Abstract
Plants have evolved a plethora of responses to cope with phosphate (Pi) deficiency, including the transcriptional activation of a large set of genes. Among Pi-responsive genes, the expression of the Arabidopsis phospholipase DZ2 (PLDZ2) is activated to participate in the degradation of phospholipids in roots in order to release Pi to support other cellular activities. A deletion analysis was performed to identify the regions determining the strength, tissue-specific expression, and Pi responsiveness of this regulatory region. This study also reports the identification and characterization of a transcriptional enhancer element that is present in the PLDZ2 promoter and able to confer Pi responsiveness to a minimal, inactive 35S promoter. This enhancer also shares the cytokinin and sucrose responsive properties observed for the intact PLDZ2 promoter. The EZ2 element contains two P1BS motifs, each of which is the DNA binding site of transcription factor PHR1. Mutation analysis showed that the P1BS motifs present in EZ2 are necessary but not sufficient for the enhancer function, revealing the importance of adjacent sequences. The structural organization of EZ2 is conserved in the orthologous genes of at least eight families of rosids, suggesting that architectural features such as the distance between the two P1BS motifs are also important for the regulatory properties of this enhancer element.
Keywords: enhancer element, gene regulation, phosphate deprivation, phospholipase, root
Introduction
Phosphorus, as a major component of fundamental biomolecules such as ATP, nucleic acids, and phospholipids, influences virtually every developmental and biochemical process in plants. It is also a central metabolic regulator of many cellular processes including energy transfer, modulation of protein activity, and carbon and nitrogen metabolism. Phosphate (Pi), the assimilable form of phosphorus, can be abundant in some soils; however, because of its low mobility in the soil solution and its rapid conversion to organic forms that are not readily available for plant uptake, Pi is often a limiting factor for plant growth in both natural and agricultural ecosystems. As a response to this limitation, plants have evolved a series of developmental, biochemical, and symbiotic adaptive strategies (Lynch, 1995; Raghothama, 1999). At the morphological and developmental level, the strategies involving the root system include changes in growth, branching, and root hair elongation (López-Bucio et al., 2003; Sánchez-Calderón et al., 2005). Additionally, the biochemical strategies include the transcriptional induction of genes encoding high-affinity Pi transporters, the activation of alternative pathways that reduce the need for high-energy phosphate bonds, and the recycling of Pi from membrane phospholipids (Bariola et al., 1994; Baldwin et al., 2001).
Microarrays have been used to study the global transcriptional response to Pi deprivation, showing that several hundreds and up to thousands of genes are responsive to Pi (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005; Morcuende et al., 2007) and that this response is affected by sucrose and cytokinin levels (Wang et al., 2006; Müller et al., 2007). Several components of the signal transduction pathway that mediate the activation of gene expression in response to Pi availability have been described and a number of conserved cis-elements have been identified in the promoters of Pi-responsive genes. The P1BS element (GNATATNC) is a DNA motif present in the promoters of a large subset of Pi-responsive genes in Arabidopsis and corresponds to the binding site of the transcription factor PHR1, a member of the Arabidopsis MYB-CC gene family. The P1BS element is also bound by other members of this family, such as PHR1-LIKE1. Loss-of-function mutations of both genes result in a significant decrease in the induction of a large number of genes responsive to Pi availability and reveal a partial functional redundancy between these transcription factors (Rubio et al., 2001; Bustos et al., 2010). Several other Arabidopsis transcription factors such as WRKY75, ZAT6, and MYB62 have been reported to influence the expression of Pi-responsive genes, suggesting that a complex interaction of transcription factors and their cognate cis-acting elements is involved in the transcriptional regulation of the Pi-starvation response in plants (Devaiah et al., 2007a,b, 2009).
Pi deprivation leads to changes in membrane lipid composition: a decrease in the total content of phospholipids such as phosphatidylcholine, phosphatidylethanolamine, and phosphatidylglycerol, which together constitute about 30% of the total Pi storage molecules in plants, is coupled with an increased rate of synthesis of non-phosphorous lipids, such as the galactolipid digalactosyldiacylglycerol and the sulpholipid sulphoquinovosyldiacylglycerol, presumably to preserve membrane integrity and functionality (Essigmann et al., 1998; Kelly and Dörmann, 2002; Andersson et al., 2003). Synthesis of digalactosyldiacylglycerol and sulphoquinovosyldiacylglycerol requires diacylglycerol, and two alternative pathways have been proposed for the production of this compound from phosphatidylcholine and phosphatidylethanolamine: direct hydrolysis by phospholipase C or a two-step reaction involving phospholipase D (PLD) and a phosphatidic acid phosphatase (Awai et al., 2001; Dörmann and Benning, 2002; Kelly and Dörmann, 2002).
Two members of the PLD gene family, PLDZ1 and PLDZ2, have been shown to be involved in phospholipid remodelling in roots as a response to Pi deficiency. The expression of these genes is strongly induced in Pi-deprived plants both in the shoot and root and the increase in expression of PLDZ2 is regulated at the transcriptional level. Furthermore, expression of the GUS reporter gene under the control of the PLDZ2 promoter shows that it contains regulatory sequences able to direct Pi-responsive gene expression (Nakamura et al., 2005; Cruz-Ramírez et al., 2006).
This work reports a detailed characterization of the promoter region of PLDZ2 gene. In silico analysis shows that it contains five copies of the P1BS motif, whose presence has been associated with the Pi-responsive gene expression of the Arabidopsis Pi transporter genes AtIPS1 and AtPht1;4, the barley Pht1;1, and the Medicago truncatula MtPT1 and MtPT2 (Rubio et al., 2001; Schünmann et al., 2004; Xiao et al., 2006; Karthikeyan et al., 2009). Deletion analysis of the PLDZ2 promoter led to the discovery and characterization of a 65-bp Pi-responsive enhancer element (here named EZ2) containing two P1BS motifs. This enhancer element confers gene expression properties similar to those of the PLDZ2 promoter in terms of the response to Pi starvation and the effect of sucrose and cytokinin levels. Mutation analysis of EZ2 shows that the P1BS elements are necessary but not sufficient for the enhancer function, revealing the importance of adjacent sequences. The structural organization of EZ2 is conserved in the orthologous genes of at least eight families of rosids, suggesting that the distance between the two P1BS motifs is also important for the regulatory properties of this enhancer element.
Materials and methods
Plant material and growth conditions
WT and transgenic lines generated in this study are in the Col0 ecotype background. Seeds from all lines were surface sterilized with 20% (v/v) bleach for 10 min and then germinated and grown under limiting (0 mM) or sufficient (1 mM) NaH2PO4 concentrations on 0.1 MS medium agar plates supplemented with 0.5% sucrose.
PLDZ2 promoter deletion series
In silico analyses of the PLDZ2 promoter region were done using the Signal Scan Search feature of the PLACE database (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999). After, a series of deletions were generated from genomic Arabidopsis DNA by PCR amplification using different primers (Supplementary Table S1, available at JXB online). The PCR products were cloned by recombination in the vector pDONR221 using a GATEWAY BP kit (Invitrogen). They were then transferred into the destination vector pKGWFS7 by recombination using a GATEWAY LR kit to generate transcriptional fusions and drive GFP-GUS expression. To produce the EZ2::GUS construct, the fragment between positions –782 bp and –717 bp relative to the start codon of the PLDZ2 promoter was ligated to the –46 35S minimal promoter generating the minimal Pi-responsive enhancer. A 65-bp oligonucleotide containing the sequence of the enhancer, flanked by KpnI and HindIII sites, was synthesized. The complementary chain was generated using the Klenow fragment of DNA polymerase, digested with KpnI and HindIII, and cloned in pBS 46S carrying the 35S minimal promoter.
