Abstract
Endemic strains of Legionella pneumophila sequence type 1 (ST1), in particular the ST1/Paris pulsotype, are dispersed worldwide and represent about 10% of culture-proven clinical cases of Legionnaires' disease in France. The high rate of isolation of this strain from both clinical and environmental samples makes identification of the source of infection difficult during epidemiological investigations. The full-length genome sequence of this strain was recently determined, and it revealed the presence of a CRISPR/cas complex. The aim of this study was to develop and evaluate a spoligotyping tool based on the diversity of this CRISPR locus that would allow the accurate subtyping of the L. pneumophila serogroup 1 ST1/Paris pulsotype. The CRISPR loci of 28 L. pneumophila ST1/Paris pulsotype isolates were sequenced, and 42 different spacers regions were characterized. A membrane-based spoligotyping method was developed and used to determine the subtypes of 406 L. pneumophila isolates, including 233 with the ST1/Paris pulsotype profile that were collected in France from 2000 to 2011. A total of 46 different spoligotypes were detected, and 41 of these were specifically identified in the ST1/Paris pulsotype isolates. In 27 of 33 epidemiological investigations, the environmental source of contamination was confirmed by comparing spoligotypes of clinical isolates with those of environmental isolates. With an index of discrimination of 79.72% (95% confidence interval, 75.82 to 83.63), spoligotyping of the L. pneumophila ST1/Paris pulsotype has the potential to be a useful complementary genotyping tool for discriminating isolates with undistinguishable pulsed-field gel electrophoresis (PFGE) and ST genotypes, which could help to identify environmental sources of infection.
INTRODUCTION
Legionella spp. are ubiquitous bacteria present in natural and artificial water systems. Inhalation of Legionella spp. in aerosolized water droplets from contaminated water sources is known to cause a type of pneumonia called Legionnaires' disease (LD). In the event of an LD outbreak, the successful outcome of an epidemiological investigation can help prevent further cases by rapidly identifying and containing the source of contamination.
Legionella pneumophila is responsible for more than 90% of the cases of LD, and serogroup 1 alone accounts for almost 85% of cases (9, 23). Diagnosis of LD can be made by serology, direct immunofluorescence, PCR, urinary antigen detection, or culturing of clinical specimens; almost 20% of confirmed LD cases are detected by culture (2). Epidemiological analyses based on pulsed-field gel electrophoresis (PFGE) and/or sequence-based typing (SBT) of clinical isolates of L. pneumophila serogroup 1 have been used to classify isolates as sporadic, epidemic, or endemic (1). A strain is considered endemic when several isolates of an identical genotype are responsible for several epidemiologically unrelated cases of LD. Among the endemic strains of L. pneumophila serogroup 1, sequence type 1 (ST1) strains are among the most prevalent, in particular the ST1/Paris pulsotype. This endemic type was responsible for 8.2% of French culture-proven cases of LD from 1995 through 2006 (1, 10, 15). ST1/Paris pulsotype isolates have also been detected in clinical and environmental samples from several other countries around the world, including Switzerland, Italy, Spain, Sweden, the United States, Japan, Senegal, and Canada (1, 4). The high isolation rate of this strain in clinical and environmental samples makes it difficult, and frequently impossible, to identify the environmental source of an infection during epidemiological investigations.
Recent studies have demonstrated the value of using the diversity of CRISPR spacers as genotyping markers for several pathogenic agents, and spoligotyping tools have been successfully developed for this purpose (14, 17, 19, 20).
The aim of this study was to design the first spoligotyping tool for subtyping L. pneumophila ST1/Paris pulsotype isolates and to evaluate its performance and efficiency on a collection of clinical and environmental isolates from France.
(These results were presented in part as an oral communication at the EWGLI Meeting in 2010 and as a poster at the FEMS Microbiology Congress in 2011.)
MATERIALS AND METHODS
Strains and growth conditions.
