Abstract
In Drosophila collections of green fluorescent protein (GFP) trap lines have been used to probe the endogenous expression patterns of trapped genes or the subcellular localization of their protein products. Here, we describe a method, based on nonoverlapping, highly specific, shRNA transgenes directed against GFP, that extends the utility of these collections to loss-of-function studies. Furthermore, we used a MiMIC transposon to generate GFP traps in Drosophila cell lines with distinct subcellular localization patterns, which will permit high-throughput screens using fluorescently tagged proteins. Finally, we show that fluorescent traps, paired with recombinant nanobodies and mass spectrometry, allow the study of endogenous protein complexes in Drosophila.
A commonly used method for visualizing endogenous proteins without the need for specific antibodies is the “protein trap” approach (Gossler et al. 1989; Morin et al. 2001; Clark et al. 2011). Protein trapping relies on transposons that harbor a fluorescent or epitope tag flanked by splice acceptor and donor sites. If such a transposon is inserted into an intron, then its tag is spliced into the open reading frame (ORF) of the trapped gene. This approach has been used successfully in a number of model organisms, including Drosophila, where several collections of green fluorescent protein (GFP)-trapped fly lines have become available over the years (Morin et al. 2001; Clyne et al. 2003; Buszczak et al. 2007; Quiñones-Coello et al. 2007; Rees et al. 2011; http://flytrap.med.yale.edu; http://www.flyprot.org).
GFP traps have mainly been used to study the endogenous expression patterns of trapped genes or the subcellular localization of their protein products. Here, we show that the GFP tag can also be used to interfere with gene function by RNAi-mediated knockdown of the tagged transcripts. This method, which we refer to as “tag-mediated loss-of-function,” addresses major shortcomings of the classical RNAi approach in which gene-specific sequences are targeted. Furthermore, we show that the GFP tag can be used to efficiently purify endogenous protein complexes for mass spectrometric analysis using recombinant nanobodies against GFP. Finally, we screen for mCherry traps in Drosophila cultured cells and describe several lines with mCherry expression in specific subcellular patterns for use in high-throughput screening.
Materials and Methods
Fly strains
The following protein traps were described in (Buszczak et al. 2007): dlg1–GFP (CC01936), Spt6–GFP (CA07692), Cp1–GFP (CC01377), Pabp2–GFP (CC00380). Additional protein traps used in this study are listed in Supporting Information, Figure S1. nanos-GAL4 was described by Van Doren et al. (1998), and matα4-tub-GAL4 (also known as mat67) is a gift from D. St Johnston. EGFP-shRNA constructs 1, 2 and 3 were generated by annealing the oligos listed in Table S2 and cloning them into the pVALIUM20 or pVALIUM22 vectors (Ni et al. 2011) using NheI and EcoRI. Transgenic flies were established using the attP40 and attP2 landing sites (Groth et al. 2004; Markstein et al. 2008). The Spt6-specific shRNA (HMS00364) was obtained from the TRiP at Harvard Medical School.
Fluorescently tagged baits were immunoprecipitated from embryos expressing a GFP-tagged par-6 rescue construct in a par-6 null mutant background (referred to as par-6GFP; Wirtz-Peitz et al. 2008) or from sgg–YFP (CPTI-002603) obtained from the Drosophila Genetic Resource Center, and w− embyros were used as controls.
All tag-mediated loss-of-function experiments in the germline were controlled by driving EGFP-shRNAs in the background of the heterozygous GFP trap. The observed phenotypes were invariably wild type, indicating that the targeted genes are neither haplo-insufficient nor subject to transitive RNAi effects; i.e., targeting the GFP exon does not give rise to secondary siRNAs directed against other regions of the transcript (Roignant 2003). Embryos analyzed in Figure 2B were from the following cross: par-6GFP; mat67/UAS–EGFP-shRNA × par-6GFP/Y; UAS–EGFP-shRNA. The par-6GFP; mat67 stock served as the control in Figure 2A.
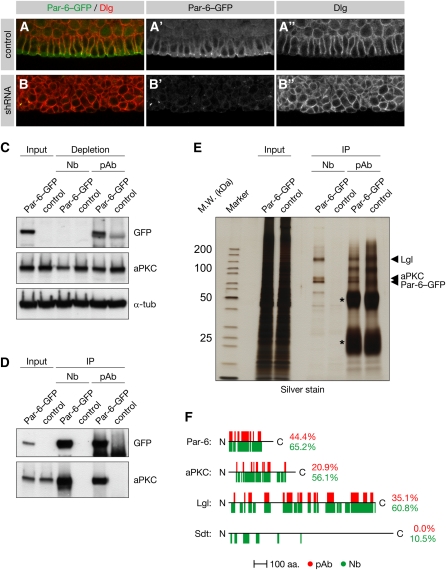
Figure 2 .