The different mutant versions of the enhancer element were obtained using the same strategy, with specifically designed oligonucleotides as templates (Supplementary Table S1). All transcriptional fusions were transformed into Agrobacterium tumefaciens and used for obtaining Arabidopsis transgenic lines following the modification to the floral dip Arabidopsis transformation protocol reported by Martinez-Trujillo et al. (2004). At least 15 independent transgenic lines were produced for each construct and tested by GUS histochemical assays for low Pi responsiveness. Three representative lines covering the range of expression levels for each PLDZ2 deletion construct were selected for detailed characterization.
Protein extraction and fluorometric GUS assays
GUS activities of Arabidopsis transgenic lines were determined in seedlings 10 days after germination (dag). Plant tissues were ground in GUS extraction buffer, and 2 μg protein was used for the fluorometric assay. Protein extracts were incubated in 2 mM 4-methylumbelliferyl-β-D-glucuronide, 50 mM KPO4, pH 7.0, 0.1 % Sarkosyl, 0.1% Triton X-100, 10 mM β-mercaptoethanol, and 10 mM EDTA for 2 h, followed by analysis with a fluorometer (Hoefer Scientific Instruments).
Histochemical localization of GUS expression
Histochemical analysis of GUS activity was performed in GUS reaction buffer (0.5 mg/ml of 5-bromo-4-chloro-3-indolyl-β-D-glucuronide in 100 mM sodium phosphate, pH 7.0). Seedlings were incubated overnight at 37 °C. After the GUS reaction, seedlings were cleared by the method described by Malamy and Benfey (1997) and representative plants were observed and recorded by Nomarski optics in a Leica DMR microscope.
Real-time quantitative reverse-transcription PCR expression analysis
Total RNA of 10 dag seedlings of Col0 and phr1 grown in P-limiting (P–) or P-sufficient (P+) conditions were isolated using an RNAeasy kit (Qiagen). cDNA templates for PCR amplification were prepared from all samples by using reverse specific primers (Supplementary Table S1) and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Each reaction contained cDNA template from 30 μg total RNA, 1× SYBR Green PCR Master Mix (Applied Biosystems), and 500 nM forward and reverse primers. Real-time PCR was performed in an ABI PRISM 7500 sequence detection system (Applied Biosystems) under the following thermal cycling conditions: 10 min at 95 °C and 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s. For quantitative reverse-transcription PCR, relative transcript abundance was calculated and normalized with respect to actin to minimize variation in cDNA template levels, with the Col0 P+ sample acting as calibrator with a nominal value of 1. The data shown represent mean values obtained from at least three independent amplification reactions. All calculations and analysis were performed using 7500 Software v2.0.1 (Applied Biosystems) and the 2−ΔΔCt method with a relative quantification confidence set at 95 % (Livak and Schmittgen, 2001). Amplification efficiency (0.6611–1.0834) for the primer sets was determined by amplification of cDNA dilution series (1:5). Specificity of the reverse-transcription PCR products was followed by a melting curve analysis with continual fluorescence data acquisition during the 65–95 °C melt.
Reverse-transcription PCR expression analysis
Plantlets carrying the PLDZ2::GUS and EZ2::GUS constructs were used. Total RNA was isolated from seedlings grown for 7 dag in P-limiting conditions and transferred to Murashige and Skoog liquid medium supplemented with Pi (1 mM) for 30 min, 60 min, 3 h, 6 h, 12 h, 24 h, and 48 h. Total RNA was extracted by using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized using 100 ng of total RNA with SuperScript III reverse transcriptase, according to the manufacturer’s instructions. Specific primers were used for PLDZ2 and GUS transcript detection (Supplementary Table S1). Amplification reactions were performed under the following conditions: 94 °C for 3 min and 28 cycles at 94 °C for 30 sec, 60 °C for 30 sec, and a final extension step at 72 °C for 30 sec. Amplification reactions for ACTIN2 used 25 cycles.
Sucrose and benzilaminopurine analysis
Seedlings of PLDZ2::GUS and EZ2::GUS were germinated and grown for 7 days under P-sufficient and P-limiting conditions, and transferred to Murashige-Skoog liquid medium with (P+) and without Pi (P–), supplemented with different concentrations of sucrose (0, 0.5, 1, 3, and 5%) or benzilaminopurine (BAP; 10−7, 10−6, 10−5 M). Seedlings were harvested after 48 h and GUS activity determined by the fluorometric assay.
Sequences and sequence alignments
The promoter region and the predicted amino acid sequence of the Arabidopsis PLDZ2 gene (AT3G05630) were obtained from the Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org). A BLAST search using the amino acid sequence was performed in the database Phytozome (http://www.phytozome.net) in order to identify putative orthologues of PLDZ2 and obtain the promoter region sequences. The locus names in Phytozome for these orthologous genes are as follows: 477854 (Arabidopsis lyrata), Glyma20g38200 (Glycine max), Medtr1g100110 (Medicago truncatula), 30128.t000330 (Ricinus comunis), Cucsa.090930 (Cucumis sativus), cassava4.1_031321m (Manihot esculenta), POPTR_0013s01380 (Populus trichocarpa), Eucgr.H03262 (Eucalyptus grandis), ppa000572m (Prunus persica), orange1.1g.001225m (Citrus sinensis), and clementine0.9_000905m (Citrus clementina). The sequence of the putative orthologue of PLDZ2 in Phaseolus vulgaris is part of the Bean Genome Sequencing Project (PhasIbeAm Team) and was kindly provided by Dr. Alfredo Herrera-Estrella. Sequence searches and alignments were performed with the Lasergene software package (DNASTAR, Madison, WI, USA), with manual adjustment when necessary.
Results
The PLDZ2 promoter contains regulatory regions that influence the response to Pi deprivation
PLDZ2 has been defined as a Pi-responsive gene based on the strong induction shown in Pi-deprived seedlings and the reversion of this response after a short-term resupply of Pi, indicating that this gene is involved in direct responses to Pi availability (Cruz-Ramírez et al., 2006; Morcuende et al., 2007). It has been previously reported that a 1,232-bp fragment of the PLDZ2 promoter region is capable of directing low-Pi-inducible expression of the GUS reporter gene in transgenic Arabidopsis plants (Cruz-Ramírez et al., 2006). To determine whether the GUS reporter gene fusion displays a reversible induction similar to the endogenous PLDZ2 gene, the current study analysed the effect of Pi resupply on the level of PLDZ2 and GUS transcripts in PLDZ2::GUS transgenic seedlings germinated and grown for 7 days in medium lacking Pi. Reverse-transcription PCR analysis showed that the decrease of both PLDZ2 and GUS transcripts is evident 1 h after transfer to Pi-sufficient medium and reaches a similar level to that detected in seedlings grown continuously in high-Pi medium 48 h after the transfer (Supplementary Fig. S1). These results showed that changes in the transcript abundance of PLDZ2 in response to the Pi status are largely controlled at the transcriptional level and that the promoter fragment used for the gene construct contains the cis-acting elements responsible for the Pi-regulated expression of this gene.
