Abstract
Noisy bistable dynamics in gene regulation can underlie stochastic switching and is demonstrated to be beneficial under fluctuating environments. It is not known, however, if fluctuating selection alone can result in bistable dynamics. Using a stochastic model of simple feedback networks, we apply fluctuating selection on gene expression and run in silico evolutionary simulations. We find that independent of the specific nature of the environment–fitness relationship, the main outcome of fluctuating selection is the evolution of increased evolvability in the network; system parameters evolve toward a nonlinear regime where phenotypic diversity is increased and small changes in genotype cause large changes in expression level. In the presence of noise, the evolution of increased nonlinearity results in the emergence and maintenance of bistability. Our results provide the first direct evidence that bistability and stochastic switching in a gene regulatory network can emerge as a mechanism to cope with fluctuating environments. They strongly suggest that such emergence occurs as a byproduct of evolution of evolvability and exploitation of noise by evolution.
Keywords: environmental fluctuations, gene regulatory network, noise, phenotypic variability, stochastic switching
Introduction
Organisms are able to display diverse phenotypes under a given environment. This ability is first described in higher organisms as a bet-hedging strategy (Cohen, 1966; Philippi, 1993), but is increasingly found to be common at the cellular level, where it is underlined by stochastic switching among different phenotypic states (Rao et al, 2002; Raser and O’Shea, 2005; Samoilov et al, 2006; Losick and Desplan, 2008; López-Maury et al, 2008; Raj and van Oudenaarden, 2008). In microbes, stochastic switching is demonstrated in several phenotypic traits including persistence to antibiotics (Balaban et al, 2004) and lactose metabolism (Novick and Weiner, 1957; Ozbudak et al, 2004; Robert et al, 2010) in Escherichia coli, sporulation (Veening et al, 2008) and DNA uptake competence (Maamar et al, 2007) in Bacillus subtilis, and galactose metabolism in yeast (Acar et al, 2005). By allowing a certain fraction of the population to display a distinct phenotype, stochastic switching can allow populations to survive environmental changes (Balaban et al, 2004; Visco et al, 2010) and adapt faster to a new environment (Blake et al, 2006; Kashiwagi et al, 2006; Acar et al, 2008). On the other hand, the heterogeneity in the population can incur a fitness cost as some fraction of its members would always be maladaptive in a given environment. Theoretical studies have shown that such potential fitness costs can be balanced under environmental fluctuations, resulting in a net fitness gain from stochastic switching under a range of fluctuation rates and fitness costs (Thattai and van Oudenaarden, 2004; Salathé et al, 2009; Gaál et al, 2010; Liberman et al, 2011). Further, switching rates can evolve to be in tune with environmental fluctuation rates so as to optimize the associated fitness tradeoffs (Kussell and Leibler, 2005; Kussell et al, 2005). In line with these theoretical findings, experimental evolution implementing different environmental fluctuation rates can be used to select for higher or lower rates of stochastic switching (Stomp et al, 2008; Beaumont et al, 2009).
Despite these findings, evolution of molecular mechanisms leading to stochastic switching remains unexplained. Theoretical studies to date presume the existence of stochastic switching and focus on the evolution of the rate of switching between maladapted and adapted states either directly (Thattai and van Oudenaarden, 2004; Kussell and Leibler, 2005; Salathé et al, 2009; Gaál et al, 2010; Visco et al, 2010; Liberman et al, 2011) or thorough mutations affecting noise levels (Ribeiro, 2008). Thus, it is not clear if and how fluctuating selection alone could drive the emergence of molecular implementation of stochastic switching at the single cell level. It is generally believed that stochastic switching requires the interplay of noise and bistable dynamics in a gene regulatory network (Dubnau and Losick, 2006); bistability gives rise to two distinct states of gene expression, and noise in gene expression can then lead some cells to be tipped to one state from the other. Theoretical and experimental studies indicate that such bistable dynamics underlie the phenotypic switching seen in E. coli persister cell formation (Rotem et al, 2010) and lactose metabolism (Ozbudak et al, 2004; Robert et al, 2010), B. subtilis DNA uptake competence (Maamar et al, 2007) and sporulation (Fujita et al, 2005), and yeast galactose metabolism (Acar et al, 2005). Further, a specific synthetic implementation of a bistable gene network in E. coli is shown to display stochastic switching, and enables an adaptive response to environmental changes (Kashiwagi et al, 2006).
Here, we analyze the molecular evolution of bistability and noise in simple gene regulatory networks incorporating feedback loops and capable of displaying bistability. Starting with parameters that result in a monostable system without feedback, we run evolutionary simulations under fluctuating environments that select for different optimal gene expression levels. We find that such fluctuating selection results in the emergence and maintenance of bistability only in presence of noise and under a limited range of environmental fluctuation rates. We show that bistability emerges due to selection for increased nonlinearity, which renders the system more evolvable and allows faster adaptation to fluctuating environments. In the absence of noise, the system still evolves higher evolvability by attaining nonlinear dynamics near the bistable regime; however, the evolution of bistability is not observed. These findings suggest that bistability and consequent stochastic switching in gene regulatory networks allowing for feedback loops, evolves only in the presence of noise and as a byproduct of selection for increased evolvability.
Results
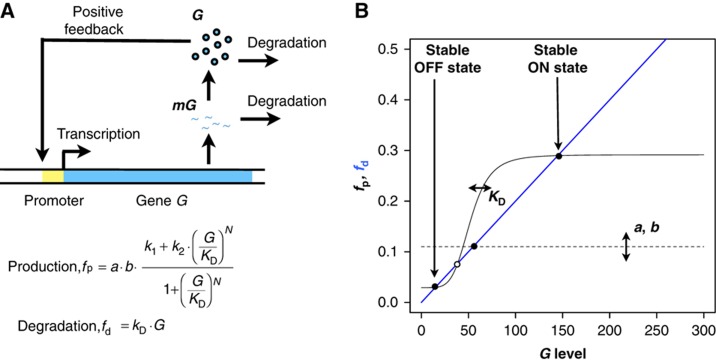
In order to study the evolution of molecular mechanisms underpinning stochastic switching in microbes, we first develop a model of the simplest genetic regulatory network that can display bistability (see Materials and methods). In this system, a single gene G regulates its own expression by acting as a transcription factor binding to its own cis-regulatory module (Figure 1A). As experimentally shown (Becskei et al, 2001; Isaacs et al, 2003; Kaufmann et al, 2007), the resulting nonlinearity from such feedback regulation can exhibit bistability, that is, two distinct and stable states of expression levels separated by an unstable state acting as a threshold (Figure 1B, open circle). Noise in gene expression can then result in protein levels stochastically reaching above the threshold of the bistable system. When this happens, the feedback regulation ensures that protein levels converge to a high value (the ON state). In contrast, when protein levels drop below threshold, protein levels can converge to a low value (OFF state). Under the right conditions, in particular when the expression level in the OFF state is close to the threshold, noise-enabled stochastic switching could maintain a heterogeneous population of cells in the ON and OFF states. This would translate to phenotypic diversity when these two states correspond to distinct phenotypes (e.g., gene G encodes for a master transcription regulator controlling downstream genes).
Figure 1.
(A) Cartoon representation of the model incorporating a single gene auto-regulatory network. (B) Steady-state analysis of the system showing the production (fp, in black) and degradation (fd, in blue) curves for G. The dashed and solid black lines correspond to parameter values a=1.0, b=1, KD=50 and N=0 (the starting condition for evolutionary simulations) and a=0.31, b=4.7, KD=52 and N=5 (mean parameters resulting form one of the simulations under v=0.05), respectively. The solid and open circles indicate the stable and unstable steady states of the system.
