Abstract
Gram-negative bacteria have evolved several secretory pathways to release enzymes or toxins into the surrounding environment or into the target cells. The type II secretion system (T2SS) is conserved in Gram-negative bacteria and involves a set of 12 to 16 different proteins. Components of the T2SS are located in both the inner and outer membranes where they assemble into a supramolecular complex spanning the bacterial envelope, also called the secreton. The T2SS substrates transiently go through the periplasm before they are translocated across the outer membrane and exposed to the extracellular milieu. The T2SS is unique in its ability to promote secretion of large and sometimes multimeric proteins that are folded in the periplasm. The present review describes recently identified protein–protein interactions together with structural and functional advances in the field that have contributed to improve our understanding on how the type II secretion apparatus assembles and on the role played by individual proteins of this highly sophisticated system.
Keywords: type II secretion system (T2SS), secreton, pseudopilus, secretin, protein–protein interaction
1. Introduction
Gram-negative bacteria are surrounded by a dual membrane structure establishing an interface between the environment and the interior of the cells. The two membranes are separated by an aqueous periplasmic space containing a rigid peptidoglycan layer. This cell envelope constitutes a highly selective barrier for uptake and release of various compounds. Gram-negative bacteria have evolved several highly specialized secretory pathways to release proteins into their surrounding environment. Among them, the type II secretion pathway is a two-step process dedicated to the secretion of folded and/or oligomeric exoproteins. This ability to secrete large molecules is extremely valuable and is achieved by a sophisticated molecular nano-machine embedded in the bacterial envelope called the secreton.
The type II secretion pathway is conserved in Gram-negative bacteria [1] with prevalence in bacterial pathogens of plants (Pseudomonas fluorescens, Erwinia or Xanthomonas species), animals (Aeromonas hydrophila) and humans (Klebsiella oxytoca, Pseudomonas aeruginosa, Vibrio cholerae or Legionella pneumophila) [2–5]. The number of proteins secreted via the T2SS by any given organism is variable and ranges from one, in the case of K. oxytoca [6], to more than ten in P. aeruginosa [7], V. cholerae [8] or L. pneumophila [9]. The functions of these proteins are extremely diverse and include toxins [10,11], surface-associated virulence factors [12,13], cytochromes [14] and a broad range of enzymes that hydrolyse macromolecules such as lipids, polysaccharides and proteins [15].
(a). Genetic organization
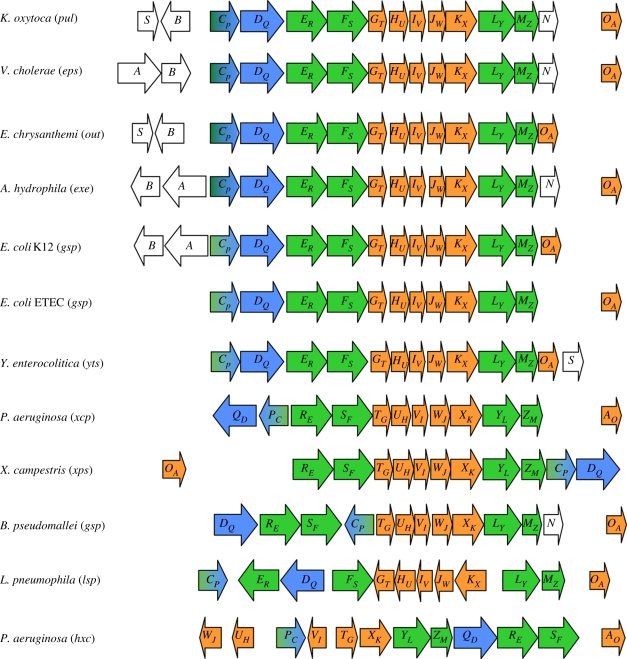
Typical type II secretion systems (T2SSs) are encoded by a set of 12 to 16 gsp (general secretion pathway) genes organized into large operons including the conserved ‘core’ genes denoted gspCP to OA and in some bacterial species extra gsp genes such as gspAB, gspN or gspS (figure 1). Because a different nomenclature is used for Pseudomonas and non-Pseudomonas T2SSs, the alternative gene or protein nomenclature is indicated throughout the review. For example, in GspER the ‘R’ refers to the Pseudomonas XcpR T2SS component, which is reciprocally called XcpRE. Apart from rare exceptions, mutation in any gsp gene prevents secretion and causes accumulation of the exoproteins