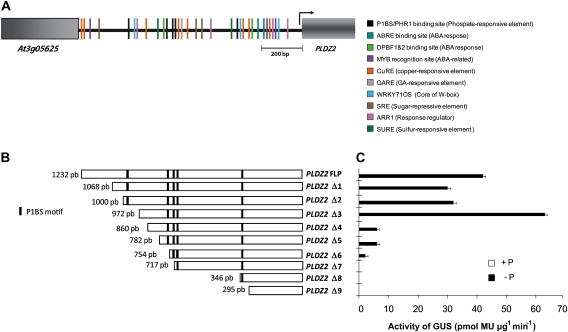
In silico analysis of the promoter fragment showed that it contains five copies of the P1BS motif (GNATATNC; Fig. 1A). To gain further insight into the relative importance of these elements or any other region for proper promoter function in response to Pi availability, a set of nine 5′ deletions of the 1,232-bp PLDZ2 promoter fragment was constructed with 5'-end points at positions –1,068 (Δ1), –1,000 (Δ2), –972 (Δ3), –860 (Δ4), –782 (Δ5), –754 (Δ6), –717 (Δ7), –346 (Δ8), and –259 (Δ9) relative to the start codon (Fig. 1B). The PLDZ2 promoter deletions were generated by PCR using specific primers (Supplementary Table S1), sequenced, and inserted upstream of the GUS gene in the plant promoter probe vector pKGWFS7 (Karimi et al., 2002) and used to produce Arabidopsis transgenic lines. At least ten independent transgenic lines were obtained for each promoter construct and the Pi-regulated expression of the reporter gene was determined for at least three lines (Supplementary Table S2). The results for one representative line for each construct are presented in the following experiments.
Fig. 1.
Deletion analysis of the PLDZ2 promoter. (A) Structure of the 1,232 bp PLDZ2 promoter showing potential transcription binding DNA motifs. (B) PLDZ2 full-length promoter and end-points of progressive promoter deletions. Promoter lengths are indicated with respect to the start codon. Positions of putative P1BS elements are shown as black vertical lines. (C) GUS specific activity determined by fluorometric assays for a representative line of each promoter deletion construct. Values indicate the mean activity obtained from four independent replicate reactions. SD for each value is shown. (This figure is available in colour at JXB online.)
To test the capacity of the different PLDZ2 promoter fragments to respond to Pi availability, 10-day-old seedlings germinated in either high (1 mM) or low (0 mM) Pi medium were analysed for GUS activity by a quantitative fluorometric assay (Fig. 1C). As shown in Supplementary Table S2, under Pi-limited conditions the complete PLDZ2 promoter displayed an average of 1,000-fold induction relative to high-Pi conditions. All promoter versions presented a very low level of GUS activity in high-Pi medium. Deletion constructs –1,068 (Δ1), –1,000 (Δ2), and –972 (Δ3) did not significantly decrease the low-Pi-inducible expression of the GUS reporter gene when compared to the complete promoter, even though one of the P1BS elements had been removed in construct –972 (Δ3) (Fig. 1C and Supplementary Table S2). Constructs –860 (Δ4) and –782 (Δ5), both containing four P1BS elements, presented a significantly lower level of GUS activity in low-Pi medium compared to longer promoter fragments; however, they still showed over 100-fold induction relative to high-Pi conditions. Deletion –754 bp (Δ6), which still contains three P1BS motifs, directed a reduced level of expression in comparison to previous deletions, although it was still reproducibly induced by low Pi. The remaining constructs –717 (Δ7), –346 (Δ8), and –259 (Δ9), which contain two, one, and no P1BS elements respectively, did not show significant levels of GUS activity in either high or low Pi. These results showed that different regions of the PLDZ2 promoter play distinct roles in regulating Pi-responsive gene expression.
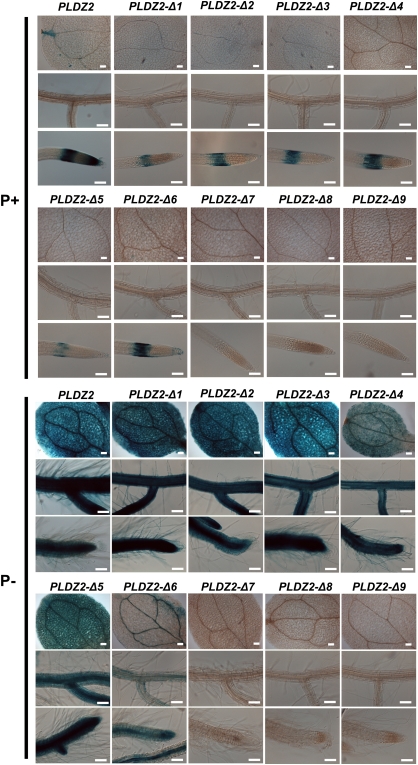
Histochemical GUS analysis of the above-mentioned transgenic lines was performed to determine the promoter regions that influence the tissue-specific expression driven by the full-length PLDZ2 promoter. Ten-day-old seedlings from each line were examined and representative images are shown in Fig. 2. When grown under high-Pi conditions, the transgenic lines harbouring the full-length PLDZ2 promoter presented GUS staining only in the root tip, particularly in the meristematic and elongation zones, whereas in low Pi a strong signal was detected in all tissues of cotyledons and roots. Deletion constructs –1,068 (Δ1), –1,000 (Δ2), and –972 (Δ3) showed patterns of expression very similar to that displayed by the full-length promoter both under low- and high-Pi conditions, except for the loss of GUS staining in the meristematic zone under high-Pi conditions. Promoter versions –860 (Δ4) and –782 (Δ5) also showed GUS expression in the root elongation zone when grown in high Pi, and the pattern of staining observed under low-Pi conditions was similar to that of the full-length promoter, albeit the intensity was comparatively lower in the cotyledons (Fig. 2). Deletion construct –754 (Δ6) in high-Pi medium displayed a expression pattern similar to that of longer promoter versions; however, even though it still showed induction under low-Pi conditions, the staining observed in the root was notably lower and that detected in the cotyledons was restricted to the vascular tissue. Shorter versions of the PLDZ2 promoter did not show any detectable GUS staining even after long incubation times, regardless of the Pi conditions (Fig. 2). Altogether, these results suggested that the region between positions –782 and –717 contains elements that are important not only for the low-Pi-inducible expression but also for the spatial expression pattern directed by the PLDZ2 promoter.
Fig. 2.
