Abstract
The Shisa family of single-transmembrane proteins is characterized by an N-terminal cysteine-rich domain and a proline-rich C-terminal region. Its founding member, Xenopus Shisa, promotes head development by antagonizing Wnt and FGF signaling. Recently, a mouse brain-specific Shisa protein CKAMP44 (Shisa9) was shown to play an important role in AMPA receptor desensitization. We used sequence similarity searches against protein, genome and EST databases to study the evolutionary origin and phylogenetic distribution of Shisa homologs. In addition to nine Shisa subfamilies in vertebrates, we detected distantly related Shisa homologs that possess an N-terminal domain with six conserved cysteines. These Shisa-like proteins include FAM159 and KIAA1644 mainly from vertebrates, and members from various bilaterian invertebrates and Porifera, suggesting their presence in the last common ancestor of Metazoa. Shisa-like genes have undergone large expansions in Branchiostoma floridae and Saccoglossus kowalevskii, and appear to have been lost in certain insects. Pattern-based searches against eukaryotic proteomes also uncovered several other families of predicted single-transmembrane proteins with a similar cysteine-rich domain. We refer to these proteins (Shisa/Shisa-like, WBP1/VOPP1, CX, DUF2650, TMEM92, and CYYR1) as STMC6 proteins (single-transmembrane proteins with conserved 6 cysteines). STMC6 genes are widespread in Metazoa, with the human genome containing 17 members. Frequently occurrences of PY motifs in STMC6 proteins suggest that most of them could interact with WW-domain-containing proteins, such as the NEDD4 family E3 ubiquitin ligases, and could play critical roles in protein degradation and sorting. STMC6 proteins are likely transmembrane adaptors that regulate membrane proteins such as cell surface receptors.
Keywords: Shisa-like proteins, WBP1/VOPP1, CX and DUF2650, TMEM92, CYYR1, transmembrane adaptors
1. Introduction
Adaptor or scaffold proteins are important components with diverse roles in cell signaling networks [1-2]. They are usually non-enzymes that contain modular domains and/or linear peptide motifs to mediate protein-protein interactions and/or protein-membrane associations [3-4]. Modular domains frequently found in adaptor proteins include SH3 [5] and WW domains [6] that recognize proline-rich peptide motifs, SH2 [7] and PTB [8] domains that recognize motifs with phosphorylated residues, PDZ domains [9] that bind specific C-terminal peptides, and PH domains [10] that recognize phosphoinositide lipids. Adaptor proteins regulate diverse aspects of cell surface receptor functions ranging from transducing signals to downstream components, attenuating signals in negative feedback loops, to the trafficking and recycling of receptors. An example of an adaptor protein is β-arrestin [11] that regulates G-protein-coupled receptor (GPCR) signaling. Agonist-bound GPCRs are subject to phosphorylation followed by β-arrestin recruitment, resulting in the blockage of receptor-G protein coupling and thus receptor desensitization [12]. Furthermore, β-arrestin-bound GPCRs are targeted for clathrin-mediated endocytosis to promote their degradation or recycling. In addition to receptor desensitization, β-arrestins also serve as scaffold molecules that mediate interactions between GPCRs and downstream effectors in G protein-independent signaling events [13]. Like most characterized adaptor proteins, β-arrestins are soluble proteins residing in the cytosol. Some adaptors also have transmembrane domains. One group of transmembrane adaptor proteins (TRAPs), such as LAT, PAG and TRIM, are key components in immunoreceptor signaling [14].
A recent study showed that overexpression of a brain-specific type I transmembrane protein CKAMP44 (cysteine-knot AMPAR modulating protein, 44 kD) enhanced AMPA receptor desensitization [15], suggesting that it is an adaptor protein. CKAMP44, also named Shisa9, is a member of the Shisa protein family. The founding member of this family is the Xenopus Shisa protein (designated later as Xenopus Shisa1) that can promotes head formation in Xenopus development [16]. During amphibian embryonic development, the Spemann organizer produces a number of secreted proteins, such as Noggin, Chordin and Cerberus, that antagonize BMP and Wnt signaling to promote the formation of anterior neuroectoderm (head) and induce dorsoventral differentiation in a non-cell-autonomous fashion [17-18]. On the other hand, Xenopus Shisa1 promotes head formation cell-autonomously by retention of Wnt [19-20] and FGF [21] receptors in the endoplasmic reticulum (ER) to inhibit their maturation [16].
Several vertebrate homologs of the original Xenopus Shisa1 protein have been cloned and studied, with implicated roles in development, cancer and apoptosis. For example, Xenopus Shisa2 was shown to function in segmental patterning during somitogenesis [22]. Expression of Shisa2 in chicken embryos exhibited an anterior pattern, suggesting its role in development [23]. High expression levels of human Shisa2 were found in certain cancer cell lines and its overexpression increased cell growth and invasion [24]. Five mouse homologs of Shisa (Shisa1–5) were shown to have distinct developmental expression patterns, and mouse Shisa2 was also shown to be an antagonist of Wnt and FGF signaling [25]. Mouse Shisa5, also named Scotin, was independently identified as a downstream pro-apoptotic protein of the P53 family proteins [26-28].
Shisa proteins have not been characterized outside vertebrates, and their evolutionary origin remains unclear. Using sensitive sequence similarity searches, we found distant homologs of Shisa proteins (called Shisa-like proteins) within and beyond vertebrates. Noticeably, we found EST evidence of Shisa-like genes in Porifera, suggesting a Metazoa origin for them. Further computational analysis suggests that Shisa and Shisa-like proteins belong to a large and diverse group of predicted single-transmembrane proteins that have undergone large expansions in certain metazoan lineages. These proteins possibly play various roles in regulating the functions of membrane proteins such as cell surface receptors.
2. Materials and methods
2.1. Sequence similarity searches against the non-redundant protein database
PSI-BLAST [29] was used to search for homologs of the Shisa family starting with the mouse Shisa2 protein (NCBI gene identification (gi) number: 21703918; range: 23–137, which excludes the predicted signal peptide and C-terminal low complexity region) against the NCBI non-redundant (nr) protein database (e-value inclusion cutoff: 1e-4). To perform transitive searches, the protein hits found by PSI-BLAST were grouped by BLASTCLUST (with the score coverage threshold (-S, defined as the bit score divided by alignment length) set to 1, length coverage threshold (-L) set to 0.5, and no requirement of length coverage on both sequences (-b F)) and a representative sequence from each group was used to initiate new PSI-BLAST searches. In practice, we noticed that some of the distant Shisa homologs could not be found with e-values less than 1e-4 when composition-based statistics were used. To increase the coverage of Shisa homologs, composition-based statistics was disabled (-t 0) in separate transitive PSI-BLAST searches while the the e-value inclusion cutoff was set to be more stringent (1e-6 in the first three rounds of transitive PSI-BLAST and 5e-6 in the fourth round of transitive PSI-BLAST searches, these values were selected based on trials of different cutoff values and manual inspections of profile corruption). The results were manually checked and no false positives were found in the first three rounds of transitive PSI-BLAST searches. A few false positives found in the fourth round of transitive PSI-BLAST searches were removed (false positives were judged by the lack of the cysteine motif C*C*CC*CC in Shisa-like proteins and the fact that PSI-BLAST searches starting from them could not reciprocally find Shisa or Shisa-like proteins). A similar approach was used to search for homologs of the WBP1/VOPP1, CX, DUF2650, TMEM92 and CYYR1 family proteins. The HHpred web server [30] was used for profile-against-profile-based similarity searches using several members found by PSI-BLAST against the Pfam [31] and human proteome databases.
2.2. Sequence similarity searches against EST and genome databases
To increase the coverage of homology detection for Shisa-like proteins and WBP1/VOPP1 homologs, especially in lophotrochozoans where not many protein records were available, we used the NCBI TBLASTN web server to search against the EST database (e-value cutoff: 10) starting with several Shisa and Shisa-like proteins. For each TBLASTN hit, the longest translated amino acid sequence segment covering the hit region and without stop codons was obtained. Sequences without the motif ‘C*C*CC*CC’ were discarded. The remaining sequences were subject to multiple sequence alignment by MAFFT [32]. The resulting alignment was manually inspected to remove likely false positives (e.g., some cysteine-rich fragments with a lot more cysteines than in Shisa-like proteins). Sequence redundancy was reduced by keeping one sequence for any group of ESTs that appear to have been transcribed from the same gene.
Translated sequences of several plant and Plasmodium vivax ESTs were found to show high similarity to Shisa or Shisa-like proteins. However, these ESTs cannot find high similarity hits in the whole genome sequences of the corresponding plant or Plasmodium species. Instead, the transcripts of Plasmodium EST sequences (e.g., gi: 260703421) show high similarity (>99%) to a human Shisa5 protein. Two plant ESTs (gi: 76604539 from Euphorbia esula and gi: 119017255 from Manihot esculenta) show high similarity (99% sequence identity) to an EST from a Platyhelminth species (gi: 84601851 from Schmidtea mediterranea). Therefore, the ESTs from plants and Plasmodium were considered as contaminations.
