Abstract
Exchange of macromolecules between the nucleus and cytoplasm is a key regulatory event in the expression of a cell’s genome. This exchange requires a dedicated transport system: (1) nuclear pore complexes (NPCs), embedded in the nuclear envelope and composed of proteins termed nucleoporins (or “Nups”), and (2) nuclear transport factors that recognize the cargoes to be transported and ferry them across the NPCs. This transport is regulated at multiple levels, and the NPC itself also plays a key regulatory role in gene expression by influencing nuclear architecture and acting as a point of control for various nuclear processes. Here we summarize how the yeast Saccharomyces has been used extensively as a model system to understand the fundamental and highly conserved features of this transport system, revealing the structure and function of the NPC; the NPC’s role in the regulation of gene expression; and the interactions of transport factors with their cargoes, regulatory factors, and specific nucleoporins.
ALTHOUGH considered “simple,” its amenability to molecular and genetic interrogation has established baker’s yeast as an outstanding model system for cell biologists. Moreover, in the context of the Eukaryota, Saccharomyces cerevisiae is closely related to humans (both being members of the opisthokonts). Thus, interrogation of the fundamental biology of yeast has proven to be not only comparatively facile, but also highly relevant to human biology, both morphologically and mechanistically. Indeed, yeast has remained at the forefront of studies on the nucleus—the defining characteristic of eukaryotes—for several decades.
Eukaryotic chromosomes are housed within the nucleus, which is delimited by the two parallel membranes of the nuclear envelope (NE). The evolution of this physical barrier endowed eukaryotes with a critical control mechanism segregating the sites of gene transcription and ribosome biogenesis from the site of protein synthesis. This compartmentalization allows cells to strictly coordinate numerous key cellular processes, but it also presents cells with the challenge of selectively managing the transport of a bewildering number of proteins and RNAs between the nucleus and cytoplasm. This is accomplished by the presence of “nuclear pores,” which arise at points where the inner and outer NE membranes conjoin to form circular channels across the nuclear envelope. Within these pores sit large proteinaceous complexes, appropriately named nuclear pore complexes (NPCs), which, in conjunction with soluble transport factors, govern all biomolecular transport into and out of the nucleus. Beyond this fundamental control of transport, the NPC has adopted a host of other activities by acting as a spatial landmark or anchor site for many of the machineries that directly control gene activity and transcriptional processing (reviewed in Ahmed and Brickner 2007; Hetzer and Wente 2009). As a transporter, it must allow small molecules to pass as freely, prevent most macromolecules from crossing, and permit the quickest possible passage of selected macromolecules bidirectionally across the NE. As an anchor, it must allow free communication between the attached control machineries and the chromatin or transcripts that they regulate without hindering nuclear transport. One can thus also consider the NPC as a major way station in eukaryotes, interacting with and regulating DNA, RNA, and membranes and communicating between the cytoplasm, nucleoplasm, and ER lumen. Because of this, the subject of the nuclear pore complex and nuclear transport is a huge one, far beyond the scope of any single review. Our aim here is therefore to give an overview, including references to many excellent reviews that detail particular areas of study.
Structure and Composition of the NPC
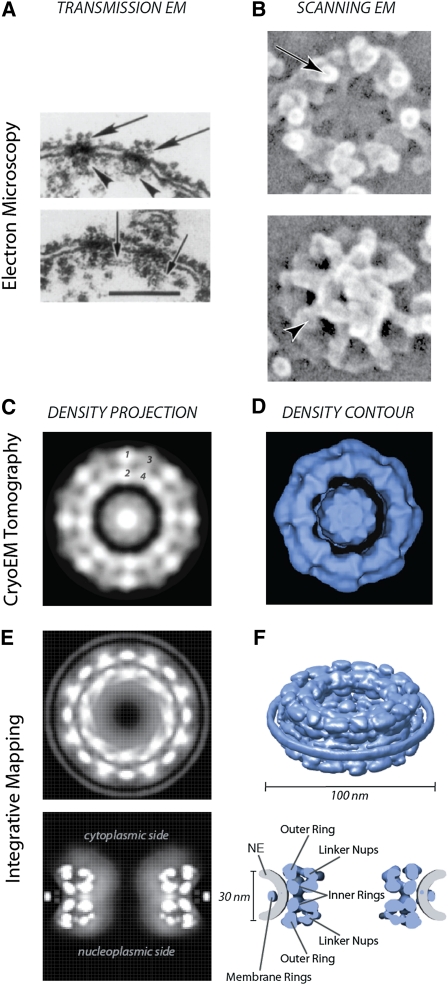
Tomographic electron cryomicroscopy and high-resolution scanning electron microscopy on rotary-shadowed specimens have shown that the yeast NPC shares its overall architectural features with those studied in other eukaryotes, although it is somewhat smaller, being ∼100 nm in diameter and ∼40 nm in height as compared with ∼130 nm × ∼80 nm for its vertebrate counterpart (Yang et al. 1998) (Figure 1). The core of the NPC consists of an octagonally symmetrical cylinder, the axis of which lies perpendicular to the plane of the NE. This core is made of coaxial inner, outer, and membrane rings surrounding a central channel (or “central transporter”) of ∼40-nm diameter through which virtually all nucleocytoplasmic trafficking occurs (membrane proteins excepted). The circular membrane of the nuclear pores actually passes between the membrane ring and the outer/inner rings, thus anchoring the NPC firmly into the NE. Eight short filaments can be seen to project from the core into the cytoplasm, and, similarly, eight filaments extend ∼50 nm into the nucleoplasm, where they conjoin distally to form a structure said to resemble a “basket” on the nuclear face of the NPC (Fahrenkrog et al. 1998; Yang et al. 1998; Kiseleva et al. 2004) (Figure 1). Starting in 1990, researchers took advantage of yeast genetic screens as well as cross-reacting monoclonal antibodies made against vertebrate NPCs to identify the first NPC components (termed nucleoporins or “Nups”) in yeast. Genetic and biochemical methods then steadily and rapidly filled in the list of yeast Nups (reviewed in Wente and Rout 2010), ultimately culminating in the yeast NPC being the first to have its composition cataloged (Rout et al. 2000). Nevertheless, the precise definition of what constitutes a “Nup” remains somewhat arbitrary, as many proteins that associate with the NPC do so transiently, with varying dwell times, and some NPC-associated proteins also extend their functions and localizations beyond the NPC (Arib and Akhtar 2011).
Figure 1 .
Visualizing the yeast NPC. (A) Transmission EM transverse sections of the NE revealing cytoplasmic filamants (large arrows), nuclear baskets (arrowheads), and interbasket connections (small arrows) (Rout and Blobel 1993). (B) Scanning EM showing a bird’s eye view of cytoplasmic filaments (top, arrow) and the nuclear basket (bottom, arrowhead) (Kiseleva et al. 2004). (C) En face slice of the mass density distribution from a cryoEM map of the yeast NPC (Yang et al. 1998). Two equivalent peaks per spoke unit are seen for the outer rings (1–3) and the inner rings (2–4) (see Figure 2). (D) En face surface-rendered view from a cryoEM map of the yeast NPC (Yang et al. 1998). (E) Projections of Nup mass density, derived from the combined Nup localization volumes (Alber et al. 2007b). (F) The structured nucleoporin domains of the NPC, represented by a density contour approximated to the combined volume of the 456 nucleoporins composing the NPC (Alber et al. 2007b).
The sheer size and flexibility of the NPC make it difficult to fully solve its molecular architecture by conventional techniques. Therefore, an orthogonal approach has been taken; large and diverse sets of proteomic data were amassed and a computational method for using these data was developed to define the relative positions and proximities of the yeast NPC’s constituent proteins. A corresponding average protein density map represents the position of every Nup with a precision of ∼5 nm, sufficient to resolve the molecular organization of the entire NPC (Alber et al. 2007a,b) (Figure 2A). The resulting map agrees with complementary data in both yeast and vertebrates (reviewed in Strambio-de-Castillia et al. 2010).
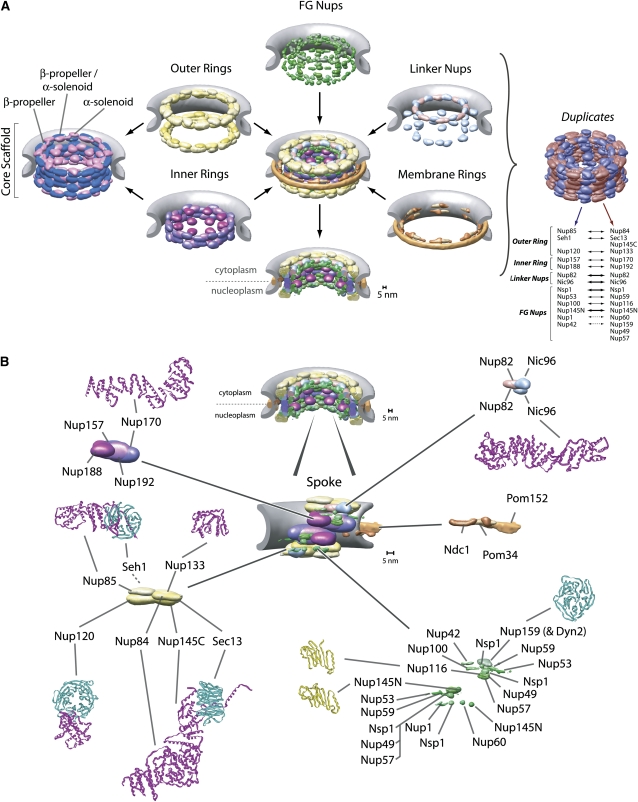
Figure 2 .
(A) Major structural features of the yeast NPC (based on the architectural map of Alber et al. 2007a,b); see main text for details. (B) Map of protein positions in the yeast NPC (based on the architectural map of Alber et al. 2007a,b), with examples of the atomic structures of pieces of Nups where known: Nic96 (2RFO) (Schrader et al. 2008), Nup84/Nup145C/Sec13 (3IKO) (Nagy et al. 2009), Nup85/Seh1 (3EWE) (Brohawn et al. 2008), Nup116 (2AIV) (Robinson et al. 2005), Nup120 (3F7F) (Seo et al. 2009), Nup133 (3KFO), Nup145N (3KEP) (Sampathkumar et al. 2010), Nup159 (1XIP) (Weirich et al. 2004), and Nup170 (3I5P) (Whittle and Schwartz 2009).
Overall Composition
Computational fold predictions and biochemical domain mapping have analyzed the fold composition of every Nup (Devos et al. 2004, 2006; Dokudovskaya et al. 2006), showing that the NPC is surprisingly simple in terms of fold composition, consisting of a few, highly repetitive fold types (Figure 3). This finding, supported by recently solved crystal structures for several Nups (Figure 2B) (see below), indicates that the bulk of its structure has evolved through extensive gene duplication from a simple precursor set of only a few proteins. Indeed, each spoke can be divided into two parallel columns, in which every Nup in the first column carries a similarly positioned homolog in the adjacent column (Figure 2A). This pattern has been interpreted as resulting from at least one (and likely more) ancient genome duplication events, which gave rise to the two columns composing each spoke. S. cerevisiae is one of several related yeast that have undergone a whole-genome duplication with subsequent gene loss (Wolfe and Shields 1997; Kellis et al. 2004). Yeast-specific homologous Nup pairs such as Nup116p/Nup100p, Nup157p/Nup170p, and Nup53p/Nup59p, which exist only as single proteins in vertebrate NPCs (Cronshaw et al. 2002), are segregated pairwise into the two columns in each spoke and so enhance the partitioning of Nups to these columns, although the functional reason for maintaining these proteins as homologous pairs in yeast is still not clear (Figure 2). Most of the motifs, domains and even domain arrangements, and Nup types found in yeast NPCs are also found in vertebrate NPCs, and even in the NPC from the most divergent eukaryote studied so far (Trypanosoma; member of the kinetoplastida), albeit with some variations (DeGrasse et al. 2009). This implies strongly that the basic structural elements of the NPC are conserved across all eukaryotes and that the NPCs of the last common ancestor of all eukaryotes had most of the major attributes of modern NPCs. It also means that studies of the yeast NPC are reasonably representative of the Eukaryota as a whole and are relevant for medically significant organisms, including ourselves.
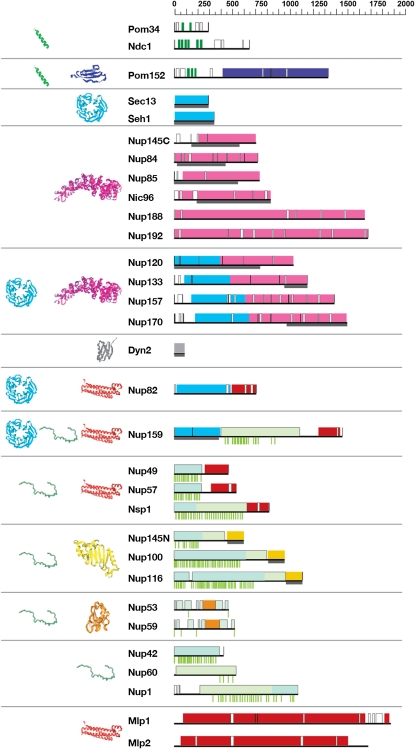
Figure 3 .
Map of major fold-type positions in the yeast nucleoporins [adapted from Devos et al. (2006)]. Here, for the sake of clarity, we define Nups as proteins that appear stoichiometrically and stably associated with the NPC. The sequence of each yeast Nup is represented to scale as a thin black horizontal line. Predicted transmembrane helices are shown in dark green, cadherin domains are in dark blue, coiled coils and α-helical coils are shown in red. β-Propellers are shown in cyan, α-solenoid domains are in magenta, the autoproteolytic domain is in yellow, and the RRM is shown in orange. Unstructured regions are shown by an empty box, except for the FG-repeat regions, which are colored blue-green for low-DERK (Asp, Glu, Arg, Lys) regions and light green for high-DERK regions; the position of each FG repeat is shown as a short green vertical line below each horizontal black sequence line. Representative models are shown on the left of the Nup domains and are colored according to the fold type. Dark-gray bars below each horizontal sequence line mark the position of crystal structures solved for yeast Nups (e.g., N-terminal region of Nup159). Position of folds is based on Devos et al. (2004, 2006); and position and type of FG repeat is based on Yamada et al. (2010).
Membrane Ring
The first class of NPC components is the set of three membrane proteins, called “poms,” which compose the membrane ring forming a distinct subcomplex in the NPC (Alber et al. 2007b; Onischenko et al. 2009) (Figure 2). All are predicted to carry transmembrane α-helices, which likely help to anchor the NPC in the NE, interact with core components and one another, and have functional redundancy perhaps as membrane domain anchors and stabilizers (Chial et al. 1998; Miao et al. 2006). This is consistent with a network of interactions formed between these three proteins (Onischenko et al. 2009). The first to be discovered and largest pom is Pom152p, a type II transmembrane protein (Wozniak et al. 1994; Tcheperegine et al. 1999). Secondary structure predictions reveal numerous repetitive motifs in the lumenal domain that are strongly indicated to form a series of cadherin folds (Devos et al. 2006) (Figure 3). This cadherin domain is predicted to form homophilic binding interfaces (Bryant and Stow 2004) and likely explains the oligomeric lumenal ring. Much less is known about Pom34. It is a small protein containing two transmembrane helices and two small domains both facing into the core scaffold. As yet, no function has been assigned to Pom34—even cells lacking both Pom34 and Pom152 are viable—and no nonfungal homolog has yet been found. However, like Pom152, Pom34 genetically interacts with several core scaffold proteins in ways that suggest these poms’ functions partially overlap (see below) (Madrid et al. 2006; Miao et al. 2006). Ndc1p (nuclear division cycle 1) is also a pom but was named differently because it was actually first characterized due to the effect of one of its mutants on the assembly of the spindle. Indeed, it turns out that, like several other Nups, Ndc1p plays at least two roles in the cell: in the case of Ndc1p, one in the NPC and one in the spindle pole body (SPB)—the mitotic spindle organizer. In yeast, both macromolecular assemblies are embedded in the NE, and it seems that Ndc1p helps to insert and attach both into their respective nuclear pores. Ndc1p has (at least) six transmembrane helices and a carboxy-terminal domain that interacts with the core scaffold and other poms (Alber et al. 2007b; Onischenko et al. 2009) and has confirmed homologs in both Schizosaccharomyces pombe [i.e., cut11, also with known roles in spindle assembly and an NPC component (West et al. 1998)] and metazoa [i.e., NDC1 (Lau et al. 2004; Mansfeld et al. 2006; Stavru et al. 2006)]. Dissecting the NPC function of Ndc1p from its SPB function has been difficult, but several lines of genetic (Lau et al. 2004) and molecular biological (Onischenko et al. 2009) evidence suggest that Ndc1p plays an important role, with the help of the other poms, in NPC assembly (see also below).