Effect of Pi availability on the tissue-specific expression of the PLDZ2 promoter and deletion derivatives. Arabidopsis seedlings of a representative line harbouring PLDZ2::GUS and truncated constructs grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)
PHR1 participates in the regulation by Pi availability of the PLDZ2 promoter
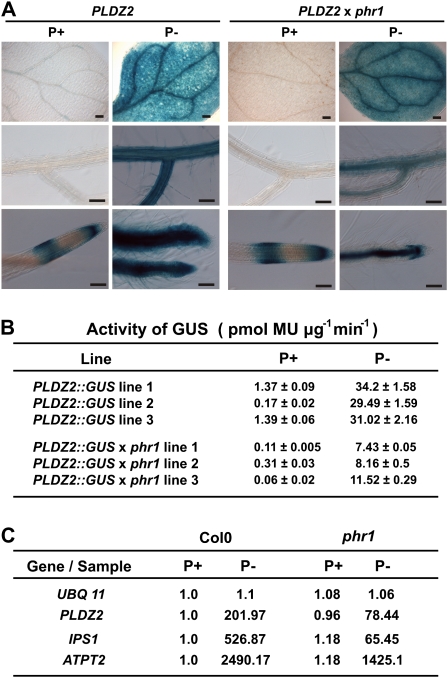
PHR1 is a key regulatory component of the stress responses to Pi deficiency in Arabidopsis. It binds to a motif designated P1BS, which is present in the promoters of many Pi-starvation-induced genes (Rubio et al., 2001). Although the PLDZ2 promoter contains five P1BS motifs, the deletion analysis showed that the two P1BS elements most proximal to the transcription start site are not sufficient for conferring low-Pi responsiveness, whereas the most distal element seems to be dispensable for promoter activity. To determine whether PHR1 participates in the regulation of PLDZ2, presumably through binding to its cognate sequences in the promoter region, the PLDZ2::GUS gene construct was introduced into the Arabidopsis phr1 mutant background. This was achieved through genetic crosses of the wild-type transgenic lines with the phr1 mutant. The results for three lines homozygous for both the phr1 mutation and the gene construct are shown in Fig. 3. Histochemical GUS staining showed that the spatial expression pattern driven by the PLDZ2 promoter in both the phr1 mutant and the wild-type backgrounds was essentially the same, except that staining in roots of phr1 seedlings was restricted to the vascular cylinder when growing in low-Pi medium (Fig. 3A). When GUS activity was determined by the fluorometric assay, it was found that low-Pi responsiveness of PLDZ2::GUS in the phr1 mutant background was reduced about 3–4-fold compared to that observed in the wild type (Fig. 3B). This reduction in the transcriptional activation of PLDZ2::GUS in the phr1 mutant background under low-Pi conditions was similar to that observed for the endogenous PLDZ2 transcript as determined by quantitative reverse-transcription PCR (Fig. 3C). A similar effect of the loss-of-function mutation of PHR1 was observed for the low-Pi-induced expression of IPS1 and AtPT2 (also known as AtPht1;4), genes that are reported as responsive to Pi starvation and whose promoters are known to possess P1BS motifs (Nilsson et al., 2007; Bustos et al., 2010) (Fig. 3C).
Fig. 3.
Expression of PLDZ2::GUS in Arabidopsis Col0 and phr1. (A) Tissue-specific expression of PLDZ2::GUS in 10-day-old seedlings grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. (B) GUS specific activity of PLDZ2::GUS in Col0 and phr1 seedlings grown under P-sufficient and P-limited conditions. (C) Quantitative reverse-transcription PCR analysis of PLDZ2, IPS1, and ATPT2 in Col0 and phr1. Expression levels are reported as relative expression of the corresponding gene in Col0 grown under Pi-sufficient conditions. UBQ 11 was used as a non-responsive control. (This figure is available in colour at JXB online.)
The PLDZ2 promoter region contains a Pi-responsive enhancer element
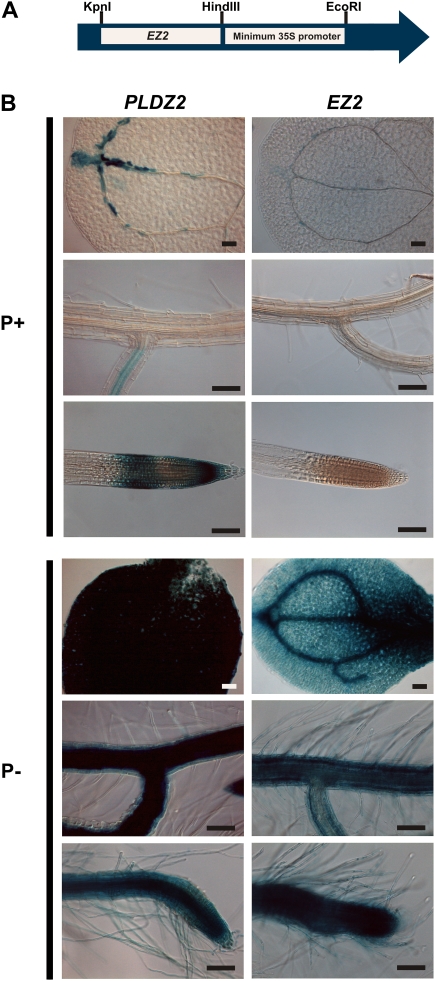
The results of deletion analysis identified the region between positions –782 and –717 as important for the low-Pi responsiveness of the PLDZ2 promoter. It was interesting to determine whether this region contained some type of Pi-responsive enhancer element. With this aim, this study constructed a chimeric promoter consisting of the 65-bp sequence from positions –782 to –717 of the PLDZ2 promoter fused to an inactive –46 cauliflower mosaic virus 35S minimal promoter and inserted it upstream of the GUS reporter gene. The resulting construct was denominated EZ2::GUS (Fig. 4A). Transgenic Arabidopsis plants harbouring the construct were produced and evaluated for GUS activity and responsiveness to Pi availability. GUS histochemical assays were carried out to determine the spatial pattern of expression directed by the EZ2 element. When seedlings were grown in high-Pi medium, expression was detected in the vasculature of cotyledons, a pattern similar to that observed for the complete promoter; however, unlike that observed for PLDZ2 promoter, under these conditions no GUS activity was detected in the root tip of EZ2::GUS plants (Fig. 4B). A considerable increase in GUS staining was observed in the vasculature and mesophyll cells of cotyledons and all root tissues of EZ2::GUS plants when grown in low-Pi medium (Fig. 4B). Quantitative fluorometric GUS assays showed that even though the GUS activity detected in EZ2::GUS plants grown in low-Pi medium was significantly lower to that observed in PLDZ2::GUS plants, the former still displayed an average of 100-fold induction relative to high-Pi conditions (Supplementary Table S3).
Fig. 4.
Pi-starvation responsiveness of EZ2, an enhancer element present in the PLDZ2 promoter. (A) Structure of a chimeric promoter composed of a PLDZ2 Pi-responsive enhancer inserted upstream of the –46 35S minimal promoter to direct expression of GUS (EZ2::GUS). (B) Arabidopsis seedlings of a representative line harbouring PLDZ2::GUS and EZ2::GUS constructs grown for 10 days in media supplemented with 1 mM (P+) or 0 mM (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)
To determine whether this EZ2 enhancer reflected the regulatory properties of the complete PLDZ2 promoter in response to Pi deficiency, this study investigated the kinetics of induction of GUS activity in PLDZ2::GUS and EZ2::GUS transgenic lines grown in low-Pi medium. Under these conditions, GUS activity increased noticeably in both transgenic lines at 4 dag, reaching a maximum between days 8 and 10 and declining in both lines by 12 dag (Supplementary Fig. S2).
Taken together, these results showed that EZ2, the enhancer element located between positions –782 and –717 of the promoter region of PLDZ2 gene, is capable of conferring low-Pi-inducible expression to a heterologous minimal promoter in a manner similar to the complete PLDZ2 promoter.