To explore the evolution of stochastic switching and bistability under fluctuating environments, we evolve an asexual population of virtual unicellular organisms, each embedding a stochastic model of the system (stochastic phenotype from now on) shown in Figure 1 (see Materials and methods). To understand the role of noise in the evolution of bistability, we also run evolutionary simulations where cells implement a deterministic version of this model (deterministic phenotype from now on). In each case, the parameters of the system that are subject to mutation are a, the scaling factor for the rate of transcription, b, the average number of proteins produced per transcript (i.e., the average size of protein bursts), N, the parameter controlling the strength of the auto-regulatory feedback and KD, the parameter controlling the threshold level of the dynamics resulting from this feedback. Changes in these parameters are biologically plausible and can result from point mutations in promoter regions and/or transcription factors regulating gene expression (Blake et al, 2006; Murphy et al, 2007). Fitness of individuals is determined according to the environment. For our main simulations, we consider a binary relationship between environment and fitness, where either low (in low environment, Elow) or high (in high environment, Ehigh) level of expression results in optimal fitness (see Equation 6). This scenario could arise whenever G encodes a protein that conveys high fitness in a certain environment, but whose activity is detrimental or highly costly in another. In additional simulations, discussed below, we also consider variations on the modeling of the simple feedback network, an alternative feedback circuit and alternative environment–fitness couplings. In the main simulations, we consider that the environment switches stochastically at a certain probability v per generation. At the start of the simulation, the population is homogenous with initial system parameters set to give a monostable system that lacks the auto-regulatory feedback loop (see Materials and methods).
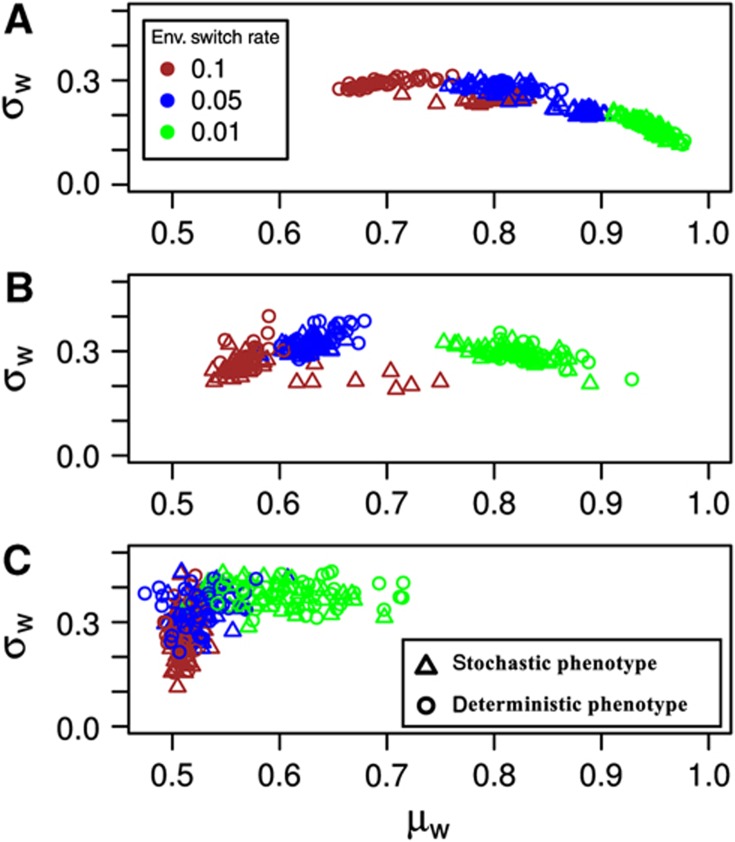
Under all the environmental fluctuation rates we consider, we find that evolution leads to significant improvements over the initial fitness and results both in an increase in the mean of population fitness averaged over 5000 generations, μw and a decrease in its variance, σw (Figure 2). The extent of these fitness improvements depends closely on the mutation rate and the rate of environmental fluctuations. Interestingly, the presence of noise (i.e., stochastic versus deterministic phenotypes) only improves fitness under intermediate ranges of environmental fluctuation rates with the exact range depending also on the mutation rate. To better understand this general trend in the improvement of fitness and its molecular basis, we quantify steady-state system behavior, resulting from the mean parameter values from the individual evolved populations (see Equation 5). Summarized in Figure 3 and Supplementary Figure S1, this analysis suggest evolution of three different strategies under three representative fluctuation rates (slow, intermediate and fast). Under slowly switching environments (v=0.001), the system evolves monostable dynamics with steady-state level of G being either very high or very low (Supplementary Figure S1C). This is because the system spends a relatively long time in either Ehigh or Elow and behaves as if it is under stable selection to adapt to one of these environments. When environments fluctuate very fast (v=0.5), the period of selection under Ehigh or Elow is so short that the best strategy seems to be to set G at a level that corresponds to a reasonable fitness value in both environments. For the particular fitness function used in the simulations shown in Figure 3, this corresponds to G=50 (see Equation 6) and all simulations have evolved parameters that gave a steady-state value close to this (Supplementary Figure S1A). These two strategies that evolved for dealing with slow and fast environmental fluctuations are interestingly independent of the presence or absence of noise in the system. In other words, evolution under these environmental fluctuation rates neither exploited nor was affected by noise.
Figure 2.
Mean fitness of the population averaged over the last 5000 generations of the evolutionary simulations, μw, versus its variance, σw. Open circles and triangles represent results from simulations embedding a deterministic and stochastic phenotype, respectively. Simulations with different environmental fluctuation rates, v, are color coded as indicated by the legend. Results from additional simulations with smaller and higher values of v are omitted to achieve clarity. (A–C) Results from simulations run with mutation rate set to 0.01, 0.001 and 0.0001, respectively.
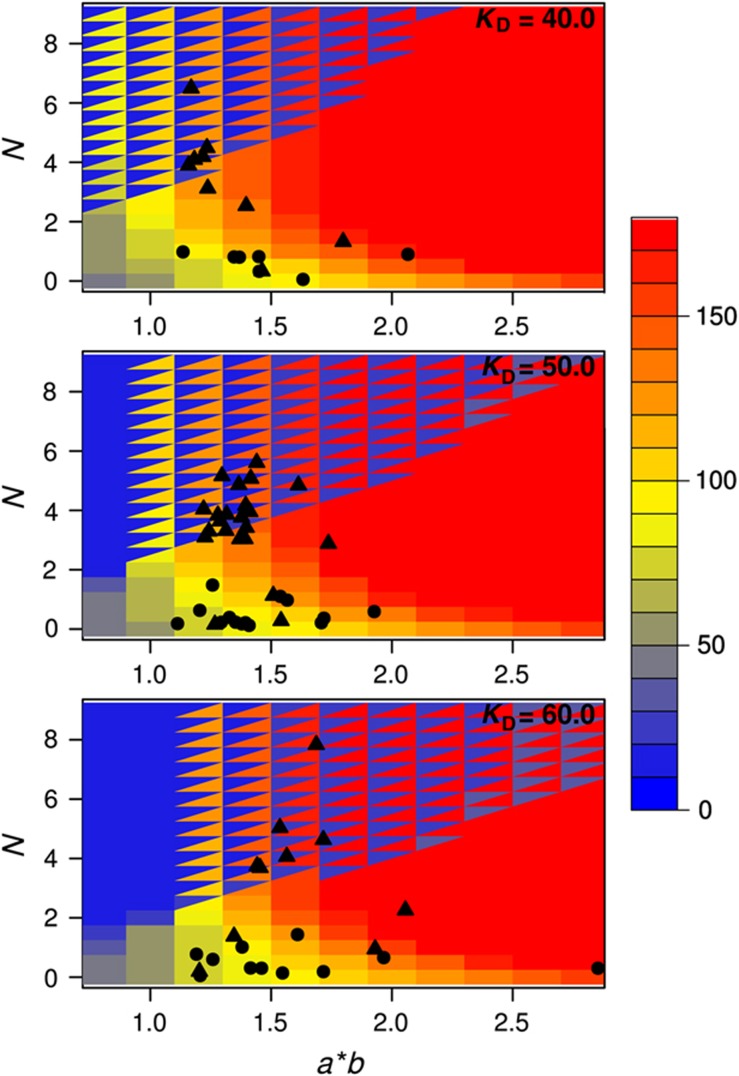
Figure 3.