Using the GFP tag to probe gene function and protein–protein interactions in the embryo. (A and B) Postgastrulation embryos expressing a GFP-tagged par-6 rescue construct in a par-6 null mutant background were stained for Discs-large (Dlg) and GFP. The embryos were derived from female germlines expressing an EGFP-shRNA (B) or from control germlines (A). (C–F) Par-6–GFP was immunoprecipitated from embryonic extract using either anti-GFP nanobodies (Nb) or anti-GFP polyclonal antibodies (pAb). w− embryos not expressing Par-6–GFP were used as a control. (C) Western blot analysis of the extract before and after immunoprecipitation of Par-6–GFP (Input and Depletion, respectively). (D) Western blot analysis of the immunoprecipitates. (E) Silver stain of the immunoprecipitates. Asterisks indicate IgG heavy and light chains. (F) Peptide coverage maps of the Par-6 bait and select co-immunoprecipitated proteins. The peptides were obtained by LC-MS/MS after in-solution digestion of the immunoprecipitates prepared using either nanobodies (green) or polyclonal control antibodies (red). Percentages indicate the overall peptide coverages of the proteins. Note that the polyclonal antibodies used in this experiment were coupled to beads and different from those used in C–E (see Materials and Methods).
Immunofluorescence
The following antibodies were used: rabbit anti-Vasa (1:500, Santa Cruz, sc-30210), rabbit anti-GM130 (Abcam, ab30637), rabbit anti-Anillin (1:1300, a gift of Tim Mitchison), rat anti-troponin H (1:500), mouse anti-1B1 (1:2, Developmental Studies Hybridoma Bank), rabbit anti-cleaved caspase (1:100, Cell Signaling), rabbit anti-GFP (1:500, Abcam, ab6556), mouse anti-Dlg (1:100, Developmental Studies Hybridoma Bank, 4F3). Immunohistochemistry in ovaries was performed as previously described (Neumüller et al. 2008). Embryos were fixed in 4% (v/v) formaldehyde in PBS/heptane, devitellinized using heptane/methanol, and blocked in 2% (v/v) NGS in PBS, 0.1% (v/v) Triton X-100. Images were acquired on either a Leica TCS SP2 or a Zeiss LSM 710 confocal microscope.
Immunoprecipitation from embryos and mass spectrometry
Overnight collections were extracted with lysis buffer (25 mM Tris–HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% [v/v] NP-40, 5% [v/v] glycerol, 1 mM DTT, 1× Halt protease and phosphatase inhibitor cocktail [Thermo Scientific]) and debris was removed by centrifuging twice at 1200 × g for 5 min. Extracts were cleared by incubation with agarose resin (Thermo Scientific) for 1 hr at 4°, followed by centrifugation at 15,000 × g for 15 min. Immunocomplexes were formed by incubation for 2 hr at 4° with the following antibodies: anti-GFP nanobodies coupled to agarose beads (10 μl of packed beads per IP; ChromoTek, GFP-Trap_A), rabbit anti-GFP antibodies (5 μl per IP; used in Figure 2, C–E; Invitrogen, A6455) precipitated using Protein A/G agarose (Thermo Scientific), rabbit anti-GFP antibodies (1 μl per IP; used in Figure S2, A and B; Abcam, ab6556), rabbit anti-GFP antibodies coupled to sepharose beads (10 μl of packed beads per IP; used in Figure 2F, Figure S2, C and D; Abcam, ab69314). The immunocomplexes were washed four times with lysis buffer, eluted in IgG Elution Buffer (Thermo Scientific), and neutralized using 100 mM Tris–HCl (pH 9). The eluates were Western blotted using standard protocols or stained using the PageSilver silver staining kit (Thermo Scientific). The following antibodies were used in Western blotting: rabbit anti-GFP (1:500, Abcam, ab6556), rabbit anti-PKCζ (1:500, Santa Cruz, sc-216), mouse anti-α-tubulin (1:1000, Sigma-Aldrich, T6199).
For mass spectrometry the immunocomplexes were washed three times with 10 mM Tris–HCl (pH 7.5) prior to elution to remove detergent. The neutralized eluate was reduced with 5 mM DTT for 30 min at 56° and alkylated with 15 mM iodacetamide for 30 min at RT. The alkylation reaction was quenched with 15 mM DTT. The alkylated eluate was digested with 1 μg of sequencing grade modified Trypsin (Promega) in 2 mM CaCl2 overnight at 37°. The digest was purified on PepClean C-18 spin columns (Thermo Scientific) according to the manufacturer’s instructions, dried in a SpeedVac, and reconstituted in 10 μl of 2% (v/v) acetonitrile, 0.1% formic acid, of which 4 μl was injected into the mass spectrometer. LC-MS/MS analysis was performed essentially as described (Dephoure and Gygi 2011). Peptide spectral matches were filtered to a 2% false discovery rate (FDR).
Immunoprecipitation from cell lines and mass spectrometry
Cells were extracted by incubation in lysis buffer (10 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.5% [v/v] NP-40, 1 mM PMSF, 1× complete protease inhibitor cocktail [Roche]) for 30 min on ice. The extract was cleared by centrifugation at 20,000 × g for 10 min at 4°. mCherry was immunoprecipitated using anti-DsRed nanobodies coupled to agarose beads (ChromoTek, RFP-Trap_A) according to the manufacturer’s instructions. The immunoprecipitate was prepared for mass spectrometry and analyzed by LC-MS/MS essentially as described (Sowa et al. 2009; Dephoure and Gygi 2011). Peptide spectral matches were filtered to a 1% FDR.