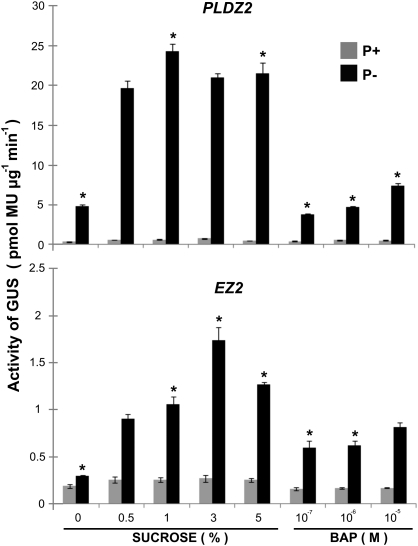
The low-Pi responsiveness of the PLDZ2 promoter and the EZ2 element is modulated by sucrose and cytokinins
It has been previously shown that the Pi-starvation response is affected by sucrose and cytokinins (Martín et al., 2000; Hou et al., 2005; Karthikeyan et al., 2007; Müller et al., 2007; Lei et al., 2011). Sucrose or photosynthates are required to activate the expression of Pi-responsive genes, and cytokinins act as antagonists of this response. To test whether the low-Pi-induced expression directed by the complete PLDZ2 promoter and the EZ2 enhancer element is similarly affected by these compounds, this study tested the effect of sucrose and the cytokinin BAP on the level of GUS activity of PLDZ::GUS and EZ2::GUS seedlings. For this purpose, plants were grown for 7 days in either low- or high-Pi medium and then transferred to the same medium supplemented with different concentrations of either sucrose or BAP for 2 additional days. When grown in high-Pi medium, no significant increase in GUS activity was observed, independently of the concentration of sucrose or BAP added to the medium (Fig. 5). However, when grown on medium lacking Pi, the increase in the level of GUS activity was dependent on the concentration of sucrose for both gene constructs. When different concentrations of sucrose were added to the low-Pi medium, the increase in GUS activity for both the PLDZ2::GUS and EZ2::GUS constructs was significantly higher compared to that observed in medium containing no sucrose (Fig. 5). It is interesting to note that when no sucrose is added, PLDZ2::GUS still showed about 15-fold induction in response to Pi starvation, while EZ2::GUS showed no significant difference in the level of GUS activity between high- and low-Pi conditions, which indicated that the low-Pi responsiveness of this enhancer element is strongly dependent on sucrose supply.
Fig. 5.
Effect of sucrose and benzilaminopurine (BAP) on the expression of PLDZ2::GUS and EZ2::GUS. Seven-day-old PLDZ2::GUS and EZ2::GUS seedlings grown in 1 mM (P+) or 0 mM Pi (P–) solid medium were transferred into liquid medium with the same concentration of Pi supplemented with different concentrations of sucrose (0, 0.5, 1, 3, and 5%) or BAP (10−7, 10−6, and 10−5 M). Seedlings were harvested after 48 h of treatment for fluorometric GUS assay. Error bars represent SD of four replicates. Asterisks indicate significant differences with respect to 0.5% sucrose group (P < 0.05; ANOVA and Tukey analyses).
Since the addition of sucrose was necessary for both gene constructs to achieve high levels of GUS activity, medium containing 0.5% sucrose was used to observe the negative effect of cytokinins. Thus, 0.5% sucrose should be considered the control condition when analysing the effect of BAP in Fig. 5. Addition of different concentrations of BAP resulted in almost 80% reduction of GUS activity in PLDZ2::GUS seedlings, whereas a maximum reduction of about 30% was observed in EZ2::GUS seedlings after treatment with this cytokinin.
These results showed that the transcriptional activation directed by the PLDZ2 promoter in response to Pi starvation requires sucrose to achieve high levels of induction and that this activation is negatively affected by the addition of cytokinins. Interestingly, these compounds also modulated the low-Pi-induced expression directed by the EZ2 enhancer, indicating that the signalling pathways for Pi deficiency, sucrose supply, and cytokinin production converge in this 65-bp regulatory element.
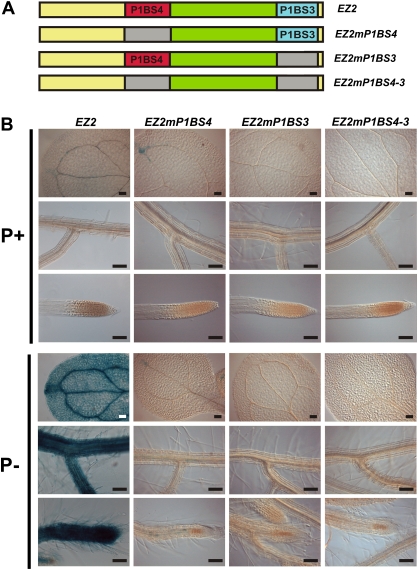
Two P1BS elements are necessary for the activity of EZ2
Since the EZ2 element contains two P1BS motifs, it was important to determine whether both sites were necessary for the enhancer activity of this promoter region. The P1BS motifs present in EZ2 were numbered according to their positions relative to the start codon in the context of the complete PLDZ2 promoter: P1BS3 (starting at position –727) and P1BS4 (starting at position –761). Diverse versions of the EZ2 element were constructed in which P1BS3, P1BS4, or both were eliminated by mutation of the consensus site (Fig. 6A). Interestingly, GUS activity detected by the histochemical (Fig. 6B) and fluorometric (Supplementary Table S3) assays showed that elimination of either of the two P1BS motifs completely abolished the activity of EZ2, suggesting that both are required to have a functional enhancer element. As expected, elimination of both P1BS sites also resulted in an inactive enhancer element (Fig. 6B and Supplementary Table S3).
Fig. 6.
Two P1BS motifs are necessary for the function of the EZ2 enhancer. (A) Diagram showing the structure of the EZ2 enhancer element and its derivatives with mutations in P1BS4 (EZ2mP1BS4), P1BS3 (EZ2mP1BS3), or both (EZ2mP1BS4–3). (B) Seedlings grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)
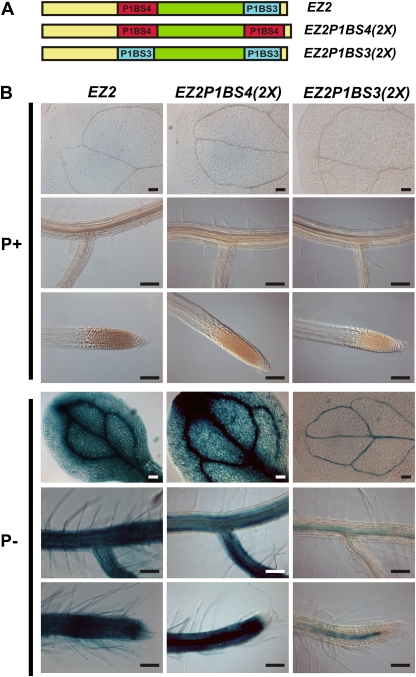
Although differing by one nucleotide, the sequences of both P1BS4 (GAATATTC) and P1BS3 (GGATATTC) have been previously reported to be binding sites for PHR1 (Rubio et al., 2001). To determine whether these two P1BS motifs are functionally equivalent, this study generated versions of EZ2 in which either the P1BS4 or P1BS3 elements were duplicated, denominated EZ2P1BS4(2X) and EZ2P1BS3(2X), respectively, while the rest of the sequence remained unchanged (Fig. 7A). Seedlings of transgenic lines carrying either of the constructs showed induction of GUS activity in response to Pi starvation. However, plants harbouring the EZ2P1BS4(2X)::GUS gene construct showed stronger and broader spatial distribution of GUS staining than the native enhancer. Quantitative determination of GUS activity revealed that this construct conferred on average 3-fold higher levels of GUS activity than EZ2 under low-Pi conditions (Fig. 7B and Supplementary Table S3). On the other hand, EZP1BS3(2X)::GUS seedlings showed weak GUS staining mainly in vascular tissues of cotyledons and roots and an average of 4-fold lower level of GUS activity compared to EZ2 (Fig. 7B and Supplementary Table S3). These results showed that although both P1BS4 and P1BS3 are sufficient to direct low-Pi-inducible expression, they are not equivalent in terms of the transcriptional strength conferred by each motif to the modified versions of EZ2.
Fig. 7.