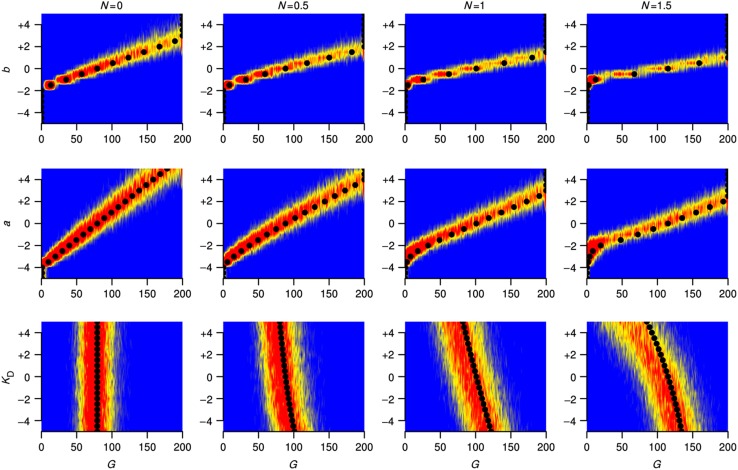
The steady-state behavior of the system with the mean values of parameters as averaged over the last 5000 generations of the evolutionary simulations. The different panels give the phase plane of the system (i.e., steady-state level of G versus system parameters N, a and b) for different values of the parameter KD as shown on each panel. On each panel, the level of G is color coded, where locations with split colors indicate bistable regime with two distinct steady-states levels of G. Solid circles and triangles correspond to results from simulations embedding a deterministic and stochastic phenotype, respectively. Results shown are from simulations with v=0.05 and mutation rate set to 0.01. Note that to achieve the mapping of the results on the phase plane, the mean value of KD obtained from each simulation is rounded to the nearest tenth. Only results with a KD value rounded to the interval 40–60 are shown for clarity and are representative of results with other KD values (in particular, with respect to values of N).
Under intermediary fluctuation rates (v=0.05) system parameters mostly evolved to values that resulted in an intermediary level of G, especially in those simulations that resulted in high fitness (Figure 3 and Supplementary Figure S1B). In the hypothetical fluctuation-free deterministic phenotype, this seems to be achieved by system dynamics that is underpinned by a non-zero N and that places steady-state expression on a ridge in the phase plane that is roughly equidistant to both low and high levels of G (black circles in Figure 3). Simulations with the stochastic phenotype took this approach of ‘being at the edge’ one step further and have predominantly evolved system parameters resulting in bistability (black triangles in Figure 3). It is important to note that in both simulations with the deterministic and stochastic phenotypes, simulations resulting in the highest fitness values (blue outliers in Figure 2A) have evolved non-zero N corresponding to nonlinearity in protein production (Supplementary Figure S1B). In the stochastic case, this nonlinearity was more pronounced and resulted in bistability.
While suggestive, these analyses based on the population average of systems parameters over generations might give misleading conclusions about systems dynamics of genotypes in each generation. Further, they do not provide detailed insight on the evolutionary dynamics, raising the question about why these simulations resulted in the evolution of nonlinearity and bistability in gene expression dynamics. A possible explanation comes from considering the selection process under such intermediary fluctuation rates. Cells experience long enough selection periods under Ehigh and Elow but also frequent environmental change, such that they need to be capable of quickly shifting their steady-state expression level. In the absence of signaling and other feedback mechanisms, such as those seen spatial epigenetic control (Kelemen et al, 2010), this can only be achieved through mutations. Thus, cells evolving under intermediary rates of environmental fluctuations are under selection for achieving abrupt changes in expression levels as fast as possible, that is, with fewest number of mutations. Such ability of a system to maximize its rate of adaptation either by decreasing number of mutations needed for adaptation or by increasing their beneficial effects is broadly referred to as evolvability (Wagner and Altenberg, 1996). Thus, we can hypothesize that the seeming evolution of nonlinearity in the dynamics of the system depicted in Figure 1 confers on it an increased evolvability.
To test this hypothesis and gain a better insight on the evolutionary dynamics, we run additional simulations under a deterministically switching environment and monitored evolvability and the distribution of parameters over individuals in the population. We concentrated this analysis on the intermediary environmental fluctuation rates, where stochastic and deterministic phenotypes gave different results (v=0.05), and run simulations with an environmental epoch duration of 20 generations. We defined evolvability as the ratio of the normalized fitness change (i.e., adaptation) over the sum of relative changes in parameters (i.e., genetic shift) (see Materials and methods and Equation 8). We also considered an alternative approach and quantified adaptation time as a proxy for evolvability (see Materials and methods).
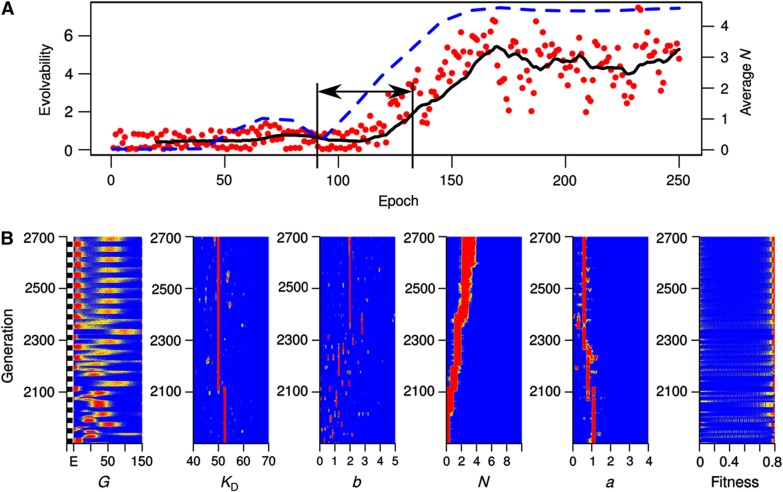
In line with the analysis of average parameter values, these simulations showed evolution of larger values of N both in simulations with deterministic and stochastic phenotypes. Interestingly, we found that such evolution of higher values of N are associated with an increase in evolvability (Figure 4A; Supplementary Figure S2) and a decrease in adaptation time (Supplementary Figures S3A and S4). Importantly, this effect is visible in simulations with the stochastic phenotype well before the emergence of bistability (Figure 4). These findings suggest that evolution of higher N is selected for its ability to confer higher evolvability (i.e., faster adaptation) in environments with intermediary fluctuation rates. To further support this finding, we run additional simulations with fixed N. We found that increasing values of N allow populations to adapt faster to environmental fluctuations (Figure 5; Supplementary Figure S5). Importantly, this trend is mostly independent of the presence or absence of noise and is visible before the bistable regime, indicating that it is mainly the increasing nonlinearity in system dynamics that confers a higher evolvability to the system.