in the periplasm. The genetic organization of the T2SS clusters is remarkably conserved. However, in some species, the position of the gspCPDQ genes is peculiar (figure 1). In the P. aeruginosa xcp cluster, these two genes form an operon divergent from the operon containing the gspER-MZ genes. In Xanthomonas campestris, the gspCPDQ genes are found after gspMZ at the end of the gsp operon (figure 1). Exceptions are with the T2SS genes in Burkholderia pseudomallei and L. pneumophila, where gspCP and gspDQ are not next to each other. Finally, it should be noted that the whole P. aeruginosa hxc (for homologous to xcp) cluster [16] has a radically different organization of its genes. This T2SS is used by P. aeruginosa for the secretion of a single exoprotein, the alkaline phosphatase LapA, and the hxc genes are expressed in phosphate-limited growth conditions [16]. The specificity of the P. aeruginosa Hxc system versus the more general Xcp pathway is probably not linked with growth in phosphate starvation conditions since phosphate-regulated phospholipases (PlcH, –N and –B) [17,18], or alkaline phosphatase, PhoA [19], are all secreted via the Xcp machinery. Interestingly, Durand et al. [20] recently identified specific Hxc phenotypes suggesting the existence of two T2SS subtypes called T2aSS and T2bSS to which, Xcp and Hxc, respectively, belong. Indeed, the authors propose that the secretion process of the Hxc T2bSS of P. aeruginosa involves a pseudopilus whose structure and stability may differ from the one commonly found in Xcp and other known T2aSSs. A second T2SS called Stt has also recently been identified in Erwinia chrysanthemi (now called Dickeya dadantii) where it involves cell-surface targeting of a non-conventional T2SS substrate, PnlH, possessing a non-cleavable Tat-dependant amino-terminal targeting signal [21].
Figure 1.
Genetic organization of the T2SS clusters. The name of each T2SS gene cluster is shown in brackets beside the name of the bacterial species. Each gene is represented by an arrow and ‘core’ genes present in all T2SS clusters are represented in colour. The gspER, FS, LY and MZ genes encoding components of the inner membrane platform are shown in green; the gspGT, HU, IV, JW and KX genes encoding pseudopilins and gspOA gene encoding the prepilin peptidase are shown in orange; the gspDQ gene encoding the secretin is shown in blue; the gspCP gene encoding the trans-periplasmic protein is represented in shaded green and blue tones because GspCP is a component of the inner membrane surface interacting with secretin. The gspA, B, N and S genes that are not considered to be core components of the T2SS are represented in white.
(b). The type II secretion pathway
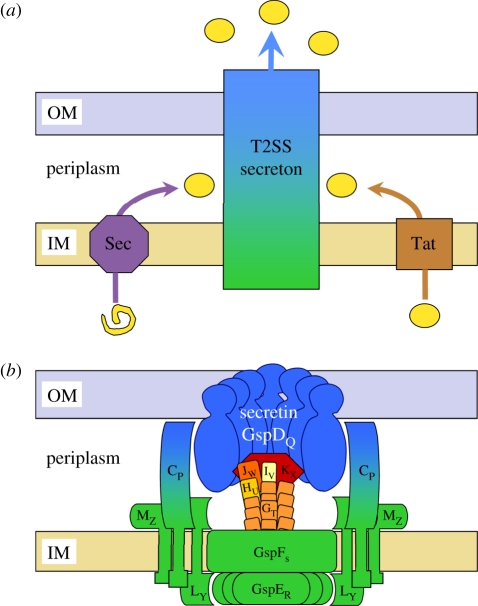
Exoproteins that use the T2SS are secreted into the extracellular medium by a two-step process in which the proteins are exported across the cytoplasmic membrane and released into the periplasm before being transported across the outer membrane (OM) (figure 2a). Exoproteins requiring cytoplasmic folding are exported through the inner membrane (IM) by the Tat export pathway, while translocation of unfolded protein precursors through the IM goes via the Sec export system [5,17]. In a second step, the folded exoproteins, transitorily localized in the periplasm, are translocated across the OM in a T2SS-dependent manner, thus involving the trans-envelope supramolecular complex, called the secreton, made of the different Gsp proteins.
Figure 2.