Core Scaffold: Inner and Outer Rings
The second class of NPC components comprises the core scaffold proteins. This scaffold is composed of an interlocking lattice of roughly a dozen evolutionarily conserved structural proteins that link together to form a core layer giving the NPC shape and strength (Figure 2). Fold composition analyses revealed that the core scaffold consists of Nups composed of only two fold types in three arrangements: consisting almost entirely of a b-propeller fold, or almost entirely of a-solenoid-like/helix-turn-helix repeat folds, or a b-propeller followed by an a-solenoid-like domain (Figure 3). These fold types together are characteristic of components of the clathrin, COPI and COPII membrane vesicle-coating complexes (reviewed in Field and Dacks 2009), and related complexes such as the intraflagellar transport complex (Jekely and Arendt 2006) and the HOPS/CORVET complexes (Nickerson et al. 2009). The latter β-propeller/α-solenoid combination is particularly characteristic of this family, as these protein types interlock in a variety of related ways into a lattice forming their vesicle coats. These similarities were recently further underscored by atomic structures solved for several core scaffold Nups (Figure 2B) (Boehmer et al. 2003; Berke et al. 2004; Hsia et al. 2007; Jeudy and Schwartz 2007; Brohawn et al. 2008; Debler et al. 2008; Schrader et al. 2008; Brohawn and Schwartz 2009; Leksa et al. 2009; Nagy et al. 2009; Seo et al. 2009; Whittle and Schwartz 2009) as well as clathrin (reviewed in Owen et al. 2004) and COPII and COPI components (Stagg et al. 2006; Fath et al. 2007; Lee and Goldberg 2010; reviewed in Stagg et al. 2007, 2008). This fold similarity, analyzed initially in yeast, led to the “protocoatomer hypothesis,” which proposes that a simple membrane-curving module, made primarily from β-propeller and α-solenoid folds, was a common ancestor for NPCs and coated vesicles that originated in the precursors to the ancient last ancestor common to all eukaryotes (Devos et al. 2004, 2006; Alber et al. 2007a,b). This protcoatomer gave these ancestors the ability to generate internal membrane systems by invagination of the plasma membrane and then to manipulate and elaborate these systems, eventually leading to the evolution of the ER, Golgi, and nucleus that characterize modern eukaryotes (Field and Dacks 2009).
The core scaffold somewhat resembles a vesicle coat, as it forms a discrete layer completely following the curve of the pore membrane, effectively coating it (Figure 2). Thus, it defines the size of the central tube/transporter of the NPC and the height of the NPC core, and all other Nups and poms are attached to either the inner or the outer face of this coat (Figure 1). Biochemical studies have shown that the core scaffold Nups compose several subcomplexes that appear to function as “building blocks” during NPC assembly and can even exchange with a soluble pool in mature NPCs (Lutzmann et al. 2002; D’Angelo et al. 2006; Makio et al. 2009). Morphologically, the scaffold is made of two inner rings, sandwiched between and interconnected with two outer rings, such that the nuclear and cytoplasmic halves of the NPC have one inner and one outer ring each (Figure 2). Although there is still some debate on the matter, a consensus remains that in both yeast and vertebrates these inner rings are compositionally, as well as morphologically, distinct (Tran and Wente 2006; Alber et al. 2007b).
Four large Nups, each just under 200 kDa in size, compose the inner rings: Nup188p and Nup192p (primarily composed of α-solenoid-like folds) and the homologous Nup170p and Nup157p (Figure 2) (made from the clathrin-like pattern of an amino-terminal β-propeller followed by an α-solenoid-like domain), proteins proposed long ago to be core components of the NPC (Aitchison et al. 1995b). The inner rings are adjacent to each other at the equator of the NPC in the same plane as the three poms of the membrane ring with which they interact extensively to anchor the core scaffold to the NE (Alber et al. 2007b; Onischenko et al. 2009). Numerous mutations in all four proteins demonstrate extensive genetic interaction networks with each other and with the membrane ring components, which likely underscore the functional importance of the inner ring in being a keystone of the core scaffold and in anchoring the NPC to the pore membrane (Aitchison et al. 1995b; Tcheperegine et al. 1999; Miao et al. 2006). Moreover, on both the nuclear and cytoplasmic sides of each spoke, one copy of the Nup Nic96p is anchored through Nup192p and a second copy through Nup188p, linking the inner ring to the other internal structures of the NPC (Figure 2) (see below).
Structurally, the most extensively studied set of core scaffold proteins are the seven Nups that compose the yeast outer rings, first identified and characterized by the Hurt laboratory; Nup133p, Nup120p, Nup145Cp, Nup85p, Nup84p, Seh1p, and Sec13p form a discrete complex that can be biochemically isolated and is termed the Nup84 complex (Figure 2B) (Siniossoglou et al. 1996, 2000; Lutzmann et al. 2002; Flemming et al. 2009). Importantly, the evolutionary link between NPCs and vesicle-coating complexes is supported by the fact that Sec13p is shared with the Sec13/31 COPII vesicle-coating complex (Siniossoglou et al. 1996; Salama et al. 1997; Devos et al. 2004). Moreover, just like the inner-ring proteins, all Nup84 complex proteins are formed almost entirely by a β-propeller fold (Seh1p, Sec13p), an α-solenoid-like fold (Nup85p, Nup84p, Nup145Cp), or an N-terminal β-propeller and a carboxy-terminal α-solenoid-like fold (Nup133p, Nup120p), again common to vesicle-coating complexes (Devos et al. 2004, 2006) (Figure 2B and Figure 3). Excitingly, crystal structures primarily from the Blobel and Schwartz laboratories have begun to piece this complex together at the atomic level (Figure 2B) (Hsia et al. 2007; Brohawn et al. 2008; Debler et al. 2008; Brohawn and Schwartz 2009; Leksa et al. 2009; Nagy et al. 2009; Seo et al. 2009; Whittle and Schwartz 2009). Electron microscopy studies have shown that the Nup84 complex formed an extended Y structure (Siniossoglou et al. 2000; Lutzmann et al. 2002; Kampmann and Blobel 2009), and pioneering work from the Hurt laboratory reconstituted this complex from bacterially expressed proteins and showed that the two short arms of this Y are composed, respectively, of Nup120p and Nup85p+Seh1p, while Nup133p, Nup84p, and Nup145Cp/Sec13p form the main stalk (Siniossoglou et al. 2000; Lutzmann et al. 2002). There is evidence that this complex is flexible (Kampmann and Blobel 2009), perhaps reflecting the known flexibility of the NPC in response to changes in NE shape and during its assembly (see below).
Few interactions have been found between the components of the Nup84 complex and the rest of the NPC, although it connects with the inner rings through, for example, a Nup157p–Nup120p connection (Lutzmann et al. 2005; Alber et al. 2007a,b). Mutations in any of these seven Nups are often characterized by temperature sensitivity, messenger RNA (mRNA) and pre-ribosomal export problems, and aberrant NPC assembly. In particular, a so-called “clustering” phenotype was first described in mutants of the Nup84 complex components in which the NPCs can be seen to cluster into one or a few patches in the NE (Doye et al. 1994; Aitchison et al. 1995a; Heath et al. 1995; Li et al. 1995; Pemberton et al. 1995; Goldstein et al. 1996; Siniossoglou et al. 1996). That outer-ring Nup mutants cause mislocalization of otherwise reasonably functional NPCs in the plane of the NE may point to a role for this structure in keeping the NPC stably located in the pore membrane. The outer rings are strategically placed at the point where the pore membrane joins the coplanar outer and inner NE membranes, and it seems reasonable that a major role for them is to ensure the smooth transition of the pore membrane into the inner and outer NE membranes (Figure 2) (Alber et al. 2007b).
Phenylalanine-Glycine Nups
It was the field’s catalog of the composition of the yeast NPC that led to a surprise. No homologs of mechanochemical proteins or NTPases of any kind that could physically drive a gating process were found as components of the NPC. Instead, strikingly, the cataloging revealed that over one-third of Nups in the NPC shared a highly characteristic repetitive motif, consisting of multiple repeated phenylalanine-glycine (FG) pairs spaced by ∼20 mainly polar amino acids (Figure 3). Although these proteins are found in all eukaryotes studied, once again they were first sequenced from yeast (Hurt 1988; Davis and Fink 1990; Nehrbass et al. 1990), and the yeast “FG Nups” remain the best studied. On the basis initially of work in yeast, two flavors of FG Nups were described: FxFG Nups typified by Nsp1p and GLFG Nups typified by Nup100p (on the basis of the typical sequence of their FG repeat), the former having some charged amino acids in their spacers and the latter having relatively uncharged spacers (reviewed in Rout and Wente 1994). These two flavors of FG repeat also appear to be conserved (Figure 3), although by examination of orthologs in syntenic yeasts it was shown that the spacer sequences between each of the FG repeats evolved more rapidly than did other Nups (Denning and Rexach 2007), a situation common to the FG-repeat regions of all eukaryotes (DeGrasse et al. 2009). On the basis of various physical measurements of purified and bacterially expressed proteins, a consensus has emerged that the FG-repeat regions of FG Nups take on a natively unfolded structure both in vitro and in vivo, such that they form long, disordered flexible filaments (Denning et al. 2003; Denning and Rexach 2007; Lim et al. 2006a,b, 2007a, 2008; Patel et al. 2007; Yamada et al. 2010); the lack of structural constraints therefore likely explains the lower evolutionary constraints on sequence conservation of these repeat regions (Denning and Rexach 2007).
FG Nups also usually carry small structured domains, which serve to anchor them to the NPC (Alber et al. 2007b). These are generally predicted to be coiled-coil or α-helical by fold analysis and comparison to vertebrate homologs (Melcak et al. 2007), in addition to β-sandwich and (oddly) RNA recognition motif (RRM) folds (Figure 3) (Devos et al. 2006). Nup82p and Nic96p seem to serve as linkers to attach many of the FG Nups to the core scaffold (mainly through the inner-ring Nups), such that the inner surface of the scaffold is lined with FG Nups whose filamentous FG-repeat regions fill the central channel and extend into the nucleoplasm and cytoplasm (Figure 2). On both the cytoplasmic and nucleoplasmic sides of each spoke two copies of Nic96p carry the FG Nups Nsp1p, Nup57p, and Nup49p and another two copies form interactions to additional copies of Nsp1p, such that these FG Nups face both the nucleus and the cytoplasm. At the cytoplasmic side, Nup82p associates with Nsp1p as well as with the cytoplasmically facing FG Nups Nup159p, Nup116p, Nup100p, and Nup42p (Grandi et al. 1995; Belgareh et al. 1998; Bailer et al. 2000, 2001; A. K. Ho et al. 2000; Rout et al. 2000; Alber et al. 2007a,b). There are also the FG Nups Nup145Np, Nup1p, and Nup60p found on the nucleoplasmic side, connecting mainly to the inner-ring Nups. In addition, Nup53p and Nup59p are attached to Nup170p and Nic96p, and both face the pore membrane (Figure 2). The latter two Nups may also belong to the class of FG Nups, as they can bind transport factors and carry degenerate FG-repeat regions that are predicted to be natively unfolded (Marelli et al. 1998; Fahrenkrog et al. 2000b; Lusk et al. 2002; Makhnevych et al. 2003; Alber et al. 2007b). One FG Nup, Nup145p, uniquely cleaves itself in half at its Phe605-Ser606 peptide bond to produce two separate Nups, Nup145Np (carrying the FG-repeat region and the autoproteolytic β-sandwich domain at its new carboxy-terminus) and Nup145Cp (a mainly α-solenoid-like protein that forms a major component of the Nup84 complex) (Figure 3) (Wente and Blobel 1994; Teixeira et al. 1997, 1999; Rosenblum and Blobel 1999). Although not essential in yeast (Emtage et al. 1997), this cleavage event appears conserved in vertebrates (Fontoura et al. 1999; Hodel et al. 2002; Sun and Guo 2008). Nup145Cp and Nup145Np remain linked as a dynamic complex such that Nup145Np can shuttle between the NPC and the nuclear interior, as does its vertebrate counterpart (Griffis et al. 2002; Ratner et al. 2007). The entirety of Nup145Np is highly conserved with the homologous yeast nucleoporins Nup100p and Nup116p, neither of which undergoes autoproteolysis as they lack a homologous counterpart for Nup145Cp. It seems that lineage-specific gene duplications of an ancestral Nup145N-like gene gave rise to the truncated versions Nup100p and Nup116p, and likely other FG Nups also originated from such, sometimes more ancient, duplication events (Mans et al. 2004; Devos et al. 2006).
Collectively, the anchored FG Nups form the business end of the NPC, as the FG-repeat domains form low-affinity, high-specificity interactions with transport factors involved in active transport through the NPC and so actually form the selective barrier in and around the central tube by providing the binding sites for transport factors that facilitate their exchange across the NE while excluding the passage of macromolecules not destined for nucleocytoplasmic transport. In one sense, the NPC can be considered a framework that provides the correct positioning of the FG repeats, flanking and filling the central tube while defining the upper diameter of the central tube and the cargoes that transit through it (Figures 1 and 2) (Rout and Aitchison 2001; Rout et al. 2003; Hetzer and Wente 2009; Walde and Kehlenbach 2010).
Cytoplasmic Filaments and Nuclear Basket
While not as morphologically prominent as their vertebrate counterparts, both cytoplasmic filaments and nuclear baskets have been seen to project from the yeast NPC (Figure 1) (Kiseleva et al. 2004). Nup159p, Nup82p, and Nup42p seem to contribute to the cytoplasmic filaments (Kraemer et al. 1995; Hurwitz et al. 1998; Strahm et al. 1999; Rout et al. 2000; Alber et al. 2007b) and function in the last stages of export from the NPC (below). Curiously, the protein Dyn2p, a light chain component of the dynein microtubule motor, binds to Nup159p and helps form a rigid filamentous structure that may stiffen the cytoplasmic filament projecting it out from the core scaffold (Stelter et al. 2007). Yeast lack an obvious homolog of the vertebrate Nup358, which is believed to produce the more prominent cytoplasmic filaments in the latter (Wu et al. 1995; Matunis et al. 1998). In vertebrates, the bulk of the nuclear basket seems to be made of Tpr (Cordes et al. 1997; Hase et al. 2001; Frosst et al. 2002; Krull et al. 2004; Qi et al. 2004). Tpr is a conserved ∼200-kDa protein made mainly of extensive coiled-coil domains that dimerize into long rods forming the basket struts. Two Tpr homologs, Mlp1p and Mlp2p, exist in yeast and localize to the region of the nuclear basket (Strambio-de-Castillia et al. 1999). Unlike metazoa, no lamina lies interwoven between NPCs beneath the NE, but both Mlp1p and Mlp2p spread out along the inner face of the NE to form a delicate network interconnecting yeast NPCs, although they are excluded where the dense crescent of the nucleolus presses against the NE (Strambio-de-Castillia et al. 1999). Mlp2p is additionally associated with the SPB (Niepel et al. 2005). In yeast, Mlp2p is the result of the specific genome duplication; however, a spindle organizer-specific copy of Tpr homologs has been independently reinvented several times in evolution for reasons that are still unclear (Jimenez et al. 2000; DeGrasse et al. 2009). Overall, a bewildering array of functionalities have been ascribed to the Mlp network, including roles in recruitment of transport factors, late processing of transcripts, and epigenetic regulation of gene expression, as will be discussed below.
Shuttling Nucleoporins
The definition of nucleoporins becomes more difficult when one considers the dynamics of some of the classically defined nucleoporins. Nup2p, for example, was defined as a nucleoporin as early as 1993 on the basis of its localization to the NPC; however, fluorescence microscopy, subcellular fractionation, and experiments monitoring its dynamics in vivo (Dilworth et al. 2001) revealed that Nup2p actually cycles on and off the nuclear basket and in this sense behaves more like a soluble transport factor. Similarly, Yrb2p (yeast ran binding protein 2; a.k.a. Nup36p) is primarily nuclear, but contains FG repeats, yet it only transiently associates with the NPC (Floer and Blobel 1996). So far, all such rapidly “shuttling” nucleoporins belong to the FG Nup family. As well as an FG-repeat region, both Nup2p and Yrb2p carry a consensus Ran-binding motif and may have a role in promoting the disassembly of transport cargos; in this way, shuttling nucleoporins may act as mediators between the stationary and soluble phases of transport (see below) (Dilworth et al. 2001, 2005; Gilchrist et al. 2002). In vertebrates, the dynamics of Nups have been comprehensively examined, revealing varying half-lives of each Nup on the NPC, and it seems likely that this will be borne out in yeast (Tran and Wente 2006). For example, Nup145Np has a localization that is biased to, but not exclusively on, the nuclear face, while Nup116p and Nup100p are similarly biased to the cytoplasmic face (Suntharalingam and Wente 2003). This variation complicates efforts to define a “stoichiometry” for components of the NPC, as any number for these more dynamic Nups will be an average of what may be a stochastic variation in Nup number and location. We expect that, the closer we look, the more difficult it will be to consider the NPC an autonomous structure; rather, perhaps it should be considered a dynamic assembly of proteins which to varying degrees, cycle between the stationary and soluble phases during transport and assembly, and functionally link the NPC to numerous other dynamic cellular activities (see below).