Figure 4.
(A) Evolvability and population mean of N over environmental epochs for a sample simulation with stochastic phenotype and implementing deterministic environmental fluctuations. The environment switches every 20 generations and mutation rate is set to 0.01. Evolvability is defined as the ratio of the normalized fitness change (i.e., adaptation) over the sum of relative changes in parameters (i.e., genetic shift). The red points show the actual evolvability data, calculated for each epoch, while the black line gives its moving average over epochs. The blue dotted line gives population average of N over epochs. (B) The distribution of individual parameters, fitness and expression level of G over the population and over generations. Each row on each panel encodes a distribution for a specific quantity (as indicated at the bottom of the panel) and for the specific generation shown on the y axis. The distributions are shown as a heat map ranging from red (highest density) to blue (lowest density). Environments Elow and Ehigh are indicated as black and white bars on the y axis of the left-most panel.
Figure 5.
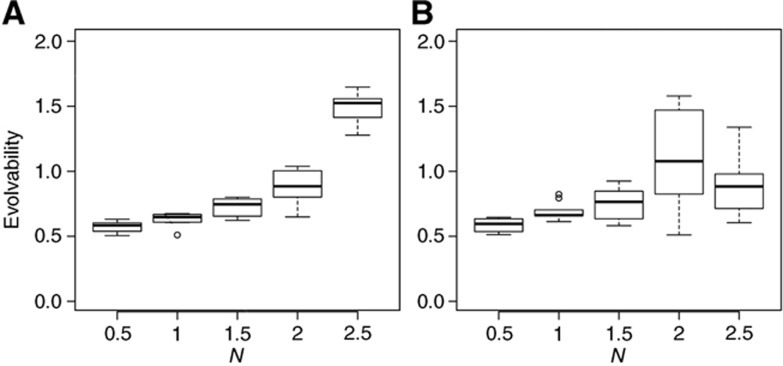
Boxplots showing a summary of the distribution of evolvability from 10 simulation runs for each fixed value of N as shown on the x axis. Note that at N=2.5, we have a bistable system. In these simulations, the other parameters of the system were free to evolve as before. The environment is deterministic and switches every 20 generations. Mutation rate is set to 0.01. Evolvability is defined as the ratio of the normalized fitness change (i.e., adaptation) over the sum of relative changes in parameters (i.e., genetic shift). (A) Results from simulations with stochastic phenotype. (B) Results from simulations with deterministic phenotype.
To better understand how higher N is linked to an increase in evolvability, we performed a systematic study of phenotypic effects of mutations in other parameters under different values of N. This analysis revealed that increasing N results in an increase in the diversity of gene expression level, and in the phenotypic effects of mutations in a, b and KD (Figure 6). The latter effect was also evident in systems encoded by the average parameters obtained from our original simulations. These display significant phenotypic shift with as few as two mutations (Supplementary Figure S6). We note that the relation of increasing N and increasing mutational effects is a direct result of how N changes the shape of the production curve fp (e.g., see Supplementary Figure S1). The former effect of increasing diversity with increasing N can be understood in light of a positive correlation between noise and nonlinearity that is also shown to exist in certain signaling motifs (Shibata and Fujimoto, 2005). To better understand how increasing noise results from increasing N in the feedback circuit, we derived an analytical expression for the protein level (see Supplementary information). This showed that, up until bistability emerges, increasing N (while keeping all other parameters of the model fixed) does not strongly affect the peak of the steady-state distribution of the protein levels, but results in a widening of it (Supplementary Figure S7A). Thus, increasing N results in higher noise level (Supplementary Figure S7B), while the mean protein level remains mostly unaltered. Exploiting this uncoupling between the effects of N on noise and on the mean protein level, we then asked if solely the increase in noise can provide a fitness advantage in either of the two environments. Fixing all other parameters of the model, we calculated the fitness of the resulting genetic system in both environments for increasing levels of noise through an increase in N (see Supplementary information). We found that increasing noise levels, while the mean protein level remains mostly unchanged, is beneficial under the fitness scheme given in Equation (6) and in the ‘low’ (‘high’) environment when mean protein levels are below (above) 50 (Supplementary Figure S8). This result also extends to an alternative scheme for coupling fitness and protein levels (see below and Supplementary Figure S16). Taken together, these results indicate that increasing N (and thus noise) is beneficial for adaptation when mean protein levels are far from optimal. This finding is in line with previous findings, which showed a beneficial effect of noise under a convex fitness function (Zhang et al, 2009).
Figure 6.
Analysis of mutational effects of parameters on the phenotype (i.e., level of protein G) in genotypes with increasing value of N. This analysis shows that as N increases, so does (i) the diversity in the expression level of G and (ii) the phenotypic effects of a given mutational increment in other parameters. Rows from top to bottom show phenotypic effects of mutations in parameters b, a and KD, respectively. Columns from left to right show genotypes with increasing value of N as indicated at the top. The x axis on each panel gives the level of protein G, while the y axis gives the deviation in a particular system parameter based on its mutational increment in the evolutionary simulations (see Materials and methods). The initial parameter is the middle point of the y axis (i.e., zero deviation) and the upper and lower halves of each panel correspond to mutational deviations from that value. The basal values used on each panel are the same; b=1.8, a=0.8 and KD=50 (different basal values gave similar qualitative results as those shown). For each value of the parameter given on the y axis, we run 1000 independent simulations for 10 generations and encoded in color the number of simulations that converged to the corresponding level of G indicated on the x axis. Solid dots correspond to simulations run with the deterministic phenotype.
These analyses strongly suggest that increasing N is selected for in our simulations, due to its positive fitness effects that arise from an increased noise (i.e., increased heterogeneity in the population) and increased effects of mutations. We have observed both of these effects in the evolutionary simulations with stochastic and deterministic phenotypes (Figure 4B; Supplementary Figure S3B). First, we find that as large values of N evolve, adaptation to a new environment occurs very fast and with few mutations (Figure 4B, generations 2100–2160). Furthermore, simulations with the deterministic phenotype show a clear pattern of increased phenotypic movement with mutations after the evolution of larger N; while expression level of G stays close to 50 and shifting slightly above and below it with every environmental epoch, it moves to more extreme values (achieving higher fitness) after the evolution of larger N (Supplementary Figure S3B, generations 0–300 versus generations 400–1000). Second, as N evolves to larger values, we observe a general increase in the phenotypic diversity in the population (Figure 4B, generations before versus after generation 2100). This increase in diversity adds on top of that resulting from the mutational diversity around the wild type (i.e., quasi species), which is evident in both the runs with the stochastic and deterministic phenotypes and is mainly driven by changes in the parameter b (e.g., see corresponding panels in Figure 4B and Supplementary Figure S3B). We note, however, that mutational diversity in all parameters is lost in the stochastic phenotype after the emergence of bistability. This is because bistability mostly alleviates the need for diversity by resulting in stochastic switching; two distinct phenotypes expressed from the same genotype (see Figure 4B, generations >2400). Subsequent mutations can then tune the noise level and the size of basins of attractions in the bistable system to achieve maximal fitness and minimal adaptation time. The former process can be seen to a certain extent in Figure 4B.