To further investigate the species distribution of Shisa subfamilies, TBLASTN was used to search against the NCBI whole genomes, whole-genome shortgun reads and high throughput genomic sequences. BLAT searches were performed in the UCSC genome bioinformatics server [33] against the lamprey genome sequences and TBLASTN searches were performed against the elephant shark genome sequences [34].
2.3. Pattern-based searches in eukaryotic proteomes
To find proteins with a similar domain structure to Shisa-like proteins, we performed pattern-based searches against a number of eukaryotic proteomes sampling various taxa (Apis mellifera, Arabidopsis thaliana, Branchiostoma floridae, Caenorhabditis elegans, Ciona intestinalis, Dictyostelium discoideum, Drosophila melanogaster, Homo sapiens, Hydra magnipapillata, Monosiga brevicollis, Naegleria gruberi, Nematostella vectensis, Paramecium tetraurelia, Saccharomyces cerevisiae, Saccoglossus kowalevskii, Tetrahymena thermophila, Tribolium castaneum and Trichoplax adhaerens). The proteins from these genomes were downloaded from the NCBI genome database. For each proteome, we predicted TMs for its proteins using Phobius [35] and selected those proteins with a single predicted TM. For these predicted single-TM proteins, we identified proteins with a ‘C*C*CC*CC’ pattern (matching the regular expression of ‘C.{2,40}C.{2,40}CC.{2,40}CC’) in the sequence segment N-terminal to the predicted TM and excluding any predicted signal peptide. The results were manually inspected to select proteins with a cysteine-rich domain similar to Shisa-like proteins. We found that Shisa and Shisa-like proteins, as well as some other proteins such as WBP1 and TMEM92, usually have a short N-terminal segment before the predict TM (less than 100 residues) with no more than eight cysteines in the N-terminal segment before the predict TM. On the other hand, some proteins identified by pattern match searches have a much longer N-terminal region before the predicted TM with much more numbers of cysteines than the six cysteines in the ‘C*C*CC*CC’ motif (one example is gi: 4557251, ADAM metallopeptidase domain 10). We thus only considered those proteins with no more than eight cysteines in the N-terminal segment as potentially related to Shisa and Shisa-like proteins. The results of pattern-based searches in selected eukaryotic proteomes are available in Supplementary Figure S14.
To expand the searches against other eukaryotes, we downloaded RefSeq sequences in the following organism categories: fungi, invertebrate, plant, protozoa, vertebrate_mammalian and vertebrate_other (these categories are defined in ftp://ftp.ncbi.nlm.nih.gov/refseq/release). We used the same pattern-based searches on these protein datasets. The results are shown in Supplementary Figure S15.
2.4. Sequence alignment and phylogenetic reconstruction
The multiple sequence alignment for each protein family was made by PROMALS [36]. These alignments were then improved by manual curation. The MOLPHY package [37] was used for phylogenetic reconstruction based on the alignment of Shisa proteins. The JTT amino acid substitution model [38] was used in MOLPHY. The local estimates of bootstrap percentages were obtained by the RELL method [39] (-R option in the ProtML program of MOLPHY). A phylogenetic reconstruction of WBP1/VOPP1 proteins was conducted in the same way.
3. Results
3.1. The Shisa family proteins in vertebrates
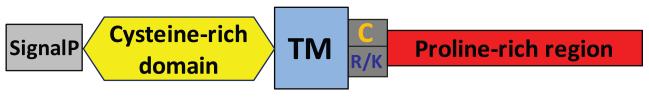
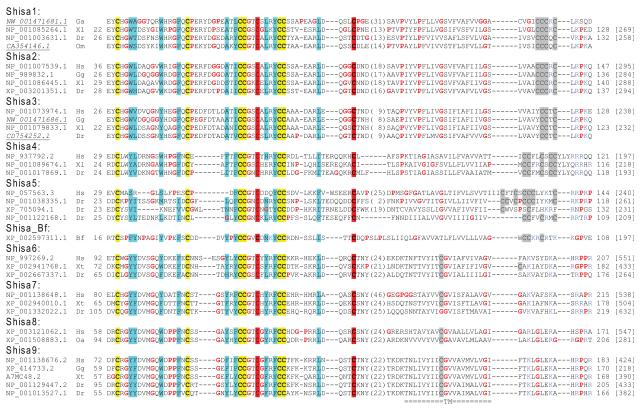
A previous study reported five vertebrate Shisa subfamilies (Shisa1–5) that exhibit varying taxonomy distributions in mammals, chicken, Xenopus and zebrafish [25]. Four additional Shisa subfamilies (Shisa6–9) were found in vertebrates in sequence similarity searches by PSI-BLAST [29] (see Materials and methods). They have a common domain architecture (Figure 1) characterized by a predicted signal peptide, an N-terminal cysteine-rich domain with eight conserved cysteines [40] exhibiting a distinct pattern (‘C*C*CC*C*CC*C’, ‘*’ represents a stretch of amino acid residues, Figure 2), a predicted transmembrane segment (TM) and a proline-rich C-terminal region. For Shisa proteins from subfamilies 1–5, several cysteines residues are present near the C-termini of their predicted TMs (Figure 2), possibly providing sites for lipid modifications such as palmitoylation to facilitate their localization to specific membrane microdomains [41]. Positively charged residues frequently occur near the C-termini of their predicted TMs and may help determine the topology of Shisa proteins [42]. They are predicted to be type I transmembrane proteins with their proline-rich, mostly disordered C-terminal regions residing in the cytosol.
Figure 1. Domain structure of Shisa and Shisa-like proteins.
These proteins usually have a signal peptide (labeled as ‘SignalP’), an N-terminal cysteine-rich domain with a distinct cysteine pattern (‘C*C*CC*C*CC*C’ for Shisa proteins and ‘C*C*CC*CC’ for Shisa-like proteins), a single predicted transmembrane segment (labeled as ‘TM’) and a C-terminal proline-rich low complexity region predicted to be largely disordered. Cysteines (labeled as ‘C’) and positively charged residues (labeled as ‘R/K’) frequently occur at the C-terminal end of the predicted TM.
Figure 2. Multiple sequence alignment of Shisa proteins.
The cysteine-rich domains and transmembrane regions of representative Shisa proteins (with a ‘C*C*CC*C*CC*C’ pattern) from nine vertebrate Shisa subfamilies (Shisa1-9) and one sequence from Branchiostoma floridae (Shisa_Bf) are included in this alignment NCBI accession numbers are shown. A few proteins were derived from genome or EST sequences (accession numbers in italic and underlined). Starting and ending residues numbers for sequences with protein accession numbers are shown before and after the sequences respectively. Protein lengths are shown in brackets. Conserved cysteines are shaded in yellow or red (the additional two cysteine positions compared to Shisa-like proteins). Non-charged residues in positions with mainly hydrophobic residues were shaded in cyan. Prolines and glycines are shown in red letters. Cysteines within and after the predicted transmembrane segments are shaded in gray. Arginines and lysines, occurring frequently C-terminal to the predicted transmembrane segments, are shown in blue letters. Species name abbreviations shown after the accession numbers are as follows: Bf, Branchiostoma floridae; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Om, Oncorhynchus mykiss; Oa, Ornithorhynchus anatinus; Xt: Xenopus tropicalis; and Xl, Xenopus laevis.
Seven of the nine Shisa subfamilies (Shisa2–7 and Shisa9) have orthologous proteins in mammals and zebrafish (Figure 2), suggesting that they were present in the last common ancestor of vertebrates. The evolutionary origin of the first reported Shisa in Xenopus laevis (belonging to the Shisa1 subfamily) [16] has been unclear [25]. In a previous study Shisa1 was not identified in amniotes, and only one putative ortholog was found in zebrafish [25]. We found EST evidence of Shisa1 in another fish Oncorhynchus mykiss (gi: 24599333). Moreover, we found genomic sequences harboring putative Shisa1 genes in birds and the elephant shark Callorhinchus milii (Figure 2 and Supplementary Figure S1), suggesting that the Shisa1 subfamily, despite apparent absence from mammals, has a deeper and wider distribution in vertebrates than previously thought [25]. On the other hand, the Shisa8 subfamily is restricted to be in mammals (including one in the basal mammalian species Ornithorhynchus anatinus, gi: 149457717). Shisa8 appears to be closely related to Shisa9 (Figure 3) and could originate from a mammalian-specific gene duplication of Shisa9. Alternatively, the Shisa8 gene could be present in the last common ancestor of vertebrates, but was lost in vertebrate lineages other than mammals. Subfamily-specific gene duplications or gene losses were observed. For example, Xenopus appears to lack a Shisa5 ortholog while zebrafish possess several Shisa5 proteins (Figure 2). Xenopus also possesses a distantly related Shisa homolog with a pair of cysteines mutated (Supplementary Figure S1, discussed below).