NPC Assembly
NPCs are not static structures. They are assembled, and their components appear to be capable of turning over during the NPC’s lifetime. In many organisms, NPCs disassemble upon NE breakdown at the beginning of mitosis or meiosis and reassemble coordinately with the NE around the newly segregated chromosomes at its end. However, yeast has a “closed” mitosis in which the NE remains intact, such that the NPCs remain assembled throughout the life cycle of the cell and negate the need for NE and NPC disassembly—in sharp contrast to the elaborate mitotic nuclear disassembly and reassembly processes seen in metazoans (Suntharalingam and Wente 2003). Careful analyses of serially sectioned yeast confirmed that NPC assembly occurs continuously throughout the entire cell cycle with a typical haploid NE containing between ∼70 NPCs just after mitosis to ∼140 NPCs in late anaphase (Winey et al. 1997). How this assembly occurs is still unclear, despite much work in both yeast and vertebrate model systems, with most of that work in metazoan cells (because researchers generally studied the synchronized assembly of NPCs in mitosis), and some of the details are only just beginning to emerge (as reviewed in Fernandez-Martinez and Rout 2009; Hetzer and Wente 2009). Nevertheless, the processes of NPC and NE assembly—and the reasons why some species opt for a closed mitosis while other related species opt for variants of an open mitosis [compare the ascomycetes Saccharomyces and Aspergillus (De Souza et al. 2004; Osmani et al. 2006; Liu et al. 2009)], remain somewhat mysterious. Work in vertebrate cell-free systems has established, finally, that new NPCs are indeed inserted de novo into the NE (rather than, e.g., “budding off” from existing NPCs) (D’Angelo et al. 2006). In yeast, it is primarily genetic approaches that have given some of these insights. As a yeast cell grows, the nucleus also grows in volume and the NE enlarges its surface area, during which time new NPCs are inserted into the NE (Winey et al. 1997). Although not proven, it seems likely that this process in yeast is similar to interphase NPC assembly in vertebrates, which has been shown to occur through de novo assembly of precursor building blocks recruited from both the nucleoplasm and cytoplasm into the regions of the NE between pre-existing NPCs (D’Angelo et al. 2006). The continued assembly of the NPC and NE throughout the yeast cell cycle has been used as a basis for genetic screens, selecting for mutants that caused mislocalization of tagged Nups. Initially, mutants in various Nups produced phenotypes that (if not lethal) gave a puzzling collection of different phenotypes that were difficult to interpret in terms of NPC assembly. Some made the NPCs cluster (above), whereas others led to herniations of the NE extending over the cytoplasmic face of NPCs to seal them (Wente and Blobel 1993, 1994). However, more recent approaches have given more interpretable phenotypes. By using a photoconvertable Dendra tag in cells blocked and then released in NPC assembly, it was shown that some pre-assembly Nup complexes congregate on both the inner and the outer membranes of the NE, including cytoplasmic-facing Nups on the cytoplasmic face of the NE and nucleoplasmic/basket Nups on the nuclear face, whereas symmetrically disposed Nups were found to accumulate on both NE faces (Makio et al. 2009; Onischenko et al. 2009). These pre-assembly complexes might correspond to the discrete complexes found to compose the NPC, such as the Nup84 complex (see above). Targeting of these pre-assembly Nups to the NE seems to require certain soluble transport factors normally used to chaperone and power the transport of cargoes through the NPC (see below), as genetic screens for conditional mutants in NPC assembly identified Ran, RanGEF, RanGAP, and Ntf2 (see Figure 4 and below) (Ryan and Wente 2002; Ryan et al. 2003, 2007). The karyopherin (Kap) Kap95p was also identified in these screens, and another karyopherin, Kap121p, seems to aid Nup53p in assembling into a complex with Nup170p (Lusk et al. 2002).
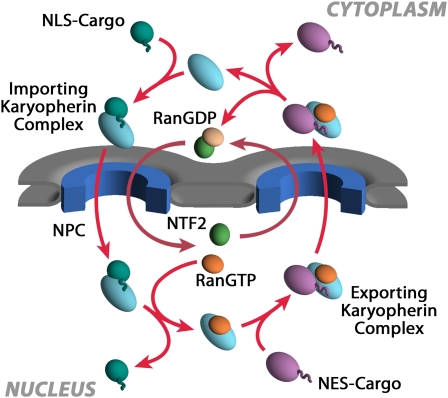
Figure 4 .
The nuclear transport cycle for karyopherins and their cargoes. See Fig. 5 legend and main text for details.
Interestingly, these mutants correspond to two key components of the cargo-carrying transport factor pathways, namely the β-karyopherins Kap95p and Kap121p and Ran cycle components [Ran, RanGAP, RanGEF, and Ntf2 (responsible for transporting RanGDP into the nucleus)] (Lusk et al. 2002; Ryan and Wente 2002; Ryan et al. 2003, 2007). The reasons for the functional associations between NPC assembly and transport factors are still being elucidated, but similar connections have been seen in vertebrates (D’Angelo et al. 2006). In yeast, Kap121p has been proposed to target Nup53p to the NPC, where it is attached to the core scaffold component Nup170p. Indeed, recent work has revealed the importance of the core scaffold to the early stages of NPC assembly. Thus, when the C-terminal domain of Nup170p is overexpressed, what appear to be intermediates of NPC assembly accumulate both in the cytoplasm and at the NE (Flemming et al. 2009). Similarly, in strains lacking both Nup53p and its paralog Nup59p, depletion of Nup170p or either of two transmembrane nucleoporins that connect with Nup170p—Pom152p or Pom34p—also caused the accumulation of such intermediates in yeast cells (Onischenko et al. 2009).
For an NPC to be inserted into the intact NE, both the inner and the outer NE membranes must approach at a site and fuse to give rise to the pore membrane, upon which the core scaffold and the rest of the NPC can then assemble. It is curious, therefore, that two of the three poms (Pom152p and Pom34p) are not essential and so are dispensable for NPC assembly and that all three poms (including Ndc1p) are not required for NPC assembly in the closely related fungi, Aspergillus (Liu et al. 2009). Taken together, this suggests that there must be other transiently or dynamically associating membrane proteins that play key roles in initiating the NPC assembly process and fusion of the inner nuclear membrane (INM) and outer nuclear membrane (ONM) to form the pore membrane.
Indeed, there has been a growing cadre of proteins that, while not strictly Nups, play a key role in yeast NPC assembly. As well as Ran, Ran cofactors, and the Kaps (above; Lusk et al. 2002; Ryan et al. 2003, 2007), the two yeast reticulons Rtn1p and Rtn2p and their interacting partner Yop1p have been implicated in NPC assembly (Dawson et al. 2009). Rtns and Yop1/DP1 proteins can deform and mold membranes, having been shown to have roles in both dynamically restructuring and maintaining tubular ER (De Craene et al. 2006; Voeltz et al. 2006; Hu et al. 2008) and, in metazoans, even in postmitotic NE shaping (Anderson and Hetzer, 2008b). Reticulons have a segment that can insert into one leaflet of a membrane, which may promote or induce membrane curvature (Oertle et al. 2003; De Craene et al. 2006; Voeltz et al. 2006; Shibata et al. 2008); indeed, they are depleted in regions of flat membrane, such as the NE between NPCs, and are found to concentrate in curved membrane regions such as tubular ER (De Craene et al. 2006; Voeltz et al. 2006; Anderson and Hetzer, 2008a,b). The apparent absence of these proteins in the mature NPC suggests that they play only a transient role at the beginning of the assembly process, perhaps helping the first NE membrane curving and fusion step to make the pore membrane. Similarly, the NE/ER proteins Apq12p and Brr6p are genetically linked to each other and are necessary for normal NPC assembly and distribution. This work indicates that both proteins are involved in maintaining lipid homoeostasis in the ER, which is necessary for proper NPC insertion and distribution in the NE (Scarcelli et al. 2007; Hodge et al. 2010).
Another candidate NPC assembly factor is Pom33p, isolated in a genetic screen for genes that are essential in cells lacking Nup133p (Chadrin et al. 2010). The transmembrane protein Pom33p and its paralog Per33p are found in both the ER and the NE, although Pom33p shows a preferential dynamic localization at NPCs. Pom33, but not Per33, genetically interacts with Nup84 complex components and the interacting proteins Nup170p and Ndc1p and physically forms a direct complex with Rtn1p (Chadrin et al. 2010). These data, plus the fact that depletion of both Nup170 and Pom33 significantly impaired assembly of NPCs, point to a role for Pom33p in NPC assembly or maintenance of the NE (Chadrin et al. 2010). Pom33p thus potentially links the reticulon membrane bending and manipulation machinery with the assembling NPC, which together possibly either help the transmembrane nucleoporins during the initial membrane fusion event required for the start of NPC assembly or facilitate the stabilization of the nascent nuclear pore. Following this initial pore formation, assembly to form the mature NPC must proceed extremely rapidly, as no naturally occurring intermediates have been found.
Of course, the NPC core scaffold is composed almost entirely of homologs of vesicle-coating proteins, whose function is to mold and fuse membranes into curved vesicles. On the basis of this similarity it has been suggested that the Nup84 complex and the Nup170 inner-ring complex (which interacts directly with poms) could be directly involved, after recruitment to the NE, in forming a coat somewhat like those in coated vesicles that produces the nascent pore membrane and pinches the inner and outer NE membranes together in a manner analogous to pinching off a curved vesicle (reviewed in Fernandez-Martinez and Rout 2009; Hetzer and Wente 2009).
In summary, there appear to be several main steps to NPC assembly. Initially, accessory factors collaborate with transmembrane and inner-ring NPC components to accumulate on both the inner nuclear membrane and the outer nuclear membrane to warp the latter into a fused pore. The recruitment of the inner-ring components would recruit the outer-ring components, permitting the assembly of the entire membrane-coating core scaffold in the pore (Alber et al. 2007b). Rapid association of the remaining FG Nups, other NPC components, and the nuclear basket would then complete the process. However, this sequence of events remains strictly speculative, and much remains to be understood about the mechanism of NPC assembly in yeast or in any other eukaryote.
It seems possible that other NE-associated structures share at least some aspects of the NPC’s assembly process. Curiously, Nup60p and Pom152p are also required for the assembly and repair of the SPB (Greenland et al. 2010). Recall that the pore membrane component, Ndc1p, has been shown to be a shared component of both the NPC and the SPB and is required for the assembly of both (Chial et al. 1998; Lau et al. 2004). The functional connection between the SPB and NPC is underscored by the putative nuclear basket component Mlp2p, which may associate with Nup60 (Zhao et al. 2008) and connects to both NPCs and SPBs (Niepel et al. 2005). It seems that several proteins are found at both locales, raising the possibility that, as both NPCs and SPBs are inserted into a membranous grommet formed from the fusion of the inner and outer nuclear membranes, there are some commonalities in their assembly mechanisms.
Turnover of NPCs
No repair mechanism, as such, has been found for the NPC. Rather, it seems that a combination of some pre-emptive replacement of components by constant turnover and dilution of “old” NPCs by new ones through cell growth and division are the tactics taken to rejuvenate the NPC population in a growing yeast population. The turnover rates of yeast Nups are not yet precisely known, although certainly some FG Nups exchange very quickly (Dilworth et al. 2001; Tran and Wente 2006). Moreover, there is some uncertainty about how “old” and “new” NPCs are partitioned between the mother and the daughter cells upon budding. While evidence was originally presented that the old NPCs are retained preferentially in mother cells, potentially ensuring that the daughters receive a fresh supply of new NPCs (Shcheprova et al. 2008), more recent work indicates instead that new and old NPCs partition equally between mother and daughter at mitosis (Khmelinskii et al. 2010, 2011). As NPC segregation and turnover have direct implications for aging studies (Kaeberlein 2010), this and related topics will doubtless be areas of intensive future investigation.
Soluble Phase of Transport: Transport Signals and Carriers
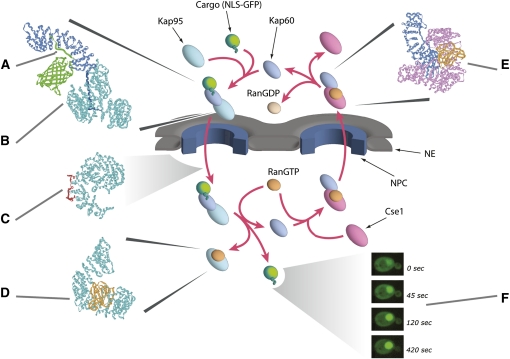
While NPC-mediated gating does not require an energy input, nucleocytoplasmic transport and the accumulation of cargoes in the nucleus and cytoplasm are driven by the formation and maintenance of concentration gradients across the NE by GTPases and ATPases in the nucleoplasm and cytoplasm (Figures 4 and 5) (reviewed in Rout et al. 2003; Strambio-de-Castillia et al. 2010). Moreover, as is typical for protein sorting throughout eukaryotic cells, proteins synthesized in the cytoplasm that are destined for the nucleus carry targeting signals [generally termed nuclear localization signals (NLSs)] that are recognized by soluble receptors, which mediate their transport. The first transport factors to be identified and purified to homogeneity were karyopherin α, karyopherin β, and a small Ras-like GTPase called Ran. Through classic biochemical fractionation combined with in vitro import assays, these proteins were purified to homogeneity from mammalian systems and shown to mediate transport of reporter proteins carrying an NLS from the SV40 large T antigen (reviewed in Pemberton and Paschal 2005; Wente and Rout 2010). Because the yeast genome had recently been completed, it was then a straightforward matter to identify orthologs in yeast (reviewed in Wozniak et al. 1998), and soon work on the mechanisms of nucleocytoplasmic transport was progressing in both yeast and mammalian systems. These studies established that karyopherin α (Kap60p in yeast) binds to the NLS and that karyopherin β (Kap95p in yeast) enhances (or stabilizes) the interaction and in turn binds to FG-repeat-containing nucleoporins. Ran-GTP provides an important source of energy to the reaction by binding to karyopherin β as it enters the nucleus with karyopherin α and cargo in tow, releasing the cargo to the nucleoplasm (Figures 4 and 5). Importantly, the versatility of yeast as a model system rapidly led to complementary approaches and insights beyond those immediately possible in mammalian systems.
Figure 5 .
The transport cycle of Kap60 and Kap95 is shown diagrammatically in the center, with relevant atomic structures shown in the surroundings. (A) The extended NLS attached to a GFP reporter [green; PDB 1EMA (Ormo et al. 1996)] binds to a long region on the inside of the Kap60 superhelix [dark blue; PDB 1EE5 (Liker et al. 2000)], made of alternating α-helical turns. (B) The characteristic superhelical solenoid of Kap95 (light blue), made of alternating α-helical turns in a related fashion to Kap60, forms a spiral with two surfaces. The inner surface wraps around the extended N-terminal IBB domain of Kap60, which links it tightly to Kap95 [PDB 1QGK (Cingolani et al. 1999)]. (C) As Kap95 passes through the NPC, it interacts with FG Nups. The repeated Phe residues on the FG-repeat region (red) insert into complementary repeated pockets formed from the crevices between adjacent α-helical repeats, all along the outer surface of Kap95’s spiral [PDB 2BPT (Liu and Stewart 2005)]. By transferring between the multiple FG repeats in the NPC, Kap95—together with Kap60 and its NLS-GFP cargo—cross the NPC. (D) In the nucleus, binding of RanGTP (orange) to Kap95 [PDB 2BKU (Lee et al. 2005)] causes a conformational change in the latter, which releases Kap60, and, in doing so, Kap60 is made to release its NLS cargo into the nucleoplasm. In either its Ran bound or free form, Kap95 can bind to FG Nups and thereby cross the NPC to continue the transport cycle. (E) Kap60 is exported from the nucleus by the RanGTP-bound form of the karyopherin Cse1 [magenta; PDB 1WA5 (Matsuura and Stewart 2004)]. In this state, the IBB domain is held tightly against the side of Kap60, inhibiting NLS binding. Both Kap60 and RanGTP are once again held to the inner surface of the Cse1 spiral, leaving the outer surface free to interact with FG repeats and carry the complex through the NPC out of the nucleus. Once in the cytoplasm, GTP on Ran is hydrolyzed to form RanGDP, causing the complex to dissociate. Kap60 remains bound to its IBB even when free in the cytoplasm, but binding to an NLS exposes the IBB and allows Kap95 to bind, initializing another round of import. (F) As a result of the import cycle, NLS-GFP accumulates in the nucleus over time, shown here by fluorescence microscopy (Timney et al. 2006).