While we observed evolution of larger N in all simulations under intermediary rates of environmental fluctuations, we also observed—mainly in simulations with the deterministic phenotype—that a large value of N can also be lost again after its emergence (Supplementary Figures S2 and S4). Such loss of nonlinearity could be explained by the fact that in the deterministic phenotype, the effect of increasing N is limited to only a change of phenotypic effects of mutations in other parameters, and that mutations in N themselves also change the expression level of G even slightly. In particular, every time the environment switches so to favor lower G, mutations that decrease N can increase in frequency. In contrast, in the stochastic phenotype, the associated phenotypic diversity with higher N ensures that the population can move to a lower G level without necessarily drifting out of the nonlinear regime. In addition, the presence of noise in the stochastic phenotype results in a positive fitness effect (Supplementary Figure S8), and potentially reduces the efficiency of selection (Wang and Zhang, 2011). Both effects would allow easier maintenance of larger values of N in the stochastic phenotype model, even under long stretches of stable environments. In other words, increased diversity provides a type of robustness against mutations that decrease N. Eventually, further mutations can lead N to reach above a certain threshold and result in the emergence of bistability. This results in stochastic switching and provides the system with the perfect solution to the particular fluctuating environment we implement. Such high fitness associated with the stochastic switching ‘strategy’ under the fluctuating environment then ensures stable maintenance of large N and bistable dynamics. In line with this view, we find that in evolutionary simulations starting with an initially large value of N, bistability is maintained under a broad range of environmental fluctuation and mutation rates (Supplementary Figure S9).
In summary, these findings strongly indicate that the fitness benefit of having a larger N (i.e., nonlinear fp) comes from an increased evolvability, rather than from noise-enabled stochastic switching per se. Such a key role for selection for higher evolvability under intermediate fluctuation rates could also explain the general trend we observe in Figure 2 with regards to fitness difference between simulations with stochastic and deterministic phenotypes. The fitness difference between these simulations is significant only when the best strategy to cope with the environmental fluctuation is to evolve a nonlinear fp, in which case simulations with the stochastic phenotype can achieve higher fitness by evolving bistability. As can be seen from Figure 2, this situation arises not only for a specific range of environmental fluctuation rates but rather for a specific combination of mutation rate and environmental fluctuation rate. This is in line with the previous findings on the evolution of evolvability (Meyers et al, 2005; Stomp et al, 2008; Tsuda and Kawata, 2010), and is a manifestation of the fact that selection for higher evolvability only becomes significant when generation of mutations and their time to fixation are in line with the duration of selection under a specific environment. Population genetic models of simple systems suggest that both higher mutation rate and higher rates of fluctuating selection can decrease the time to fixation of beneficial and neutral mutants (Ota and Kimura, 1972; Kimura, 1980), thus potentially allowing for selection of systems that can generate more of these types of mutations (Tsuda and Kawata, 2010). In our simulations, and in nature, these effects of mutation rate and fluctuating selection have to be in balance for the evolution of evolvability as suggested before (Meyers et al, 2005).
How general are these findings? In particular, could selection for incremental increase in N operate in other feedback circuits, under different modeling choices or under other environment–fitness relationships? To address this question, we run several additional simulations. First, we confirmed the robustness of the above results for a variety of alternative modeling choices in our original feedback model. We found that higher N and consequently higher evolvability evolves when we allow all of the model parameters to evolve, when feedback is modeled with an alternative form to that given in Equation (1), and finally when we consider a coupling between gene expression dynamics and growth (see Supplementary information and Supplementary Figure S10). Of these alternative modeling choices, the last one is particularly interesting as both protein and mRNA levels in bacteria are found to relate to growth rate in intricate ways (Klumpp et al, 2009). While such a coupling can itself lead to bistable gene expression when a constitutively expressed gene has a direct effect on growth rate (Klumpp et al, 2009), our simulations indicate that it might not have a bearing on the evolution of higher nonlinearity in a feedback-based gene regulatory system under fluctuating environments. It can be envisioned, however, that a constitutively expressed gene that alters growth under certain environments could act as the precursor to the feedback motif we consider here, by providing a primary increase in heterogeneity (and thus potentially in evolvability).
Second, we run additional simulations with a model that captures another commonly observed gene regulatory motif; the double negative feedback loop. This regulatory motif is well studied in the λ phage, where it underlies the lysis-lysogeny decision (Ptashne, 2004), and an engineered version of it is experimentally shown to display bistability in bacteria (Gardner et al, 2000). We developed a mathematical model of this system using the same approach as in the original model (see Supplementary information and Supplementary Figure S11A). Using this double negative feedback model, we run five evolutionary simulations for 5000 generations under a deterministically switching environment switching every 20 generations (same conditions as above). All of these simulations resulted in the evolution of bistability underpinned by higher values of N in the two feedback loops. In all these simulations, we confirmed the positive relation between nonlinearity and evolvability (Supplementary Figure S11B and C). This suggests that this positive relation could be a generic feature in a diverse set of gene regulatory motifs that involve feedback loops.
Finally, we considered alternative fitness–environment couplings and run simulations with the simple feedback model. We first relaxed the assumption of a highly nonlinear relation between the level of G and fitness. Using a more linear relationship (Hill coefficient in Equation (6) set to 2), we found that the main results remain unaltered (Supplementary Figure S12). Next, we relaxed the assumption of a sigmoidal fitness relation altogether and considered that fitness is given by a normal distribution, where the optimal fitness in a given environment corresponds to a specific level of G and where any deviations from this level are detrimental to fitness (Equation 7). Under such an environment–fitness relationship, we assumed that evolutionary fluctuations correspond to changes in the optimal level of G (i.e., the mean of the normal distribution). Biologically, this scenario might be more broadly applicable and simply assumes that each of the different environments an organism encounters requires a different optimal level of G. Within this scenario we considered two types of environmental fluctuations: (i) environments fluctuating between two specific optimal mean values (high and low) and (ii) environments fluctuating randomly between optimal mean values in a wide range.
We find that evolutionary simulations under these scenarios still result in the improvement of fitness (Supplementary Figure S13) and in the evolution of increased nonlinearity (Supplementary Figures S14 and S15). Interestingly, we find that the presence of noise is mostly detrimental under this type of environment–fitness relationship (Equation 7), resulting in the deterministic phenotype evolving higher fitness solutions compared with the stochastic phenotype. This result can be understood considering the normal distribution of fitness encoded by Equation (7), which in the presence of noise inhibits attaining optimal fitness as also observed in theoretical analysis of the effects of noise in metabolism (Wang and Zhang, 2011). Although in our simulations the parameters are free to evolve to minimize noise, the system cannot do so freely as it is also under selection to achieve a specific level of G. When the normal distribution of fitness has a wider variance, this detrimental effect of noise and consequently the difference between the fitness resulting from stochastic and deterministic phenotypes is reduced (Supplementary Figure S13). Under the environment–fitness relationship given by Equation (7), the evolution of bistability is much more limited and is only observed in few cases under the scenario where environment fluctuates between two specific optimal values of G (Supplementary Figure S14).
Both sets of results are in line with the above-described relation of noise, N and fitness. In our model, increasing N results in an increase in noise (Supplementary Figure S7), which then can have a positive effect on noise under a convex fitness function (Supplementary Figure S8). While reducing the Hill coefficient in Equation (6) does not alter these dynamics significantly, using a normal distribution for fitness (as in Equation 7) results in a fitness function that is concave near optima. This limits the positive fitness effects of increasing noise (see Supplementary Figure S16). While the effect of increasing N on other mutations is maintained, this can limit the evolution of N to high values and emergence of bistability as observed in the simulations. Taken together, these results suggest that evolution of increased evolvability (through evolution of nonlinear system dynamics), but not the evolution of bistability, might occur under broad fitness–environment relationships.