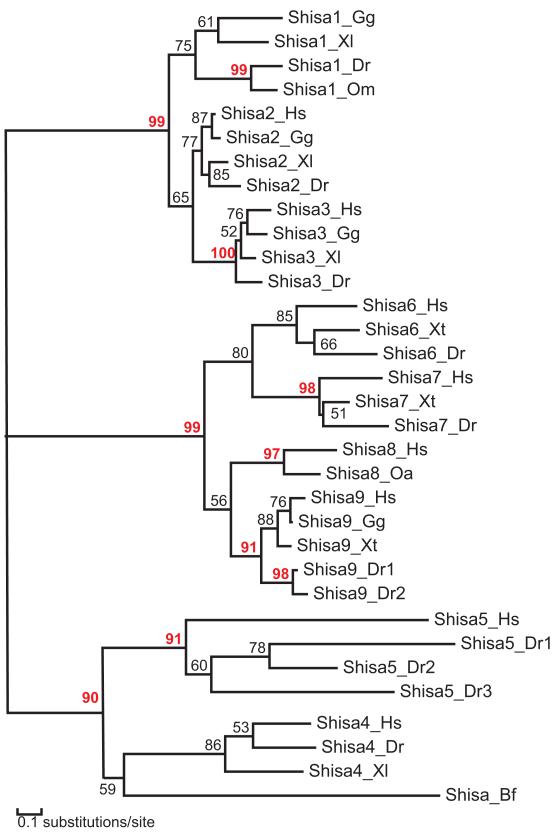
Figure 3. A phylogenetic tree of Shisa proteins by MOLPHY.
Protein names are shown as Shisa subfamily names followed by species name abbreviations defined in Figure 2. Branch support values above 90 are shown in red numbers.
A phylogenetic tree reconstructed by the MOLPHY program [37] supports three major clades among the nine Shisa subfamilies (Shisa1–3, Shisa4–5 and Shisa6– 9, Figure 3). Subfamilies Shisa1–3 form a monophyletic group with a good bootstrap support (Figure 3). Compared with other Shisa subfamilies, they share several sequence features such as longer insertions between the second and third conserved cysteines in the cysteines-rich domain (Figure 2) and a ‘PxxxP’ signature at the beginning of their predicted TMs (Figure 2). The cysteine patterns at the end of their predicted TMs can serve as subfamily-specific signatures, with ‘CCC[KQ]C’, ‘CCCRC’ and ‘YCCTC’ motifs in Shisa1, Shisa2 and Shisa3 respectively. Subfamilies Shisa6–9 are more closely related to each other than to other Shisa subfamilies (Figure 3). Unlike other subfamilies, Shisa6–9 lack cysteines near the C-termini of their predicted TMs (Figure 2), suggesting a loss of lipid modification sites.
We found genomic and/or EST evidence of several Shisa family members in the basal vertebrate lamprey and several Chondrichthyes species (cartilaginous fishes) (Supplementary Figure S1). Among other deuterostomes, only one Shisa protein was found in the amphioxus Branchiostoma floridae (Figure 2), which belongs to the clade containing subfamilies Shisa4–5 (Figure 3). The presence of a Shisa protein in B. floridae suggests that Shisa proteins probably originate in the ancestor of chordates.
3.2. FAM159 and KIAA1644: two Shisa-like protein families distantly related to Shisa proteins
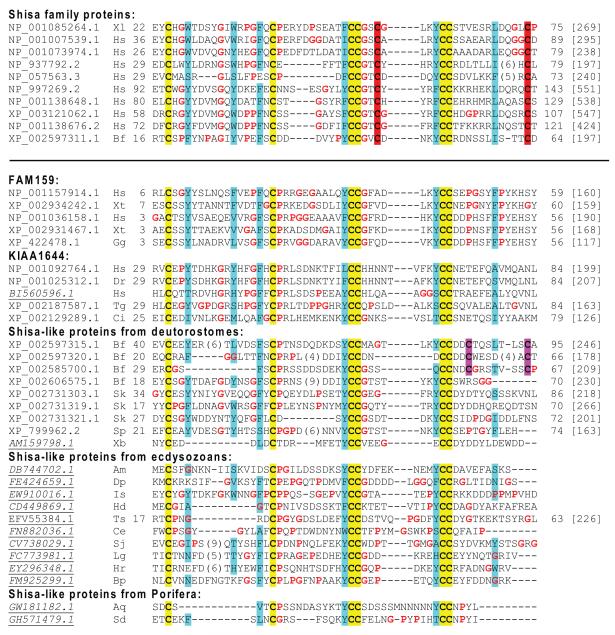
PSI-BLAST searches detected two protein families FAM159 and KIAA1644 with vertebrate members that are distantly related to the Shisa family proteins (Figure 4 and Supplementary Figure S2). Proteins in these families share a similar domain architecture to Shisa proteins (Figure 1), except that FAM159 proteins lack a predicted signal peptide. Functions of these families have not been characterized. Two copies of FAM159 proteins exist in vertebrates (FAM159A and FAM159B, Supplementary Figure S2). While FAM159 proteins were only found in vertebrates, KIAA1644 family members were present in vertebrates and tunicates such as Ciona intestinalis, suggesting an Olfactore origin of KIAA1644 proteins. There are also two KIAA1644 copies in amniotes (Figure 4 and Supplementary Figure S2).
Figure 4. Multiple sequence alignment of cysteine-rich domains from Shisa proteins and Shisa-like proteins.
This alignment is formatted and colored in the same way as in Figure 2. Shisa proteins and Shisa-like proteins are separated by a horizontal line. Species name abbreviations shown after the accession numbers are as follows: Aq, Amphimedon queenslandica; Am, Apis mellifera; Bp, Brachionus plicatilis; Bf, Branchiostoma floridae; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Dr, Danio rerio; Dp, Daphnia pulex; Gg, Gallus gallus; Hr, Helobdella robusta; Hs, Homo sapiens; Hd, Hypsibius dujardini; Is, Ixodes scapularis; Lg, Lottia gigantea; Sk, Saccoglossus kowalevskii; Sj, Schistosoma japonicum; Sp, Strongylocentrotus purpuratus; Sd, Suberites domuncula; Tg, Taeniopygia guttata; Ts, Trichinella spiralis; Xt, Xenopus tropicalis; Xl, Xenopus laevis and Xb, Xenoturbella bocki.
The cysteine-rich domains in FAM159 and KIAA1644 contain six highly conserved cysteines (‘C*C*CC*CC’, ‘*’ represents a stretch of amino acid residues) as compared to eight conserved cysteines in the Shisa family proteins (Figure 4). The extra two cysteines in the Shisa family proteins lie in between the two ‘CC’s and after the second ‘CC’, respectively, giving rise to a signature of ‘C*C*CC*C*CC*C’ (the extra cysteines underlined). Most Shisa homologs outside vertebrates (Figure 4 and discussed below) show the six-cysteine pattern similar to KIAA1644 and FAM159 while lacking the extra cysteine pairs in vertebrate Shisa proteins. Therefore, it is likely that Shisa proteins have evolved the extra pair of cysteines that possibly form a new disulfide bond to increase the stability of the cysteine-rich domain. To differentiate the proteins based on their cysteine patterns, we refer to the Shisa homologs with six conserved cysteines and without the specific cysteine pairs in Shisa proteins as ‘Shisa-like proteins’. Several positions with mainly hydrophobic residues are also conserved among Shisa and Shisa-like proteins (Figure 4), supporting the inferred homologous relationships between them. Most noticeably, each of the two ‘CC’ signatures is often preceded by an aromatic residue (tyrosine being the most frequent) (Figure 4).
3.3. Shisa-like proteins in deuterostomes other than vertebrates
In addition to FAM159 and KIAA1644 mainly found in vertebrates, we detected Shisa-like proteins in other deuterostomes by PSI-BLAST. Noticeably, Shisa-like genes have undergone large expansion in the cephalochordate B. floridae [43] and the hemichordate Saccoglossus kowalevskii [44], which possess at least 15 and 14 copies of these genes, respectively (Supplementary Figure S2). Most of the B. floridae Shisa-like proteins have an extra pair of cysteines after the second ‘CC’ giving rise to a pattern of ‘C*C*CC*CC*C*C’ (the extra cysteines underlined). Such a pattern is not observed in other deuterostome Shisa-like proteins, suggesting they originated and expanded in the amphioxus. In contrast, only one Shisa-like protein was found in the echinoderm Strongylocentrotus purpuratus [45]. We also identified EST evidence of several Shisa-like proteins with tandem cysteine-rich domains in S. kowalevskii, Xenoturbella bocki [46], and two tunicates Molgula tectiformis and Botryllus schlosseri (Supplementary Figure S3).
3.4. Shisa-like proteins in protosotomes
PSI-BLAST searches identified Shisa-like proteins from several protostomes such as insects, nematodes, and flatworm (Platyhelminthes) species from the Schistosoma genus. TBLASTN searches against EST databases using these proteins as queries revealed that Shisa-like proteins are widely distributed in various protostomes, including both ecdysozoans and lophotrochozoans.