It was clear that not all proteins destined for the nucleus contain an NLS typified by SV40 large T antigen. The diversity of cargoes and complexes that traverse the NPC is huge, ranging from proteins to RNAs and ribonucleoproteins (RNPs), including mRNPs and ribosomes, to viruses. Analysis of the yeast genome revealed family transport factors structurally related to karyopherin β (and more distantly to karyopherin α). Other eukaryotes studied, even the most evolutionarily divergent, seem to retain this same family of Kaps (DeGrasse et al. 2009; Mason et al. 2009). Members of the β-Kap family are generally large (molecular weight of 100–125 kDa) proteins that share ∼20% sequence identity with each other. Each is typified by the presence of up to ∼20 HEAT repeats (amphipathic helix-loop-helix motifs) that form a large helical solenoid (with a single extended hydrophobic core) (Figure 5) (Cansizoglu et al. 2007). There are apparently 14 Kaps in S. cerevisiae and at least 19 Kaps in humans (Stewart 2003), all of which differentially bind different classes of nuclear transport signals, FG-repeat nucleoporins, and Ran; unlike the Kap60p:Kap95p dimer, all other β-Kaps bind directly to their cargos (Figure 4).
Karyopherins responsible for importing cargoes are often called importins, and exporters are called exportins. The direction of transport for each karyopherin is dictated by its differential interaction with cargoes and Ran (Figure 4). In cells, Ran primarily exists in two forms: in the nucleus, Ran is maintained in its GTP-bound form by a GTP exchange factor (RanGEF; RCC1, Prp20p, or Srm1p in yeast); this protein is chromatin bound, thus signaling to the nucleocytoplasmic transport system the position of the nucleoplasm by virtue of generating a cloud of RanGTP around it. In contrast, the Ran GTPase-activating protein (RanGAP) is localized to the cytoplasm, so that Ran in the cytoplasm predominates in the GDP form. Karyopherins exploit this property during transport. As mentioned above, during an import cycle, Kaps bind to their cargoes in the cytoplasm, and when they reach the high Ran-GTP in the nucleus, are induced to release their cargoes. In contrast, exportin binding to cargoes is enhanced by the formation of a trimeric complex that includes Ran-GTP. Thus, as this complex meets the RanGAP in the cytoplasm, the GTP is hydrolyzed and the complex falls apart. Indeed, the direction of karyopherin-mediated transport through the NPC can be reversed by inversion of the Ran gradient (Nachury and Weis 1999). Most karyopherins are thought to be recycled to their original compartments empty, but in a few instances they are believed to chaperone another cargo on their return journey (Figure 4).
Studies of prototypical interactions among constituents of these transport pathways have shed considerable light on the structural basis of transport (Figure 5). In the classical pathway, the NLS binds to a long region on the inside of the Kap60 superhelix, made of alternating α-helical turns. Kap95p, which is also made of alternating α-helical turns, forms a spiral with two surfaces, and the inner surface wraps around an extended N-terminal domain of Kap60p (Figure 5) [a.k.a the importin β-binding (IBB) domain] (Cingolani et al. 1999). The interaction of Kap95p with FG Nups is mediated as the repeated Phe residues on the FG-repeat regions (see below) insert into complementary repeated pockets formed from the crevices between adjacent α-helical repeats all along the outer surface of Kap95p’s spiral. RanGTP binds to Kap95p (Lee et al. 2005) on the inner surface of Kap95’s amino-terminal solenoid spiral, which causes conformational changes that lead to release of Kap60p (and cargoes) (Figure 5).
During Kap60p export, Kap60p and RanGTP are bound to the inner surface of the Cse1p spiral (Matsuura and Stewart 2004). In this form, the IBB domain is held tightly against the side of Kap60p, inhibiting NLS binding and leaving the outer surface free to interact with FG repeats and thereby carrying the complex through the NPC out of the nucleus (Figure 5). Once in the cytoplasm, the RanGTP hydrolyzes to RanGDP, causing the complex to dissociate. Kap60p remains bound to its IBB even when free in the cytoplasm, but binding to an NLS exposes the IBB and allows Kap95p to bind, initializing another round of import.
Karyopherins and Their Cargoes
The apparent presence of a family of karyopherins, and the knowledge that there are numerous classes of cargoes that must be transported across the NPC, led researchers to begin to identify cargoes for each of the karyopherins. Again, yeast has been a tremendous model system for investigating this fundamental question. The mainstay approach for doing so has been to take advantage of homologous recombination techniques to genomically tag karyopherins with an epitope tag (like protein A) and to isolate the Kap and its associated cargoes (Aitchison et al. 1996). Genetic perturbations of the Kap genes have then been used to explore the consequences with respect to the potential cargo. This approach was first applied to Kap104p to establish that it is responsible for importing a subclass of RNA-binding proteins (Nab2p and Nab4p/Hrp1p) (Aitchison et al. 1996). These proteins are major mRNA-binding proteins essential for mRNA processing and export (Anderson et al. 1993). They appear to accompany the mRNA out of the nucleus, and upon reaching the cytoplasm, they are recycled for another round by Kap104p (Lee and Aitchison 1999). Mtr10p/Kap111p also appears dedicated to this essential function; it imports Npl3p, another essential mRNA biogenesis factor. Interestingly, while both Kaps import essential proteins, neither is essential (under the same conditions) by itself. This suggests that Kaps must have the capability to compensate for one another and bind to their cargoes with some promiscuity. This was first made obvious upon examination of Kap123p. Deletion of Kap123 is virtually without phenotypic consequences in laboratory strains of yeast. Yet, Kap123p is perhaps the most abundant of the Kaps in yeast, conserved throughout the Eukaryota, and it binds to Lys-rich NLSs shared by a host of ribosomal proteins and ribosome assembly factors, which leads to their import into the nucleus prior to their assembly into ribosomes (Rout et al. 1997;Leslie et al. 2002; Timney et al. 2006)—an essential process if ever there was one! Indeed, a host of genetically interacting Kaps appear to be involved in the import of proteins critical to ribosome assembly (e.g., Kap108p/Sxm1p, Kap119p/Nmd5p, Kap121p/Pse1p) (Rosenblum et al. 1997; Rout et al. 1997; Sydorskyy et al. 2003; Caesar et al. 2006), and it has been shown explicitly that, in the absence of Kap123p, Kap121p can bind to Kap123p substrates and import them into the nucleus (Rout et al. 1997).
Perhaps it is not surprising that structurally related Kaps can bind to structurally related NLSs, but it also appears that Kaps can recognize more than one type of NLS. For example, while Kap121p was originally shown to bind to noncanonical Lys-rich NLSs (Rout et al. 1997; Kaffman et al. 1998b; Leslie et al. 2002), it, like Kap104p, also imports proteins through rg-NLSs, which are reminiscent of structurally distinct RNA-binding motifs (Dreyfuss et al. 1993; Lee and Aitchison 1999; Leslie et al. 2004) characterized by repeats of Arg and Gly amino acid residues. Moreover, multiple cargo domains exist in Kap114p, and it has been proposed that this Kap is capable of importing multiple cargoes simultaneously (Hodges et al. 2005).
Although some of the cargoes for many Kaps have been defined, there are an estimated 1500–2000 proteins that transit the NPC during their life cycle, and as a field, we have identified only a handful of the cargoes that they each recognize. So, while it has been proposed many times that evolution has likely exploited their overlapping specificities and potential complexity to regulate classes of cargoes by regulating the karyopherins, it remains for the field to more comprehensively define Kap-cargo complexes to demonstrate how much this is the case and to fully appreciate how they may have done so.
Protein export from the nucleus is mediated by at least three β-karyopherins. The first (“classic”) nuclear export signal was defined in vertebrate cells in HIV-Rev protein. Rev binds specifically to unspliced and singly spliced HIV mRNA and ensures that it is exported efficiently. Studies to define this process identified a short leucine-rich region within Rev that is necessary and sufficient for nuclear export. This sequence is recognized by the karyopherin Xpo1/Crm1 (Stade et al. 1997). As it turns out, there are many proteins that contain variants of the prototypical sequence and are exported by Xpo1p. These include the proteins of the 40S and 60S preribosomal subunits (J. H. Ho et al. 2000; Stage-Zimmermann et al. 2000; Moy and Silver 2002); numerous transcriptional or signaling proteins (Ferrigno et al. 1998; Jensen et al. 2001; Menezes et al. 2004; Chang et al. 2006; Martin et al. 2006; Azevedo et al. 2007; Pelaez et al. 2009); key regulators of the cell cycle [Cdc14p (Bembenek et al. 2005)], which control exit from mitosis; and certain small RNAs (Gallardo et al. 2008; Thomson and Tollervey 2010). Xpo1p/Crm1p is also, at least indirectly, required for normal mRNA production and export (Feng et al. 1999; Strasser et al. 2000; Hammell et al. 2002; Dong et al. 2007).
Msn5p has also been shown to act as a nuclear export factor, exporting phosphorylated nuclear transcription factors (Kaffman et al. 1998a; DeVit and Johnston 1999; Gorner et al. 2002; Queralt and Igual 2003; Durchschlag et al. 2004; Ueta et al. 2007), the HO endonuclease (Bakhrat et al. 2008), and Whi5p, the yeast ortholog of Rb (Taberner et al. 2009). A consensus nuclear export signal (NES) for Msn5p has been elusive, but its preference for phosphorylated proteins suggests a role for regulated export. Indeed, regulation of transport provides an exquisite mechanism to control gene expression. Perhaps the best-characterized example of such regulation in yeast comes from studies of Pho4p. Pho4p is a transcription factor that induces the transcription of phosphate-responsive genes. When cells lack phosphate, Pho4p is imported into the nucleus by Kap121p. However, in the presence of excess phosphate, Pho4p is phosphorylated adjacent to its NLS, inhibiting Kap121p binding and consequently its import. In addition, phosphorylation at two different sites promotes the factor’s nuclear export(Kaffman et al. 1998b; Komeili and O’Shea 1999). A similar shuttling activity has been described for Tor1p (a target of the immunosuppressant rapamycin), which is a protein kinase that controls growth in response to nutrients through the regulation of diverse cellular processes in the cytoplasm and nucleus. Interestingly, PolIII transcription is also regulated by transport; Maf1p, a global inhibitor of PolIII, is exported by Msn5p when cells are shifted to conditions that favor growth and high PolIII activity (Towpik et al. 2008).
Msn5p appears to be the most versatile of the Kaps. It has also been shown to import proteins into the nucleus (Yoshida and Blobel 2001), and both Los1p and Msn5p have been shown to be capable of exporting transfer RNAs (tRNAs) from the nucleus. Both appear to bind double-stranded RNA directly (Shibata et al. 2006), and (at least) Los1p appears to play a proofreading role, ensuring that its tRNA substrates are appropriately structured prior to their export to the cytoplasm (although splicing per se appears not to be proofread by Los1p binding (Arts et al. 1998; Lipowsky et al. 1999; Cook et al. 2009; Hopper et al. 2010). The coupling of transport to function of the cargo is certainly not limited to the tRNA example; import Kaps such as Kap114p, Kap104p, and Mtr10p release their cargoes in the nucleus in concert with their cargoes binding to DNA and RNA. In effect, nuclear-binding sites compete with Kaps for their cargoes upon import, suggesting a mechanism for controlled release and intranuclear targeting.
Competition as a Major Factor in Nuclear Transport
Similarly, competition effects play a major role in the behavior of nuclear transport. Using an in vivo assay and by manipulating the amounts, types, and affinities of Kaps and cargos, it was shown that import rates in vivo are governed in a straightforward manner by the concentrations of Kaps and their cargo and the affinity between them and that the main limiting factor for import (accounting for the fact that nuclear accumulation of transported cargo was much slower than expected) was the poor ability of Kaps and cargoes to find each other in the cytoplasm in a background of overwhelming nonspecific competition. In other words, the key rate-limiting step of the transport cycle is not transiting through the NPC itself, but is instead the formation of the Kap/cargo complex within the cell’s crowded environment (Timney et al. 2006). The importance of competition seems to extend to the mechanism of the NPC. A recent computational model indicated how the selectivity of the NPC could be enhanced by the exclusion of nonspecific molecules by specific ones, due to competition for binding sites and limited space inside the channel. By using recombinant purified full-length yeast FG Nups and transport factors, it was shown that FG Nup-functionalized nanopores behave as a nanoselective filter, reproducing key features of trafficking through the NPC. It was also confirmed that competition between transport factors and nonspecific proteins is a major factor in the transport mechanism (Jovanovic-Talisman et al. 2009).
Not Just Karyopherins: RNA Export
Many small RNAs, such as small nuclear RNA and tRNAs, are exported using the same RanGTP-powered karyopherin-dependent pathways used by exporting proteins, although some karyopherins seem to specialize in this function, such as the tRNA exporter Los1 (reviewed in Köhler and Hurt 2007). However, the process is considerably more complicated for mRNAs and ribosomal RNAs. All such large RNAs are transcribed and assembled into RNP complexes, and each RNP can be considered an intermediate along an assembly line as proteins flit on and off the assembling structures to trim and assemble the RNA. Consider the ribosome. Over 200 proteins are believed to be involved in the assembly of ribosomes as they mature during their complex biogenesis. Similarly, each mRNA is assembled into an RNP (mRNP) particle involving a series of complex assembly intermediates, which associate with each species of RNA in a dynamic fashion to allow for precise transcript maturation (Fatica and Tollervey 2002; Hopper and Phizicky 2003; Vinciguerra and Stutz 2004).
Surprisingly, mRNA uses a Kap- and Ran-independent mechanism for export (Santos-Rosa et al. 1998; Katahira et al. 1999). Moreover, mRNP assembly and export involve strict surveillance mechanisms to ensure that only fully mature and functional RNPs are transported to the cytoplasm (Palancade et al. 2005; Schmid and Jensen 2008; Skruzný et al. 2009). In addition, there are many different species of mRNA, each potentially with its own particular maturation pathway. This is a topic that has been both extensively researched and comprehensively reviewed, so we will only summarize these findings briefly and refer the reader to these reviews (Rondon et al. 2010; Stewart 2010; Rodriguez-Navarro and Hurt 2011).
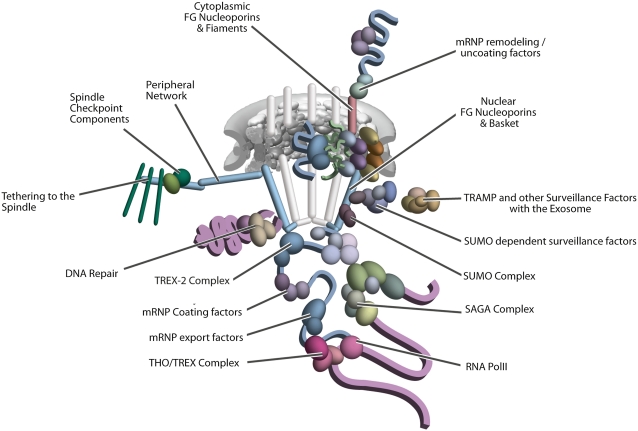
Upon being processed and packaged into mRNP particles in the nucleus, the non-Kap transport factors Mex67p and Mtr2p associate with and chaperone the mRNP through the NPC (Figure 6). These factors are co-transcriptionally recruited to the maturing mRNP by a highly coordinated process that couples transcription, post-transcriptional processing, mRNP assembly, and docking to the nuclear basket, beginning in many cases even before the nascent transcript has left the gene; and once again, research in yeast has pioneered much of our understanding of these processes. Thus, co-transcriptional recruitment of the THO/TREX complex to the nascent mRNA of intron-containing transcripts ultimately leads both to correct 3′-end processing and to recruitment of (among other proteins) Yra1p, which in turn recruits the Mex67p-Mtr2p heterodimer (reviewed in Rodriguez-Navarro and Hurt 2011) (Figure 6). Another complex, TREX-2, associates with the mRNP at the NPC, apparently assuring that it is correctly packaged for its journey across the NPC. The Mex67p-Mtr2p heterodimer, by binding both the mRNP and the FG Nups, mediates the actual mRNP transport event, which can be surprisingly rapid; recent work in vertebrates has suggested that the actual translocation event, even for a multi-megadalton mRNP complex, lasts only milliseconds (Grunwald and Singer 2010; Mor et al. 2010). After transiting the NPC’s central channel, the mRNP encounters Dbp5p, an ATP-driven RNA helicase tethered to the cytoplasmic filament protein Nup159p. Regulated by Nup42p-tethered Gle1p and the small molecule IP6 (inositol hexaphosphate), Dbp5p’s action on the exiting mRNP serves to release the transport factors Mex67p and Mtr2p as well as mRNP proteins such as Nab2p, both actions preventing re-import of the mRNP (and thereby helping to confer directionality to export) and preparing the mRNA for translation (Figure 6) (reviewed in Carmody and Wente 2009; Rodriguez-Navarro and Hurt 2011).