Generation of mCherry cell lines
mCherry trap cassettes were amplified from pBS-KS-attB1-2-PT-SA-SD-0-mCherry, pBS-KS-attB1-2-PT-SA-SD-1-mCherry, pBS-KS-attB1-2-PT-SA-SD-2-mCherry (Venken et al. 2011) using the primers SA-M-SD-General-NotI-F (AAGGAAAAAAGCGGCCGCAGTCGATCCAACATGGCGAC) and SA-M-SD-General-EcoRI-R (CCGGAATTCAGAAGTTCAAATGGGCTTTC) and subcloned into pMiLR-attP1-2 (Venken et al. 2011), a mini-Minos transposon containing both Minos inverted repeats and two inverted attP sites for ΦC31 recombinase-mediated cassette exchange. The resulting plasmid, pGTC, had the following structure: MiL–attP1–SA–mCherry–SD–attP2–MiR.
Drosophila S2R+ cells were transfected with the pCoB plasmid and blasticidin-resistant cells were selected with 25 μg/ml of blasticidin 1 day post-transfection. Resistant cells were cotransfected with the mCherry trap vector described above (pGTC) and a Minos transposase helper plasmid (Pavlopoulos et al. 2004) in a 1:1 ratio. Three independently transfected cultures, one for each of the three reading frame constructs, were pooled 2 days post-transfection and cultured for 1 week to allow for maximal expression of the mCherry tag. Approximately 10% of the transfected cells scored positive by FACS and more than 2000 cells, selected from the top 3–5 percentile, were seeded as single cell in 384-well plates. Blasticidin-sensitive feeder cells, 5 × 106, had been added to the plates to promote the survival of these single cells. The cell clones were cultured for 4 weeks in the presence of 12.5 μg/ml of blasticidin to suppress proliferation of the feeder cells. Visible colonies were then isolated, progressively expanded to larger culture sizes, and cryopreserved. See Table S1 for a list of cell lines generated in this study. The Minos insertions were mapped by inverse PCR using 0.5 μg of genomic DNA, as described (Bellen et al. 2011).
Results
Efficient knockdown of protein traps using shRNAs directed against GFP
We sought to establish reagents for knocking down GFP-trapped genes using transgenic RNAi (Figure 1A). For this, we opted for small hairpin RNAs (shRNAs) over long double-stranded RNAs (dsRNAs) because of the consistently high efficacy reported for shRNAs in both soma and germline (Haley et al. 2008; Ni et al. 2011). In addition, despite the small size of the GFP coding sequence, shRNAs allowed us to design multiple nonoverlapping constructs. This provides a choice of constructs if any are found to induce off-target effects in a particular tissue of interest. Consequently, we designed three nonoverlapping shRNAs against the EGFP tag used in the three original Drosophila protein trap screens (Morin et al. 2001; Buszczak et al. 2007; Quiñones-Coello et al. 2007). These shRNAs are predicted to also target the EYFP derivative used in a more recent protein trap screen (Rees et al. 2011). We cloned these shRNAs into UAS vectors optimized for either somatic or germline expression (see Materials and Methods).
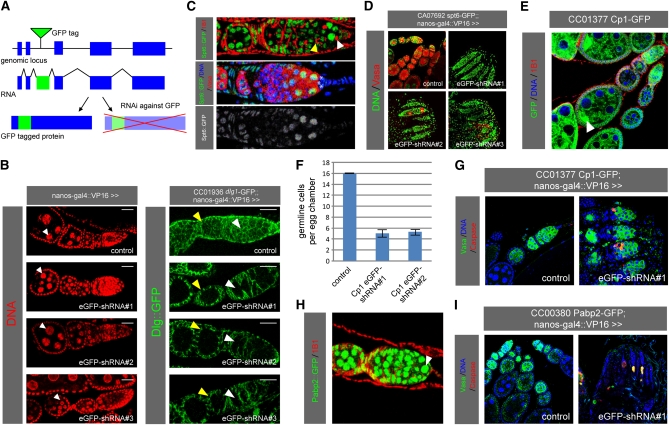
Figure 1 .
The GFP tag as a tool for stringent loss-of-function studies in vivo. (A) GFP-mediated loss-of-function strategy. (B) Individual EGFP-shRNAs were driven by nanos-GAL4 in a wild-type background (left) or in the background of the Dlg–GFP trap (right), and ovarioles were stained with DAPI. Dlg–GFP is specifically depleted in the germline (right). The arrowheads point to polyploid nurse cells (left); the yellow arrowheads point to somatic cells, and the white arrowheads point to the germline (right). The scale bar is 20 µm. (C) Germaria from the Spt6–GFP trap were stained with 1B1 or for Vasa and DAPI. The white arrowhead points to a GSC, and the yellow arrowhead points to a cystocyte (see text for details). Asterisks indicate cap cells. (D) Individual EGFP-shRNAs were driven in the background of the Spt6–GFP trap, and ovaries were stained for Vasa and DAPI. Vasa-positive germline cells are lost upon knockdown of Spt6–GFP. (E) Ovarioles from the Cp1–GFP trap were stained with 1B1 and DAPI. (F and G) Individual EGFP-shRNAs were driven in the background of the Cp1–GFP trap. (F) Quantification of germline cells per egg chamber. SEM is shown. (G) Ovarioles were stained for Vasa, cleaved caspase, and DAPI. Germline cells are lost upon knockdown of Cp1–GFP. (H) A germarium from the Pabp2–GFP trap stained with 1B1. Pabp2–GFP is ubiquitously expressed and localizes to the nucleus (arrowhead). (I) An EGFP-shRNA was driven by nanos-GAL4 in the background of the Pabp2–GFP trap, and ovaries were stained for Vasa, DAPI, and cleaved caspase. Most germline cells are lost upon knockdown of Pabp2–GFP, while the remaining cells stain positive for cleaved caspase.