Shisa-like proteins derived from ESTs were detected in the Arthropoda phylum (including species from insects, crustaceans and chelicerates) and two other ecdysozoan phyla Tardigrada and Nematoda (Supplementary Figure S4). Various EST hits in Pterygota insects were found, including one species from the Ephemeroptera order (e.g., mayflies) and quite a few species from the Neoptera orders such as Hymenoptera (e.g., bees and ants), Orthoptera (e.g., grasshoppers) and Hemiptera (e.g., true bugs) (Supplementary Figure S4). Among the Endopterygota (Holometabola) orders, Shisa-like proteins are only present in Hymenoptera and are absent from Coleoptera (e.g., beetles), Lepidoptera (e.g., butterflies and moths) and Diptera (e.g., flies) (Supplementary Figure S5). For example, no significant hits of Shisa-like sequences were found in species such as Drosophila melanogaster (a dipteran) [47], Tribolium castaneum (a coleopteran) [48] and Bombyx mori (a lepidopteran) [49] in BLAST or TBLASTN searches against NCBI RefSeq protein sequences, EST databases and whole genome sequences. Recent phylogenetic studies (e.g., [50]) supported that Hymenoptera is the sister group to other Endopterygota insects. Therefore, a single gene loss event in the last common ancestor of Endopterygota orders other than Hymenoptera could explain the lack of Shisa-like proteins in these orders (Supplementary Figure S5).
EST evidence of Shisa-like proteins was found in several lophotrochozoan phyla such as Annelida, Mollusca, Platyhelminthes and Rotifera (Supplementary Figure S4). Multiple copies of Shisa-like proteins were detected in certain lophotrochozoan species, such as Schmidtea mediterranea and Crassostrea gigas (Supplementary Figure S4). The cysteine-rich domain in most of the protostomian Shisa-like proteins has six conserved cysteines (C*C*CC*CC) similar to FAM159 and KIAA1644, while lacking the extra pair of cysteines in vertebrate Shisa proteins. A few Shisa-like proteins in Mollusca and Platyhelminthes have an extra pair of cysteines after the second CC signature, giving rise to a similar pattern (C*C*CC*CC*C*C) to some B. floridae Shisa-like proteins (Supplementary Figure S4).
EST searches retrieved five Shisa homologs in the Rotifera species Brachionus plicatilis. One of them has six cysteines in the N-terminal domain, characteristic of Shisa-like proteins (C*C*CC*CC), while the the other four sequences match the motif of vertebrate Shisa proteins with eight cysteines (C*C*CC*C*CC*C) (Supplementary Figures S1 and S4). These four B. plicatilis sequences are quite interesting as no Shisa-specific cysteine patterns (C*C*CC*C*CC*C) were detected in other protostomian species. B. plicatilis Shisa proteins have predicted TMs quite different from those observed in any subfamily of Shisa proteins from chordates (Supplementary Figure S1). Besides, two of the four B. plicatilis Shisa proteins possess two tandem cysteine-rich domains. Such tandem repeats of cysteine-rich domains were not observed in any Shisa proteins from chordates. Thus, they are unlikely to be contaminations of chordate cDNAs in the B. plicatilis EST library. In one possible evolutionary scenario, Shisa proteins (with the C*C*CC*C*CC*C pattern) were present in the common ancestor of Bilateria, but were subsequently lost in all lineages but chordates and Rotifera. A more parsimonious scenario is that B. plicatilis acquired Shisa genes through horizontal transfer from certain chordate species, and these genes have since undergone divergent evolution such as domain duplications.
3.5. Shisa-like proteins in Porifera
A TBLASTN search using the Apis mellifera Shisa-like protein (gi: 110760589) as the query against EST database found a weak hit to an EST (gi: 282462982) from the Porifera (sponge) species Amphimedon queenslandica (e-value: 0.68, with five out of the six conserved cysteines in the cysteine-rich domain and the predicted TM aligned). The translated sequence of this EST sequence has the same domain architecture as Shisa-like proteins. TBLASTN using this translated sequence as query found an EST from a different Porifera species (gi: 300475707 from Suberites domuncula, Figure 4) with an e-value of 0.002. These results suggest that Shisa-like proteins were present in the last common ancestor of metazoans.
Aside from apparent contaminations (see Materials and methods), no protein or EST evidence of Shisa-like proteins was found outside metazoans. We also did not detect Shisa-like genes in Cnidaria or Placozoa, suggesting that they were lost in these metazoan lineages.
3.6. Prediction of disulfide bond connectivity in Shisa-like proteins
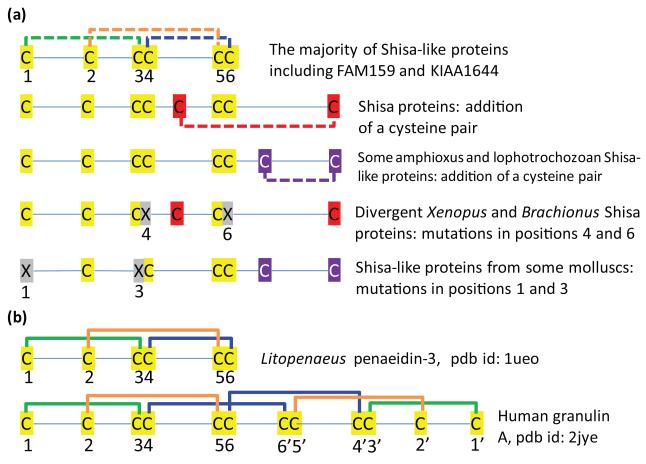
Conserved cysteines are often involved in disulfide bond formation. A newly added conserved pair of cysteines could form a disulfide bond, as in the case of Shisa proteins with eight cysteines compared to Shisa-like proteins with six cysteines. Similarly, concerted mutations in an otherwise conserved pair of cysteine positions suggest disulfide bond connectivity between them [51]. Several divergent homologs of Shisa or Shisa-like proteins have one pair of cysteines mutated, suggesting possible connectivity between the mutated positions (Figure 5). A few Mollusca Shisa-like proteins have the the first and third cysteines in C*C*CC*CC mutated, suggesting that they form a disulfide bond. In a divergent Shisa homolog from Xenopus tropicalis (gi: 113205932) and one cysteine-rich domain of a B. plicatilis Shisa protein (gi: 225249116) (Supplementary Figure S1), the fourth and sixth cysteines in the general C*C*CC*CC pattern were mutated, suggesting a disulfide bond between them. Thus the inferred cysteine connectivity for the six conserved cysteines would be 1-3, 2-5 and 4-6 (Figure 5).
Figure 5. Cysteine patterns and disulfide bond connectivity predictions for Shisa and Shisa-like proteins.
(a) The six conserved cysteines in Shisa-like proteins are labeled from 1 to 6. Cysteine pairs with inferred disulfide bond connectivity are connected by dashed lines. Mutations in the cysteine positions are marked by ‘X’s. (b) Disulfide bond connections are shown for two protein domains with known structures and containing the cysteine pattern ‘C*C*CC*CC’.
Manual inspection of the results of a regular expression search of current protein structure database revealed two structural folds with the C*C*CC*CC pattern (a few other hits also encompassing this pattern were not considered since they have a lot more conserved cysteines). One structural fold contains penaeidins, which are antimicrobial peptides found in shrimps [52] (protein databank (pdb) id: 1ueo). They have six conserved cysteines following the C*C*CC*CC pattern and exhibit a 1-3, 2-5, and 4-6 disulfide bond connectivity that coincides with the inferred connectivity pattern for Shisa and Shisa-like proteins (Figure 5). The other structural fold consists of granulin domains [53] (pdb id: 2jye), which have a palindromic cysteine pattern of ‘C*C*CC*CC*CC*CC*C*C’, with two subdomains exhibiting a mirror symmetry of structures and cysteine patterns (Figure 5). The first subdomain with a C*C*CC*CC pattern also has the 1-3 and 2-5 cysteine connectivity. The fourth and sixth cysteines of the first subdomain make a disulfide bond with the 6′ and 4′ cysteines of the second subdomain respectively, and vice versa (Figure 5). The first subdomain of granulin can also form a folding unit as its structure alone is available [54] (pdb id: 1g26). In this case, the fourth and sixth cysteines in this subdomain are spatially close and can potentially form a disulfide bond. It should be noticed that penaeidin and the first subdomain of granulin have different structural folds despite sharing the same cysteine pattern and similar connectivity, suggesting that sequences with a C*C*CC*CC pattern, such as Shisa-like proteins and the WBP1/VOPP1 proteins (described below) could also adopt different 3-dimensional structural folds. No statistically significant sequence similarities were found between Shisa-like proteins and proteins with known structures. Structural determination of the cysteine-rich domains in Shisa and Shisa-like proteins could reveal possible structural similarities and homologous relationships to other proteins.
3.7. Additional protein families with Shisa-like domain architectures - defining the STMC6 proteins
We searched a number of eukaryotic proteomes for predicted single-transmembrane proteins that possess an N-terminal cysteine-rich domain with a C*C*CC*CC pattern similar to the one in Shisa-like proteins (see Materials and methods). Several additional families were uncovered, including WBP1/VOPP1, CX, DUF2650, TMEM92, and CYYR1 (Figure 6 and Supplementary Figures S6–S11). Together with Shisa and Shisa-like proteins, we refer to them as STMC6 proteins (single-transmembrane proteins with conserved 6 cysteines) based on the shared cysteine pattern and the common domain structure.