Reference strains used in this study were L. pneumophila Paris CIP107629 (ST1/Paris pulsotype) and L. pneumophila 130b ATCC BAA-74 (non-ST1/non-Paris pulsotype). All other clinical (257) and environmental (149) L. pneumophila isolates were part of the collection from the French Centre National de Référence des Légionelles and were selected based on their genotypes (PFGE and SBT). Among the 406 isolates, 46 belonged to the ST1/non-Paris pulsotype (11 unrelated and 35 isolates from the same water sample), 15 to the non-ST1/Paris pulsotype (unrelated isolates), 112 to the non-ST1/non-Paris pulsotype (unrelated isolates), and 233 to the ST1/Paris pulsotype. The 233 ST1/Paris pulsotype isolates were divided into 66 unrelated isolates, several sets of environmental isolates where members of each set were from the same water samples from three different environments (9, 10, and 3 isolates), 24 isolates from one patient's sputum, and 121 clinical and related environmental isolates from 33 epidemiological investigations (Fig. 1A). Three passages of the Paris CIP 107629 strain (after 1, 3, and 31 subcultures) were also used to assess the stability of the spoligotype markers. All isolates were grown on standard BCYEα medium (Oxoid, Dardilly, France) at 37°C.
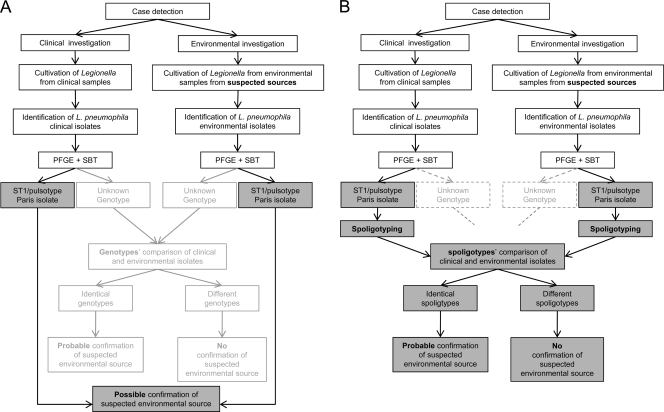
Fig 1.
Flowcharts of L. pneumophila typing during epidemiological investigations without (A) and with (B) spoligotyping of ST1/Paris pulsotype isolates.
CRISPR research in the genome.
CRISPR sequences in the Legionella genomes were identified using CRISPRFinder software (12).
Distribution of CRISPR loci.
The presence of a CRISPR locus in each strain was assessed by PCR using primers DR_F (5′-TTAGATGAGGGATTATTG-3′) and DR_R (5′-AAATCCAACCACTGAAAC-3′), which were designed in the direct repeat region of the CRISPR (DR), followed by agarose gel electrophoresis. The primers were designed using the sequence of the Paris CIP107629 reference strain (accession no. NC_006368).
Sequencing of the CRISPR locus.
The CRISPR locus was amplified by a pair of primers framing the locus: Crispr_F (5′-TGTTGATTTCAGGGTATCGA-3′) and Crispr_R (5′-ACAGAGAATAAAGTATGGAGT-3′). The CRISPR loci of the first set of 14 isolates (ST1/Paris pulsotype) were sequenced by Cogenics (Beckman Coulter Genomics, Grenoble, France). Nucleotide sequences for 19 supplementary CRISPR loci (14 of the ST1/Paris pulsotype and 5 of types other than the ST1/Paris pulsotype) were obtained with the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) using a PCR primer walking strategy (primers are available upon request). Sequence chromatograms were obtained with an ABI3730XL automated sequence analyzer (Applied Biosystems). Sequence analysis was performed using BioNumerics version 6.5 (Applied-Maths, Sint-Martens-Latem, Belgium). All of the CRISPR loci sequences are available on the Institut Pasteur website: http://www.pasteur.fr/recherche/genopole/PF8/crispr/CRISPRDB.html.
Spoligotyping tool.