To validate the efficacy of these constructs, we used nanos-GAL4 to drive the shRNAs in the background of a GFP trap inserted in the discs large 1 (dlg) gene (Figure 1B). Germline-specific expression of the EGFP-shRNAs in females resulted in the loss of Dlg–GFP signal in the germline, whereas the cortical signal remained unaffected in the somatic follicle cells. To extend these observations, we tested a total of 12 additional GFP traps and consistently observed germline-specific loss of GFP signal upon EGFP-shRNA expression using nanos-GAL4 (Figure S1A; data not shown). Importantly, expression of these EGFP-shRNAs in the germline using either nanos-GAL4 or matα4-tub-GAL4 had no adverse effect on germline development as judged from normal differentiation of germaria and egg chambers as well as normal fertility (Figure 1B; data not shown). We conclude that our EGFP-shRNA lines are effective and specific in knocking down GFP-trapped genes in the female germline.
Tag-mediated loss-of-function reveals a role for Spt6 in germline stem cell maintenance
To showcase the utility of these shRNAs in genetic screening, we used them to knock down homozygous viable GFP traps available from the Carnegie collection (Buszczak et al. 2007) that showed strong GFP signal in female germline stem cells (GSCs) (Figure S1A; data not shown). Among these, CA07692 (an insertion in Spt6) was expressed at variable levels throughout oogenesis (Figure 1C). The Spt6–GFP signal overlapped with the nuclear DAPI signal, consistent with a recent study implicating Spt6 in transcriptional elongation in Drosophila (Ardehali et al. 2009). The 1B1 antibody, which labels spectrosomes and fusomes, and the Vasa antibody, which labels all germline cells, allowed us to correlate Spt6–GFP expression with specific cell types in the germarium. We found that Spt6–GFP was strongly expressed in GSCs adjacent to the stem cell niche (identified as Vasa-positive cells containing a single spectrosome), while lower expression levels were observed in the transit-amplifying cystocytes (distinguished by the presence of a fusome) (Figure 1C).
To assess whether Spt6 is required in GSCs, we used nanos-GAL4 to drive individual EGFP-shRNAs in the background of the Spt6–GFP trap. In contrast to wild-type ovaries, where Vasa-positive germline cells are detected in egg chambers of all stages, tag-mediated knockdown of Spt6–GFP using three nonoverlapping EGFP-shRNAs resulted in the depletion of Vasa-positive cells from the germline with only somatic cells remaining in the ovaries (Figure 1D). This, together with the finding that knockdown of Spt6 did not trigger apoptosis in the germline (data not shown), suggests that Spt6 is required for GSC maintenance, which is consistent with recent findings that transcriptional elongation is an important regulatory event in stem cell homeostasis (Bai et al. 2010; Neumüller et al. 2011). We corroborated these data using a gene-specific shRNA against Spt6, which caused a similar phenotype when expressed in the germline (Figure S1C).
Tag-mediated loss-of-function identifies genes required for germ cell survival
Another trap identified in our tag-mediated loss-of-function screen was CC01377, an insertion in Cysteine proteinase-1 (Cp1), which is expressed at all stages of oogenesis and accumulates predominantly at the fusome in germaria and in the oocytes of developing egg chambers (Figure 1E). Knockdown of Cp1 using two independent EGFP-shRNAs resulted in egg chambers with fewer Vasa-positive germline cells compared to wild type (Figure 1, F and G; Figure S1D). To identify the underlying cause, we stained for caspase cleavage as a marker for apoptosis. Whereas cleaved caspase-positive cells were rarely observed in the wild-type germline, we detected a high frequency of apoptotic germline cells upon Cp1 knockdown (Figure 1G). The highest incidence of caspase cleavage was observed in cystocytes (region 2 of the germarium), but apoptotic cells were also detected at later stages of oogenesis. Interestingly, apoptotic cells were not detected in region 1 of the germarium, consistent with our observation that ovaries still contained germline cells in 4-day-old flies.