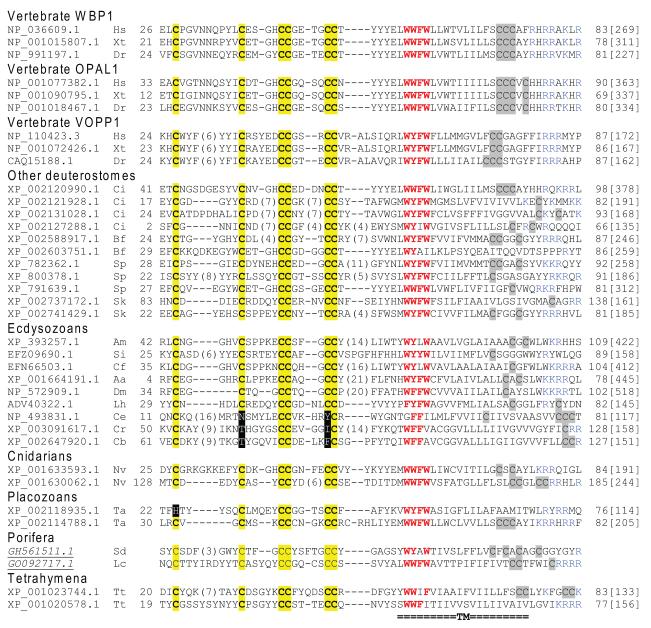
Figure 6. Multiple sequence alignment of WBP1/VOPP1 family and its homologous proteins.
This alignment includes aligned regions of the cysteine-rich domains and predicted transmembrane segments (labeled TM at the bottom). The sequences are denoted by NCBI accession numbers. EST-derived proteins have italic and underlined accession numbers. Starting residue numbers for sequences with protein accession numbers are shown before the sequences. Conserved cysteines are shaded in yellow. Mutations in conserved cysteines are shaded in black. Aromatic residues in the four positions with mainly aromatic residues are in red letters. Cysteines near the C-terminal ends of predicted transmembrane segments are shaded in gray. Arginines and lysines near the C-terminal ends of the predicted TMs are shown as blue letters. Species abbreviations shown after the accession numbers are as follows: Aa, Aedes aegypti; Am, Apis mellifera; Bf, Branchiostoma floridae; Cb, Caenorhabditis briggsae; Ce, Caenorhabditis elegans; Cr, Caenorhabditis remanei; Cf, Camponotus floridanus; Ci, Ciona intestinalis; Dr, Danio rerio; Dm, Drosophila melanogaster; Hs, Homo sapiens; Lh, Latrodectus hesperus; Lc, Leucetta chagosensis; Nv, Nematostella vectensis; Sk, Saccoglossus kowalevskii; Si, Solenopsis invicta; Sp, Strongylocentrotus purpuratus; Sd, Suberites domuncula; Tt, Tetrahymena thermophila; Ta, Trichoplax adhaerens and Xt, Xenopus tropicalis.
Sequence similarities among the WBP1/VOPP1, TMEM92, CX and DUF2650 families were found by PSI-BLAST searches with statistical supports, suggesting that members of these four families are homologous. For example, a CX family member (gi: 268555046) found a WBP1/VOPP1 family member (gi: 169234886) with an e-value of 7e-5 in the seventh PSI-BLAST iteration (e-value inclusion threshold: 1e-4, no composition-based statistics). Another CX family member (gi: 253683499) found a TMEM92 protein (gi: 73966363) with an e-value of 1e-5 in the third PSI-BLAST iteration (e-value inclusion threshold: 1e-4, no composition-based statistics). A DUF2650 member (gi: 115532536) found a TMEM92 member (gi: 269954664) with an e-value of 8e-04 in the seventh PSI-BLAST iteration (e-value inclusion threshold: 1e-4, no composition-based statistics). Distant homologous relationships among WBP1/VOPP1, CX, DUF2650 and TMEM92 were also supported by the sensitive profile-profile-based HHpred searches [30]. For example, an HHpred search using a CX protein (gi: 17557266; range: 132–191, covering the cysteine-rich domain and the predicted TM) as the query against the Pfam and human proteome databases found the Pfam domain DUF2650, the human VOPP1 protein and the human TMEM92 protein with probability scores of 96.38, 91.98 and 86.89, respectively. An HHpred search using a TMEM92 protein (gi: 253683499; range 28–85, covering the cysteine-rich domain and the predicted TM) as the query against the Pfam and human proteome databases found Pfam domains WBP-1 and DUF2650 with probability scores of 96.75 and 93.62, respectively.
The WBP1/VOPP1 family proteins include vertebrate proteins WBP1 (WW domain-binding protein 1) [55], its close homolog OPAL1 (outcome predictor in acute leukemia 1) [56], and a less similar protein VOPP1 (Vesicular Over-expressed in cancer Prosurvival Protein 1) [57-58] (Figure 6). These proteins have a sequence motif of four consecutive positions with mainly aromatic residues in their predicted TMs (Figure 6, Supplementary Figures S6 and S7). The WBP1/VOPP1 family proteins in Caenorhabditis remanei and Caenorhabditis briggsae have been expanded (Supplementary Figure S6). The second and fifth cysteines of the C*C*CC*CC signature in these nematode proteins are mutated (C*X*CC*XC), suggesting a disulfide bond connection between them (Supplementary Figure S6). Coupled with concerted mutations in the fourth and sixth cysteines in some of the CX family members (C*C*CX*CX, Supplementary Figure S8), these proteins were inferred to have 1-3, 2-5 and 4-6 disulfide bond connectivity, identical to the predicted cysteine connectivity pattern of Shisa and Shisa-like proteins (Figure 5). Some WBP1/VOPP1 proteins (e.g., gi: 268567772 from C. briggsae and gi: 291234472 from S. kowalevskii, Supplementary Figure S6) have more than one cysteine-rich domains before their predicted TMs.
Similar to Shisa/Shisa-like proteins, most members in WBP1/VOPP1, CX, DUF2650, TMEM92 and CYYR1 families have cysteines and positively charged residues near the C-termini of their predicted TMs, suggesting that they are type I transmembrane proteins that can potentially undergo lipid modifications. WBP1, OPAL1 and VOPP1 proteins have CCCAϕ (ϕ, an aromatic residue), CCCVC and CCGAG signatures near the C-termini of their predicted TMs, respectively (Supplementary Figure S6). A phylogenetic reconstruction of WBP1/VOPP1 proteins suggests that WBP1 and OPAL1 proteins form a major group (group 1 in Supplementary Figure S12, the bootstrap support is low at 61). OPAL1 proteins, being closely related to WBP1, probably result from a gene duplication of WBP1 in vertebrates (Supplementary Figure S12). Vertebrate VOPP1 proteins together with a number of invertebrate proteins form a second major group (group 2 in Supplementary Figure S12, the bootstrap support is 84), although the relative positions of many invertebrate proteins do not have strong bootstrap supports. Insect and nematode sequences form long branches in the tree (Supplementary Figure S12), reflecting elevated evolutionary rates in these lineages.
WBP1/VOPP1 proteins are widespread in Metazoa (Supplementary Figure S13). Outside metazoans, they are only found in the ciliate Tetrahymena thermophila based on both protein and EST evidence (Figure 6 and Supplementary Figure S7). CX and DUF2640 are nematode-specific families, while TMEM92 appears to be mammalian-specific. The other family CYYR1 (cysteine and tyrosine rich protein 1) is vertebrate-specific [59]. Weak sequence similarities were found between CYYR1 and Shisa /Shisa-like proteins by PSI-BLAST, suggesting that they are evolutionarily related. For example, A PSI-BLAST search (e-value inclusion threshold 1e-4, with no composition-based statistics) starting with a zebrafish Shisa protein (gi: 192453554) identified a chicken CYYR1 protein (gi: 118083813) with an e-value of 1e-4 in the eighth iteration.
4. Discussion
4.1. Homology among the STMC6 proteins?
Homology inference for STMC6 proteins was complicated by the biased amino acid composition in the cysteine-rich domains, the predicted TM regions and the C-terminal proline-rich low complexity regions. To reduce the chance of finding false hits, we removed the predicted signal peptides and the C-terminal low complexity regions. The remaining regions covering the cysteine-rich domains and predicted TMs were subject to PSI-BLAST searches under more strict e-value cutoffs (1e-4 or lower) than default (2e-3 for the local blastpgp program or 5e-3 in online server). Shisa and Shisa-like proteins form a clear homologous group, as they can be readily linked by PSI-BLAST under stringent settings. On the other hand CYYR1can only be linked to Shisa proteins under more relaxed PSI-BLAST settings (disabling of composition-based statistics). WBP1 and VOPP1 belong to another clear homologous group, while CX, DUF2650 and TMEM92 can be linked to WBP1/VOPP1 proteins under less stringent PSI-BLAST settings (disabling of low complexity statistics and with higher e-value inclusion cutoffs). HHpred search results also support the distant homologous relationships among WBP1/VOPP1, CX, DUF2650 and TMEM92.