Figure 6 .
Diagrammatic representation of mRNA export, adapted from Strambio-de-Castillia et al. (2010). The SAGA complex is recruited to the promoter of a subset of inducible genes and promotes their transcription. SAGA and the NPC-associated TREX-2 complex may help the genes move to the vicinity of the NPC. The nascent transcripts recruit shuttling mRNA-coating factors, THO, TREX, and, subsequently, the mRNA export factors Mex67p and Mtr2p, resulting in the formation of an export-competent mRNP (Rodriguez-Navarro and Hurt 2011); the association of the maturing mRNPs with components of the nuclear basket is strengthened in preparation for nuclear translocation, while nuclear basket-associated TRAMP and exosome complex-associated mRNP surveillance mechanisms ensure that the mRNP is correctly assembled for export (Fasken and Corbett 2009). After translocation through the NPC, the release of mRNA export factors from mRNPs is induced by the combined action of Dbp5p and Gle1p, which are docked to NPC cytoplasmic filaments via interaction with Nup42p and Nup159p, respectively, and are thought to act as mRNP-remodelling factors (Carmody and Wente 2009). It is presumed that this process drives the directionality of mRNP export while at the same time priming mRNAs for translation initiation.
Ribosomal subunit export is still a little less well characterized, and while clearly differing from both protein and mRNP export, oddly shares elements from both (Hage and Tollervey 2004; Zemp and Kutay 2007; Henras et al. 2008; Lo and Johnson 2009). Three transport factors have been implicated in yeast in the export of the large ribosomal subunit: Mex67p-Mtr2p (Yao et al. 2007), Crm1p (which docks to the large subunit adaptor protein Nmd3p (J. H. Ho et al. 2000; Gadal et al. 2001), and the noncanonical receptor Arx1p (Bradatsch et al. 2007; Hung et al. 2008). Fusions of Mex67p, Los1p, Mtr2p, Cse1p, or Msn5p to Nmd3p, lacking its Crm1p-dependent NES, all function in export, suggesting that there may not be a fundamental requirement for any specific export receptor for the large subunit, in contrast to the specific export factors required for mRNAs (Lo and Johnson 2009). There is some evidence that up to a dozen α-solenoid proteins, possibly resembling Kaps, such as Rrp12p and Nog1p, may also aid ribosomal subunit export (Oeffinger et al. 2004; Pertschy et al. 2007). The export of 40S subunits is still poorly understood, but may be somewhat simpler than for the 60S subunit (Zemp and Kutay 2007; Maggi et al. 2008; Perreault et al. 2008; Carron et al. 2011). Two different yeast nonribosomal proteins, Dim2p and Ltv1p, have been proposed to function as adapters for Crm1p-mediated 40S export in yeast (Seiser et al. 2006; Vanrobays et al. 2008). Both are late-acting 40S biogenesis factors that shuttle between the nucleus and the cytoplasm.
Mechanism of Nuclear Transport
The molecular details are beginning to emerge as to exactly how the NPC mediates the active exchange of selected macromolecules while excluding all others, although this remains a subject of vigorous debate. Nevertheless, certain basic features of the NPC as a transport machine are generally accepted.
First, the NPC defines a tube of defined width and height that connects between the nucleoplasm and cytoplasm. These dimensions delimit the upper size of the transport cargos, defined in vertebrate NPCs as ∼35 nm, and, on the basis of morphological maps, are likely to be similar in yeast and other eukaryotes (reviewed in Strambio-de-Castillia et al. 2010). Second, the tube is lined with FG-repeat regions contributed by the ∼160 copies of the different types of FG Nups anchored in and around this tube; work in yeast indicated that no ATPases or GTPases are needed as components of the NPC, such that the NPC does not appear to open and shut as a physical gate, but rather behaves as a “virtual” one (Figures 1 and 2) (Rout et al. 2000, 2003; Peters 2009). Thus, the power for transport is generated in the nucleoplasm and cytoplasm, and the NPC is chiefly responsible for selectivity. On the basis of mapping and deletion mutagenesis experiments, an affinity gradient of FG-binding sites between the nuclear and cytoplasmic faces of the NPC also does not seem to be essential for nuclear transport in yeast (Rout et al. 2000; Strawn et al. 2004). As the general architecture, distribution, and composition of the FG-repeat regions are similar throughout the eukaryote (see above), the mechanism of gating is likely conserved. The FG-repeat regions do not appear to fold into permanent secondary or tertiary structures, and indeed it is likely that they never form such structures. Rather, they appear highly flexible, allowing them both to assume many possible conformations and to dynamically switch between those conformations. Because they are unfolded, the FG-repeat regions fill a volume many times that of a folded protein of the same size. This means that they can extend tens of nanometers from their anchor point, such that the central tube is flanked by, and filled with, filamentous FG repeats, accounting for the “cloud” of filaments seen to surround the yeast NPC by electron microscopy (Fahrenkrog et al. 2000a; Kiseleva et al. 2004). Another advantage of disordered filaments as binding sites is that only a little protein is needed to fill a lot of volume—a very economical way of having a small amount of protein generate a huge binding site. As stated above, transport factors bind FG-repeat regions, and it is through this binding that they are allowed selective passage through the central channel. Regardless of their differing atomic structures, it seems that all transport factors carry numerous copies of surface-accessible hydrophobic pockets into which several of the F residues of an FG-repeat region can bind (see above). These appear to have low affinity and rapid exchange rates, although as there are several such interactions per transport factor (at least 14 in the case of the karyopherin transport factor Cse1), the avidity of transport factors for FG Nups is expected to be high (Isgro and Schulten 2005, 2007). In a sense, then, the FG repeats can be thought of as antennae, reaching out in a cloud of binding sites many tens of nanometers from the nuclear and cytoplasmic faces of NPCs to efficiently funnel transport factors and their associated cargoes into the NPC, while generating a zone of exclusion for nonspecific materials around the NPC (Figure 1) (Rout and Aitchison 2000, 2001; Rout et al. 2000, 2003; Macara 2001).
How does it actually work? We still do not know, but attempts have been made to describe the basic physical principles of NPC-mediated gating although this has been done without a detailed description of FG Nup behavior, considering only the consensus features of the NPC and making some basic physical assumptions, thus treating the NPC as a narrow hole lined with binding sites and allowing that molecules access and transit this hole through normal diffusion. It has been shown that a narrow channel filled with FG-repeat regions presents a significant barrier to passage across the NPC, such that the probability of a macromolecule translocating through the channel is low. However, transient trapping by a macromolecule that can bind to the FG repeats (such as a transport factor) increases the probability of that molecule remaining in the central channel and thus enhances its transport through the channel (Zilman et al. 2007, 2010). Such explanations are similar to those applied successfully to account for the transport properties of other channels (e.g., Berezhkovskii and Szabo 2005; Berezhkovskii and Bezrukov 2005). More elaborate analyses consider some of the proposed biophysical properties of the FG-repeat regions or invoke others (e.g., Bickel and Bruinsma 2002; Kustanovich and Rabin 2004). Molecular dynamics simulations are also beginning to shed considerable light on the likely behaviors of FG-repeat regions in the NPC (e.g., Miao and Schulten 2009, 2010), but the sheer complexity of computationally simulating this system remains a significant challenge. However, the fact that a narrow hole filled with a selective polymer is, in principle, all that is needed at the NPC for gating has been demonstrated by chemical analogs (Caspi et al. 2008) and, importantly, by a nanochannel filled with FG-repeat regions from yeast that exhibited selective passage of transport factors over control proteins and even transport of a cargo-carrying karyopherin (Jovanovic-Talisman et al. 2009). In this system, gating was exhibited without any other proteins, including an energy-regenerating system; thus gating in principle requires only the FG-repeat regions.
Although perhaps not needed to understand many of the basic principles of NPC-mediated gating, a full understanding of transport will ultimately require an understanding of how the FG Nups behave at the molecular level. Indeed, in the absence of any folded structure, the question of the physical form and behavior of the FG repeats in and around the NPC’s central channel comes down to what is the precise balance of the FG-repeat regions’ intramolecular and intermolecular forces. Cohesive forces have been measured between at least some types of FG-repeat regions and even within individual repeat regions (Krishnan et al. 2008). Conversely, repulsive forces resulting from entropic exclusion—the tendency of the Brownian motion of a disordered polymer to sweep away other molecules from its vicinity—has also been measured for FG-repeat regions (Lim et al. 2006b). The distribution of FG-repeat types has recently been cataloged and characterized extensively, showing that the FxFG (and similar) repeat regions are characterized by being highly charged and in vitro adopt dynamic, extended-coil conformations whereas the low-charge-content GLFG regions have been reported to form more globular, collapsed coil configurations in vitro (Yamada et al. 2010). Nsp1p, Nup159p, Nup1p, Nup60p, and Nup2p carry mainly charged (FxFG) regions while Nup100p, Nup116p, Nup145Np, Nup57p, Nup49p, and Nup42p carry mainly uncharged regions; however, many of these FG nucleoporins, although predominantly featuring one type, actually have both types of regions next to each other, such as Nsp1p, which has an ∼190-amino-acid GLFG-like amino-terminus followed by an ∼430-amino-acid region that is canonically FxFG-like (Yamada et al. 2010). The extended regions would tend to push away from each other and are predicted to be more mobile, while the more compact regions would tend to be cohesive and less mobile (Ader et al. 2010; Yamada et al. 2010). Thus, if intermolecular attraction forces dominate—even rigid, amyloid-like interactions as measured in vitro for these proteins—then the FG repeats would form a gel; such a “hydrogel” barrier has been proposed, with the central channel filled with a gel of FG Nups cross-linked by intermolecular cohesion, excluding nonspecific molecules by sieving but through which the transport factors dissolve a tunnel by binding to the FG repeats and thereby unzipping them (Ribbeck and Gorlich 2002; Frey et al. 2006; Frey and Gorlich 2007; Mohr et al. 2009; Ader et al. 2010). If repulsive forces such as entropic exclusion dominate, then the central channel is filled with a polymer brush. The repulsive forces exclude nonspecific molecules but can be nullified by the binding forces of transport factors (Lim et al. 2006b, 2007a,b, 2008; Rout et al. 2000, 2003). If intra- and intermolecular cohesions are modulated by entropic exclusion effects, then various “hybrid” models are possible. One such model is the “reduction in dimensionality” (“oily spaghetti”) model that suggests that the FG-repeat regions are more compact, such that they form a layer around the inner walls of the central channel (Macara 2001; Peters 2005). Because they bind, transport factors can enter this layer, allowing them access to the entire central channel, while nonbinding molecules can access only the narrow middle region of the channel, which is devoid of FG repeats. In addition, in vitro measurements led to the suggestion that the reversible binding of transport factors causes the FG repeats to collapse and open a path (Lim et al. 2006b, 2007b, 2008). Similarly, it has been proposed that the more compact and more elongated FG-repeat regions are arranged in such a way as to form an organized but highly dynamic structure, forming a tubular density within the central region of the NPC (Yamada et al. 2010), and perhaps accounting for the “central transporter” as seen in electron micrographs and tomographic reconstructions of the yeast NPC (Yang et al. 1998; Kiseleva et al. 2004). Different transport factors would have access to different portions of this transporter, allowing them to pass by each other to cross the NPC, while entropic exclusion effects prevent nonspecific molecules from crossing (Akey 2010; Yamada et al. 2010). Certainly, there is significant evidence that the different classes of FG repeats are spatially segregated in the yeast NPC (Rout et al. 2000; Alber et al. 2007a,b).
Interestingly, by taking advantage of the molecular genetic techniques available to yeast, a set of targeted deletions were made of the FG regions of FG Nups, either singly or in numerous combinations. These experiments showed that certain of these deletion combinations influenced the transport of only particular transport factors (Strawn et al. 2004; Terry and Wente 2007), and preferences of transport factors for certain Nups has been indicated in living yeast (Marelli et al. 1998; Makhnevych et al. 2003). This preference might manifest as spatially distinguished pathways because recent EM studies in yeast have suggested that mRNPs traffic mainly through the center of the NPC’s channel whereas karyopherins pass through the channel’s periphery (Fiserova et al. 2010). Thus, the different types of FG repeats might mediate multiple but physically and functionally segregated transport pathways through the NPC, perhaps so that the passage of one transport factor through the central channel does not adversely affect another (Fiserova et al. 2010; Yamada et al. 2010). Similarly, the observed asymmetric distribution of certain FG Nups along the nucleocytoplasmic axis of the NPC (Rout et al. 2000; Alber et al. 2007b), although not required for transport (Strawn et al. 2004), may aid in biasing the directionality of transport by providing a high-affinity binding site at the far end of a transport factor’s route through the NPC, trapping a transport factor and its cargo and preventing it from returning through the NPC until the transport reaction is terminated (below) (Rout et al. 2000; Rout and Aitchison 2001; Gilchrist et al. 2002; Gilchrist and Rexach 2003; Pyhtila and Rexach 2003; Strawn et al. 2004; Zilman et al. 2007). Such an idea agrees with affinity measurements made in vitro, which suggested a correlation of the position of asymmetrically located FG Nups with their affinity for certain transport factors (Gilchrist et al. 2002; Gilchrist and Rexach 2003; Pyhtila and Rexach 2003).
While, as discussed above, FG Nups theoretically may be sufficient for gating the NPC, recent work suggests that competition by transport factors themselves also plays a key role in enhancing gating selectivity. Unladen karyopherins, for example, exchange rapidly back and forth across the NPC (Ribbeck et al. 1999), thus occupying space and binding sites in the channel. Therefore, while nonspecific molecules could also bind weakly to FG Nups and in principle pass through the channel, the exchange of transport factors across the NPC, either cargo-laden or free, would sweep these nonspecific molecules out of the central channel, the more strongly binding transport factors effectively out-competing the weaker binding nonspecific molecules. This competition effect has been both predicted theoretically (Zilman et al. 2007, 2010) and measured experimentally in an artificial yeast NPC (Jovanovic-Talisman et al. 2009). In effect, the transport factors may be acting as bouncers at the NPC gate, excluding nonspecific macromolecules and so appearing to be important components of the selectivity barrier. Clearly, competition effects cannot be ignored and likely play a significant role in the gating behavior of the NPC.
Accessory Transport and Processing Factors at the NPC
While the FG-repeat regions serve to concentrate transport factors in the central channel, thereby aiding their nucleocytoplasmic exchange, various more peripherally localized Nups also harbor specific and dynamic binding sites for various accessory transport factors—proteins required to modulate the efficiency, directionality, or energetics of transport but which do not themselves carry cargoes across the NPC (Figure 6). At first, some of these factors were identified as potential Nups, for example, both Gle1p and Gle2p in yeast, but all of these factors are also found free in the cytoplasm and nucleoplasm. The function of these binding sites seems to be to produce a concentrated cloud of the factors around the entrances of the NPC to make the various processes of cargo loading and release more efficient (reviewed in Strambio-de-Castillia et al. 2010; Wente and Rout 2010). Like the shuttling Nups and gatekeeping transport factors, then, these clouds blur the distinction between Nup and transport factor and underscore how the environment in and around the NPC is maintained to be very different from the surrounding nucleoplasm and cytoplasm.
A major factor in the efficiency of cellular processes is the rate at which macromolecules in a given process can interact. As the cell size goes up, so the rate of diffusion of a macromolecule across the cell exponentially decreases (Fick’s law), potentially decreasing the efficiency of cellular processes. This effect can be offset by increasing the local concentration of macromolecules that are part of the same process either by compartmentalizing them (as in eukaryotic internal membranes) or, as here, by providing anchored binding sites. The binding sites for accessory transport factors found on the yeast NPC seem less numerous and diverse than those on vertebrate NPCs; the vertebrate Nup358, carrying up to half a dozen such sites (reviewed in Wente and Rout 2010), is absent in yeast. This may be because vertebrate cells are usually significantly larger than yeast cells.