The CRISPR spacers were amplified using oligonucleotides Dig-DR_F (5′-Dig-TTAGATGAGGGATTATTG-3′) and DR_R (5′-AAATCCAACCACTGAAAC-3′), which are complementary to the DR; the forward primer was 5′ digoxigenin labeled. Probes were designed as amino-linked oligonucleotides synthesized with a C6 spacer (Table 1), and these were diluted to the appropriate concentrations for membrane preparation with 0.5 M NaHCO3 (9). Biodyne C membrane (Pall Biosupport, Ann Arbor, MI) was activated by incubation in 16% (wt/vol) 1-ethyl-2-(3-dimethylaminopropyl) carbodiimide (Sigma-Aldrich, Saint Quentin Fallavier, France) for 10 min at room temperature. Following a brief wash with deionized water, 150 μl of each probe solution was applied in a line by using a miniblotter system (enhanced chemiluminescence [ECL] multiprobe; GE Healthcare Life Science, Saclay, France). After 5 min of incubation at room temperature, probe solutions were removed from the membrane by aspiration. The membrane was inactivated by incubation in 100 mM NaOH for 10 min at room temperature and washed briefly with deionized water before 5 min of incubation with 2× SSPE (0.36 M NaCl, 20 mM NaH2PO4, 2 mM EDTA [pH 7.7])–0.1% sodium dodecyl sulfate (SDS) at 60°C. The membrane was then incubated in 20 mM EDTA for 15 min at room temperature and stored at 4°C until use.
Table 1.
Spoligotyping oligonucleotide probes
| Probe | Sequence |
|---|---|
| spacer1 | TAGATATAAAAAGATTAAATCTTCTAGCGCACAT |
| spacer2 | TCACTACTCCTGAAGGTTATAATTTTTGCTATAA |
| spacer3 | TTCGAATACAATCCTAGTGTCTCTGTGTGAATTAAG |
| spacer4 | CAGGCACTGGTTCACTAGACACTGTAACATCTAT |
| spacer5 | CAATAACAAGCGAGCCTTTTGTACTAGAAGGTTTA |
| spacer6 | CTACCAGTTAATCGTAACTCAATCTCTTTTTCAA |
| spacer7 | ATAGAATACATAAGTGCAAATTATTAAATGTTAC |
| spacer8 | TGAATGTAGAAACCAGATGCCACGAATTATTAGA |
| spacer9 | TTTGTATAAACGTTCTGATATGACTTAGGTAATCT |
| spacer10 | CTAACCTGATTGCTCAACAAATAATGCTATTGGC |
| spacer11 | TCACTTTAGGCCAACGCCGATCCTCCGCTTCGAA |
| spacer12 | CTCACATCTTACCCTCAGGGCGGATATTGTGAATC |
| spacer13 | ATTAATAATATTTTAGAAGATTGGCACATAATA |
| spacer14 | TGACGCAAAGGATTTATTAAAAACGCCTTGTAAT |
| spacer15 | ATTTTACCTTTTAACACATATTGATAGGCGT |
| spacer16 | TCCATAACTGAAACGTCCTTATGCCTCAACATAATG |
| spacer17 | GAATTTGTCGGCCGCATAGACCGCTTTTATCAAA |
| spacer18 | CTATTGCAAGCTAGTTTGATCGTGTTATTATAAGAA |
| spacer19 | TGACAAACGTTTGTTTTTTAGACACAACACTAAAAG |
| spacer20 | CCTGAAAACCCGCCACAACCCGCGCCAGACTTGAA |
| spacer21 | ACCAAGTCGAAACAACATACCGAGACCGTGTTGA |
| spacer22 | TACATTGTTACGTTCATTTCACTCAGTTTTTCATA |
| spacer23 | AGCAATAACCCAAAGTTTCGCGCGCGTGCGCGGG |
| spacer24 | TTATAACATCGGGATGGCGGTTTATTGGTTAAGTAAC |
| spacer25 | TTCTTTTTTCAGATTTCATTTCCTTTTCCTTGTG |
| spacer26 | TTTTTATCGTAAGCTTGATTAACTAGACATCTACTA |
| spacer27 | GTTTTAATTTAATCATTATTGCTTCCTTATTAA |
| spacer28 | TAAGATATTGGTCGATTGTTTGCAAGACGTCATCAAA |
| spacer29 | CTAAATCAGCAGTCATCAAAACCCATATGATTGT |
| spacer30 | TTCTCAGAATGGGAACGTTACACATCATATT |