A third trap identified in our screen is CC00380, an insertion in polyA-binding protein II (Pabp2). Tag-mediated knockdown of Pabp2 using two independent EGFP-shRNAs resulted in almost complete loss of germline cells (Figure 1I and Figure S1D). The few remaining germline cells showed signs of nuclear fragmentation (data not shown) and stained positive for cleaved caspase (Figure 1I), suggesting that Pabp2 is required for cell survival across the germline. Consistent with this, Pabp2–GFP was uniformly expressed in the germline (Figure 1H). Our findings are also consistent with a study reporting that flies mutant for a hypomorphic allele of Pabp2 are sterile and contain degenerating egg chambers arresting at stage 8 of oogenesis (Benoit et al. 2005). The stronger phenotype observed upon tag-mediated knockdown most likely reflects a null or strongly hypomorphic phenotype. Taken together, our tag-mediated knockdown screen revealed a requirement for both Cp1 and Pabp2 in cell survival in the germline, with Cp1 fulfilling this function mostly at the cystocyte stage of oogenesis.
Tag-mediated loss-of-function in the embryo
Our functional analysis of Spt6, Cp1, and Pabp2 in the female germline indicates that tag-mediated knockdown is a powerful strategy by which to study phenotypes with a high degree of stringency. Besides inducing phenotypes in the germline, shRNAs have been used to deplete the maternal contribution of proteins required in embryogenesis (Ni et al. 2011). To test if our EGFP-shRNAs are effective in inducing embryonic phenotypes we turned to par-6, a gene required for the establishment of epithelial polarity. While embryos derived from par-6 homozygous mutant germline clones (par-6GLC) show an embryonic lethal defect in cell polarity, the zygotic mutant mostly survives to the larval stage owing to the maternal contribution of Par-6 protein (Petronczki and Knoblich 2001).
We used matα4-tub-GAL4 to drive one of our EGFP-shRNAs in par-6 mutant females expressing a GFP-tagged genomic rescue construct (par-6GFP) (Wirtz-Peitz et al. 2008) and analyzed the resulting embryos using antibodies against Dlg and GFP. The ectodermal epithelium of control embryos was organized as a monolayer, and its constituent cells showed a columnar morphology (Figure 2A). In contrast, epithelial cells in RNAi embryos had rounded up and formed an irregular, multilayered, epithelium (Figure 2B), as has been reported for par-6GLC embryos (Petronczki and Knoblich 2001). Furthermore, in control embryos Dlg was confined to the basolateral cell cortex, whereas Par-6–GFP was localized to the subapical region (Figure 2A). In RNAi embryos lacking Par-6–GFP, Dlg was localized all around the cell cortex except in the outermost cells (Figure 2B). Therefore, the EGFP-shRNA directed against par-6–GFP closely phenocopies the epithelial defects in par-6GLC embryos.
Using fluorescently tagged genes to isolate endogenous protein complexes
Par-6 assembles a well-defined complex with two other proteins, atypical protein kinase C (aPKC) and Lethal (2) giant larvae (Lgl) (Betschinger et al. 2003). This led us to explore if GFP-tagged fly lines could be used to analyze endogenous protein complexes when combined with recently commercialized high-affinity GFP antibodies raised in llamas (Rothbauer et al. 2006, 2008). These so-called nanobodies lack light chains, which allows the heavy chain’s variable domain to be cloned and purified to high titer from a recombinant source. Thus, these antibodies promise to combine the high antigen-binding capacity of polyclonal sera with the specificity of monoclonal antibodies.
To test this we used anti-GFP nanobodies to immunoprecipitate Par-6 from par-6GFP embryos. Western blotting showed that Par-6–GFP was immunoprecipitated and its binding partner aPKC was co-immunoprecipitated from these embryos by both nanobodies and a polyclonal control serum (Figure 2D). However, in contrast to the control antibodies, the nanobodies completely depleted Par-6 from the extract after as little as 60 min of incubation and even partially depleted its binding partner aPKC (Figure 2C and Figure S2, A and B). Although Par-6–GFP is only weakly expressed as inferred from its comparatively faint fluorescence (data not shown), the Par complex containing Par-6, Lgl, and aPKC was clearly detected on a silver-stained gel in immunoprecipitates from ∼200 μl of packed embryos (Figure 2E). In addition, these proteins were readily identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after in-solution digestion of the immunoprecipitate (Figure 2F and File S1). These proteins were also identified when Par-6–GFP was immunoprecipitated using agarose-coupled polyclonal control antibodies, albeit with significantly lower peptide coverages. In fact, Stardust, which so far could be shown to bind Par-6 only when both proteins were overexpressed in cultured cells (Hurd et al. 2003), was specifically identified in the nanobody immunoprecipitate but not in the polyclonal control immunoprecipitate (Figure 2F and File S1).
To assess the performance of anti-GFP nanobodies with a different bait we turned to a YFP trap in shaggy (Rees et al. 2011), the Drosophila ortholog for glycogen synthase kinase 3 and a core subunit of the Armadillo destruction complex (Macdonald et al. 2009). Silver staining of the nanobody immunoprecipitate revealed a double band (Figure S2C), which in-solution digestion and LC-MS/MS identified as Shaggy and its well-characterized interactor Axin (Figure S2D). Although both proteins were also identified using polyclonal control antibodies, the nanobodies recovered a higher amount of bait protein with less background, resulting in superior peptide coverage. Together, these experiments indicate that GFP traps or GFP-tagged genomic rescue constructs can be used in conjunction with anti-GFP nanobodies to interrogate the Drosophila interactome with high sensitivity and stringency.