Sequence similarity search methods such as PSI-BLAST and HHpred did not link the group of Shisa/Shisa-like/CYYR1 to the group of WBP1/VOPP1/CX/DUF2650/TMEM92 with statistically significant scores. However, these proteins share common sequence features including a similar domain architecture (Figure 1), a cysteine-rich domain with the general C*C*CC*CC pattern, and frequently a C-terminal low complexity region (Supplementary Figures S6–S11) with PY motifs (discussed below). These features collectively tend to argue that all these families of STMC6 proteins are evolutionarily related. Future structural determination of the cysteine-rich domains of these families can provide insights into the possible homologous relationships among STMC6 proteins. For example, a shared structural fold and the same disulfide bond connectivity would favor the proposed common evolutionary origin of STMC6 proteins.
4.2. Divergent evolution of STMC6 proteins
Several types of recurring evolutionary events were observed for STMC6 proteins. First, gene expansion by duplications is a frequent scheme in the evolution of different STMC6 families, resulting in quite a number of STMC6 genes in some metazoan genomes. For example, the human genome has 17 STMC6 genes (eight copies of Shisa; two copies of FAM159, two copies of KIAA1644, three copies of WBP1/VOPP1, one TMEM92, and one CYYR1). Large gene expansions of Shisa-like genes were observed in deuterostomes such as B. floridae and S. kowalevskii (Supplementary Figure S2). High gene copy numbers of WBP1/VOPP1, CX and DUF2650 families represent a prominent feature in some nematode genomes (Supplementary Figures S6 and S8–S11). Duplicated STMC6 gene products could evolve new functions and potentially regulate the functions of different membrane proteins in a variety of ways (discussed below).
Second, domain-level duplications resulting in tandem cysteine-rich domains occurred occasionally and independently in several STMC6 families, including Shisa-like proteins from a few deuterostomes (Supplementary Figure S3), two B. plicatilis Shisa proteins (Supplementary Figure S4), and some WBP1/VOPP1 proteins from C. briggsae and S. kowalevskii (Supplementary Figure S6). Such domain-level duplication events support that the cysteine-rich domain is an independent evolutionary and folding unit in STMC6 proteins. Duplicated cysteine-rich domains could contribute to extra binding sites for their interaction partners.
Third, at the amino acid residue level, concerted mutations of the conserved cysteines were found independently in a few Shisa and Shisa-like proteins (Figure 5), some of the WBP1/VOPP1 proteins in nematodes (Supplementary Figure S7), and some CX family proteins (Supplementary Figure S8). These mutations suggest the 1-3, 2-5 and 4-6 disulfide bond connectivity in the general C*C*CC*CC pattern of the cysteine-rich domains in STMC6 proteins. On the other hand, the extra cysteine pairs in Shisa proteins and some B. floridae and lophotrochozoan Shisa-like proteins could form an additional disulfide bond. Structural studies of the cysteine-rich domains in STMC6 proteins would be required to validate these disulfide bond connectivity predictions based on co-varying positions.
4.3. Functions of STMC6 proteins
The STMC6 proteins, especially WBP1/VOPP1 and Shisa-like proteins, are widespread in Metazoa (Supplementary Figure S13), yet not much functional information is available for most of them except for Xenopus Shisa1 [16] and mouse Shisa9/CKAMP44 [15]. WBP1 is one of the first WW domain-interacting proteins identified more than a decade ago [55, 60]. The C-terminal region of WBP1 has several PY motifs ([LP]PxY) that are supposed to interact with WW domains. However, no experimental studies have been performed to study the cellular functions of WBP1 ever since. High gene expression of OPAL1, a close homolog of WBP1, was originally indicated as an outcome predictor of acute lymphoblastic leukemia (ALL) [61-62], although a later multivariate analysis has shown that its expression may not be an independent prognostic feature for ALL [56]. VOPP1 (previously named ECOP for EGFR-Coamplified and Over-expressed Protein), a distantly related WBP1 homolog, was implicated in several types of human cancers as a pro-survival protein [57, 63-64]. VOPP1 was found to localize in cytoplasmic vesicles [58]. Its knockdown induces apoptosis that is likely related to oxidative cellular injury [65]. Despite these sporadic studies, the specific functional roles of members in the WBP1/VOPP1 family are not understood. Members from the CX, DUF2650 and TMEM92 families also lack experimental studies and detailed information about their cellular functions.
Like WBP1, the majority of STMC6 proteins contain PY motifs ([LP]PXY) in their C-terminal proline-rich, mainly disordered regions (Supplementary Figures S1, S2, S4 and S6–S11). PY motifs are the most common WW-domain-interacting motifs [6]. WW domains often occur in tandem in a protein. Consistently, two or more PY motifs have been observed in quite a number of STMC6 proteins, suggesting they could interact with multiple WW domains from a single protein. One important class of proteins with tandem WW domains are the NEDD4 family HECT-domain-containing E3 ubiquitin ligases [66]. The NEDD4 family proteins, such as Rsp5 from yeast and the mammalian proteins NEDD4, NEDD4-2, SMURF1, SMURF2 and ITCH, target a variety of proteins for ubiquitination to facilitate their degradation by the proteasome and the lysosome or play roles in their sorting to specific subcellular locations [66]. The presence of an N-terminal C2 domain facilitates membrane localization of the NEDD4 family proteins, and thus many of their targets are transmembrane proteins including ion channels such as epithelial sodium channel (EnaC) [67] and cell surface receptors such as TGF-β [68]. The interactions between the NEDD4 family proteins and their targets are often mediated by the WW domains in the NEDD4 family proteins and the PY motifs in the targets [66].
Some transmembrane proteins do not possess the PY motifs themselves, but are still targeted by the NEDD4 family proteins through interactions with PY-motif-containing adaptor proteins. One example is TGF-β, which degradation can be mediated by SMAD7 [68]. Several transmembrane adaptors with PY motifs have also been identified, such as LAPTM5 [69-70] and NDFIP proteins [71-72], that can promote the degradation of transmembrane proteins. The presence of PY motifs in STMC6 proteins suggests that it could play similar roles. In one simplified model, STMC6 proteins could interact with target membrane proteins and recruit the NEDD4 family E3 ligases to ubiquitinate the targets and promote their degradation (Figure 7a). In this way, the STMC6 proteins act as down-regulators of cell surface expression of the target proteins. Alternatively, competitive binding of STMC6 proteins to NEDD4 members can cause up-regulation of cell surface expression of transmembrane proteins that are otherwise targeted by NEDD4 members, as proposed by some researchers [73-74] (Figure 7b).
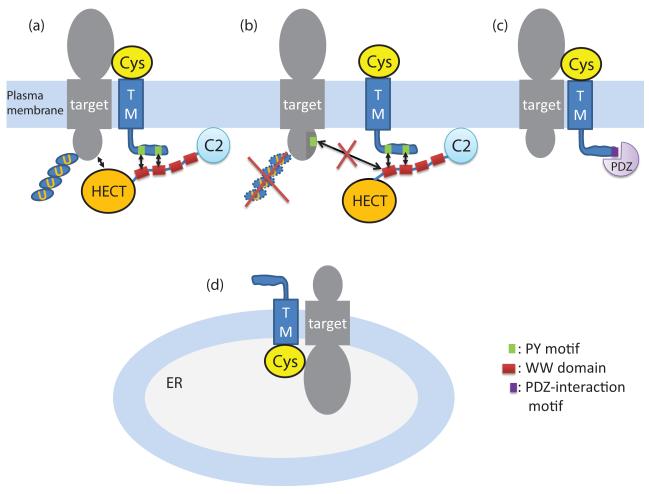
Figure 7. Putative mechanisms of STMC6 proteins.
This diagram depicts four putative mechanisms of STMC6 proteins in regulating other transmembrane proteins. (a). The STMC6 protein can decrease cell surface expression of a target transmembrane protein by recruiting a NEDD4 family E3 ubiquitin ligase to ubiquitinate the target, possibly resulting in its degradation or relocalization. (b). The STMC6 protein can enhance cell surface expression of a target transmembrane protein by competitively interacting with the WW domains of a NEDD4 family E3 ubiquitin ligase and thus blocking the interaction between the E3 ligase and the target. (c). The STMC6 protein Shisa9 (CKAMP44) desensitizes the AMPA receptor by interacting with it. Shisa9 can also potentially interact with PDZ-domain-containing proteins via a C-terminal motif. (d). The STMC6 protein Shisa1 from Xenopus retains receptors in the ER and inhibits their cell surface expression.
PY motifs are present in STMC6 members from the basal metazoan branches, such as Shisa-like proteins from Porifera (Supplementary Figure S4) and WBP1/VOPP1 members from Porifera, Placozoa and Cnidaria (Supplementary Figure S7), suggesting that their interactions with WW-domain containing proteins existed in the ancestor of metazoans.
