any defects in glucose induction or glucose repression of gene expression caused by deleting these genes (either singly, doubly, or in combination with a reg1 mutation). The function of the N terminus of Reg1, the portion that is similar to Ykr075 and Yor062, is not known. This part of Reg1 is required neither for interaction with the Glc7 phosphatase nor for interaction with Snf1 (57), though it is required for the full repression of SUC2 (but not ADH2) expression (17). The pattern of transcriptional regulation of YKR075C (repression by Rgt1 in the absence of glucose and repression by Mig1 and Mig2 at high levels of glucose) is similar to those of HXT2 and HXT4, which encode high- and intermediate-affinity glucose transporters, respectively, leaving a relatively narrow window of glucose concentrations within which these genes are maximally expressed (43, 53). By contrast, the YKR075c paralogue YOR062c is expressed mainly in the presence of high levels of glucose (Table 4). This pattern of regulation suggests that Ykr075 operates under conditions of low levels of glucose and that Yor062 operates when glucose levels are high.
The protein encoded by YGL157W is similar to oxidoreductases with dihydroflavanol 4-reductase activity (Gene Ontology Consortium, SGD; http://www.geneontology.org/). A yeast mutant lacking all four dihydroflavanol 4-reductase-like proteins (Ygl157, Ygl039, Gre2/Yol151w, and Ydr541) has been constructed (14), but no phenotype of the mutant was reported. The substrate(s) of Ygl157 is unknown, but its ortholog, Gre2, catalyzes the reduction of methylglyoxal (10), a by-product of glycolysis. Another Rgt1 target, YNL234W, encodes a protein that is similar to the globins and contains a functional heme-binding domain (59). Other transcription factors in addition to Rgt1 must control the expression of YNL234W, since the gene is also activated by stress (e.g., nitrogen shortage and heat shock) (59). The role of these two proteins, if any, in glucose signaling or metabolism remains to be determined.
Perhaps the most significant insight gained from our gene expression profiling results is the revelation of an intricate intra- and interpathway regulatory circuit connecting two glucose sensing and signaling pathways that cause glucose repression and glucose induction of gene expression (Fig. 2). There are feedback components as well as feedforward components of this regulatory circuit. In the glucose induction pathway, STD1 expression is feedback regulated by Rgt1; glucose inhibits Std1 function by stimulating its degradation (44a; V. Brachet, unpublished), and glucose induces STD1 expression through the Rgt2/Snf3-Rgt1 signaling pathway (Table 6 and Fig. 2). Thus, STD1 expression is increased at the same time that Std1 levels are decreasing in response to glucose. This regulation should serve to dampen glucose induction of gene expression. It should also provide for a rapid reestablishment of repression when glucose is depleted. Std1 may also play a role in the glucose repression pathway, because it interacts with and regulates Snf1 (36).
In contrast to STD1, its paralogue MTH1, which has an overlapping function, is feedforward regulated; glucose stimulates the proteasome-mediated degradation of Mth1 (19) while also reducing MTH1 expression via repression mediated by Mig1 and Mig2 (and Mig3) (Table 6) (62). This regulation should reinforce the inhibitory effect of glucose on Mth1 function and ensure maximal glucose induction of Rgt1-repressed genes. MTH1 expression is also activated on galactose by Gal4 (54). As a result, in the absence of glucose (when Rgt1 is repressing HXT expression), Mth1 seems to be the major promoter of Rgt1 repression function.
The expression of a third component of the glucose induction pathway, SNF3, is repressed by high levels of glucose through the action of Mig1 and Mig2 (and Mig3) (47, 52). This finding reflects the likely function of Snf3 as a high-affinity glucose sensor (a sensor of low levels of glucose). Glucose repression of MIG1 expression mediated by Mig1 itself (43, 71), in collaboration with Mig2 (and Mig3) (Table 8), constitutes a feedback regulatory circuit, whereby synthesis of the repressor is decreased under conditions in which it is active. The effect of this regulation is to dampen Mig1-mediated glucose repression, possibly to avoid overrepression, and conceivably to enable a more rapid recovery from repression when glucose becomes depleted from the medium.