P1BS motifs in EZ2 have different properties. (A) Diagram showing the structure of the EZ2 enhancer element and its derivatives containing either two copies of P1BS4 [EZP1BS4(2X)] or P1BS3 [EZP1BS3(2X)] motifs. (B) Seedlings grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)
Sequence elements other than the P1BS motifs are important for EZ2 functionality
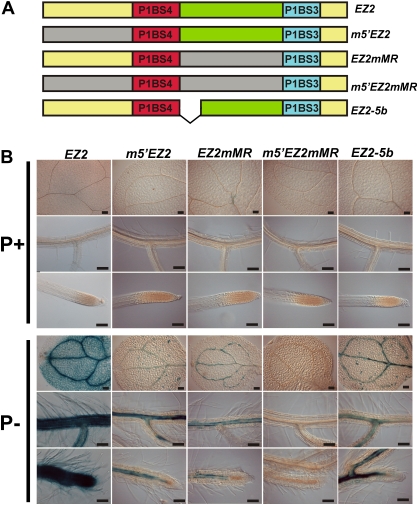
Although the P1BS motifs are the most conspicuous regulatory elements present in EZ2, it was important to determine whether additional sequences constitute functional components of this enhancer element. With this aim, this study produced transgenic Arabidopsis plants harbouring mutant versions of EZ2 in which the 5' region adjacent to the P1BS4 motif (m5'EZ2), the spacer sequence between the two P1BS elements (EZ2mMR), or both (m5'EZ2mMR) were replaced by randomly generated sequences lacking palindromes and with no homology to the corresponding native sequences (Fig. 8A). In high-Pi conditions, none of the constructs showed any GUS activity detectable by histochemical staining in either cotyledons or roots (Fig. 8B). Under low-Pi conditions, both the m5'EZ2 and EZ2mMR versions presented GUS staining in the vascular tissue of cotyledons and roots, although the intensity of the staining was much lower than that observed for the native enhancer element. Fluorometric GUS assays provided quantitative data confirming these observations, showing that both constructs present about 5-fold lower activity than the EZ2 element. Even though the GUS activity was comparatively lower than that observed for the native element, it is noteworthy that both altered versions of EZ2 were still able to display a significant response to Pi deficiency (Supplementary Table S3). In the same conditions, no GUS activity could be detected for m5'EZ2mMR in either high or low Pi media (Fig. 8B and Supplementary Table S3).
Fig. 8.
Role of the sequences adjacent to the P1BS motifs in the function of the EZ2 enhancer. (A) Diagram showing the structure of the EZ2 enhancer element and its mutant derivatives where deletions and substitutions were introduced. The region upstream of P1SB4 (m5'EZ2), the spacer sequence between P1BS4 and P1BS3 (EZ2mMR), or both the upstream and spacer sequences (m5'EZ2mMR) were replaced by random DNA sequences and the distance between the P1BS sites was reduced by 5 bp (EZ2-5b). (B) Seedlings grown for 10 days in media supplemented with 1 mM (P+) or 0 mM Pi (P–), subjected to histochemical GUS assays, and photographed using Nomarsky optics. The name of each construct is indicated at the top of each set of three images, which show the expression pattern for cotyledons, lateral roots, and root apical meristems from top to bottom, respectively. Bars, 100 μm. (This figure is available in colour at JXB online.)
The arrangement of DNA motifs present in EZ2 shows characteristics of a composite regulatory element. In a composite element, there are usually distance constraints between neighbouring sites, which facilitates specific protein–protein interactions between transcription factors. In some cases, even a small difference in the distance between binding motifs results in dramatic changes in the regulatory properties of the element (Makeev et al., 2003). To determine whether the distance between the P1BS motifs in EZ2 is important for the activity of this element, this study produced a construct in which the spacer length was reduced by 5 bp (EZ2-5b; Fig. 8A). As observed for other constructs, this version of the enhancer element showed no GUS activity detectable by histochemical staining when seedlings were grown in high-Pi medium, whereas when grown under Pi-limiting conditions, EZ2-5b seedlings presented GUS staining in the vascular tissue of cotyledons and roots. The intensity of this staining was perceptibly lower than that observed for the native EZ2 element (Fig. 8B). Quantitative measurements of GUS activity confirmed this observation and showed that this construct presents a reduction in activity more than 3-fold compared to the EZ2 element (Supplementary Table S3).
Collectively, these results showed that sequences in the flanking regions of the two P1BS motifs are important to determine the functionality and transcriptional strength of EZ2. These results also suggested that proper spacing between the P1BS motifs is an important structural feature affecting the activity of the EZ2 element.
The effect of sucrose supply on the transcriptional activation directed by the EZ2 mutant versions was also tested (Supplementary Fig. S3). As mentioned above, the low-Pi responsiveness of the native EZ2 element was strongly dependent on sucrose supply, denoted by an increase of GUS activity when 0.5% sucrose was added to the low-Pi medium. This induction of GUS activity was even stronger when 3% sucrose was used. Responsiveness to Pi starvation of mutant version m5'EZ2mMR was completely impaired regardless of the sucrose concentration in the medium, and a very small induction was observed for mutant versions EZ2mMR and EZ2-5b when sucrose was added. Interestingly, GUS activity in seedlings harbouring the mutant version m5'EZ2 increased in both high- and low-Pi conditions in the presence of sucrose, although the response to Pi starvation was still significantly higher.
These results suggested that a negative element may be involved in the response to sucrose which, when eliminated, increases the expression of the EZ2 enhancer element in the presence of sucrose even under conditions of sufficient Pi.
Structural organization of EZ2 is phylogenetically conserved
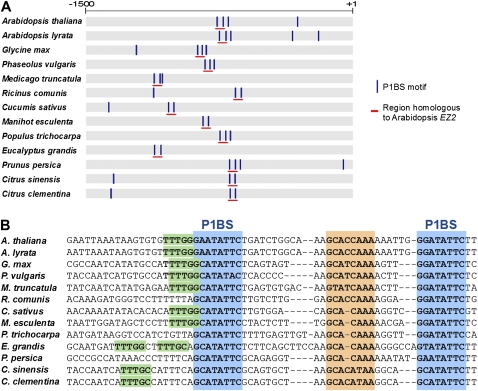
Mutation analysis of EZ2 showed that this enhancer element functions as a cis-regulatory module, since it is an arrangement of adjacent cis-acting elements that operate collectively to implement a higher-order regulatory function. This arrangement has been demonstrated to function independently in gene regulation as a Pi-starvation-responsive enhancer. It was interesting to determine whether this element has been maintained as a single arrangement in evolution, providing further support of its functional significance. The predicted amino acid sequence of PLDZ2 was used with BLAST in the Phytozome website to search for homologous protein sequences in the database and retrieve the genomic sequences of orthologous genes. This study examined the presence and distribution of P1BS motifs in the promoter regions of these genes (Fig. 9A), and segments that contained two P1BS motifs in an arrangement similar to that observed in EZ2 were used for alignment. Interestingly, the modular arrangement observed in EZ2 is conserved in the promoter regions of genes orthologous to PLDZ2 from 11 genera belonging to eight families of rosids (Fig. 9B). Promoters of PLDZ2 orthologues from monocots and other dicot clades were also analysed, but this arrangement of two P1BS motifs was not found outside of rosids. The alignment of orthologous sequences revealed not only the conservation of the P1BS motifs, all of them matching the consensus sequence GNATATNC, but also the conservation of the distance between these elements, with a length varying between 21 and 28 bp. A conserved motif in the spacer region with the sequence GCACAAA or GCAYCAAA and a motif in the 5′ region of EZ2 with the sequence TTTGG or TTTGC could also be identified (Fig. 9).
Fig. 9.