Some STMC6 proteins do not contain PY motifs in their C-terminal disordered regions, such as Shisa-like protein families FAM159 and KIAA1644 (Supplementary Figure S2) and many nematode members of the WBP1/VOPP1, CX and DUF2650 families (Supplementary Figures S6, S8 and S9). STMC6 proteins without PY motifs may have evolved new functions that do not depend on the interactions with WW-domain-containing proteins. Functional divergence after gene duplications is evidenced for the Shisa family proteins in vertebrates, which have nine subfamilies with divergent C-terminal regions among them (Supplementary Figure S1). Most members of Shisa subfamilies 3–8 contain PY motifs in their C-terminal regions (Supplementary Figure S1), suggesting that they could potentially serve as adaptor proteins to regulate the ubiquitination of other target proteins through the interactions with the NEDD4 family proteins. On the other hand, most members of Shisa subfamilies 1, 2 and 9 lack PY motifs (Supplementary Figure S1), suggesting that they do not target other proteins for ubiquitination and degradation. The mouse Shisa9 protein (CKAMP44) functions to desensitize AMPA receptors in the plasma membrane [15] (Figure 7c). The Xenopus Shisa1 protein plays a role in embryonic development via the retention of Wnt and FGF receptors in the endoplasmic reticulum [16] (Figure 7d). Therefore, existing experimental results suggest that Shisa1 and Shisa9 are both adaptor proteins that regulate the localization or function of receptors. However, their targets, subcellular localizations and cellular functions are different. New functions for the Shisa paralogs could be acquired by evolving new linear sequence motifs in the C-terminal low complexity region. For example, it has been observed that mouse Shisa9 contains a type 2 PDZ-domain interacting signature ([ST]x[VLI]) at the C-terminus [15]. Examination of other Shisa members revealed that such a PDZ-interacting motif is a conserved feature in subfamilies Shisa2 and Shisa6–9, while it is lacking in subfamilies Shisa3–5 (Supplementary Figure S1).
4.4. Comparison of STMC6 proteins with a class of immunoreceptor-related transmembrane adaptor proteins
Several transmembrane adaptor proteins (TRAPs), such as LAT, NTAL, PAG and SIT, play important roles in mediating the functions immunoreceptors [14]. These adaptor proteins do not interact with receptors, but function as downstream targets of immunoreceptor-activated tyrosine kinases. Phosphorylated TRAPs then serve as scaffolds to recruit other proteins to regulate downstream signaling. These immunoreceptor-related TRAPs and STMC6 proteins are all single-transmembrane proteins with cytosolic C-terminal regions that contain linear peptide motifs. However, all characterized immunoreceptor-related TRAPs have very short N-terminal regions before their TMs [14], while STMC6 proteins possess an N-terminal cysteine-rich domain that could interact with the ectodomains of receptors or other transmembrane proteins. In this sense, STMC6 proteins are similar to some receptor-associated proteins that also possess an ectodomain for mediating protein-protein interactions, such as T-cell-receptor-associated CD3 components [75] and B-cell-receptor associated CD79 components [76]. STMC6 proteins could also interact with receptors or other transmembrane proteins via their predicted TMs. For WBP1/VOPP1 proteins (Figure 6), the conserved motif with several aromatic residues in their predicted TMs could serve as interaction sites for other transmembrane proteins. Future experimental investigations are required to identify the interaction partners of STMC6 proteins, which could help reveal their cellular functions.
5. Conclusions
We catalogued a large and diverse group of STMC6 proteins that include the Shisa/Shisa-like, WBP1/VOPP1, CX, DUF2650, TMEM92, and CYYR1 family proteins. STMC6 members are widespread in Metazoa, yet with scarce experimental data. They share a domain structure characterized by an N-terminal cysteine-rich domain with a distinct cysteine pattern, a single predicted transmembrane segment, and a proline-rich C-terminal region. Most STMC6 proteins possess PY motifs in their C-terminal regions, suggesting that they could interact with WW-domain-containing proteins such as the NEDD4 family E3 ubiquitin ligases and play important roles in protein sorting and degradation. These proteins are likely transmembrane adaptors that regulate the functions of other transmembrane proteins such as cell surface receptors, as exemplified by Xenopus Shisa1 and mouse Shisa9/CKAMP44. Future experimental studies on STMC6 proteins could reveal new functional aspects of receptor regulation.
Supplementary Material
Highlights.
Several Shisa proteins play important roles in development and receptor regulation.
STMC6 transmembrane proteins possess a Shisa-like cysteine-rich domain.
STMC6 families also include WBP1/VOPP1, CX, DUF2650, TMEM92 and CYYR1.
PY motifs were frequently identified in the C-termini of STMC6 proteins.
STMC6 proteins are likely adaptors for membrane protein sorting and degradation.
Acknowledgements
We would like to thank Lisa Kinch for critical reading of the manuscript. This work was supported by Howard Hughes Medical Institute, National Institute of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG).
Footnotes
Competing interests None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pawson T, Scott JD. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- [2].Zeke A, Lukacs M, Lim WA, Remenyi A. Trends Cell Biol. 2009;19:364–374. doi: 10.1016/j.tcb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pawson T. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- [4].Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Front Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- [5].Kaneko T, Li L, Li SS. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- [6].Macias MJ, Wiesner S, Sudol M. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- [7].Pawson T, Gish GD, Nash P. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- [8].van der Geer P, Pawson T. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- [9].Saras J, Heldin CH. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- [10].Lemmon MA. Biochem Soc Symp. 2007:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- [12].Ferguson SS. Pharmacol Rev. 2001;53:1–24. [PubMed] [Google Scholar]
- [13].Luttrell LM, Gesty-Palmer D. Pharmacol Rev. 2010;62:305–330. doi: 10.1124/pr.109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Horejsi V, Zhang W, Schraven B. Nat Rev Immunol. 2004;4:603–616. doi: 10.1038/nri1414. [DOI] [PubMed] [Google Scholar]
- [15].von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H. Science. 2010;327:1518–1522. doi: 10.1126/science.1184178. [DOI] [PubMed] [Google Scholar]
- [16].Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- [17].De Robertis EM. Mech Dev. 2009;126:925–941. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Niehrs C. Trends Genet. 1999;15:314–319. doi: 10.1016/s0168-9525(99)01767-9. [DOI] [PubMed] [Google Scholar]
- [19].Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- [20].MacDonald BT, Tamai K, He X. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dorey K, Amaya E. Development. 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nagano T, Takehara S, Takahashi M, Aizawa S, Yamamoto A. Development. 2006;133:4643–4654. doi: 10.1242/dev.02657. [DOI] [PubMed] [Google Scholar]
- [23].Hedge TA, Mason I. Int J Dev Biol. 2008;52:81–85. doi: 10.1387/ijdb.072355th. [DOI] [PubMed] [Google Scholar]
- [24].Zhu Y, Tsuchida A, Yamamoto A, Furukawa K, Tajima O, Tokuda N, Aizawa S, Urano T, Kadomatsu K. Nagoya J Med Sci. 2008;70:73–82. [PubMed] [Google Scholar]
- [25].Furushima K, Yamamoto A, Nagano T, Shibata M, Miyachi H, Abe T, Ohshima N, Kiyonari H, Aizawa S. Dev Biol. 2007;306:480–492. doi: 10.1016/j.ydbio.2007.03.028. [DOI] [PubMed] [Google Scholar]
- [26].Bourdon JC, Renzing J, Robertson PL, Fernandes KN, Lane DP. J Cell Biol. 2002;158:235–246. doi: 10.1083/jcb.200203006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Terrinoni A, Ranalli M, Cadot B, Leta A, Bagetta G, Vousden KH, Melino G. Oncogene. 2004;23:3721–3725. doi: 10.1038/sj.onc.1207342. [DOI] [PubMed] [Google Scholar]