Extending the protein trap approach to Drosophila cell lines
Although in vivo approaches are strongly preferred for most Drosophila studies, cell lines remain a powerful system for high-throughput RNAi screening. To generate cell lines with fluorescent proteins that label distinct subcellular features for use in such applications, we developed a protein trap approach for Drosophila S2 cells using the MiMIC transposon. MiMIC is a Minos-based protein trap featuring a mutagenic marker cassette containing a splice acceptor site followed by a triple stop (Venken et al. 2011). The cassette is flanked by two inverted ΦC31 integrase attP sites that allow its replacement with a sequence of interest by recombinase-mediated cassette exchange (RMCE). As depicted in Figure 3A, we replaced the mutagenic marker cassette with a fluorescent protein trap consisting of the mCherry coding sequence in one of three frames flanked by splice acceptor and donor sites.
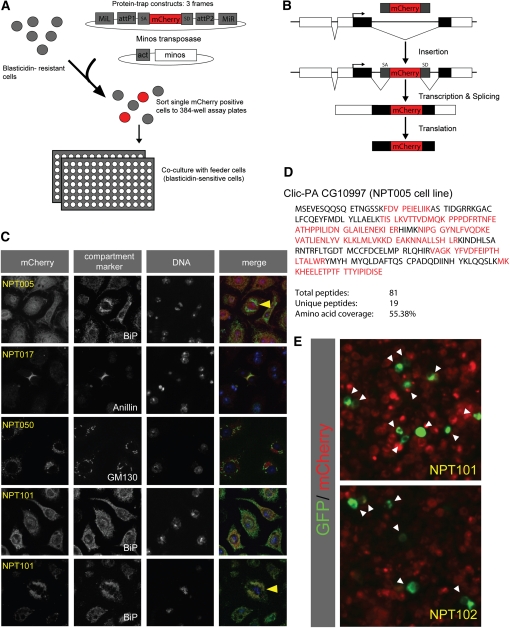
Figure 3 .
GFP trapping in Drosophila cell lines. (A) The strategy used to establish GFP-trapped cell lines. The Minos transposon harboring the mCherry trap cassette and a Minos transposase helper plasmid were cotransfected into blasticidin-resistant Drosophila S2R+ cells. Single mCherry-positive cells were FACS-sorted and cocultured with blasticidin-sensitive feeder cells in 384-well plates. (B) Schematic for the expression of an mCherry fusion protein from a trapped gene. Splice acceptor (SA) and splice donor (SD) sites mediate the integration of the mCherry coding sequence into the endogenous transcript. (C) Drosophila S2R+ cells expressing mCherry traps in specific subcellular patterns. Cells were stained for the indicated markers and DAPI. Anillin labels the cleavage furrow; BiP (Drosophila Hsc70-3) labels the endoplasmic reticulum; GM130 labels the Golgi. NPT005 (Clic)–RFP colocalized with BiP at most stages of the cell cycle, but least in mitotisis. Colocalization of NPT101–mCherry was independent of the cell cycle. Arrowheads point to mitotic cells. (D) NPT005–RFP was immunoprecipitated using anti-DsRed nanobodies and analyzed by LC-MS/MS after in-solution digestion of the immunoprecipitate. The amino acid sequence for the top-ranked protein hit (Clic) is shown, and the identified peptides are labeled red. (E) Examples of ΦC31-mediated cassette exchange in the S2R+ trap lines NPT101 and NPT102 using an attB–EGFP–attB cassette (arrowheads point to GFP-positive cells).
This modified MiMIC element was mobilized into random positions in S2R+ cells using a transiently transfected Minos transposase (Metaxakis et al. 2005). As part of a pilot screen we FACS-sorted more than 2000 mCherry-positive cells and established approximately 50 stable clones from single cells (Figure 3A and Table S1). In a number of these lines mCherry assumed a distinct subcellular distribution, indicative of a successful trapping event (Figure 3, B and C, and Table S1). For example, cell line NPT017 showed a cell-cycle-dependent localization of mCherry, being distributed diffusely in the cytoplasm at interphase and relocalizing to the cleavage furrow at cytokinesis. In cell line NPT005, by contrast, mCherry was localized to cytoplasmic puncta, which partially overlapped with BiP, a marker for the endoplasmic reticulum (Otero et al. 2010; Figure 3C).
To identify the gene trapped in NPT005 we purified the mCherry-tagged protein using anti-DsRed nanobodies followed by LC-MS/MS of the in-solution digested immunoprecipitate. This identified chloride intracellular channel (Clic) as the highest-ranked protein hit (Figure 3D), and inverse PCR-based mapping of the MiMIC insertion site confirmed Clic as the trapped gene (data not shown). Using the same strategy we identified atx2 and gap1 as the genes trapped in NPT022 and NPT102, respectively (Table S1; data not shown).