Discussion
This analysis shows that the main and broadly applicable effect of fluctuating selection on the evolution of gene regulatory networks that allow for a feedback loop is selection for specific system dynamics that confer an increased evolvability. Increased evolvability mainly results from evolution of system parameters controlling the feedback loop, into a nonlinear regime, where both phenotypic diversity and the amount of phenotypic shift caused by individual mutations are increased. We find that under a specific but biologically plausible form of fluctuating environment, selection for having system parameters in such a nonlinear regime results in the emergence of bistability and the associated stochastic switching. Under a switch-like environment, bistability offers immediate adaptation by allowing expression of two distinct phenotypes from the same genotype and offers additional ‘points of operation’ for evolution, such as the size of basins of attractions for the steady states.
Under the evolutionary scenario considered here and in the gene regulatory motifs involving a feedback loop, the beneficial effects of bistability are realized only under specific environment–fitness relationships and in the presence of noise. Consequently, we find that noise is a necessary but not sufficient condition for the evolution of bistability through fluctuating selection under these conditions. Once bistability arises, noise-induced stochastic switching can give rise to additional fitness benefits, such as survival under advert conditions (Balaban et al, 2004). Furthermore, the rate of switching can be tuned to environmental fluctuation rates (Kussell and Leibler, 2005) via changes altering system dynamics and/or noise level (Ribeiro, 2008). Noise is an inevitable feature of cellular systems (Lestas et al, 2010) and could itself be under positive or negative selection (van Hoek and Hogeweg, 2007; Tănase-Nicola and ten Wolde, 2008; Zhang et al, 2009; Wang and Zhang, 2011). Thus, other selective forces that lead to higher/lower noise might also enhance/inhibit the evolution of bistability and nonlinearity in gene regulation as indicated for example in the lactose metabolism of E. coli (van Hoek and Hogeweg, 2007).
We find that for the gene regulatory networks considered here, evolution of both evolvability and bistability are limited to a specific combination of mutation and environmental fluctuation rates. This is in line with other observations reporting the evolution of higher evolvability (Meyers et al, 2005; Stomp et al, 2008; Tsuda and Kawata, 2010) and indicates a need for a balance between environmental fluctuation rate and mutation rate so that the latter can lead to a selection for higher evolvability (Meyers et al, 2005). In our simulations, evolvability leads to the emergence of bistability in the gene regulatory network and interestingly, we find this bistability to be maintained in subsequent evolution under a broader range of environmental fluctuation and mutation rates (Supplementary Figure S9). These findings do not contradict earlier studies, demonstrating a fitness benefit for stochastic switching under a wide range of environmental fluctuation rates (Thattai and van Oudenaarden, 2004; Kussell and Leibler, 2005; Kussell et al, 2005; Salathé et al, 2009; Gaál et al, 2010; Visco et al, 2010; Liberman et al, 2011), but show that those results cannot be interpreted as fluctuating selection to be a general mechanism for leading to molecular evolution of stochastic switching.
It is important to emphasize that the increased evolvability we observe in these simulations is based solely in system dynamics; a shift into a nonlinear regime results in larger phenotypic jumps from a given mutation and also to an increased phenotypic diversity. These two effects can essentially be described as changing the genotype–phenotype mapping in such a way to increase evolvability and endowing the system the ability to adapt to environmental shifts quickly. Similar mechanisms of evolvability, but that draw mainly on mutational biases and result from structural and sequence-based features have been described in both gene (Crombach and Hogeweg, 2008; Tsuda and Kawata, 2010) and logic circuits (Parter et al, 2008) and also in promiscuous proteins (Aharoni et al, 2005), viral RNA (Burch and Chao, 2000) and TATA-box containing genes (Landry et al, 2007). Our results extend this list of observed evolvability in biological systems and could explain the observed stochasticity and nonlinearity in the regulation of genes whose fitness effects are directly coupled with the environment, including antitoxin–toxin proteins, metabolic enzymes and master transcriptional regulators (Ozbudak et al, 2004; Fujita et al, 2005; Robert et al, 2010; Rotem et al, 2010). We note that interestingly, some of the epigenetic and genetic mechanisms other than feedback regulation that control the expression of environmentally relevant genes in a number of microbes (van Der Woude and Baumler, 2004) can display high levels of nonlinearity and bistability (Sedighi and Sengupta, 2007), supporting the hypothesis that high nonlinearity might be a general strategy to achieve high evolvability in the regulation of environmental genes.
Materials and methods
Single gene positive-feedback loop model
In order to capture the underlying stochasticity of gene regulation, we constructed a positive-feedback model using the stochastic chemical kinetic framework (Gillespie, 2007). The model describes the regulation of a gene G by its own protein product G. In particular, we consider that G can positively regulate the transcription of G by binding to its cis-regulatory module. This type of regulation is common in biology and a synthetic implementation of it has experimentally verified its potential for achieving bistability so to give rise to two distinct steady-state levels of protein amount (Becskei et al, 2001; Isaacs et al, 2003). The model consists of two species corresponding to the protein (G) and mRNA (mG) and four reaction processes: transcription, translation, mRNA degradation and protein degradation (Figure 1A). One molecule of mG is synthesized from G via the transcription reaction whose propensity is given by:
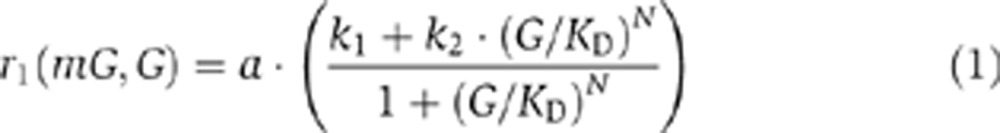 |
where the parameters k1 and k2 are arbitrarily set to 0.02 and 0.2, respectively, while parameters a, KD and N are free to evolve. This production function is based on the equilibrium statistical thermodynamic model of transcriptional regulation (Ackers et al, 1982), which is used frequently for modeling the dynamics of gene expression processes. This approach represents transcription dynamics based on the probabilities of the configuration of the promoter and operator sites in thermodynamic equilibrium. In the Supplementary information, we consider an alternative model that uses the Hill function as the production term.
One molecule of G is synthesized using mG as a template through the translation reaction while one molecule of mG is removed through the mRNA degradation reaction. The propensity functions of these reactions are, respectively, given by:
 |
 |
where the parameter k4 is set to 0.1 and parameter k3 is free to evolve. At the start of evolutionary simulations, we set k3=0.1 resulting in b=k3/k4=1, which indicates that on average one copy of protein will be synthesized per transcript. The other reaction process in our model is the protein degradation where one molecule of G is removed from the system. The propensity function of this reaction is given by:
 |
where the parameter k5 is set to 0.002. See also Supplementary information for an alternative implementation of these parameters, considering their coupling to growth rate (i.e., fitness), and also for simulations that allow all of the parameters to evolve.
In the continuous-deterministic framework, setting all the kinetic law functions to zero allows us to solve for the steady-state values of mG and G. In particular, using the G* notation for the steady-state value of G, we can derive the following relation at steady state:
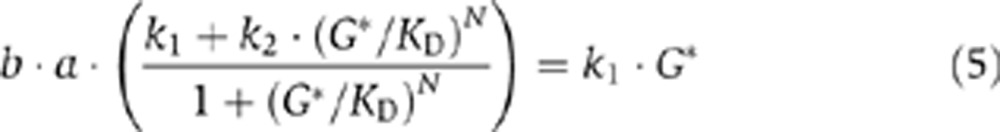 |
where the left and right sides denote the production (fp) and degradation (fd) functions for G, respectively. This reveals that, with an appropriately high value of N (i.e., a high-binding cooperativity of G to the promoter of the gene G), we can have three distinct and unique steady-state values for G, making the system bistable (Figure 1B).