| spacer31 | TTAAGTATACGTTTCTTTGGGCTATTAGTTCTAA |
| spacer32 | TTCTCATATTTTAAAACAATAGAGTAATTCATTTTA |
| spacer33 | AATCTTAATTTATGCGCCTTACCTTCTGCTTCATC |
| spacer34 | TATACTTAGACAATATCATCATTTCCTATGTTTCGA |
| spacer35 | GCTATTCCAGATAGGTAGTTGTTTAGAGCATTTTGT |
| spacer36 | TTCGATACTCCTTAGCGGTACTTGCTAAAGTAGTT |
| spacer37 | AAATAATTCTTGTGACTCACGTGCCGCCATTTGAA |
| spacer38 | TAGGAATTGATTGGGGTAACGCCATCGCCATAGAAG |
| spacer39 | TTACTTCATGACCCGGACATTTTACTAAAAAAT |
| spacer40 | TCCTATACCTTGTCACAATTTCCTCATATGGCTC |
| spacer41 | CATAATTGGGAATTGGTGTGAAATGCTCACCGTCCG |
| spacer42 | TTAATTGCGCCAGAAACAGCACCATTTATGGTTACAGC |
Before the hybridization assay, membranes were incubated in 2× SSPE–0.1% SDS for 5 min at 60°C and then inserted into the miniblotter apparatus such that the lines of the previously applied oligonucleotides were perpendicular to the sample lanes. Residual liquid was removed by vacuum aspiration. For the hybridization assay, a 40-μl volume of the PCR product was diluted in 300 μl of 2× SSPE–0.1% SDS and boiled for 10 min and cooled on ice. For each sample, 150 μl of diluted PCR product was added to one slot of the miniblotter in duplicate. The entire miniblotter apparatus was then incubated at 60°C. After 1 h, the samples were removed by vacuum aspiration for at least 1 min. The membrane was removed from the miniblotter and washed twice with 25 ml of 2× SSPE–0.5% SDS for 10 min at 60°C.
To detect PCR-amplified spacer hybridization, the membranes were briefly washed with washing buffer (100 mM maleic acid, 150 mM NaCl [pH 7], 0.3% Tween 20), incubated for 30 min in blocking buffer (100 mM maleic acid, 150 mM NaCl [pH 7], 10% [wt/vol] blocking reagent), and then incubated another 30 min with anti-digoxigenin AP antibody (Roche Diagnostic, Meylan, France) diluted at 1/10,000 in blocking buffer. The membrane was washed twice with washing buffer during 20 min, briefly rinsed with revelation buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl), and finally revealed with the nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate. A membrane can be used several times after it has been stripped according to the manufacturer's instructions.
Index of discrimination.
Discriminating power was determined by using the index of discrimination as described by Hunter and Gaston according to the equation
where D is the index of discrimination, S is the number of different profiles, N is the total number of sampled profiles, and n is the number of isolates with the same profile (13).
RESULTS
The presence of the CRISPR locus in the 406 clinical and environmental isolates was assessed by a PCR specific for the DR region, and detection of this region was positive for all of the 233 ST1/Paris pulsotype isolates, 45 of 46 (97.8%) ST1/non-Paris pulsotype isolates, 15 of 15 (100%) non-ST1/Paris pulsotype isolates, and 12 of 112 (10.7%) non-ST1/non-Paris pulsotype isolates (Table 2).