Model of the type II secretion pathway in Gram-negative bacteria. (a) The T2SS-dependent exoproteins, shown as yellow circles, are first exported across the IM via the Sec (purple) or Tat (brown) machineries. The exoproteins are subsequently recognized and transported across the OM by the secreton (blue/green). (b) Schematic of the secreton divided into three sub-complexes. Secretin GspDQ (blue) forms a dodecameric pore in the OM through its C-terminal domain whilst its N-terminal part protrudes in the periplasm. The IM surface, composed of T2SS proteins FS, LY and MZ, and the traffic ATPase GspER are coloured green. Secretin is connected to the IM surface through the transperiplasmic protein GspCP (blue/green). The pseudopilus, mostly constituted by the GspGT major pseudopilin and capped by the minor pseudopilins GspHU, IV, JW and KX quaternary complex, is shown in orange and red. Gsp proteins are indicated by their corresponding letter.
(c). Structural organization of the secreton
Based on data obtained by many different experimental approaches, including subcellular localization, protein–protein interactions between individual components of the T2SS and resolution of protein structure, the current model for the secreton is represented by three functional sub-complexes (figure 2b). An inner membrane platform (IMP) (figure 2b, green) is composed of the GspCP, FS, LY and MZ IM proteins; the cytoplasmic traffic ATPase GspER is associated with this through an interaction with the bitopic protein GspLY [22–25]. The secreton also contains five proteins that display homologies with the type IV pilin PilA and are designated pseudopilins [26–28]. These proteins have been proposed to be involved in the formation of a fibrillar piston-like structure, the pseudopilus (figure 2b, orange/red) [29–33]. Finally, GspDQ, the OM component of the system, belongs to the secretin family and likely constitutes the channel giving T2SS substrates access to the extracellular medium (figure 2b, blue) [34,35]. Whereas the proton motive force has been shown to be involved in the translocation of T2SS substrates across the OM [36,37], GspER, which contains motifs characteristic of traffic ATPases, also contributes to energize the T2SS-dependent process [38,39] and could drive the pseudopilus through the GspDQ channel, pushing out exoproteins to the external medium [15,40,41].
(d). Cellular localization of the secreton
In P. aeruginosa the number of assembled secreton machines is thought to be relatively low and has been estimated at 50–100 secretons per cell [34]. Moreover, while results obtained in K. oxytoca and V. cholerae [42–44] with GFP-fused Gsp proteins indicate a circumferential distribution of the machinery into foci, the P. aeruginosa Xcp secreton was proposed to be polar. This was shown by adding a Lumino tag onto XcpR or XcpS or by the visualization of protease secretion with an intramolecularly quenched casein conjugate [45]. Such discrepant results could be due to artefacts related to the artificial production of the reporters used, as clearly demonstrated by Lybarger et al. [42]. Alternatively, it cannot be ruled out that cellular localization of T2SSs might vary from one species to another. Interestingly, further localization experiments of the secreton, which were performed in various gsp backgrounds, indicate that in contrast to other Gsp proteins, secretin does not need other secreton components for correct localization in the bacterial envelope, thus suggesting an assembly of the secreton from the OM [42]. This relatively new concept of molecular machines assembly from their OM secretins was also recently proposed for the type III secretion machinery [46].
In this review, we will summarize what is currently known about the individual organization of the three secreton sub-complexes briefly outlined in this introduction. We will particularly highlight new findings on solved protein structures and protein–protein interactions among and between the three sub-complexes. Finally, we will propose an integrative model for T2SS assembly and mechanism.
2. The outer membrane secretin
Secretins are members of a protein superfamily [47] involved not only in T2SS, but also in type III secretion system [48], type IV pilus assembly [49], DNA uptake and extrusion of the filamentous phage [50]. These proteins form large homo-multimers of 12–15 subunits assembled in the OM [51]. They form a ring-shaped structure with a central cavity 50–80 Å in diameter [47].
In K. oxytoca and E. chrysanthemi, the insertion of the secretin in the OM has been shown to depend on the presence of a small OM lipoprotein, the pilotin GspS [41,52]. This protein has chaperone-like properties since it is involved in the protection of the secretin from proteolytic degradation. PulS is also involved in secretin transport since in its absence, K. oxytoca secretin PulD mislocalizes to the IM [53], indicating that a lipid-anchored chaperone is required for efficient and correct insertion of the secretin into the OM. To date, genes encoding GspS members have not been found in all T2SSs (figure 1). Therefore, it cannot be ruled out that genes with low homologies, and which are not associated with the gsp cluster, could encode proteins playing the same function as GspS [54]. Alternatively, secretin transport is assisted in some species by the non-core components GspAB. Since the X. campestris ExeA directly interacts with the peptidoglycan layer, the complex may contribute to create space in the peptidoglycan mesh to allow the transport and assembly of the megadalton-sized secretin multimer in the OM [55]. Finally, some T2SS secretins do not require any specific assistance for their transport to the OM. This is the case for the liposecretin HxcQ of P. aeruginosa, which is targeted to the OM by its N-terminal lipid anchor [56]. Interestingly, it has been shown that transport to the OM of the T2SS secretin PulD is not dependent on the general Bam OM protein transport pathway [57]. It is therefore possible that secretins use the Lol lipoprotein transport route for their transport to the OM either directly for liposecretin [56] or via their pilotin [58]. Alternatively, it cannot be excluded that some secretins use the Bam pathway as was demonstrated for the Neisseria type IV pilus secretin PilQ [59].