Finally, as proof of principle for RMCE in these cell lines we successfully swapped mCherry for EGFP in NPT101 and NPT102 by cotransfection of an EGFP donor cassette along with ΦC31 integrase (Figure 3E). Taken together, these data demonstrate that the MiMIC transposon can be used to trap specific genes in Drosophila cell lines and that stable clones of such traps may be established for use in diverse applications ranging from imaging to biochemical studies.
Discussion
Advantages of tag-specific over gene-specific knockdown
Here, we have described a set of validated shRNA constructs for tag-mediated loss-of-function in GFP trap lines. This method addresses two major challenges in screens based on gene-specific RNAi constructs. First, gene-specific constructs may cause off-target effects, which manifest as false positives in RNAi screens (Ma et al. 2006; Kulkarni et al. 2006; Dietzl et al. 2007). Off-target effects can be controlled by rescue of the RNAi phenotype using an RNAi-resistant construct, but such transgenes are not readily available and need to be generated on a gene-by-gene basis (Mohr et al. 2010). By contrast, the shRNA used for tag-mediated knockdown can be expressed in the target tissue in the absence of a GFP trap to exclude the possibility of unspecific phenotypes.
Second, the efficacy of gene-specific RNAi constructs varies widely, with ineffective constructs giving rise to false negatives in RNAi screens (Dietzl et al. 2007; Booker et al. 2011). While the false-negative rate may be lowered by targeting each gene with two or more independent RNAi constructs (Mohr et al. 2010), the use of a single optimized shRNA in tag-mediated knockdown ensures consistently high efficacy. In addition, the degree of knockdown is readily verified by monitoring GFP fluorescence in the target tissue. GFP-mediated knockdown is therefore a compelling method by which to remove artifacts and ambiguity from RNAi experiments.
Strategies for isolating endogenous protein complexes
Analogous to gene-specific RNAi reagents in functional studies, biochemical studies of protein–protein interactions typically rely on protein-specific antibody reagents. Because these are time consuming and expensive to raise and recognize their protein antigen with widely variable specificity and affinity, an alternative approach is to ectopically express epitope-tagged baits, which may then be immunoprecipitated using monoclonal antibodies. However, if such constructs are not expressed at endogenous levels or in the proper spatial patterns, then this is predicted to increase the occurrence of false-positive interactors, currently a major challenge of such studies (Gingras et al. 2007). By contrast, the use of GFP traps permits the purification of endogenously expressed protein complexes using a single validated antibody reagent, thereby combining the advantages of both methods.
In addition, our data suggest that the favorable binding properties of GFP-specific nanobodies offset the low endogenous expression levels of many proteins and permit the mass spectrometric detection of even weak interactions. Interestingly, neither the interaction between Par-6 and Stardust nor the interaction between Shaggy and Axin were reported in a recent S2 cell-based co-immunoprecipitation study (Guruharsha et al. 2011), suggesting that many critical interactions are detected only by in vivo assays. Our data, together with another recent study on protein complexes purified from protein traps (Rees et al. 2011), provide an experimental framework for analyzing physical interactions between endogenous proteins in Drosophila.
Perspectives for fluorescent protein trapping in Drosophila
Although it is estimated that approximately 3200 Drosophila genes are permissive to protein trapping by conventional P-element and piggyBac transposons, only a few hundred Drosophila genes have so far been tagged with a transgenic GFP trap (Aleksic et al. 2009). Among these, three classes of traps need to be distinguished. First, insertions that disrupt protein function and are thus homozygous lethal. These traps are not amenable to tag-mediated knockdown, owing to the absence of transitive RNAi in Drosophila (see Materials and Methods), and they are similarly unsuitable for biochemical analysis because a tag that disrupts a protein’s activity may also interfere with its physical interactions. Second, insertions spliced into only some of a gene’s transcripts (Quiñones-Coello et al. 2007). Such traps have been used to analyze the distribution of cell-type-specific isoforms (Silies and Klämbt 2010), and we were able to induce germline phenotypes by tag-mediated knockdown of isoform-specific traps. At the same time, we have also encountered a number of lines in which the expression of an unaffected splice form masked the tag-mediated knockdown of the trapped transcripts (data not shown). Finally, the third class is composed of homozygous viable insertions that trap all of a gene’s transcripts and which are generally the most useful.
While the genome coverage of usable protein traps remains limited, a large-scale effort is currently ongoing to generate over 6000 insertions using the MiMIC transposon (Venken et al. 2011). Although these traps will need to be individually converted to GFP traps by RMCE, we anticipate that the methods described by us, as well as the traditional utility of the GFP tag in localization studies, will drive the conversion of a large number of these lines. At the same time, improved methods for generation of GFP knockins (Huang et al. 2009) and the generation of GFP-tagged genomic rescue constructs by individual investigators or as part of high-throughput projects (Venken et al. 2006; Ejsmont et al. 2009) are expected to increase genome coverage. Such targeted methods will also allow tagging of genes refractory to protein trapping (e.g., intronless genes). Finally, we consider the methods described here applicable to other model systems, for example, to haploid cell lines, which appear uniquely suited to gene trapping and tag-mediated knockdown (Debec 1978; Carette et al. 2009).