- [28].Zocchi L, Bourdon JC, Codispoti A, Knight R, Lane DP, Melino G, Terrinoni A. Biochem Biophys Res Commun. 2008;367:271–276. doi: 10.1016/j.bbrc.2007.12.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Soding J, Biegert A, Lupas AN. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Katoh K, Kuma K, Toh H, Miyata T. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H, Barber GP, Haussler D, Kent WJ. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kall L, Krogh A. Sonnhammer EL, Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pei J, Grishin NV. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- [37].Adachi J, Hasegawa M. Computer Science Monographs (The Institute of Statistical Mathematics) 1996;28:1–150. [Google Scholar]
- [38].Jones DT, Taylor WR, Thornton JM. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- [39].Hasegawa M, Kishino H, Saitou N. J Mol Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- [40].Katoh Y, Katoh M. Int J Mol Med. 2005;16:181–185. [PubMed] [Google Scholar]
- [41].Linder ME, Deschenes RJ. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- [42].von Heijne G. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- [43].Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- [44].Lowe CJ. Philos Trans R Soc Lond B Biol Sci. 2008;363:1569–1578. doi: 10.1098/rstb.2007.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Belle R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Geneviere AM, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallbook F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Rottinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Telford MJ. Genesis. 2008;46:580–586. doi: 10.1002/dvg.20414. [DOI] [PubMed] [Google Scholar]
- [47].Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- [48].Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ, Klingler M, Lorenzen M, Roth S, Schroder R, Tautz D, Zdobnov EM, Muzny D, Attaway T, Bell S, Buhay CJ, Chandrabose MN, Chavez D, Clerk-Blankenburg KP, Cree A, Dao M, Davis C, Chacko J, Dinh H, Dugan-Rocha S, Fowler G, Garner TT, Garnes J, Gnirke A, Hawes A, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Jackson L, Kovar C, Kowis A, Lee S, Lewis LR, Margolis J, Morgan M, Nazareth LV, Nguyen N, Okwuonu G, Parker D, Ruiz SJ, Santibanez J, Savard J, Scherer SE, Schneider B, Sodergren E, Vattahil S, Villasana D, White CS, Wright R, Park Y, Lord J, Oppert B, Brown S, Wang L, Weinstock G, Liu Y, Worley K, Elsik CG, Reese JT, Elhaik E, Landan G, Graur D, Arensburger P, Atkinson P, Beidler J, Demuth JP, Drury DW, Du YZ, Fujiwara H, Maselli V, Osanai M, Robertson HM, Tu Z, Wang JJ, Wang S, Song H, Zhang L, Werner D, Stanke M, Morgenstern B, Solovyev V, Kosarev P, Brown G, Chen HC, Ermolaeva O, Hlavina W, Kapustin Y, Kiryutin B, Kitts P, Maglott D, Pruitt K, Sapojnikov V, Souvorov A, Mackey AJ, Waterhouse RM, Wyder S, Kriventseva EV, Kadowaki T, Bork P, Aranda M, Bao R, Beermann A, Berns N, Bolognesi R, Bonneton F, Bopp D, Butts T, Chaumot A, Denell RE, Ferrier DE, Gordon CM, Jindra M, Lan Q, Lattorff HM, Laudet V, von Levetsow C, Liu Z, Lutz R, Lynch JA, da Fonseca RN, Posnien N, Reuter R, Schinko JB, Schmitt C, Schoppmeier M, Shippy TD, Simonnet F, Marques-Souza H, Tomoyasu Y, Trauner J, Van der Zee M, Vervoort M, Wittkopp N, Wimmer EA, Yang X, Jones AK, Sattelle DB, Ebert PR, Nelson D, Scott JG, Muthukrishnan S, Kramer KJ, Arakane Y, Zhu Q, Hogenkamp D, Dixit R, Jiang H, Zou Z, Marshall J, Elpidina E, Vinokurov K, Oppert C, Evans J, Lu Z, Zhao P, Sumathipala N, Altincicek B, Vilcinskas A, Williams M, Hultmark D, Hetru C, Hauser F, Cazzamali G, Williamson M, Li B, Tanaka Y, Predel R, Neupert S, Schachtner J, Verleyen P, Raible F, Walden KK, Angeli S, Foret S, Schuetz S, Maleszka R, Miller SC, Grossmann D. Nature. 2008;452:949–955. [Google Scholar]
- [49].Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Su J, Wang X, Zhang Z, Wu Q, Li J, Zhang Q, Wei N, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan F, Huang X, Li Y, Fang L, Li D, Sun Y, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Ye J, Chen H, Zhou Y, Liu B, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Wong GK, Yang H. Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- [50].Meusemann K, von Reumont BM, Simon S, Roeding F, Strauss S, Kuck P, Ebersberger I, Walzl M, Pass G, Breuers S, Achter V, von Haeseler A, Burmester T, Hadrys H, Wagele JW, Misof B. Mol Biol Evol. 2010;27:2451–2464. doi: 10.1093/molbev/msq130. [DOI] [PubMed] [Google Scholar]
- [51].Rubinstein R, Fiser A. Bioinformatics. 2008;24:498–504. doi: 10.1093/bioinformatics/btm637. [DOI] [PubMed] [Google Scholar]
- [52].Yang Y, Poncet J, Garnier J, Zatylny C, Bachere E, Aumelas A. J Biol Chem. 2003;278:36859–36867. doi: 10.1074/jbc.M305450200. [DOI] [PubMed] [Google Scholar]
- [53].Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, Bennett HP, Bateman A, Ni F. Protein Sci. 2008;17:711–724. doi: 10.1110/ps.073295308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tolkatchev D, Ng A, Vranken W, Ni F. Biochemistry. 2000;39:2878–2886. doi: 10.1021/bi992130u. [DOI] [PubMed] [Google Scholar]
- [55].Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, Kelly JW, Sudol M. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- [56].Holleman A, den Boer ML, Cheok MH, Kazemier KM, Pei D, Downing JR, Janka-Schaub GE, Gobel U, Graubner UB, Pui CH, Evans WE, Pieters R. Blood. 2006;108:1984–1990. doi: 10.1182/blood-2006-04-015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Eley GD, Reiter JL, Pandita A, Park S, Jenkins RB, Maihle NJ, James CD. Neuro Oncol. 2002;4:86–94. doi: 10.1093/neuonc/4.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baras A, Moskaluk CA. J Mol Histol. 2010;41:153–164. doi: 10.1007/s10735-010-9272-8. [DOI] [PubMed] [Google Scholar]
- [59].Vitale L, Casadei R, Canaider S, Lenzi L, Strippoli P, D’Addabbo P, Giannone S, Carinci P, Zannotti M. Gene. 2002;290:141–151. doi: 10.1016/s0378-1119(02)00550-4. [DOI] [PubMed] [Google Scholar]
- [60].Chen HI, Sudol M. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mosquera-Caro M, Helman P, Veroff R, Shuster J, Martin S, Davidson G, Potter J, Harvey R, Hromas R, Andries E, Atlas S, Wilson C, Ar K, Xu YX, Chen IM, Carroll A, Camitta B, Willman C. Blood. 2003;102:4A–4A. [Google Scholar]
- [62].Carroll WL, Bhojwani D, Min DJ, Raetz E, Relling M, Davies S, Downing JR, Willman CL, Reed JC. Hematology Am Soc Hematol Educ Program. 2003:102–131. doi: 10.1182/asheducation-2003.1.102. [DOI] [PubMed] [Google Scholar]
- [63].Baras A, Yu Y, Filtz M, Kim B, Moskaluk CA. Oncogene. 2009;28:2919–2924. doi: 10.1038/onc.2009.150. [DOI] [PubMed] [Google Scholar]
- [64].Park S, James CD. Oncogene. 2005;24:2495–2502. doi: 10.1038/sj.onc.1208496. [DOI] [PubMed] [Google Scholar]
- [65].Baras AS, Solomon A, Davidson R, Moskaluk CA. Lab Invest. 2011 doi: 10.1038/labinvest.2011.70. [DOI] [PubMed] [Google Scholar]
- [66].Ingham RJ, Gish G, Pawson T. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- [67].Eaton DC, Malik B, Bao HF, Yu L, Jain L. Proc Am Thorac Soc. 2010;7:54–64. doi: 10.1513/pats.200909-096JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- [69].Ouchida R, Kurosaki T, Wang JY. J Immunol. 2010;185:294–301. doi: 10.4049/jimmunol.1000371. [DOI] [PubMed] [Google Scholar]
- [70].Ouchida R, Yamasaki S, Hikida M, Masuda K, Kawamura K, Wada A, Mochizuki S, Tagawa M, Sakamoto A, Hatano M, Tokuhisa T, Koseki H, Saito T, Kurosaki T, Wang JY. Immunity. 2008;29:33–43. doi: 10.1016/j.immuni.2008.04.024. [DOI] [PubMed] [Google Scholar]
- [71].Mund T, Pelham HR. EMBO Rep. 2009;10:501–507. doi: 10.1038/embor.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Howitt J, Putz U, Lackovic J, Doan A, Dorstyn L, Cheng H, Yang B, Chan-Ling T, Silke J, Kumar S, Tan SS. Proc Natl Acad Sci U S A. 2009;106:15489–15494. doi: 10.1073/pnas.0904880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Konstas AA, Shearwin-Whyatt LM, Fotia AB, Degger B, Riccardi D, Cook DI, Korbmacher C, Kumar S. J Biol Chem. 2002;277:29406–29416. doi: 10.1074/jbc.M203018200. [DOI] [PubMed] [Google Scholar]
- [74].Ohzono C, Etoh S, Matsumoto M, Nakayama KI, Hirota Y, Tanaka Y, Fujita H. Biol Pharm Bull. 2010;33:951–957. doi: 10.1248/bpb.33.951. [DOI] [PubMed] [Google Scholar]
- [75].Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Cold Spring Harb Perspect Biol. 2010;2:a005140. doi: 10.1101/cshperspect.a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Neuberger MS, Patel KJ, Dariavach P, Nelms K, Peaker CJ, Williams GT. Immunol Rev. 1993;132:147–161. doi: 10.1111/j.1600-065x.1993.tb00841.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.