Sequence comparison of the PLDZ2 promoter with orthologous promoter regions. (A) Schematic representation of the promoter region of orthologues for PLDZ2 showing the location of the P1BS motifs (vertical lines) and the region corresponding to the EZ2 element (horizontal lines). Scale represents location in bp relative to the start codon. (B) Sequence comparison of the EZ2 element with similar regions found in orthologous promoter sequences. P1BS motif and other conserved sequences are shaded. Gaps (–) were included for clarity of the alignment. (This figure is available in colour at JXB online.)
EZ2 is functional in a heterologous system
It has been shown that several plant species have in common a large group of genes inducible by Pi deprivation. In the case of common bean (Phaseolus vulgaris), it has been reported that PHR1 also plays an important role in the regulation of Pi-responsive genes (Valdés-López et al., 2008), and this work shows that EZ2 is conserved in the promoter region of the gene orthologous to PLDZ2 in bean. To test whether the PLDZ2 promoter and the EZ2 enhancer are functional and maintain Pi-responsive regulatory properties in a plant species other than Arabidopsis, this study used the transgenic hairy root system of common bean and introduced the constructs PLDZ2::GUS and EZ2::GUS. In both cases, no GUS activity was detected in roots cultivated in Pi-sufficient liquid medium. In contrast, GUS staining in the transgenic hairy roots for both constructs was significantly induced when cultivated in liquid medium lacking Pi (Supplementary Fig. S4). These data showed that the molecular mechanism that regulates the Pi-responsive activity of the EZ2 enhancer element is functionally conserved between Arabidopsis and bean.
Discussion
The hydrolysis of phospholipids is one of the biochemical mechanisms that plants activate during Pi deprivation to recycle Pi from storage compounds, leading to a decrease of this membrane lipids and the concomitant increase of non-phosphorous lipids like digalactosyldiacylglycerol (Essigmann et al., 1998; Kelly and Dörmann, 2002; Andersson et al., 2003). The PLDZ2 gene encodes a PLD that participates actively in the hydrolysis of phospholipids to release Pi and provide diacylglycerol for galactolipid synthesis under Pi starvation. It has been previously shown that the expression of PLDZ2 is specifically regulated by Pi availability at the transcriptional level (Cruz-Ramírez et al., 2006).
The P1BS motif is the cognate sequence of PHR1, and this element has been found to be over-represented in the promoters of a large subset of Pi-responsive genes, demonstrating the importance of this cis-regulatory element in mediating Pi-starvation responses (Bustos et al., 2010). The PLDZ2 promoter contains five P1BS motifs; therefore these sequences were the ideal candidates for elements determining the regulatory properties of this promoter. Since it has been reported that not all P1BS motifs present in a promoter are equally relevant for Pi-starvation responses (Bustos et al., 2010), deletion analysis of the PLDZ2 promoter was performed to determine the relative importance of the different P1BS motifs or any other region in the response to Pi deficiency. The deletion analysis shows that the promoter region between positions –972 and –860 is important in determining the promoter strength, but it is dispensable for its ability to respond to low-Pi conditions. Interestingly, in the region between positions –782 and –717 there are two closely associated P1BS motifs, and removal of this segment results in a complete loss of responsiveness to low Pi, suggesting these motifs or additional regulatory elements contained in that region are important in mediating the induction of PLDZ2 in response to Pi deprivation. Thus, according to this analysis, the most distal P1BS element relative to the start codon is dispensable for the Pi-responsive promoter function and the two most proximal are not sufficient to confer transcriptional activation. This indicates that not all P1BS motifs present in the PLDZ2 promoter have profound effects on its transcriptional activity. The importance of PHR1 in mediating Pi-starvation responsiveness was demonstrated introducing the PLDZ2::GUS construct into the phr1 mutant background, where the induction of gene expression displayed by PLDZ2 was reduced significantly. It is clear from this result that PHR1 has an important role in the regulation of the transcriptional activation of PLDZ2 via elements found in the promoter region, presumably the P1BS motifs. Nevertheless, it is very likely that additional factors participate in the Pi-deficiency response of PLDZ2, since the low-Pi-inducible expression is not completely abolished in the mutant background. This could reflect partial gene redundancy, since PHR1 belongs to a gene family with over a dozen members, some of which display similar DNA-binding properties (Bustos et al., 2010; Castrillo et al., 2011). In fact, it has recently been reported that another member of the PHR1 gene family, PHR1-LIKE1, also plays an important and partially redundant role in the control of plant responses to low-Pi conditions, demonstrated by the additive or synergistic effects of mutations in these two genes on the transcriptional responsiveness to Pi starvation (Bustos et al., 2010).
The results from the deletion analysis allowed the identification of EZ2, a 65-bp element located between positions –782 and –717. Gain-of-function analysis of EZ2 showed that this sequence acts as an enhancer element and resembles the regulatory properties of the complete PLDZ2 promoter, displaying a significant low-Pi-inducible expression and a spatial and temporal expression pattern similar to the full-length promoter. This enhancer element is also responsive to compounds known to modulate low-Pi-induced gene expression, namely sucrose and cytokinins. When no sucrose is added to the low-Pi medium, the PLDZ2 promoter is still able to direct a significantly high level of induction, which increases when the medium is supplemented with sucrose. In contrast, EZ2 displays a stronger dependence on sucrose supply, since no significant induction is observed in the absence of exogenous sucrose. The transcriptional activation directed by both the complete promoter and the enhancer element in low-Pi conditions is modulated negatively by cytokinins. These results show that EZ2 is not only necessary but sufficient to direct low-Pi-induced expression and that sucrose and cytokinin responsiveness also reside in the cis-acting motifs that constitute this 65-bp enhancer element.
EZ2 contains two closely associated P1BS motifs (here designated P1BS3 and P1BS4) whose presence is absolutely necessary for the enhancer function. Both elements match the consensus P1BS motif and differ only in one nucleotide. Despite this apparently small difference, these motifs are not functionally equivalent in terms of the strength conferred when duplicated in the context of EZ2. When duplicated, P1BS4 is a stronger motif directing an even higher transcriptional activation than the native EZ2 enhancer. P1BS4 is a perfect palindromic sequence, which is not the case for P1BS3. It is possible that PHR1 and related transcription factors show higher-affinity binding to a palindromic sequence, thus improving transcription efficiency. Recently, Bustos et al. (2010) reported a similar arrangement of two P1BS motifs in the promoter of the Pi-starvation responsive gene IPS1, where these cis-elements are separated by 26 bp, as in EZ2. However, despite their apparent structural similarities, these arrangements show very different properties. In the case of IPS1, it was shown that only one of the two P1BS motifs is necessary for the Pi-starvation responsiveness of this gene, while in EZ2 two motifs are required. Although Bustos et al. (2010) also found the two elements are not functionally equivalent, in IPS1 the core sequence of both P1BS motifs is identical, thus indicating that the observed differences are mainly due to the effect of adjacent sequences. In contrast, for EZ2 the functional differences between the two P1BS motifs can be attributed to differences in their core sequences. This indicates that features other than the sole presence of two P1BS motifs with a 26-bp spacing determine the ability of EZ2 to function as a regulatory unit.