Starting from the nuclear face of the NPC, the nuclear basket has been implicated by numerous experiments to interact directly with exporting ribonucleoprotein complexes (Figure 6). The best characterized of these interactions to date is between the major basket protein Mlp1p and the essential yeast RNP-binding protein Nab2p, which regulates poly(A) tail length and is important for mRNA nuclear export. Nab2p’s N-terminal ∼100 amino acids interact with the nuclear basket component, Mlp1p, helping to dock mRNPs as they export the nucleus. The atomic structure of this N-terminal region has been solved and is typical of a PWI fold found in numerous RNA-binding proteins (Grant et al. 2008). Another protein on the nuclear face of the NPC and associated with the nuclear basket is Nup1p, which is involved in a number of interactions with transport factors. As well as being an FG Nup docking site (above), Nup1p carries a high-affinity binding site for the Kap95/Kap60 complex in its last ∼40 amino acids; it is believed that this site increases the translocation efficiency of Kap95p import complexes across the NPC by trapping them there prior to Ran-mediated dissociation (Gilchrist et al. 2002; Pyhtila and Rexach 2003). Nup60p may play a similar role, having high-affinity sites for the Kap95/Kap60 complex and Kap123p. Nup60p also carries a binding site for Nup2p, and the recruitment of this mobile Nup to a Kap95/Kap60 complex binding site may aid in dissociating the cargo from this complex and in recycling of Kap60p and Kap95p to the cytoplasm (Denning et al. 2001). In the symmetrical core of the NPC, the non-FG Nups of the core scaffold may also interact with transport factors to aid their passage across the NPC. The Nup84 complex, and in particular Nup85p within it, appears to be able to bind the RNA export factor Mex67p and to help it mediate RNA export (Yao et al. 2008). Another possible binding site for RNA is in the ∼100-amino-acid RRM-like domains of Nup53p and its homolog Nup59p, although as yet no RNA-binding function for these has been assigned in vivo (Devos et al. 2006). Indeed, many RNA-binding and -processing proteins are found on the cytoplasmic side of the NPC. Thus, a putative ∼40-amino-acid redundant RNA-binding domain has been assigned to the homologs Nup116p, Nup100p, and Nup145Np (Fabre and Hurt 1994), and Nup116p contains a binding site for the conserved poly(A)+ RNA export factor Gle2p. The Gle2-binding site (GLEBS) of Nup116p, at residues 110–166, is in the middle of the FG-repeat region, although it is not found in the same region of either Nup145Np or Nup100p; although removal of the GLEBS from Nup116p is normally deleterious, placing it instead in the same site in Nup100p results in a fully viable strain (Bailer et al. 1998). The precise role of Gle2p in RNA export is still unclear, but seems to be required for mRNP export under conditions of heat shock and has been implicated in the nuclear export of the small ribosomal subunit (Takemura et al. 2004). Moreover, as mentioned above, the RNA helicases Dbp5p and Gle1p are attached to the cytoplasmic filaments and mediate release mRNP proteins such as Mex67p and Nab2p from the mRNPs exiting the NPC and drive mRNP transport by restructuring them and helping the long RNP complexes to roll out of the NPC, thereby imparting directionality to the process (Figure 6) (Snay-Hodge et al. 1998; Tseng et al. 1998; Tran et al. 2007; Fan et al. 2009).
Balancing the Books
As has been discussed (Rout et al. 2003), the action of the NPC in transport can be likened to that of an enzyme in a biochemical reaction. Enzymes function as catalysts, lowering the activation energy of a reaction by creating transition states with lowered energy. In this way, they accelerate the rate of transition between substrate and product. Like an enzyme, the facilitation of nucleocytoplasmic exchange by NPCs also works by lowering the activation barrier, but instead of a chemical reaction, they catalyze the movement of macromolecules across the NE. In the NPC, the energy barrier is overcome by the binding energy of specific transport factors, but, as in catalysis, this binding should be neither too weak nor too strong (Dill and Bromberg 2003). As with a catalyst, this process does not necessarily favor transport in any particular direction. Hence, unladen transport factors can diffuse back and forth across the NE far faster than similarly sized nonbinding macromolecules. However, to provide unidirectional transport, other cues are needed. In the case of nuclear import, NLS cargo accumulates in the nucleus because it becomes “trapped” there; while the cargo is allowed to enter the nucleus by virtue of its attachment to a Kap, the high concentration of RanGTP in the nucleus causes its dissociation from the Kap and so prevents its re-exit from the nucleus. Because Kap-cargo complexes constantly form in the cytoplasm and are constantly broken apart in the nucleus, there is a Kap-cargo complex concentration gradient that drives the complexes into the nucleus (Rout et al. 2003). Similarly, export Kaps bind RanGTP and their NES cargoes in the nucleus and diffuse down a concentration gradient into the cytoplasm where RanGAP dissociates the complexes and releases their cargoes. This builds a cytoplasmic concentration of free Kaps and RanGDP. These free Kaps can rapidly exchange across the NPC (Ribbeck et al. 1998), allowing them to diffuse between the nucleus and cytoplasm and search for new cargoes (Macara 2001). Thus, the hydrolysis of GTP maintains the diffusion gradients that ultimately force cargoes to concentrate on one or the other side of the NE (Figure 4). The accumulation of other kinds of cargos in the cytoplasm or nucleoplasm must be driven by analogous concentration gradients; for example, mRNA export is likely driven by the formation of mRNP complexes in the nucleus and their ATP-powered helicase-driven disassembly in the cytoplasm.
Regulation of Transport by the NPC
As an understanding of the principles governing transport across the NPC has emerged, so has our understanding of the regulation of transport. As discussed above, transport is clearly regulated at the level of transport factor recognition and binding in the soluble phase of transport. However, the NPC itself also regulates transport. Earlier work in vertebrate cells indicated that the functional diameter of the channel changes as cells progress through the cell cycle (Feldherr et al. 1984). In yeast, the potential for NPC-regulated transport has been most extensively studied in the context of Nup53p. In contrast to the FG motifs present on numerous Nups, which bind somewhat indiscriminately to multiple members of the β-karyopherin family with low affinity, Nup53p contains a 30-amino-acid residue segment that binds specifically and with relatively high affinity to Kap121p. Detailing this interaction has led to a model in which cell-cycle-dependent phosphorylation of Nup53p causes molecular rearrangements in the NPC during M phase that expose the high-affinity binding site for Kap121p and that retard Kap121p movement through the NPC (Marelli et al. 1998; Makhnevych et al. 2003). While other transport pathways function normally throughout the cell cycle, the mitosis-specific inhibition of Kap121p-mediated import and the reinitiation of import in G1 phase are predicted to regulate the subcellular distribution of molecules required for progression through mitosis. Because of the redundancy of the mulitple Kap transport pathways operating in yeast, it is predicted that only a subset of the repertoire of Kap121p cargoes would be specifically affected by this transient transport block; however, these cargoes remain to be discovered. Similarly environmental perturbations in yeast lead to differential phosphorylation of nucleoporins (Saleem et al. 2010). Morevover, phosphorylation-triggered changes in the NPC alter nuclear transport in Aspergillus nidulans (De Souza et al. 2004; De Souza and Osmani 2007), and Nup phosphorylation is correlated with mitotic NE assembly and disassembly in mammalian cells (Hetzer and Wente 2009). These results suggest that the NPC may be a suitable target for drugs designed to control the cell cycle or specific cell-cycle-regulated transport events.
Beyond Transport: The NPC as a Platform for Other Nuclear Processes
While the NPC is the nexus of communication between the nucleus and cytoplasm, its function is not limited to macromolecular movement transport. The unique positioning of NPCs has also provided evolution with the opportunity to exploit this structure as a scaffold or positional beacon. It is no great surprise, then, that NPCs have been implicated in a host of nuclear processes.
The nucleus is a complex, highly organized subcellular compartment, and the chromatin contained within the organelle is not uniformly distributed. In yeast and metazoans alike, heterochromatin often appears concentrated at the nuclear periphery under the nuclear envelope. In many eukaryotes, heterochromatin is readily observed as electron-dense material in electron micrographs. In yeast, regions of silent chromatin associated with the nuclear periphery include telomeres and the mating-type loci (Cockell and Gasser 1999), and silencing of these domains requires peripheral localization. Indeed, repositioning normally repressed telomere proximal genes to distal positions along chromosomes leads to their inappropriate expression (Maillet et al. 1996), and physically tethering transcriptionally active genes to the nuclear periphery induces gene silencing (Andrulis et al. 1998). It is proposed that the peripheral domain of the nucleus is rich in Sir proteins—silencing factors that promote deacetylation of histones and alter chromatin structure (Gotta et al. 1996; Maillet et al. 1996). Thus, genes that are positioned near telomeres, which themselves are generally tethered to the periphery, are exposed to high concentrations of histone deacetylases and are silenced, while genes distal to telomeres are potentially active. How chromosomal regions come into association with the periphery has been the subject of some debate, but NPC proteins are certainly central to the picture (Figure 6); the basket proteins such as Nup60p, Mlp1p, and Mlp2p, and the mobile nucleoporin, Nup2p, function in the repression of the HMR locus and other subtelomeric genes (Galy et al. 2000; Feuerbach et al. 2002; Dilworth et al. 2005).
In addition to their roles in the formation and maintenance of the peripheral silencing apparatus, NPC components are also linked to the activation of gene expression (Figure 6). For example, while ChIP-chip experiments have revealed that Nups associate with silent mating-type loci and subtelomeric genes, Nups have also been detected in association with active genes (Brickner and Walter 2004; Casolari et al. 2004; Cabal et al. 2006; Taddei et al. 2006; Sarma et al. 2007). NPC-associated genes have been shown to be enriched with the binding site for the transcription factor Rap1p, which, together with its coactivators Gcr1p and Gcr2p, is proposed to link transcriptional machinery to the Nup84 complex (Menon et al. 2005) in a process termed “reverse recruitment.” Furthermore, highly expressed genes appear to be recruited to the periphery when activated (Brickner and Walter 2004; Casolari et al. 2004; Cabal et al. 2006; Taddei et al. 2006; Sarma et al. 2007). Once localized to the periphery, this positioning is maintained to ensure the potential for rapid reactivation of the gene, providing an important advantage to cells exposed to changing environmental conditions. This transcriptional memory, which can persist through several yeast cell divisions, involves the incorporation of the histone variant, Htz1p, into nucleosomes. Htz1p is known to be required for the rapid and robust activation of many environmentally responsive genes (Guillemette et al. 2005; Zhang et al. 2005; Wan et al. 2009) and, in the context of the NPC, functions to retain genes at the periphery and to promote their reactivation (Brickner et al. 2007; Brickner 2009).
Further obscuring the distinction between the gene-activating and gene-repressing functions of the NPC, some NPC components, for example, Nup2p, have been shown to harbor “boundary activity.” This activity is defined by the ability of a protein, when bound to DNA, to prevent the spread of transcriptional repression into adjacent genes by a mechanism that, at least in the case of Nup2p, involves chromosome association with the NPC (Ishii et al. 2002). In this case, the NPC appears to both prevent and promote gene expression, and the nuclear periphery appears to define different territories—both gene activating and repressing. Reconciling these apparent contradictions, it is attractive to think that the association of chromatin with NPCs represents a transient interaction, perhaps reflecting a role for the NPC in transitioning between states, and that these transcriptional states are further defined by chromosome movement between subnuclear territories, a model supported by observations associated with Nup2p (Dilworth et al. 2005). Nup2p has been shown to function in establishing boundaries between active and inactive genes, in transcriptional memory, and in transitioning genes between active and inactive states (Ishii et al. 2002; Dilworth et al. 2005; Brickner et al. 2007) but, unlike typical nucleoporins, Nup2p is not stably associated with NPCs. Rather, this protein transits between the nuclear interior and NPCs, suggesting that Nup2p-dependent recruitment and/or maintenance of genes at the periphery does not dictate stable NPC association. This emerging view is exemplified by recent work in higher eukaryotes, where non-NPC-associated pools of certain nuclear pore complex proteins were shown to associate with, and aid in, the expression of transcriptionally active genes deep within the interior of the nucleus (Capelson et al. 2010; Kalverda et al. 2010). Thus, a direct association with the NPC at the nuclear periphery is not a requisite element in mechanisms of gene regulation by NPC components.
The function of NPCs and nuclear basket-associated structures in chromatin organization is not limited to transcriptional regulation. The myriad of physical and genetic interactions observed between DNA repair factors, components of the Nup84 complex of the NPC and the Mlp proteins in yeast indicate that this nuclear subdomain also acts in the maintenance of telomere length and repair of DNA double-stranded breaks (Galy et al. 2000; Zhao et al. 2004; Zhao and Blobel 2005; Therizols et al. 2006; Palancade et al. 2007; Nagai et al. 2008). Taken together, this convergence of functions at NPCs—in the regulation of transcription, in the stabilization and repair of DNA ends, and in transcript quality control through mRNA surveillance—suggests that the NPC serves as a nexus to inextricably link these processes (Figure 6) (reviewed in Strambio-de-Castillia et al. 2010).
We now have an impression of the NPC as a large multimeric structure that not only plays a pivotal role in transport but also operates to facilitate nuclear organization, gene expression, and chromosome maintenance. In many cases, these functions are positionally linked to NPCs and the extended structures emanating from the nuclear basket, but it is becoming increasingly clear that NPC components exert control far beyond this nuclear microenvironment. A well-studied means by which NPCs exert far-reaching control is through regulated sequestration. The most well-understood examples of this mechanism come from the study of cell cycle regulation in yeast. The overarching theme in these mechanisms is that NPCs and their associated structures provide a scaffold at the nuclear periphery capable of acting as a switchable molecular reservoir, or sink, for proteins temporally regulated through the cell cycle. One example is the cell cycle regulation of septin sumoylation at the bud neck in yeast. Septins are sumoylated by Siz1p prior to anaphase and desumoylated by Ulp1p at cytokinesis (Johnson and Blobel 1999; Makhnevych et al. 2007). Septin sumoylation by Siz1p occurs earlier in mitosis, preceding desumoylation by Ulp1p, and is also controlled by sequestration; however, in this case, Siz1p is sequestered in the nucleus by NES masking. Phosphorylation of Siz1p during mitosis renders the protein competent for nuclear export by Msn5p/Kap142p and, as a result, Siz1p relocalizes to the cytoplasm where it mediates septin sumoylation (Makhnevych et al. 2007). Ulp1p is normally sequestered at NPCs, but is specifically released during mitosis, allowing temporally regulated desumoylation of septins at the bud neck (Takahashi et al. 2000; Makhnevych et al. 2007).
Another cell-cycle-regulated process that employs NPC-mediated sequestration is the spindle assembly checkpoint (SAC), which functions to prevent premature chromosome segregation during mitosis in cells that lack a properly formed mitotic spindle. This conserved mechanism involves the mitotic-arrest-deficient proteins, Mad1p and Mad2p, and components of the NPC and nuclear basket-associated structures (reviewed in Wozniak et al. 2010). In this example, the Mad proteins are sequestered during interphase at NPCs but transiently associate with kinetochores during mitosis until SAC has been passed (Iouk et al. 2002; Scott et al. 2005, 2009; Lee et al. 2008; De Souza et al. 2009).
Summary
Starting from the early work to define the both the yeast nuclear pore complex composition and architecture, and also the mechanism of transport factor-mediated translocation, studies in yeast have led to fundamental and far-reaching discoveries in how the NPC actively participates in transport and many other cellular processes, including chromatin silencing, gene regulation, and control of the cell cycle. In higher eukaryotes, major pathological cellular processes are associated with altered NPCs, and many viruses target components of the nucleocytoplasmic transport pathway to usurp it (Faustino et al. 2007; Chahine and Pierce 2009). Hence, nucleoporins and transport factors are key potential targets for drug therapy. But they have been almost entirely neglected so far because only recently, through much of the pioneering work in yeast, have we inventoried both the transport factors and the molecular constituents of the NPC and so have begun to gain an understanding of the critical parameters of the NPC’s roles in diverse functions. Recent results are revealing the enormous potential for how we might be able to specifically control various cellular processes by interventions targeting the NPC. Yet, much remains to be discovered to understand the molecular mechanisms underlying the numerous roles that the NPC and nuclear transport play in sufficient detail to design interventions and to predict their effects. It is clear that yeast will continue to play a central role in these discoveries.
Acknowledgments
We wish to acknowledge all members of the Aitchison and Rout laboratories for their support, Caterina Strambio de Castilla for helpful comments, and Marlene Oeffinger for critical reading of the manuscript. We are grateful for the support of the National Institutes of Health from grants U54 RR022220 and U01 GM098256 (M.R. & J.A.), R01 GM62427 (M.R.), R01 GM071329 (M.R.), R01 GM075152 (J.A.), P50 GM076547 (J.A.).