Table 2.
Distribution of CRISPR arrays and numbers of spoligotypes in different L. pneumophila genotypes
| Pulsotype and ST | No. of isolates |
No. of spoligotypes | ||
|---|---|---|---|---|
| Total | CRISPR positive | Spoligotyping positive | ||
| Paris | ||||
| ST1 | 233 | 233 | 233 | 41 |
| Non-ST1 | 15 | 15 | 14 | 4 |
| Non-Paris | ||||
| ST1 | 46 | 45 | 45 | 7 |
| Non-ST1 | 112 | 11 | 0 | 0 |
| All isolates | 406 | 304 | 292 | 46a |
Some spoligotypes were shared by isolates from the four different populations.
The sequencing of CRISPR loci from the first set of ST1/Paris pulsotype isolates allowed the identification of 42 different CRISPR spacer sequences. The spacer content of these isolates ranged from 31 to 42 nucleotides. The 42 spacers were used as probes for the spoligotyping experiments (Table 1).
Spoligotyping was performed on a set of 66 unrelated ST1/Paris pulsotype isolates to assess the ability of this method to discriminate different spoligotypes. Twenty-four different spoligotypes were obtained for these 66 isolates, with an index of discrimination of 84.71% (95% confidence interval, 78.05 to 91.37).
The stability of the marker was also investigated by spoligotyping different subcultures of the reference strain Paris CIP107629, immediately after unfreezing and after 3 or 31 subcultures. The three subcultures had identical spoligotypes that corresponded to the theoretical spoligotype deduced from the reference sequence (NC_006368).
To evaluate whether several spoligotypes could be isolated from the same sample, 24 isolates from the same sputum sample and four sets of isolates from the same water sample were assessed. All isolates from the sputum sample had identical spoligotypes, and the spoligotypes were identical for isolates from three of four environmental samples. The fourth environmental sample had three different spoligotypes among the 35 isolates.
To evaluate the performance of the method during epidemiological investigations, we used our spoligotyping approach to analyze 42 clinical isolates and 58 related environmental isolates from 33 epidemiological investigations (Table 3). Nineteen different spoligotypes were obtained. Spoligotypes of clinical and environmental isolates were concordant for 27 investigations, thus identifying the suspected contamination sources. For 5 of these investigations, the spoligotypes identified isolates that were specific to the corresponding investigation. For the other 22 investigations, the spoligotypes were also identified in other unrelated samples. For 6 investigations, the spoligotypes of the clinical isolates were different from the spoligotypes of the related environmental isolates and did not confirm the environmental source of infection.
Table 3.
Concordance of spoligotypes from clinical and related environmental isolates in 33 epidemiological investigations
| Epidemiological investigation | No. of spoligotypes (no. of isolates) |
No. of common spoligotypesa | Probable identification of environmental source | |
|---|---|---|---|---|
| Clinical isolates | Environmental isolates | |||
| 1 | 2 (6) | 3 (5) | 2 | + |
| 2 | 1 (1) | 1 (3) | 1 | + |
| 3 | 1 (3) | 1 (2) | 0 | − |
| 4 | 1 (1) | 2 (2) | 1 | + |
| 5 | 1 (1) | 1 (2) | 0 | − |
| 6 | 1 (1) | 1 (1) | 1 | + |
| 7 | 1 (2) | 1 (2) | 1 | + |
| 8 | 1 (1) | 1 (2) | 1 | + |
| 9 | 1 (1) | 1 (2) | 1 | + |
| 10 | 1 (2) | 1 (10) | 1 | + |
| 11 | 1 (1) | 1 (2) | 1 | + |
| 12 | 1 (1) | 1 (2) | 0 | − |
| 13 | 1 (1) | 1 (1) | 1 | + |
| 14 | 1 (1) | 1 (1) | 1 | + |
| 15 | 1 (1) | 1 (2) | 1 | + |
| 16 | 1 (1) | 1 (2) | 1 | + |
| 17 | 1 (1) | 1 (2) | 1 | + |