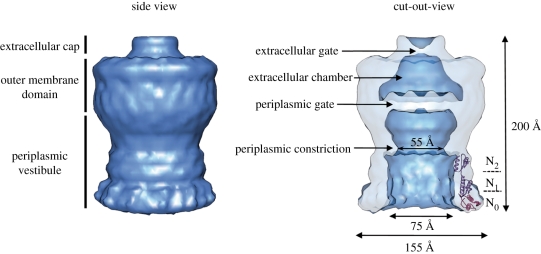
Secretin monomers are bipartite proteins ranging from 50 to 70 kDa in size. Homology among members of the secretin family resides in the C-terminal half of the protein that is important for oligomerization, whereas the N terminus is conserved only within subgroups from related transport pathways and thought to be involved in system-specific interactions [60]. Recently, Reichow et al. [61] have solved the low-resolution structure of the V. cholerae full-length GspDQ (EspDQ) secretin using cryo-electron microscopy (figure 3). The cryo-EM reconstitution of the V. cholerae secretin at 19 Å resolution suggests a dodecameric structure reminiscent of a barrel with a large internal channel containing two compartments separated by a closed periplasmic gate, a periplasmic vestibule and an extracellular chamber located in the OM (figure 3).
Figure 3.
Electron microscopy structure of T2SS secretin. Cryo-EM reconstitution of V. cholerae T2SS secretin GspDQ at 19 Å resolution (EMDB1763 and adapted from [61] by permission from Nature Publishing Group). The GspDQ cryo-EM density reveals a cylindrical channel assembly 155 Å in diameter and 200 Å in length. In side view, three domains are identified from top to bottom: the extracellular cap, the outer-membrane domain and the periplasmic vestibule domain. In a cutout view, secretin contains an extracellular chamber limited by an extracellular gate and a periplasmic gate. The vestibule domain shows a constriction which results in a narrowing of the channel diameter from 75 to 55 Å. The crystal structure of the N-terminal periplasmic subdomains N0–N1–N2 from ETEC [62] is fitted into the GspDQ periplasmic vestibule (adapted from [47] by permission from Elsevier).
The N terminus of T2SS secretins comprises four structurally independent domains, N0–N3 (figure 4). The three-dimensional structure of the N0–N1–N2 region of the enterotoxigenic Escherichia coli (ETEC) GspDQ secretin has been solved at 2.8 Å by X-ray crystallography [62]. This structure has been used to generate a 12-fold symmetrical ring, which was fitted into the density map of the full length V. cholerae EpsDQ obtained by cryo-EM [61] (figure 3). The reconstitution confirmed that the C-terminal and N-terminal regions are two structurally separate domains located in the OM and the periplasm, respectively. Since only low-resolution structures have been obtained for the pore-forming part of the secretin, the precise structural folding of this domain remains unknown. Like the majority of the bacterial OM proteins, secretins are predicted to adopt a β-barrel structure [63]. For example, the topology of the XcpQD secretin together with predictions for β-strands in the primary amino acid sequence has previously been assessed [64]. If this is the case, whether this domain is formed by one large single homomultimeric β-barrel or by the assembly of 12 individual β-barrels remains an open question. Alternatively, it is still a likely possibility that secretin pores do not form β-barrels but adopt alternative α-helical folds. Such an original fold was first identified in the E. coli capsular polysaccharides OM pore Wza [65], but is also required for insertion of the type IV secretion OM protein VirB10 in Agrobacterium tumefaciens [66] or the P. aeruginosa PelC protein [67]. The hypothesis of an α-helical fold in T2SS secretins is supported by the observation that transport of this group of proteins does not involve the Bam complex [57] which is required for insertion of OM proteins forming β-barrels [59,68].