Footnotes
Communicating editor: T. N. Davis
Literature Cited
- Ader C., Frey S., Maas W., Schmidt H. B., Gorlich D., et al. , 2010. Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. USA 107: 6281–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S., Brickner J. H., 2007. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet. 23: 396–402 [DOI] [PubMed] [Google Scholar]
- Aitchison J. D., Blobel G., Rout M. P., 1995a. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131: 1659–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., Marelli M., Blobel G., Wozniak R. W., 1995b. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131: 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Blobel G., Rout M. P., 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274: 624–627 [DOI] [PubMed] [Google Scholar]
- Akey C. W., 2010. The NPC-transporter, a ghost in the machine. Structure 18: 1230–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007a. Determining the architectures of macromolecular assemblies. Nature 450: 683–694 [DOI] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007b. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Anderson D. J., Hetzer M. W., 2008a. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Hetzer M. W., 2008b. Shaping the endoplasmic reticulum into the nuclear envelope. J. Cell Sci. 121: 137–142 [DOI] [PubMed] [Google Scholar]
- Anderson J. T., Wilson S. M., Datar K. V., Swanson M. S., 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E. D., Neiman A. M., Zappulla D. C., Sternglanz R., 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Arib, G., and Akhtar, A., 2011 Multiple facets of nuclear periphery in gene expression control. Curr. Opin. Cell Biol. 23: 346–353. [DOI] [PubMed]
- Arts G. J., Kuersten S., Romby P., Ehresmann B., Mattaj I. W., 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17: 7430–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo D., Nascimento L., Labarre J., Toledano M. B., Rodrigues-Pousada C., 2007. The S. cerevisiae Yap1 and Yap2 transcription factors share a common cadmium-sensing domain. FEBS Lett. 581: 187–195 [DOI] [PubMed] [Google Scholar]
- Bailer S. M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., et al. , 1998. Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J. 17: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer S. M., Balduf C., Katahira J., Podtelejnikov A., Rollenhagen C., et al. , 2000. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275: 23540–23548 [DOI] [PubMed] [Google Scholar]
- Bailer S. M., Balduf C., Hurt E., 2001. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol. Cell. Biol. 21: 7944–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhrat A., Baranes-Bachar K., Reshef D., Voloshin O., Krichevsky O., et al. , 2008. Nuclear export of Ho endonuclease of yeast via Msn5. Curr. Genet. 54: 271–281 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Snay-Hodge C., Pasteau F., Dagher S., Cole C. N., et al. , 1998. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell 9: 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek J., Kang J., Kurischko C., Li B., Raab J. R., et al. , 2005. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle 4: 961–971 [DOI] [PubMed] [Google Scholar]
- Berezhkovskii A. M., Bezrukov S. M., 2005. Optimizing transport of metabolites through large channels: molecular sieves with and without binding. Biophys. J. 88: L17–L19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhkovskii A., Szabo A., 2005. One-dimensional reaction coordinates for diffusive activated rate processes in many dimensions. J. Chem. Phys. 122: 14503. [DOI] [PubMed] [Google Scholar]
- Berke I. C., Boehmer T., Blobel G., Schwartz T. U., 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 167: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel T., Bruinsma R., 2002. The nuclear pore complex mystery and anomalous diffusion in reversible gels. Biophys. J. 83: 3079–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T., Enninga J., Dales S., Blobel G., Zhong H., 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 100: 981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., et al. , 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Brickner J. H., 2009. Transcriptional memory at the nuclear periphery. Curr. Opin. Cell Biol. 21: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Walter P., 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., et al. , 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Schwartz T. U., 2009. A lattice model of the nuclear pore complex. Commun. Integr. Biol. 2: 205–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U., 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. M., Stow J. L., 2004. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 14: 427–434 [DOI] [PubMed] [Google Scholar]
- Cabal G. G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., et al. , 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Caesar S., Greiner M., Schlenstedt G., 2006. Kap120 functions as a nuclear import receptor for ribosome assembly factor Rpf1 in yeast. Mol. Cell. Biol. 26: 3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., Chook Y. M., 2007. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., et al. , 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody S. R., Wente S. R., 2009. mRNA nuclear export at a glance. J. Cell Sci. 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carron C., O’Donohue M. F., Choesmel V., Faubladier M., Gleizes P. E., 2011. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 39: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., et al. , 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Caspi Y., Zbaida D., Cohen H., Elbaum M., 2008. Synthetic mimic of selective transport through the nuclear pore complex. Nano Lett. 8: 3728–3734 [DOI] [PubMed] [Google Scholar]
- Chadrin A., Hess B., San Roman M., Gatti X., Lombard B., et al. , 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189: 795–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahine M. N., Pierce G. N., 2009. Therapeutic targeting of nuclear protein import in pathological cell conditions. Pharmacol. Rev. 61: 358–372 [DOI] [PubMed] [Google Scholar]
- Chang J. S., Henry K., Geli M. I., Lemmon S. K., 2006. Cortical recruitment and nuclear-cytoplasmic shuttling of Scd5p, a protein phosphatase-1-targeting protein involved in actin organization and endocytosis. Mol. Biol. Cell 17: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H. J., Rout M. P., Giddings T. H., Winey M., 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143: 1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., Muller C. W., 1999. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399: 221–229 [DOI] [PubMed] [Google Scholar]
- Cockell M., Gasser S. M., 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9: 199–205 [DOI] [PubMed] [Google Scholar]
- Cook A. G., Fukuhara N., Jinek M., Conti E., 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- Cordes V. C., Reidenbach S., Rackwitz H. R., Franke W. W., 1997. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J., 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M. A., Anderson D., Richard E., Hetzer M., 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312: 440–443 [DOI] [PubMed] [Google Scholar]
- Davis L. I., Fink G. R., 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell 61: 965–978 [DOI] [PubMed] [Google Scholar]
- Dawson T. R., Lazarus M. D., Hetzer M., Wente S. R., 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler E. W., Ma Y., Seo H. S., Hsia K. C., Noriega T. R., et al. , 2008. A fence-like coat for the nuclear pore membrane. Mol. Cell 32: 815–826 [DOI] [PubMed] [Google Scholar]
- De Craene J. O., Coleman J., Estrada de Martin P., Pypaert M., Anderson S., et al. , 2006. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol. Biol. Cell 17: 3009–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse J. A., DuBois K. N., Devos D., Siegel T. N., Sali A., et al. , 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. P., Rexach M. F., 2007. Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell. Proteomics 6: 272–282 [DOI] [PubMed] [Google Scholar]
- Denning D., Mykytka B., Allen N. P., Huang L., Al B., et al. , 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M., 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani S. A., 2007. Mitosis, not just open or closed. Eukaryot. Cell 6: 1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A., 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14: 1973–1984 [DOI] [PubMed] [Google Scholar]
- De Souza, C. P., Hashmi, S. B., Nayak, T., Oakley, B., and Osmani, S. A., 2009 Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell. 20: 2146–2149. [DOI] [PMC free article] [PubMed]
- DeVit M. J., Johnston M., 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., et al. , 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., et al. , 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 103: 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Bromberg S., 2003. Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology. Garland Science, New York/London [Google Scholar]
- Dilworth D. J., Suprapto A., Padovan J. C., Chait B. T., Wozniak R. W., et al. , 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153: 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth D. J., Tackett A. J., Rogers R. S., Yi E. C., Christmas R. H., et al. , 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S., Williams R., Devos D., Sali A., Chait B. T., et al. , 2006. Protease accessibility laddering: a proteomic tool for probing protein structure. Structure 14: 653–660 [DOI] [PubMed] [Google Scholar]
- Dong S., Li C., Zenklusen D., Singer R. H., Jacobson A., et al. , 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V., Wepf R., Hurt E. C., 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13: 6062–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M. J., Pinol-Roma S., Burd C. G., 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Durchschlag E., Reiter W., Ammerer G., Schuller C., 2004. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J. Biol. Chem. 279: 55425–55432 [DOI] [PubMed] [Google Scholar]
- Emtage J. L., Bucci M., Watkins J. L., Wente S. R., 1997. Defining the essential functional regions of the nucleoporin Nup145p. J. Cell Sci. 110(Pt. 7): 911–925 [DOI] [PubMed] [Google Scholar]
- Fabre E., Hurt E. C., 1994. Nuclear transport. Curr. Opin. Cell Biol. 6: 335–342 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Hurt E. C., Aebi U., Pante N., 1998. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. 143: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Aris J. P., Hurt E. C., Pante N., Aebi U., 2000a. Comparative spatial localization of protein-A-tagged and authentic yeast nuclear pore complex proteins by immunogold electron microscopy. J. Struct. Biol. 129: 295–305 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Hubner W., Mandinova A., Pante N., Keller W., et al. , 2000b. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol. Biol. Cell 11: 3885–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. S., Cheng Z., Zhang J., Noble C., Zhou Z., et al. , 2009. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J. Mol. Biol. 388: 1–10 [DOI] [PubMed] [Google Scholar]
- Fasken M. B., Corbett A. H., 2009. Mechanisms of nuclear mRNA quality control. RNA Biol. 6: 237–241 [DOI] [PubMed] [Google Scholar]
- Fath S., Mancias J. D., Bi X., Goldberg J., 2007. Structure and organization of coat proteins in the COPII cage. Cell 129: 1325–1336 [DOI] [PubMed] [Google Scholar]
- Fatica A., Tollervey D., 2002. Making ribosomes. Curr. Opin. Cell Biol. 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Faustino R. S., Nelson T. J., Terzic A., Perez-Terzic C., 2007. Nuclear transport: target for therapy. Clin. Pharmacol. Ther. 81: 880–886 [DOI] [PubMed] [Google Scholar]
- Feldherr C. M., Kallenbach E., Schultz N., 1984. Movement of a karyophilic protein through the nuclear pores of oocytes. J. Cell Biol. 99: 2216–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Benko A. L., Lee J. H., Stanford D. R., Hopper A. K., 1999. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 112(Pt. 3): 339–347 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J., Rout M. P., 2009. Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 21: 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A., 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17: 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., et al. , 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4: 214–221 [DOI] [PubMed] [Google Scholar]
- Field M. C., Dacks J. B., 2009. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 21: 4–13 [DOI] [PubMed] [Google Scholar]
- Fiserova J., Richards S. A., Wente S. R., Goldberg M. W., 2010. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J. Cell Sci. 123: 2773–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D., Sarges P., Stelter P., Hellwig A., Bottcher B., et al. , 2009. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J. Cell Biol. 185: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., Blobel G., 1996. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 271: 5313–5316 [DOI] [PubMed] [Google Scholar]
- Fontoura B. M., Blobel G., Matunis M. J., 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144: 1097–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Gorlich D., 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130: 512–523 [DOI] [PubMed] [Google Scholar]
- Frey S., Richter R. P., Gorlich D., 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314: 815–817 [DOI] [PubMed] [Google Scholar]
- Frosst P., Guan T., Subauste C., Hahn K., Gerace L., 2002. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., et al. , 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F., Olivier C., Dandjinou A. T., Wellinger R. J., Chartrand P., 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27: 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin J. C., Scherthan H., Doye V., Rascalou N., et al. , 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112 [DOI] [PubMed] [Google Scholar]
- Gilchrist D., Rexach M., 2003. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J. Biol. Chem. 278: 51937–51949 [DOI] [PubMed] [Google Scholar]
- Gilchrist D., Mykytka B., Rexach M., 2002. Accelerating the rate of disassembly of karyopherin.cargo complexes. J. Biol. Chem. 277: 18161–18172 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., Snay C. A., Heath C. V., Cole C. N., 1996. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell 7: 917–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., et al. , 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., et al. , 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Emig S., Weise C., Hucho F., Pohl T., et al. , 1995. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J. Cell Biol. 130: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. P., Marshall N. J., Yang J. C., Fasken M. B., Kelly S. M., et al. , 2008. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J. Mol. Biol. 376: 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland K. B., Ding H., Costanzo M., Boone C., Davis T. N., 2010. Identification of Saccharomyces cerevisiae spindle pole body remodeling factors. PLoS ONE 5: e15426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E. R., Altan N., Lippincott-Schwartz J., Powers M. A., 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13: 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D., Singer R., 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B., Bataille A. R., Gevry N., Adam M., Blanchette M., et al. , 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage A. E., Tollervey D., 2004. A surfeit of factors: Why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 1: 10–15 [PubMed] [Google Scholar]
- Hammell C. M., Gross S., Zenklusen D., Heath C. V., Stutz F., et al. , 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase M. E., Kuznetsov N. V., Cordes V. C., 2001. Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol. Biol. Cell 12: 2433–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath C. V., Copeland C. S., Amberg D. C., Del Priore V., Snyder M., et al. , 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 131: 1677–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A. K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., et al. , 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W., Wente S. R., 2009. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. K., Shen T. X., Ryan K. J., Kiseleva E., Levy M. A., et al. , 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20: 5736–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W., 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel A. E., Hodel M. R., Griffis E. R., Hennig K. A., Ratner G. A., et al. , 2002. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell 10: 347–358 [DOI] [PubMed] [Google Scholar]
- Hodge C. A., Choudhary V., Wolyniak M. J., Scarcelli J. J., Schneiter R., et al. , 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges J. L., Leslie J. H., Mosammaparast N., Guo Y., Shabanowitz J., et al. , 2005. Nuclear import of TFIIB is mediated by Kap114p, a karyopherin with multiple cargo-binding domains. Mol. Biol. Cell 16: 3200–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Phizicky E. M., 2003. tRNA transfers to the limelight. Genes Dev. 17: 162–180 [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Pai D. A., Engelke D. R., 2010. Cellular dynamics of tRNAs and their genes. FEBS Lett. 584: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K. C., Stavropoulos P., Blobel G., Hoelz A., 2007. Architecture of a coat for the nuclear pore membrane. Cell 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., et al. , 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Hung N. J., Lo K. Y., Patel S. S., Helmke K., Johnson A. W., 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell 19: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E. C., 1988. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 7: 4323–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz M. E., Strambio-de-Castillia C., Blobel G., 1998. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc. Natl. Acad. Sci. USA 95: 11241–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T., Kerscher O., Scott R. J., Basrai M. A., Wozniak R. W., 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159: 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgro T. A., Schulten K., 2005. Binding dynamics of isolated nucleoporin repeat regions to importin-beta. Structure 13: 1869–1879 [DOI] [PubMed] [Google Scholar]
- Isgro T. A., Schulten K., 2007. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J. Mol. Biol. 366: 330–345 [DOI] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U. K., 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562 [DOI] [PubMed] [Google Scholar]
- Jekely G., Arendt D., 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28: 191–198 [DOI] [PubMed] [Google Scholar]
- Jensen T. H., Boulay J., Rosbash M., Libri D., 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Jeudy S., Schwartz T. U., 2007. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J. Biol. Chem. 282: 34904–34912 [DOI] [PubMed] [Google Scholar]
- Jimenez M., Petit T., Gancedo C., Goday C., 2000. The alm1+ gene from Schizosaccharomyces pombe encodes a coiled-coil protein that associates with the medial region during mitosis. Mol. Gen. Genet. 262: 921–930 [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G., 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic-Talisman T., Tetenbaum-Novatt J., McKenney A. S., Zilman A., Peters R., et al. , 2009. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457: 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., 2010. Lessons on longevity from budding yeast. Nature 464: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank N. M., O’Neill E. M., Huang L. S., O’Shea E. K., 1998a. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank N. M., O’Shea E. K., 1998b. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12: 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V. V., Fornerod M., 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Kampmann M., Blobel G., 2009. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 16: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser K., Podtelejnikov A., Mann M., Jung J. U., et al. , 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18: 2593–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S., 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P. J., Lorenz H., Schiebel E., Knop M., 2010. Segregation of yeast nuclear pores. Nature 466: E1. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Meurer M., Knop M., Schiebel E., 2011. Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr. Biol. 21: R17–R18 [DOI] [PubMed] [Google Scholar]
- Kiseleva E., Allen T. D., Rutherford S., Bucci M., Wente S. R., et al. , 2004. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145: 272–288 [DOI] [PubMed] [Google Scholar]
- Köhler A., Hurt E. C., 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Komeili A., O’Shea E. K., 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284: 977–980 [DOI] [PubMed] [Google Scholar]
- Kraemer D. M., Strambio-de-Castillia C., Blobel G., Rout M. P., 1995. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 270: 19017–19021 [DOI] [PubMed] [Google Scholar]
- Krishnan V. V., Lau E. Y., Yamada J., Denning D. P., Patel S. S., et al. , 2008. Intramolecular cohesion of coils mediated by phenylalanine–glycine motifs in the natively unfolded domain of a nucleoporin. PLoS Comput. Biol. 4: e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Thyberg J., Bjorkroth B., Rackwitz H. R., Cordes V. C., 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 15: 4261–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustanovich T., Rabin Y., 2004. Metastable network model of protein transport through nuclear pores. Biophys. J. 86: 2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. K., Giddings T. H., Jr, Winey M., 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell 3: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Goldberg J., 2010. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell 142: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. C., Aitchison J. D., 1999. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 274: 29031–29037 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Sterling H., Burlingame A., McCormick F., 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22: 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Matsuura Y., Liu S. M., Stewart M., 2005. Structural basis for nuclear import complex dissociation by RanGTP. Nature 435: 693–696 [DOI] [PubMed] [Google Scholar]