Our observation that the expression of the Mig2 repressor, which collaborates with Mig1, is induced by glucose through the Rgt2/Snf3-Rgt1 signaling pathway (Tables 6 and 7) provides a satisfying explanation for the fact that Mig2 function is not regulated by Snf1 (43). Indeed, glucose induction of MIG2 expression is sufficient to account for the glucose activation of Mig2 function (Table 9). The two main glucose repressors, Mig1 and Mig2, are mostly redundant (43, 66) but are regulated in different ways by different signaling pathways responding to different glucose signals, one of which operates through Snf1, is dependent on the Glc7 phosphatase and hexokinase (58, 64), and is probably based on glucose metabolism and the other of which operates through the glucose sensors to sense extracellular glucose by receptor-mediated signaling. Thus, the phenomenon of the glucose repression of gene expression is a result of outputs from two glucose signal transduction pathways: the Mig1 component regulated by the Snf1 kinase and the Mig2 (and the ancillary Mig3) component regulated at the level of their transcription by the Snf3/Rgt2-Rgt1 signaling pathway. The cross talk between the two glucose signaling pathways and the redundancy of the two main glucose repressors probably serve to integrate cellular responses to different glucose signals. Apparently, it is advantageous for the cell to employ two signaling systems to respond to its preferred sugar, but by locking the signaling pathways in a cross talking network, they respond coordinately. The dynamic structure of this glucose sensing regulatory network may serve to determine the range of its response to glucose, its robustness, and its sensitivity to environmental changes in glucose availability.
Supplementary Material
Acknowledgments
We thank Mark Watson and Kate Hamilton at the GeneChip Core Facility of the Siteman Cancer Center, Barnes-Jewish Hospital and Washington University School of Medicine, St. Louis, Mo., for performing target synthesis and hybridization to the Affymetrix oligonucleotide arrays and data analysis. We also to thank Diana Lamendola for constructing pBM4440 and our colleagues Barak Cohen, John Majors, Rob Mitra, and Priya Sudarsunam for helpful and insightful comments on the manuscript.
The study was supported by NIH grant RO1-32540.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 3.Bojunga, N., and K. D. Entian. 1999. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262:869-875. [DOI] [PubMed] [Google Scholar]
- 4.Boles, E., H. W. Gohlmann, and F. K. Zimmermann. 1996. Cloning of a second gene encoding 5-phosphofructo-2-kinase in yeast, and characterization of mutant strains without fructose-2,6-bisphosphate. Mol. Microbiol. 20:65-76. [DOI] [PubMed] [Google Scholar]
- 5.Boles, E., and C. P. Hollenberg. 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21:85-111. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Brejning, J., L. Jespersen, and N. Arneborg. 2003. Genome-wide transcriptional changes during the lag phase of Saccharomyces cerevisiae. Arch. Microbiol. 179:278-294. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 9.Carlson, M. 1998. Regulation of glucose utilization in yeast. Curr. Opin. Genet. Dev. 8:560-564. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. N., L. Porubleva, G. Shearer, M. Svrakic, L. G. Holden, J. L. Dover, M. Johnston, P. R. Chitnis, and D. H. Kohl. 2003. Associating protein activities with their genes: rapid identification of a gene encoding a methylglyoxal reductase in the yeast Saccharomyces cerevisiae. Yeast 20:545-554. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, M. C., and H. L. Chiang. 1998. Vid24p, a novel protein localized to the fructose-1,6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J. Cell Biol. 140:1347-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 14.Delneri, D., G. C. Tomlin, J. L. Wixon, A. Hutter, M. Sefton, E. J. Louis, and S. G. Oliver. 2000. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 252:127-135. [DOI] [PubMed] [Google Scholar]
- 15.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 16.Dlugai, S., S. Hippler, R. Wieczorke, and E. Boles. 2001. Glucose-dependent and -independent signalling functions of the yeast glucose sensor Snf3. FEBS Lett. 505:389-392. [DOI] [PubMed] [Google Scholar]
- 17.Dombek, K. M., S. Camier, and E. T. Young. 1993. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol. Cell. Biol. 13:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson, J. R., and M. Johnston. 1994. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics 136:1271-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 22.Guarente, L., and E. Hoar. 1984. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box.” Proc. Natl. Acad. Sci. USA 81:7860-7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 24.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 25.Hong, S. P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horak, J., J. Regelmann, and D. H. Wolf. 2002. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J. Biol. Chem. 277:8248-8254. [DOI] [PubMed] [Google Scholar]