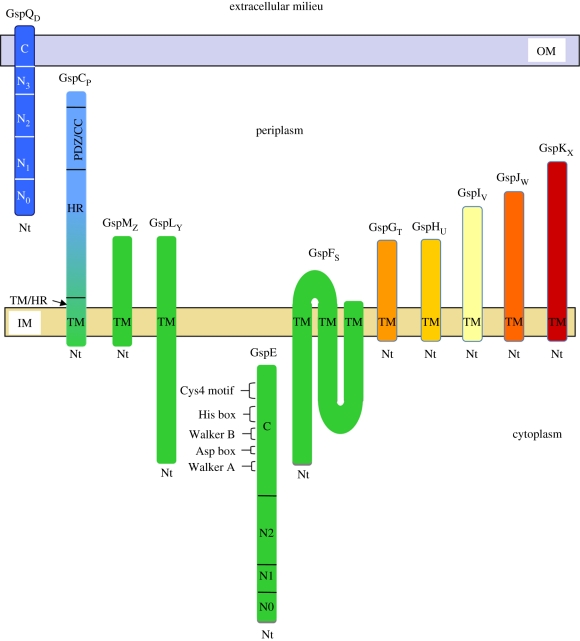
Figure 4.
Topology of the Gsp proteins. Schematic of the topology of each individual component of the secreton. The domains of the Gsp proteins discussed in the review are mentioned. The colour code used is the one generally used in all figures, i.e. blue for OM secretin, green for IMP components and orange/red for pseudopilins. The N terminus (Nt) and the transmembrane (TM) domain of each Gsp protein are indicated where appropriate. CC, coiled coil.
A recent cryo-EM study showed that the T2SS-dependent cholera toxin binds in the lower part of the periplasmic vestibule of the V. cholerae EpsDQ secretin [69]. This observation is in agreement with the previous interaction found between the T2SS secretin OutDQ and its cognate substrate PelB [41] and confirms the role of the secretin in substrate binding. How the substrate further travels through the pore is still unknown but the closed state of the channel suggests that several conformational changes might occur in the secretin core to accommodate the substrate and to trigger communication between the different chambers. In addition, and as we shall see in §3–7, other components of the secreton such as the transperiplasmic protein GspCP and the pseudopilus are also involved in this process since they also interact with the substrate [70].
3. The trans-periplasmic protein GspCP
GspCP proteins are bitopic IM proteins that are the least conserved of the T2SS components. Proteins of this family are organized into several domains including an N-terminal cytoplasmic region, a transmembrane (TM) domain, a highly conserved central periplasmic domain (homology region, HR) and a C-terminal part containing specific secondary structures such as coiled-coil domains (in P. aeruginosa and Pseudomonas alcaligenes, for instance) or PDZ domains (in K. oxytoca and E. chrysanthemi) (figure 4) [71]. GspCP was shown to be active as a dimer and self-associates by its TM domain, which is not a simple membrane anchor but plays an active role in the function of the protein [72]. The HR domain of V. cholerae GspCP (EpsCP) was shown to interact directly with the periplasmic N0 domain of the secretin EpsDQ [44,73]. This interaction was also seen in E. chrysanthemi where the interaction site of OutCP on OutDQ was localized between residues 139 and 158 of the HR domain. This peptide called OutCP-sip `secretin interacting peptide’ recognizes two different sites on the OutDQ secretin. The first one is localized on domain N0, whilst the second one sits astride domains N2 and N3 [74]. Furthermore, mapping of the interaction between XcpPC and XcpQD in P. aeruginosa showed that the N3 domain is essential for this interaction [70]. These different interaction sites between secretins and GspCs suggest that GspCP/DQ partners have evolved various strategies to interact with each other.
Several genetic data indicate that GspCP and GspDQ form a functional couple determining the specificity of the machinery. For example, in the very closely related E. chrysanthemi and E. carotovora Out systems, all genes are individually exchangeable except for outCP and outDQ [75]. Similar investigations performed comparing P. aeruginosa and P. alcaligenes also indicate that XcpPC and XcpQD are determinants of substrate specificity [76]. In order to localize the GspCP domain directly or indirectly involved in substrate specificity, chimeras between E. chrysanthemi OutCP and P. aeruginosa XcpPC domains have been generated and their ability to support secretion in P. aeruginosa tested [71]. Interestingly, XcpPC chimeras containing the TM, HR or C-terminal domain of OutCP remain functional, indicating that none of these domains play a role in substrate specificity. However, the replacement of the intermediary domain between TM and HR called TM/HR (figure 4) leads to secretion defect, suggesting that this domain plays an essential role in specificity and potentially in substrate recognition. Recently, a set of in vitro experiments has revealed direct interactions between purified XcpPC and two substrates of the Xcp T2SS, the elastase (LasB) and the lipase (LipA) [70]. Importantly, no interaction was detected using the substrate of the second P. aeruginosa T2SS (Hxc), i.e. the alkaline phosphatase (LapA). These observations revealed that the species-specificity of the T2SS mechanism is largely contributed by the exoproteins and involves GspCP and GspDQ, which directly interact with cognate substrates.