- Leksa N. C., Brohawn S. G., Schwartz T. U., 2009. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17: 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie D. M., Grill B., Rout M. P., Wozniak R. W., Aitchison J. D., 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22: 2544–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie D. M., Zhang W., Timney B. L., Chait B. T., Rout M. P., et al. , 2004. Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24: 8487–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li O., Heath C. V., Amberg D. C., Dockendorff T. C., Copeland C. S., et al. , 1995. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell 6: 401–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liker E., Fernandez E., Izaurralde E., Conti E., 2000. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 19: 5587–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R. Y., Aebi U., Stoffler D., 2006a. From the trap to the basket: getting to the bottom of the nuclear pore complex. Chromosoma 115: 15–26 [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Huang N. P., Koser J., Deng J., Lau K. H., et al. , 2006b. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103: 9512–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R. Y., Fahrenkrog B., Koser J., Schwarz-Herion K., Deng J., et al. , 2007a. Nanomechanical basis of selective gating by the nuclear pore complex. Science 318: 640–643 [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Koser J., Huang N. P., Schwarz-Herion K., Aebi U., 2007b. Nanomechanical interactions of phenylalanine-glycine nucleoporins studied by single molecule force-volume spectroscopy. J. Struct. Biol. 159: 277–289 [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Ullman K. S., Fahrenkrog B., 2008. Biology and biophysics of the nuclear pore complex and its components. Int. Rev. Cell Mol. Biol. 267: 299–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff F. R., Izaurralde E., Kutay U., Schafer S., et al. , 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L., De Souza C. P., Osmani A. H., Osmani S. A., 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84–120 complex. Mol. Biol. Cell 20: 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. M., Stewart M., 2005. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 349: 515–525 [DOI] [PubMed] [Google Scholar]
- Lo K. Y., Johnson A. W., 2009. Reengineering ribosome export. Mol. Biol. Cell 20: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Makhnevych T., Marelli M., Aitchison J. D., Wozniak R. W., 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E., 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Stangl K., Stelter P., Toth K. F., et al. , 2005. Reconstitution of Nup157 and Nup145N into the Nup84 complex. J. Biol. Chem. 280: 18442–18451 [DOI] [PubMed] [Google Scholar]
- Macara I. G., 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A. S., Mancuso J., Cande W. Z., Weis K., 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi L. B., Jr, Kuchenruether M., Dadey D. Y., Schwope R. M., Grisendi S., et al. , 2008. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 28: 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., et al. , 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Lusk C. P., Anderson A. M., Aitchison J. D., Wozniak R. W., 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115: 813–823 [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Ptak C., Lusk C. P., Aitchison J. D., Wozniak R. W., 2007. The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 177: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T., Stanton L. H., Lin C. C., Goldfarb D. S., Weis K., et al. , 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans B. J., Anantharaman V., Aravind L., Koonin E. V., 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3: 1612–1637 [DOI] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L. A., Panté N., Mall M., et al. , 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22: 93–103 [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J. D., Wozniak R. W., 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143: 1813–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Powers T., Hall M. N., 2006. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 4: 259–260 [DOI] [PubMed] [Google Scholar]
- Mason D. A., Stage D. E., Goldfarb D. S., 2009. Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 68: 351–365 [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Stewart M., 2004. Structural basis for the assembly of a nuclear export complex. Nature 432: 872–877 [DOI] [PubMed] [Google Scholar]
- Matunis M. J., Wu J., Blobel G., 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcak I., Hoelz A., Blobel G., 2007. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science 315: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Menezes R. A., Amaral C., Delaunay A., Toledano M., Rodrigues-Pousada C., 2004. Yap8p activation in Saccharomyces cerevisiae under arsenic conditions. FEBS Lett. 566: 141–146 [DOI] [PubMed] [Google Scholar]
- Menon B. B., Sarma N. J., Pasula S., Deminoff S. J., Willis K. A., et al. , 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, L., and Schulten, K., 2009 Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics. Structure 17: 449–459. [DOI] [PMC free article] [PubMed]
- Miao L., Schulten K., 2010. Probing a structural model of the nuclear pore complex channel through molecular dynamics. Biophys. J. 98: 1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Ryan K. J., Wente S. R., 2006. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics 172: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D., Frey S., Fischer T., Guttler T., Gorlich D., 2009. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28: 2541–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., Suliman S., Ben-Yishay R., Yunger S., Brody Y., et al. , 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12: 543–552 [DOI] [PubMed] [Google Scholar]
- Moy T. I., Silver P. A., 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115: 2985–2995 [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Weis K., 1999. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA 96: 9622–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., et al. , 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V., Hsia K. C., Debler E. W., Kampmann M., Davenport A. M., et al. , 2009. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc. Natl. Acad. Sci. USA 106: 17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Kern H., Mutvei A., Horstmann H., Marshallsay B., et al. , 1990. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell 61: 979–989 [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., Brett C. L., Merz A. J., 2009. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B. T., Rout M. P., 2005. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Dlakic M., Tollervey D., 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertle T., Klinger M., Stuermer C. A., Schwab M. E., 2003. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17: 1238–1247 [DOI] [PubMed] [Google Scholar]
- Onischenko E., Stanton L. H., Madrid A. S., Kieselbach T., Weis K., 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., et al. , 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273: 1392–1395 [DOI] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Nile A., Osmani S. A., 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17: 4946–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R., 2004. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 20: 153–191 [DOI] [PubMed] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., Doye V., 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell 16: 5258–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., et al. , 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 18: 2912–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. S., Belmont B. J., Sante J. M., Rexach M. F., 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96 [DOI] [PubMed] [Google Scholar]
- Pelaez R., Herrero P., Moreno F., 2009. Nuclear export of the yeast hexokinase 2 protein requires the Xpo1 (Crm1)-dependent pathway. J. Biol. Chem. 284: 20548–20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M., 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Rout M. P., Blobel G., 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault A., Bellemer C., Bachand F., 2008. Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 36: 6132–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., et al. , 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27: 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R., 2005. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic 6: 421–427 [DOI] [PubMed] [Google Scholar]
- Peters, R., 2009 Functionalization of a nanopore: the nuclear pore complex paradigm. Biochim. Biophys. Acta 1793: 1533–1539. [DOI] [PMC free article] [PubMed]
- Pyhtila B., Rexach M., 2003. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278: 42699–42709 [DOI] [PubMed] [Google Scholar]
- Qi H., Rath U., Wang D., Xu Y. Z., Ding Y., et al. , 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell 15: 4854–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E., Igual J. C., 2003. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol. Cell. Biol. 23: 3126–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner G. A., Hodel A. E., Powers M. A., 2007. Molecular determinants of binding between Gly-Leu-Phe-Gly nucleoporins and the nuclear pore complex. J. Biol. Chem. 282: 33968–33976 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Gorlich D., 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21: 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky G., Kent H. M., Stewart M., Gorlich D., 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17: 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Kutay U., Paraskeva E., Gorlich D., 1999. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol. 9: 47–50 [DOI] [PubMed] [Google Scholar]
- Robinson M. A., Park S., Sun Z. Y., Silver P. A., Wagner G., et al. , 2005. Multiple conformations in the ligand-binding site of the yeast nuclear pore-targeting domain of Nup116p. J. Biol. Chem. 280: 35723–35732 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., and Hurt, E., 2011 Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 23: 302–309. [DOI] [PubMed]
- Rondon A. G., Jimeno S., Aguilera A., 2010. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim. Biophys. Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- Rosenblum J. S., Blobel G., 1999. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96: 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum J. S., Pemberton L. F., Blobel G., 1997. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 139: 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., 2000. Pore relations: nuclear pore complexes and nucleocytoplasmic exchange. Essays Biochem. 36: 75–88 [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276: 16593–16596 [DOI] [PubMed] [Google Scholar]
- Rout M. P., Blobel G., 1993. Isolation of the yeast nuclear pore complex. J. Cell Biol. 123: 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Wente S. R., 1994. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Rout M. P., Blobel G., Aitchison J. D., 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell 89: 715–725 [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., et al. , 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T., 2003. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 13: 622–628 [DOI] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R., 2002. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., McCaffery J. M., Wente S. R., 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160: 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Zhou Y., Wente S. R., 2007. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell 18: 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. R., Chuang J. S., Schekman R. W., 1997. Sec31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol. Biol. Cell 8: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem R. A., Rogers R. S., Ratushny A. V., Dilworth D. J., Shannon P. T., et al. , 2010. Integrated phosphoproteomics analysis of a signaling network governing nutrient response and peroxisome induction. Mol. Cell. Proteomics 9: 2076–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar P., Ozyurt S. A., Do J., Bain K. T., Dickey M., et al. , 2010. Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145. Proteins 78: 1992–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., et al. , 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18: 6826–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma N. J., Haley T. M., Barbara K. E., Buford T. D., Willis K. A., et al. , 2007. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 175: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli J. J., Hodge C. A., Cole C. N., 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 178: 799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jensen T. H., 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schrader N., Stelter P., Flemming D., Kunze R., Hurt E., et al. , 2008. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell 29: 46–55 [DOI] [PubMed] [Google Scholar]
- Scott R. J., Lusk C. P., Dilworth D. J., Aitchison J. D., Wozniak R. W., 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 16: 4362–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. J., Cairo L. V., Van de Vosse D. W., Wozniak R. W., 2009. The nuclear export factor Xpo1p targets Mad1p to kinetochores in yeast. J. Cell Biol. 184: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser R. M., Sundberg A. E., Wollam B. J., Zobel-Thropp P., Baldwin K., et al. , 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H. S., Ma Y., Debler E. W., Wacker D., Kutik S., et al. , 2009. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc. Natl. Acad. Sci. USA 106: 14281–14286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S. B., Gonnet G., Barral Y., 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature 454: 728–734 [DOI] [PubMed] [Google Scholar]
- Shibata S., Sasaki M., Miki T., Shimamoto A., Furuichi Y., et al. , 2006. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 34: 4711–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., C. Voss, J. M. Rist, J. Hu, T. A. Rapoport et al., 2008 The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283: 18892–18904. [DOI] [PMC free article] [PubMed]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., et al. , 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., Lutzmann M., Santos-Rosa H., Leonard K., Mueller S., et al. , 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzný M., Schneider C., Rácz A., Weng J., Tollervey D., et al. , 2009. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 7: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge C. A., Colot H. V., Goldstein A. L., Cole C. N., 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford C. S., Guthrie C., Weis K., 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt U., Silver P. A., 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11: 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg S. M., Gurkan C., Fowler D. M., LaPointe P., Foss T. R., et al. , 2006. Structure of the Sec13/31 COPII coat cage. Nature 439: 234–238 [DOI] [PubMed] [Google Scholar]
- Stagg S. M., LaPointe P., Balch W. E., 2007. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 17: 221–228 [DOI] [PubMed] [Google Scholar]
- Stagg S. M., LaPointe P., Razvi A., Gurkan C., Potter C. S., et al. , 2008. Structural basis for cargo regulation of COPII coat assembly. Cell 134: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Hulsmann B. B., Spang A., Hartmann E., Cordes V. C., et al. , 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173: 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P., Kunze R., Flemming D., Hopfner D., Diepholz M., et al. , 2007. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9: 788–796 [DOI] [PubMed] [Google Scholar]
- Stewart M., 2003. Structural biology. Nuclear trafficking. Science 302: 1513–1514 [DOI] [PubMed] [Google Scholar]
- Stewart M., 2010. Nuclear export of mRNA. Trends Biochem. Sci. 35: 609–617 [DOI] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., et al. , 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 18: 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M. P., 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144: 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Niepel M., Rout M. P., 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11: 490–501 [DOI] [PubMed] [Google Scholar]
- Strasser K., Bassler J., Hurt E., 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R., 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6: 197–206 [DOI] [PubMed] [Google Scholar]
- Sun Y., Guo H. C., 2008. Structural constraints on autoprocessing of the human nucleoporin Nup98. Protein Sci. 17: 494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S. R., 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Sydorskyy Y., Dilworth D. J., Yi E. C., Goodlett D. R., Wozniak R. W., et al. , 2003. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23: 2042–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberner F. J., Quilis I., Igual J. C., 2009. Spatial regulation of the start repressor Whi5. Cell Cycle 8: 3010–3018 [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., et al. , 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Mizoi J., Toh E. A., Kikuchi Y., 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. 128: 723–725 [DOI] [PubMed] [Google Scholar]
- Takemura R., Inoue Y., Izawa S., 2004. Stress response in yeast mRNA export factor: reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J. Cell Sci. 117: 4189–4197 [DOI] [PubMed] [Google Scholar]
- Tcheperegine S. E., Marelli M., Wozniak R. W., 1999. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 274: 5252–5258 [DOI] [PubMed] [Google Scholar]
- Teixeira M. T., Siniossoglou S., Podtelejnikov S., Benichou J. C., Mann M., et al. , 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16: 5086–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. T., Fabre E., Dujon B., 1999. Self-catalyzed cleavage of the yeast nucleoporin Nup145p precursor. J. Biol. Chem. 274: 32439–32444 [DOI] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178: 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G. G., Genovesio A., Olivo-Marin J. C., et al. , 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E., Tollervey D., 2010. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell. Biol. 30: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney B., Tetenbaum-Novatt J., Agate D., Williams R., Zhang W., et al. , 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towpik J., Graczyk D., Gajda A., Lefebvre O., Boguta M., 2008. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 283: 17168–17174 [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R., 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tran E. J., Zhou Y., Corbett A. H., Wente S. R., 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28: 850–859. [DOI] [PubMed] [Google Scholar]
- Tseng S. S., Weaver P. L., Liu Y., Hitomi M., Tartakoff A. M., et al. , 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17: 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta R., Fujiwara N., Iwai K., Yamaguchi-Iwai Y., 2007. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E., Leplus A., Osheim Y. N., Beyer A. L., Wacheul L., et al. , 2008. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA 14: 2061–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., Stutz F., 2004. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 16: 285–292 [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A., 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Walde S., Kehlenbach R. H., 2010. The part and the whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 20: 461–469 [DOI] [PubMed] [Google Scholar]
- Wan Y., Saleem R. A., Ratushny A. V., Roda O., Smith J. J., et al. , 2009. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell. Biol. 29: 2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Berger J. M., Weis K., 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16: 749–760 [DOI] [PubMed] [Google Scholar]
- Wente S. R., Blobel G., 1993. A temperature-sensitive nup116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Blobel G., 1994. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J. Cell Biol. 125: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. R., Vaisberg E. V., Ding R., Nurse P., McIntosh J. R., 1998. cut11(+): a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell 9: 2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle, J. R., and Schwartz, T. U., 2009 Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J. Biol. Chem. 284: 28442–28452. [DOI] [PMC free article] [PubMed]
- Winey M., Yarar D., Giddings T. H., Jr, Mastronarde D. N., 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 8: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Shields D. C., 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Wozniak R. W., Blobel G., Rout M. P., 1994. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 125: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. W., Rout M. P., Aitchison J. D., 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8: 184–188 [DOI] [PubMed] [Google Scholar]
- Wozniak R., Burke B., Doye V., 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cellular and molecular life sciences. Cell. Mol. Life Sci. 67: 2215–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Matunis M. J., Kraemer D., Blobel G., Coutavas E., 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270: 14209–14213 [DOI] [PubMed] [Google Scholar]
- Yamada J., Phillips J. L., Patel S., Goldfien G., Calestagne-Morelli A., et al. , 2010. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9: 2205–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rout M. P., Akey C. W., 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1: 223–234 [DOI] [PubMed] [Google Scholar]
- Yao W., Roser D., Kohler A., Bradatsch B., Bassler J., et al. , 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Yao W., Lutzmann M., Hurt E., 2008. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J. 27: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Blobel G., 2001. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 152: 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Kutay U., 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
- Zhang H., Roberts D. N., Cairns B. R., 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Brkljacic J., Meier I., 2008. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu C. Y., Blobel G., 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167: 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilman A., Di Talia S., Chait B. T., Rout M. P., Magnasco M. O., 2007. Efficiency, selectivity, and robustness of nucleocytoplasmic transport. PLoS Comput. Biol. 3: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilman A., Di Talia S., Jovanovic-Talisman T., Chait B. T., Rout M. P., et al. , 2010. Enhancement of transport selectivity through nano-channels by non-specific competition. PLoS Comput. Biol. 6: e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]