| 18 | 1 (1) | 1 (2) | 1 | + |
| 19 | 1 (1) | 1 (2) | 1 | + |
| 20 | 1 (2) | 1 (1) | 1 | + |
| 21 | 1 (1) | 3 (3) | 1 | + |
| 22 | 1 (1) | 1 (2) | 1 | + |
| 23 | 1 (1) | 1 (1) | 1 | + |
| 24 | 1 (1) | 4 (4) | 0 | − |
| 25 | 1 (1) | 1 (1) | 0 | − |
| 26 | 1 (1) | 1 (2) | 1 | + |
| 27 | 1 (1) | 1 (2) | 1 | + |
| 28 | 1 (1) | 1 (5) | 1 | + |
| 29 | 1 (1) | 1 (3) | 1 | + |
| 30 | 1 (1) | 2 (3) | 1 | + |
| 31 | 1 (1) | 1 (1) | 1 | + |
| 32 | 1 (1) | 1 (1) | 0 | − |
| 33 | 1 (1) | 1 (3) | 1 | + |
Number of spoligotypes that were common between clinical and environmental isolates.
Finally, the other 36 isolates that did not belong to the ST1/Paris pulsotype (ST1/non-Paris pulsotype, non-ST1/Paris pulsotype, and non-ST1/non-Paris pulsotype) were also spoligotyped based upon the CRISPR locus. Positive results were obtained for 24 of these isolates, resulting in 8 different spoligotypes. For the 12 remaining isolates (10 isolates non-ST1/non-Paris pulsotype, 1 ST1/non-Paris pulsotype, and 1 non-ST1/Paris pulsotype), no hybridization was detected. In total, 305 PCR-positive isolates were tested by spoligotyping, resulting in 46 different spoligotypes and 12 negative results (Table 2; Fig. 2). The CRISPR loci from 5 of the 12 non-ST1/Paris pulsotype isolates that were negative for hybridization were sequenced. A total of 124 new spacer sequences were identified from these 5 isolates, and these spacers were all different from the 42 spacers already identified and used as probes in the newly developed ST1/Paris pulsotype spoligotyping assay.
Fig 2.
Schematic representation of all spoligotypes (1, presence of spacer; 0, absence of spacer).
To look for potentially new spacers, 14 supplementary CRISPR loci from ST1/Paris pulsotype isolates, selected based on spoligotype diversity, were fully sequenced; however, no new spacer sequences were detected. The spacer sequence content of these isolates ranged from 7 to 42 nucleotides. For these 14 isolates, the corresponding spoligotypes were identical to the theoretical spoligotype deduced from the sequences obtained. In one isolate, a single nucleotide polymorphism was identified in spacer region 8, but this polymorphism did not impair its detection in the spoligotyping experiment.
DISCUSSION
The diversity of CRISPR spacer sequences has been used as the basis for genotyping several pathogens, and these spoligotyping methods have demonstrated comparable in index of discrimination values to those of the gold standard genotyping methods, such as PFGE, multilocus sequence typing (MLST), amplified fragment length polymorphism (AFLP), or multiple-locus variable-number tandem-repeat analysis (MLVA), demonstrating the usefulness of this method for typing purposes (14, 16, 17, 20). In this study, using the genome of the Paris CIP107629 reference strain, we explored the diversity of CRISPR spacers and developed a spoligotyping tool that was able to discriminate L. pneumophila ST1/Paris pulsotype isolates that were formerly undistinguishable with PFGE and SBT (5). Using this method, 233 identical L. pneumophila ST1/Paris pulsotype strains were distinguished based upon the presence or absence of 42 specific markers that, in theory, allow detection of 242 unique combinations (4.398E12). In our set of analyzed isolates, only 46 different spoligotypes were identified, and these were determined to be of unequal distribution. This was due in large part to the selection of related isolates from epidemiological investigations; however, some unrelated isolates also displayed the same spoligotypes, suggesting an evolutionary link.