Note that while our manuscript was under review, Pastor-Pareja and Xu (2011) independently described loss-of-function phenotypes by RNAi-mediated knockdown of GFP in protein trap lines. While our approach is conceptually similar to theirs, the use of shRNA constructs rather than a long dsRNA construct lends added flexibility and stringency to this method. First, shRNAs permit the induction of phenotypes in the female germline and in the embryo, where dsRNAs have proven ineffective (Ni et al. 2011). Second, the availability of multiple nonoverlapping constructs allows phenotypes to be validated with an independent shRNA.
Supplementary Material
Acknowledgments
We thank Allan Spradling, Tim Mitchison, Frank Schnorrer, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies, Quentin Gilly, Christians Villata, and Rich Binari for technical assistance, Noah Dephoure, Robert Everley, and Steven Gygi for technical help with mass spectrometry, and Martha Evans-Holm and Joseph W. Carlson for assistance with mapping of transposon insertions. This work was supported by an European Molecular Biology Organization (EMBO) Long-Term Fellowship to R.A.N. and Human Frontier Science Program (HFSP) Long-Term Fellowships to R.A.N. and F.W.P. Y.K. is supported by the Damon Runyon Cancer Research Foundation. This work was supported in part by R01-GM067761 and R01-GM084947 to N.P. S.E.M. is supported by GM067761 with additional support from the Dana-Farber/Harvard Cancer Center. N.P. and H.B. are investigators of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: T. Schupbach
Literature Cited
- Aleksic J., Lazic R., Müller I., Russell S. R., Adryan B., 2009. Biases in Drosophila melanogaster protein trap screens. BMC Genomics 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali M. B., Yao J., Adelman K., Fuda N. J., Petesch S. J., et al. , 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., et al. , 2010. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B., Mitou G., Chartier A., Temme C., Zaessinger S., et al. , 2005. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev. Cell 9: 511–522 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A., 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422: 326–330 [DOI] [PubMed] [Google Scholar]
- Booker M., Samsonova A. A., Kwon Y., Flockhart I., Mohr S. E., et al. , 2011. False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genomics 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., et al. , 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., et al. , 2009. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326: 1231–1235 [DOI] [PubMed] [Google Scholar]
- Clark K. J., Balciunas D., Pogoda H.-M., Ding Y., Westcot S. E., et al. , 2011. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Methods 8: 506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P. J., Brotman J. S., Sweeney S. T., Davis G., 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics 165: 1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec A., 1978. Haploid cell cultures of Drosophila melanogaster. Nature 274: 255–256 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Gygi S. P., 2011. A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods 54: 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Ejsmont R. K., Sarov M., Winkler S., Lipinski K. A., Tomancak P., 2009. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6: 435–437 [DOI] [PubMed] [Google Scholar]
- Gingras A.-C., Gstaiger M., Raught B., Aebersold R., 2007. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8: 645–654 [DOI] [PubMed] [Google Scholar]
- Gossler A., Joyner A. L., Rossant J., Skarnes W. C., 1989. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science 244: 463–465 [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J.-F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B., Hendrix D., Trang V., Levine M., 2008. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 321: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A. M., Hong Y., 2009. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA 106: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T. W., Gao L., Roh M. H., Macara I. G., Margolis B., 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5: 137–142 [DOI] [PubMed] [Google Scholar]
- Kulkarni M. M., Booker M., Silver S. J., Friedman A., Hong P., et al. , 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 3: 833–838 [DOI] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L., Beachy P. A., 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363 [DOI] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X., 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A., Savakis C., 2005. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S., Bakal C., Perrimon N., 2010. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 79: 37–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W., 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Betschinger J., Fischer A., Bushati N., Poernbacher I., et al. , 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., et al. , 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8: 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero J. H., Lizák B., Hendershot L. M., 2010. Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Xu T., 2011. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted collagen IV and perlecan. Dev. Cell 21: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A., Berghammer A. J., Averof M., Klingler M., 2004. Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: quantitative and qualitative analysis of genomic integration events. Genetics 167: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J. A., 2001. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3: 43–49 [DOI] [PubMed] [Google Scholar]
- Quiñones-Coello A. T., Petrella L. N., Ayers K., Melillo A., Mazzalupo S., et al. , 2007. Exploring strategies for protein trapping in Drosophila. Genetics 175: 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees J. S., N. Lowe, I. M. Armean, J. Roote, G. Johnson, et al., 2011 In vivo analysis of proteomes and interactomes using parallel affinity capture (iPAC) coupled to mass spectrometry. Mol. Cell. Proteom.10: M110.002386. [DOI] [PMC free article] [PubMed]
- Roignant J.-Y., 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., et al. , 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3: 887–889 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., et al. , 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteom. 7: 282–289 [DOI] [PubMed] [Google Scholar]
- Silies M., Klämbt C., 2010. APC/C(Fzr/Cdh1)-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nat. Neurosci. 13: 1357–1364 [DOI] [PubMed] [Google Scholar]
- Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., A. L. Williamson, and R. Lehmann, 1998 Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–6. [DOI] [PubMed]
- Venken K. J. T., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J. T., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8(9): 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J. A., 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.