Evolutionary simulations
We consider a population of 1000 cells, each of which contains the positive-feedback genetic circuit described above. The evolutionary simulations start with a homogenous population with initial parameters set to a=1.0, b=1, KD=50 and N=0. Note that this results in a monostable system that is lacking feedback regulation (Figure 1B). We simulated the population in a fluctuating environment for 10 000 generations. The initial values of mG and G at the first generation are both set to zero. For each generation, the stochastic models are simulated using the direct method implementation of Gillespie’s stochastic simulation algorithm (Gillespie, 1977) for 2000 time units. In the deterministic case of the model, we iterate functions fp and fd to update the amount of G to calculate the steady-state level for each generation. We assume that the system reached steady state when the difference between fp and fd becomes less than 1/(k5 × c), where c=5 × 105. At the beginning of each succeeding generation, the values of mG and G are reduced to the largest integers that are smaller than mG/2 and G/2, respectively, to model cell division. We have also considered the case of relating fitness to concentration, in which case we run simulations without the cell division step described above (as concentrations can be considered to remain constant during cell division). These simulations resulted in qualitatively the same results as described in the main text (see Supplementary Figures S17 and S18).
At the end of each generation, a new population is produced from the current one using random drawing with replacement. A random individual is picked from the population and is cloned into the new population if a uniformly distributed random number (from the interval [0,1]) is below its normalized fitness. Then, it is put back into the current population and a new draw is made, the process continued until the new population contains 1000 individuals. During cell replication, mutations happen at a rate u, and are modeled as small perturbations to either one of the parameters a, k3, KD and N (with equal probability). Note that mutating k3 is equivalent to mutating the burst size, b. Mutations of a, b and KD are captured by adding A(0, 0.22), A(0, 1.02) and A(0, 5.02), respectively, where A(μ, σ2) is a normal random variable with mean μ and s.d. σ. Note that we imposed the condition that the values of a, b and KD be at least 0.01, 0.01 and 10−20, respectively. The mutation of N is captured by adding 0.5 or −0.5 at the equal probability, where the value of N is set to be ⩾0. See also Supplementary information for alternative simulations that allow all of the model parameters to evolve.
Fitness is determined as a function of the environment and the amount of G at the end of each generation. For the main simulations, we consider two environments, where either a low (Elow) or high (Ehigh) level of the protein is beneficial. The simulations start in Ehigh and subsequently the environment changes in each generation with a probability v. The fitness under the two environments is given by functions whigh and wlow:
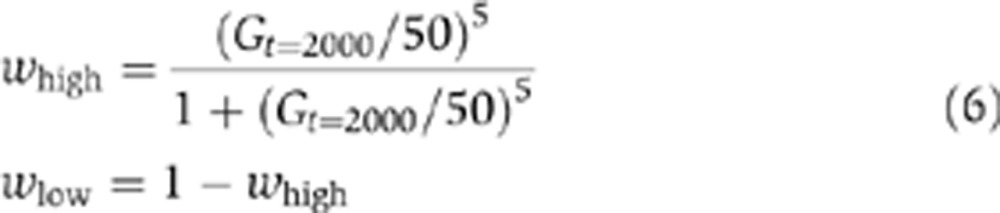 |
where Gt=2000 corresponds to the amount of G in the cell at the end of one generation. Note that Equation (6) assumes a symmetric fitness in the two environments in relation to the level of G, and assumes that the costs of producing G are negligible. Based on previous studies, we expect that such a cost, or more broadly, having an asymmetric fitness for the two environments would reduce selection for bistability and stochastic switching as indicated previously (Salathé et al, 2009).
In additional simulations, we considered alternative scenarios for environmental fluctuations and environment–fitness relations. For these simulations, fitness was given by;
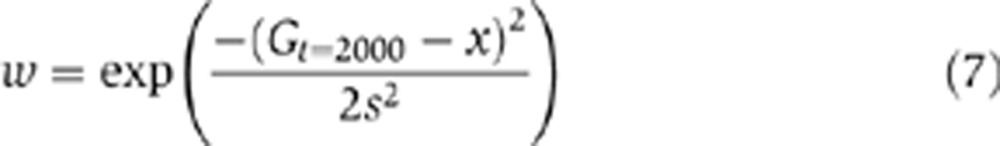 |
where Gt=2000 corresponds to the amount of G in the cell at the end of one generation as before and x and s determines the level of G corresponding to maximal fitness and the variance of fitness distribution around that level, respectively. Using this fitness function, we considered two scenarios for environmental fluctuations: (i) x changes between two specific optimal mean values corresponding to a ‘low’ (x=20) and ‘high’ (x=80) environment and (ii) x changes randomly in the interval [0,120].
All evolutionary simulations are coded in the C language. The source code for simulations that implement the single feedback loop model with all its parameters defined as evolvable and deterministic environmental switching are made available as Supplementary information. Source code of the remaining simulations, as described in the main text and the Supplementary information, can be derived from the provided code or could be obtained from the authors upon request.
Measuring evolvability
For the evolutionary simulations with a deterministically switching environment, we quantified the evolvability of a population adapting to a fluctuating environment as the ratio of the normalized fitness change over the sum of relative changes in parameters. This measure is similar to the control coefficient used in the analysis of metabolic networks (Heinrich and Schuster, 1996). The control coefficient gives the logarithmic sensitivity as the relative change of a system variable (e.g., flux) in response to a relative change in a rate. Similarly, we define evolvability as the sensitivity of the systems’ fitness, and measure it as a relative change of the average fitness arising from relative mutational shifting of the parameters over a given epoch (i.e., time window). We define relative change in fitness with respect to final rather than initial fitness, so to avoid relative change in fitness to become artificially sensitive to a small value of the initial fitness, and to account for the experimentally observed saturation in fitness as it approaches higher values (Schoustra et al, 2009; MacLean et al, 2010). Thus, evolvability is given as;
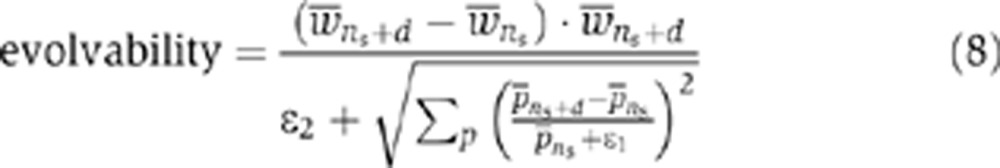 |
where 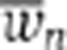 and
and  are the average of the fitness and evolvable parameter p over the population at generation n, respectively. ns indicates the first generation following an environmental switch, while d is the number of generations between the first and last generation in an environmental epoch in the evolutionary simulations with a deterministically switching environment (i.e., d=19 when epoch length is 20). We use the small correcting factors ε1=10−10 and ε2=1, to avoid division by zero. In Equation (8), the nominator gives the gain in the average fitness in a given epoch scaled by the average fitness of the last generation in that epoch (i.e., the extent of adaptation), while the denominator expresses the relative change in evolvable parameters during that same epoch (i.e., the extent of genetic change).
are the average of the fitness and evolvable parameter p over the population at generation n, respectively. ns indicates the first generation following an environmental switch, while d is the number of generations between the first and last generation in an environmental epoch in the evolutionary simulations with a deterministically switching environment (i.e., d=19 when epoch length is 20). We use the small correcting factors ε1=10−10 and ε2=1, to avoid division by zero. In Equation (8), the nominator gives the gain in the average fitness in a given epoch scaled by the average fitness of the last generation in that epoch (i.e., the extent of adaptation), while the denominator expresses the relative change in evolvable parameters during that same epoch (i.e., the extent of genetic change).