In this study, 33 L. pneumophila CRISPR loci were sequenced, 28 from ST1/Paris pulsotype isolates. A total of 42 different spacers were identified from the 28 ST1/Paris pulsotype isolates, forming the set of probes used to develop the spoligotyping method.
The stability of the spoligotype method was confirmed using the reference strain that gave similar spoligotype results after multiple subcultures.
The index of discrimination of the spoligotyping method was first evaluated on 66 unrelated L. pneumophila ST1/Paris pulsotype strains. The index of discrimination of 84.71% was quite low for a usual genotyping method but was in fact very high for isolates exhibiting identical ST and PFGE patterns. Although the index of discrimination was calculated for all 233 ST1/Paris pulsotype isolates, the index of discrimination was still low, 79.72%, despite the larger number of analyzed isolates. This was probably due to an underestimation resulting from the inclusion of related isolates.
The spoligotype diversity of L. pneumophila isolated from the same sample was investigated in four environmental samples and in one clinical sample. The spoligotypes of the isolates from the same clinical sample were similar, but different spoligotypes were observed in one of the environmental samples. The possible diversity of the population of L. pneumophila present in a single sample suggests that to cover this diversity, it will be necessary to genotype several isolates for each sample during epidemiological investigations.
For 6 of 33 epidemiological investigations, the spoligotypes of the clinical isolates were totally different from the spoligotypes determined for related environmental isolates. In these cases, either the correct sources were not investigated or the correct sources were investigated but there were not enough environmental isolates tested to cover the spoligotype diversity of the samples. Indeed, in 5 of these 6 investigations, a limited number of isolates were analyzed with only one or two environmental isolates. For the sixth investigation, several environmental isolates were analyzed, resulting in 5 different spoligotypes, all of which were different from the clinical isolates, suggesting that the source of infection was not identified. For 27 of 33 epidemiological investigations, the spoligotypes of the clinical isolates were in concordance with the spoligotypes found in the environmental isolates of the suspected sources for each investigation. As for other genotyping markers, when epidemiologically related clinical and environmental isolates shared specific patterns, there was a high level of confidence in identifying the contamination sources. This was the case for 5 investigations; however, 19 spoligotypes were identified for isolates from the 33 investigations, indicating that some spoligotypes were shared by several unrelated isolates. Despite the fact that identical spoligotypes were not strictly specific to each investigation, these investigations highlight similarities between related clinical and environmental isolates, at a sublevel compared to the gold standard methods, and reinforce the probability of identifying the suspected source of contamination.
Genomic analyses for the presence of CRISPR loci from the available Legionella genomes showed that the CRISPR locus is not present in all strains (3, 5, 6, 8, 11, 18, 21). In contrast to the consistent detection of CRISPR loci in the 233 ST1/Paris pulsotype isolates, the CRISPR locus was detected in 11 of 112 non-ST1/non-Paris pulsotype isolates, 45 of 46 ST1/non-Paris pulsotype isolates, and 15 of 15 non-ST1/Paris pulsotype isolates. The sequencing of the CRISPR locus of 5 non-L. pneumophila ST1/Paris pulsotype isolates (PCR positive but hybridization negative) revealed totally different spacer sequence content for these strains. A total of 124 new spacers were identified, showing that the negative results obtained by the spoligotyping assay were due not to technical problems but to the absence of sequence similarity between probes and spacers in these strains. The genome sequence of the reference strain 130b revealed a spacer sequence that was a combination of spacers 16 and 42 found in ST1/Paris pulsotype isolates, as well as new spacer sequences. Closely related isolates from the L. pneumophila ST1/Paris pulsotype had similar CRISPR loci, differing only by missing spacer sequences, probably lost by recombination between DR in the absence of selection pressure, while distant isolates had CRISPR loci with different spacer sequence contents.