Mutation analysis of EZ2 demonstrates that sequences in the flanking regions of the two P1BS motifs are necessary for the transcriptional strength and functionality of EZ2. When either the 5' region or the spacer sequence are mutated, a strong reduction of the gene induction in response to Pi starvation is observed but induction is still significant, suggesting these regions are important in determining the strength of the response but not Pi responsiveness. However, when both regions are mutated, EZ2 enhancer activity is completely abolished. It is not clear from these experiments whether this effect is due to an additive effect of the mutations, generating a very weak enhancer element, or whether the combined mutation of these regions has an effect on the low-Pi responsiveness of EZ2. The mutation of all flanking sequences, leaving intact P1BS4 and P1BS3, further demonstrates that these elements are necessary but not sufficient to constitute an enhancer element in the context of EZ2. The fact that a reduction in the distance between the P1BS motifs by 5 bp affects low-Pi-inducible expression suggests that proper spacing is necessary for the assembly of a functional gene-transcription activating complex (Makeev et al., 2003). This study cannot exclude the possibility that this reduction also eliminates an important regulatory element, thus affecting the function of EZ2. Sequence comparison with orthologous promoters shows that the deleted region is not evolutionarily conserved; however, the possibility remains that the eliminated sequence is important in the context of Arabidopsis EZ2. Additional experiments would be necessary to determine the architectural features, including the distance requirements that contribute to the function of EZ2.
The analysis presented here shows that EZ2 presents characteristics of a cis-regulatory module, representing the arrangement of cis-acting elements that function collectively, since mutations in any of the components affect enhancer activity. It would be expected that if this modular enhancer is functional only as a unit, its structural organization should be conserved in evolution. Indeed, EZ2 represents a modular arrangement conserved in the orthologous promoters of at least eight families of rosids, but not in other clades of dicots or in monocots. This indicates this arrangement of regulatory motifs has persisted in PLDZ promoters at least since the Early Cretaceous (108–117 million years ago), to which the origin of the rosids has been dated (Sanderson et al., 2004). The striking evolutionary conservation of the structural organization of EZ2 suggests this transcriptional enhancer is a composite element in which efficient assembly of a functional transcription complex depends on optimally spaced protein–protein interactions among the cognate DNA-binding factors. Additional motifs were found to be phylogenetically conserved in this modular arrangement: a motif with the sequence GCACAAA or GCAYCAAA located in the spacer sequence between the P1BS elements and a motif in the 5′ region of EZ2 with the sequence TTTGG or TTTGC. Even though mutations of the 5′ region and the spacer sequence affected EZ2 activity, it cannot be established at this point whether these conserved elements represent functional binding sites for transcription factors. Evolutionary conservation points to that possibility, but additional experimental evidence is needed. This study also searched for motifs reported to be associated to the arrangement of two P1BS motifs in the IPS1 promoter, namely motifs A and B (Bustos et al., 2010), but they were not present in EZ2. It is possible that interaction of PHR1 and related transcription factors with different coupling factors determine the specific regulatory properties displayed by different genes in response to Pi starvation.
The fact that this modular arrangement is conserved in evolution suggests that the molecular mechanisms determining its functionality as a Pi-responsive enhancer are also conserved. This is true in the case of common bean, where EZ2 is functional, indicating that this enhancer element and the signalling pathways mediating Pi-starvation responsiveness are functionally conserved between Arabidopsis and bean.
This work provides important understanding of the regulatory properties of the PLDZ2 promoter in terms of its responsiveness to Pi availability. Moreover, it defines EZ2 as an evolutionarily conserved, low-Pi-inducible transcriptional enhancer and establishes the relative importance of the elements that compose it in determining the enhancer functionality. The data presented here indicate that EZ2 is a small, albeit complex, regulatory unit that requires at least four DNA motifs for its enhancer activity. Because of its relatively small size and its characteristics as a native sequence, EZ2 could be a useful model to study the DNA–protein and protein–protein interactions that determine its low-Pi-induced enhancer activity.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Comparison of the expression of PLDZ2 and PLDZ2::GUS in response to Pi availability.
Supplementary Fig. S2. Temporal expression profiles of PLDZ2::GUS and EZ2::GUS.
Supplementary Fig. S3. Effect of sucrose on the low-Pi responsiveness of mutant versions of EZ2.
Supplementary Fig. S4. Expression of EZ2::GUS in transgenic bean roots.
Supplementary Table S1. Sequences of oligonucleotides used to produce deletion versions of the PLDZ2 promoter, the EZ2 enhancer versions, and the primers used for real-time and reverse-transcription PCR analysis.
Supplementary Table S2. GUS activity of PLDZ2 promoter and deletion derivatives.
Supplementary Table S3. GUS activity directed by the PLDZ2 promoter, the EZ2 enhancer, and the derivative constructs.
Acknowledgments
This work was supported in part by grants from the Howard Hughes Medical Institute (grant 55005946) and CONACyT (grant 299/43979) to L.H.-E.
References
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Letters. 2003;537:128–132. doi: 10.1016/s0014-5793(03)00109-1. [DOI] [PubMed] [Google Scholar]
- Awai K, Maréchal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 2001;98:10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG. LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiology. 2001;125:728–737. doi: 10.1104/pp.125.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Oñate-Sánchez L. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PloS ONE. 2011;6:e21524. doi: 10.1371/journal.pone.0021524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proceedings of the National Academy of Sciences USA. 2006;103:6765–6770. doi: 10.1073/pnas.0600863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology. 2007a;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Molecular Plant. 2009;2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiology. 2007b;145:147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C. Galactolipids rule in seed plants. Trends in Plant Science. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- Essigmann B, Güller S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant, Cell and Environment. 2005;28:353–364. [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Ballachanda DN, Raghothama KG. Promoter deletion analysis elucidates the role of cis elements and 5′UTR intron in spatiotemporal regulation of AtPht1;4 expression in Arabidopsis. Physiologia Plantarum. 2009;136:10–18. doi: 10.1111/j.1399-3054.2009.01207.x. [DOI] [PubMed] [Google Scholar]
- Karthikeyan A, Varadarajan D, Jain A, Held M, Carpita N, Raghothama K. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta. 2007;225:907–918. doi: 10.1007/s00425-006-0408-8. [DOI] [PubMed] [Google Scholar]
- Kelly AA, Dörmann P. DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. Journal of Biological Chemistry. 2002;277:1166–1173. doi: 10.1074/jbc.M110066200. [DOI] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiology. 2011;156:1116–30. doi: 10.1104/pp.110.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Lynch J. Root architecture and plant productivity. Plant Physiology. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Makeev VJ, Lifanov AP, Nazina AG, Papatsenko DA. Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Research. 2003;31:6016–6026. doi: 10.1093/nar/gkg799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, Del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. The Plant Journal. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce J, Herrera-Estrella L. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Molecular Biology Reporter. 2004;22:63–70. [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell and Environment. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiology. 2007;143:156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. Journal of Biological Chemistry. 2005;280:7469–7476. doi: 10.1074/jbc.M408799200. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant, Cell and Environment. 2007;30:1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes and Development. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant and Cell Physiology. 2005;46:174–184. doi: 10.1093/pcp/pci011. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Thorne JL, Wikström N, Bremer K. Molecular evidence on plant divergence times. American Journal of Botany. 2004;91:1656–1665. doi: 10.3732/ajb.91.10.1656. [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Richardson AE, Vickers CE, Delhaize E. Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiology. 2004;136:4205–4214. doi: 10.1104/pp.104.045823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant, Cell and Environment. 2008;31:1834–1843. doi: 10.1111/j.1365-3040.2008.01883.x. [DOI] [PubMed] [Google Scholar]
- Xiao K, Liu J, Dewbre G, Harrison M, Wang ZY. Isolation and characterization of root-specific phosphate transporter promoters from Medicago truncatula. Plant Biology. 2006;8:439–449. doi: 10.1055/s-2005-873053. [DOI] [PubMed] [Google Scholar]
- Wang X, Yi K, Tao Y, Wang F, Wu Z, Jiang D, Chen XIN, Zhu L, Wu P. Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant, Cell and Environment. 2006;29:1924–1935. doi: 10.1111/j.1365-3040.2006.01568.x. [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.