As an alternative measure for evolvability, we have also monitored adaptation time defined as the number of generations it takes for the mean fitness to reach 80% of the maximum possible following an environmental switching event.
Supplementary Material
The source code for one of the main evolutionary simulations discussed in the main text.
Supplementary information, supplementary figures S1-18
Acknowledgments
We thank Nicolas Buchler, Marcel Salathé, Juan Poyatos, Raul Guantes and Peter Ashwin for comments. OSS acknowledges support of Exeter University Science Strategy. HK was supported by Lane fellowship through the Ray and Stephanie Lane Center for Computational Biology at Carnegie Mellon University.
Author contributions: OSS and HK together conceived the research, developed the tools, run simulations, analyzed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 [DOI] [PubMed] [Google Scholar]
- Acar M, Mettetal JT, van Oudenaarden A (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40: 471–475 [DOI] [PubMed] [Google Scholar]
- Ackers GK, Johnson AD, Shea MA (1982) Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci USA 79: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, Tawfik DS (2005) The ‘evolvability’ of promiscuous protein functions. Nat Genet 37: 73–76 [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622. [DOI] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB (2009) Experimental evolution of bet hedging. Nature 462: 90–93 [DOI] [PubMed] [Google Scholar]
- Becskei A, Séraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ (2006) Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Burch CL, Chao L (2000) Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature 406: 625–628 [DOI] [PubMed] [Google Scholar]
- Cohen D (1966) Optimizing reproduction in a randomly varying environment. J Theor Biol 12: 119–129 [DOI] [PubMed] [Google Scholar]
- Crombach A, Hogeweg P (2008) Evolution of evolvability in gene regulatory networks. PLoS Comput Biol 4: e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R (2006) Bistability in bacteria. Mol Microbiol 61: 564–572 [DOI] [PubMed] [Google Scholar]
- Fujita M, González-Pastor JE, Losick R (2005) High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187: 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaál B, Pitchford JW, Wood JA (2010) Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics 184: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 6767. [DOI] [PubMed] [Google Scholar]
- Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81: 2340–2361 [Google Scholar]
- Gillespie DT (2007) Stochastic simulation of chemical kinetics. Annu Rev Phys Chem 58: 35–55 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Schuster S (1996) The Regulation of Cellular Systems. USA: Springer [Google Scholar]
- Isaacs FJ, Hasty J, Cantor CR, Collins JJ (2003) Prediction and measurement of an autoregulatory genetic module. Proc Natl Acad Sci USA 100: 7714–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi A, Urabe I, Kaneko K, Yomo T (2006) Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS One 1: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann BB, Yang Q, Mettetal JT, van Oudenaarden A (2007) Heritable stochastic switching revealed by single-cell genealogy. PLoS Biol 5: e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen JZ, Ratna P, Scherrer S, Becskei A (2010) Spatial epigenetic control of mono- and bistable gene expression. PLoS Biol 8: e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1980) Average time until fixation of a mutant allele in a finite population under continued mutation pressure: studies by analytical, numerical, and pseudo-sampling methods. Proc Natl Acad Sci USA 77: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Zhongge Z, Hwa T (2009) Growth rate-dependent global effects on gene expression in bacteria. Cell 139: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Kishony R, Balaban NQ, Leibler S (2005) Bacterial persistence: a model of survival in changing environments. Genetics 169: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL (2007) Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J (2010) Fundamental limits on the suppression of molecular fluctuations. Nature 467: 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman U, Cleve JV, Feldman MW (2011) On the evolution of mutation in changing environments: recombination and phenotypic switching. Genetics 187: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J (2008) Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9: 583–593 [DOI] [PubMed] [Google Scholar]
- Losick R, Desplan C (2008) Stochasticity and cell fate. Science 320: 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D (2007) Noise in gene expression determines cell fate in Bacillus subtilis. Science 317: 526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, Perron GG, Gardner A (2010) Diminishing returns from beneficial mutations and pervasive Epistasis shape the fitness landscape for Rifampicin resistance in Pseudomonas Aeruginosa. Genetics 186: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers LA, Ancel FD, Lachmann M (2005) Evolution of genetic potential. PLoS Comput Biol 1: 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KF, Balázsi G, Collins JJ (2007) Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci USA 104: 12726–12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A, Weiner M (1957) Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA 43: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Kimura M (1972) Fixation time of overdominant alleles influenced by random fluctuation of selection intensity. Genet Res 20: 1–7 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A (2004) Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740 [DOI] [PubMed] [Google Scholar]
- Parter M, Kashtan N, Alon U (2008) Facilitated variation: how evolution learns from past environments to generalize to new environments. PLoS Comput Biol 4: e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi T (1993) Bet-hedging germination of desert annuals: variation among populations and maternal effects in Lepidium lasiocarpum. Am Nat 142: 488–507 [DOI] [PubMed] [Google Scholar]
- Ptashne MA (2004) Genetic Switch: Phage Lambda Revisited. USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP (2002) Control, exploitation and tolerance of intracellular noise. Nature 420: 231–237 [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AS (2008) Dynamics and evolution of stochastic bistable gene networks with sensing in fluctuating environments. Phys Rev E Stat Nonlin Soft Matter Phys 78: 061902. [DOI] [PubMed] [Google Scholar]
- Robert L, Paul G, Chen Y, Taddei F, Baigl D, Lindner AB (2010) Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol Syst Biol 6: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ (2010) Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathé M, van Cleve J, Feldman MW (2009) Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics 182: 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoilov MS, Price G, Arkin AP (2006) From fluctuations to phenotypes: the physiology of noise. Sci STKE 2006: re17. [DOI] [PubMed] [Google Scholar]
- Sedighi M, Sengupta AM (2007) Epigenetic chromatin silencing: bistability and front propagation. Phys Biol 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra SE, Bataillon T, Gifford DR, Kassen R (2009) The properties of adaptive walks in evolving populations of fungus. PLoS Biol 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Fujimoto K (2005) Noisy signal amplification in ultrasensitive signal transduction. Proc Natl Acad Sci USA 102: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomp M, van Dijk MA, van Overzee HM, Wortel MT, Wortel MT, Sigon CA, Egas M, Hoogveld H, Gons HJ, Huisman J (2008) The timescale of phenotypic plasticity and its impact on competition in fluctuating environments. Am Nat 172: 169–185 [DOI] [PubMed] [Google Scholar]
- Tănase-Nicola S, ten Wolde PR (2008) Regulatory control and the costs and benefits of biochemical noise. PLoS Comput Biol 4: e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A (2004) Stochastic gene expression in fluctuating environments. Genetics 167: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda ME, Kawata M (2010) Evolution of gene regulatory networks by fluctuating selection and intrinsic constraints. PLoS Comput Biol 6, pii: e1000873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Woude MW, Baumler AJ (2004) Phase and antigenic variation in bacteria. Clin Microbiol Rev 17: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek M, Hogeweg P (2007) The effect of stochasticity on the lac operon: an evolutionary perspective. PLoS Comput Biol 3: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW (2008) Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci USA 105: 4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visco P, Allen RJ, Majumdar SN, Evans MR (2010) Switching and growth for microbial populations in catastrophic responsive environments. Biophys J 98: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Altenberg L (1996) Complex adaptations and the evolution of evolvability. Evolution 50: 967–976 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang J (2011) Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci USA 108: E67–E76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qian W, Zhang J (2009) Positive selection for elevated gene expression noise in yeast. Mol Syst Biol 5: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The source code for one of the main evolutionary simulations discussed in the main text.
Supplementary information, supplementary figures S1-18