The absence of a CRISPR locus and the diversity of spacer sequences in the Legionella CRISPR make it difficult to use Legionella spp. as a routine genotyping tool, as has been previously described for Mycobacterium (14). However, when required, genotyping tools based on Legionella spp. can be used to discriminate isolates into subgroups for the correct identification of environmental sources of infections during epidemiological investigations.
A previous study reported subtyping the L. pneumophila Paris pulsotype by detecting insertion sequences via RFLP experiments (RFLP-IS) (22). That study demonstrated that RFLP-IS can also be used to discriminate Paris pulsotype isolates with a discriminatory index similar to that achieved with spoligotyping. These two methods are membrane-based methods dependent on manual hybridization steps; nevertheless, as described for Mycobacterium, L. pneumophila spoligotyping could be improved by switching from membranes to microbead-based hybridization assays (7, 24). Compared to membrane-based assays, these systems allow better standardization of the assays and high-throughput analyses, which should promote the utility of spoligotyping for routinely performed L. pneumophila ST1/Paris pulsotype subtyping.
Taken together, these data demonstrate that spoligotyping can be used to efficiently discriminate L. pneumophila ST1/Paris pulsotypes and could easily be integrated into a fallback genotyping process (Fig. 1B). The L. pneumophila spoligotype marker seems to be stable after several subcultures. The diversity of spoligotypes identified in isolates from a single sample suggests that it may be necessary to test more than one isolate per sample, especially for environmental isolates.
ACKNOWLEDGMENTS
We are grateful to American journal experts for editorial assistance.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Aurell H, et al. 2003. Legionella pneumophila serogroup 1 strain Paris: endemic distribution throughout France. J. Clin. Microbiol. 41:3320–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campese C, et al. 2011. Progress in the surveillance and control of Legionella infection in France, 1998–2008. Int. J. Infect. Dis. 15:e30–e37 [DOI] [PubMed] [Google Scholar]
- 3. Cazalet C, et al. 2010. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet. 6:e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cazalet C, et al. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18:431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cazalet C, et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165–1173 [DOI] [PubMed] [Google Scholar]
- 6. Chien M, et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 7. Cowan LS, Diem L, Brake MC, Crawford JT. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, Spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. D'Auria G, Jimenez-Hernandez N, Peris-Bondia F, Moya A, Latorre A. 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doleans A, et al. 2004. Clinical and environmental distributions of Legionella strains in France are different. J. Clin. Microbiol. 42:458–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ginevra C, et al. 2008. Lorraine strain of Legionella pneumophila serogroup 1, France. Emerg. Infect. Dis. 14:673–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glockner G, et al. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298:411–428 [DOI] [PubMed] [Google Scholar]
- 12. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawrence C, et al. 1999. Single clonal origin of a high proportion of Legionella pneumophila serogroup 1 isolates from patients and the environment in the area of Paris, France, over a 10-year period. J. Clin. Microbiol. 37:2652–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu F, et al. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl. Environ. Microbiol. 77:1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mokrousov I, et al. 2009. Novel macroarray-based method of Corynebacterium diphtheriae genotyping: evaluation in a field study in Belarus. Eur. J. Clin. Microbiol. Infect. Dis. 28:701–703 [DOI] [PubMed] [Google Scholar]
- 18. Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34:281–294 [DOI] [PubMed] [Google Scholar]
- 19. Price EP, Smith H, Huygens F, Giffard PM. 2007. High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat locus of Campylobacter jejuni. Appl. Environ. Microbiol. 73:3431–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schouls LM, et al. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schroeder GN, et al. 2010. Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192:6001–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vergnes M, et al. 2011. Insertion sequences as highly resolutive genomic markers for sequence type 1 Legionella pneumophila Paris. J. Clin. Microbiol. 49:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu VL, et al. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127–128 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Andremont A, Ruimy R, Sola C. 2010. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J. Med. Microbiol. 59:285–294 [DOI] [PubMed] [Google Scholar]