- 27.Huang, P. H., and H. L. Chiang. 1997. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J. Cell Biol. 136:803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia, M. H., R. A. Larossa, J. M. Lee, A. Rafalski, E. Derose, G. Gonye, and Z. Xue. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3:83-92. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, H., K. Tatchell, S. Liu, and C. A. Michels. 2000. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:411-422. [DOI] [PubMed] [Google Scholar]
- 30.Johnston, M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15:29-33. [DOI] [PubMed] [Google Scholar]
- 31.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709-719. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J.-H., J. Polish, and M. Johnston. 2003. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol. Cell. Biol. 23:5208-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 34.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchin, S., V. K. Vyas, E. Kanter, S. P. Hong, and M. Carlson. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn, K. M., J. L. DeRisi, P. O. Brown, and P. Sarnow. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 21:916-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lafuente, M. J., C. Gancedo, J. C. Jauniaux, and J. M. Gancedo. 2000. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35:161-172. [DOI] [PubMed] [Google Scholar]
- 39.Li, F. N., and M. Johnston. 1997. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16:5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang, H., and R. F. Gaber. 1996. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 7:1953-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundblad, V., G. Hartzog, and Z. Moqtaderi. 1997. Manipulation of cloned yeast DNA, p. 13.10.1-13.10.14. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 43.Lutfiyya, L. L., V. R. Iyer, J. DeRisi, M. J. DeVit, P. O. Brown, and M. Johnston. 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150:1377-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutfiyya, L. L., and M. Johnston. 1996. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 16:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Moriya, H., and M. Johnston. Proc. Natl. Acad. Sci. USA, in press.
- 45.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 46.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neigeborn, L., P. Schwartzberg, R. Reid, and M. Carlson. 1986. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol. Cell. Biol. 6:3569-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Özcan, S., J. Dover, and M. Johnston. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Özcan, S., J. Dover, A. G. Rosenwald, S. Wölfl, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Özcan, S., and M. Johnston. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63:554-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Özcan, S., and M. Johnston. 1995. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Özcan, S., and M. Johnston. 1996. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell. Biol. 16:5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 55.Rolland, F., J. Winderickx, and J. M. Thevelein. 2002. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2:183-201. [DOI] [PubMed] [Google Scholar]
- 56.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Sanz, P., G. R. Alms, T. A. J. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz, P., A. Nieto, and J. A. Prieto. 1996. Glucose repression may involve processes with different sugar kinase requirements. J. Bacteriol. 178:4721-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sartori, G., L. Aldegheri, G. Mazzotta, G. Lanfranchi, H. Tournu, A. J. Brown, and G. Carignani. 1999. Characterization of a new hemoprotein in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:5032-5037. [DOI] [PubMed] [Google Scholar]
- 60.Scheffler, I. E., B. J. de la Cruz, and S. Prieto. 1998. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Int. J. Biochem. Cell Biol. 30:1175-1193. [DOI] [PubMed] [Google Scholar]
- 61.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wölfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 64.Tu, J., and M. Carlson. 1994. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 66.Vallier, L. G., and M. Carlson. 1994. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics 137:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Velasco, I., I. L. Calderon, and B. Andre. 2003. The AQR1 transporter mediates amino acid excretion in Saccharomyces cerevisiae. Yeast 20(Suppl.):S237. [Google Scholar]
- 68.Vyas, V. K., S. Kuchin, and M. Carlson. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, Z., and L. F. Bisson. 1996. The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast 12:1407-1419. [DOI] [PubMed] [Google Scholar]
- 70.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278:26146-26158. [DOI] [PubMed] [Google Scholar]
- 71.Zaragoza, O., O. Vincent, and J. M. Gancedo. 2001. Regulatory elements in the FBP1 promoter respond differently to glucose-dependent signals in Saccharomyces cerevisiae. Biochem. J. 359:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.