4. The ATPase of the system: GspER
A functional T2SS requires the presence of a traffic ATPase, GspER. Traffic ATPases are involved in several other transport machines such as type IV secretion, conjugation and type IV piliation systems. Structural analysis on the type IV secretion and type IV piliation ATPases indicated that they may function as dynamic hexamers [77,78]. The members of the traffic ATPase superfamily are characterized by two nucleotide-binding motifs designated Walker A and B boxes and also His and Asp boxes (figure 4) [79]. GspER proteins have a characteristic Walker box A containing the P-loop of an NTP-binding motif, and a less well-defined Walker B box in which the second conserved aspartate residue is replaced by either a glycine or an alanine. Mutation of a conserved glycine residue within the Walker A motif of GspER from P. aeruginosa, K. oxytoca, E. chrysanthemi or V. cholerae causes the bacteria to be secretion-defective, showing the important role played by this protein in the secretory process [80–83]. Mutations in the less conserved Walker B box have little or no effect on the secretion process [83]. The T2SS traffic ATPase family is distinct from other ATPases in three additional conserved regions: first the aspartate box ‘Asp Box’ between the Walker A and B boxes consisting of two short aspartate-rich motifs (figure 4), which is required for the function of GspER in the secretion process and may be involved in the formation and stabilization of the nucleotide-binding fold by interacting with Mg2+[83]; second, the His box, including two histidine residues, which is located downstream of the Walker B box, although the role of the His box in GspER function is still unknown; last, a tetracysteine (Cys4) motif that appears to be essential for function, since replacement of any of the cysteine residues by a serine within the K. oxytoca GspER leads to a large decrease in pullulanase secretion (figure 4) [15].
GspER traffic ATPases lack hydrophobic domains and exhibit the general characteristics of a cytoplasmic protein (figure 4). However, they were found to be associated with the IM through an interaction with the bitopic protein GspLY [80,84]. Results obtained from V. cholerae indicate that ATP hydrolysis by the EpsER/EpsLY complex is stimulated by acidic phospholipids, whereas the activity of EpsER alone is unaffected [85]. Further mutagenesis revealed that the membrane-proximal region of the cytoplasmic domain of EpsLY subtly controls the interaction of EpsER with the cytoplasmic membrane and influences its oligomerization, thereby stimulating its ATPase activity [85]. Other results from X. campestris have shown that XpsER oligomerization, as well as its association with XpsLY, requires ATP binding but not ATP hydrolysis, thus indicating that association between XpsER and XpsLY is needed for ATPase activity [86].
The crystal structure of a truncated V. cholerae EpsER protein lacking the N-terminal 90 residues was determined with or without the nucleotide bound [87]. These structures reveal a two-domain architecture with the five characteristic motifs of the GspER subfamily clustering around the nucleotide-binding site in the C-terminal domain. The EpsER subunits form a right-handed helical arrangement in the crystal with extensive and conserved contacts between the C and N domains of neighbouring subunits, thus suggesting that EpsER is organized as a hexameric structure. The hexameric state of GspER is confirmed by results obtained in V. cholerae and X. campestris showing that optimal ATPase activity is obtained with hexameric GspER [38,86]. The crystal structure of the N-terminal part of V. cholerae EpsER in complex with the cytoplasmic domain of V. cholerae EpsLY showed that these two proteins form a hetero-tetramer in which EpsLY forms a central dimer and EpsER binds at the periphery [88].
Amino acid sequence alignments have shown that XpsER of X. campestris contains an additional N-terminal extension not found in most other GspERs. This additional domain appears to be essential for XpsLY binding, therefore indicating that a more sophisticated interaction process between GspER and GspLY might occur within the Xps secreton of X. campestris [89]. To date, the structure of a full-length GspER has not been reported and this would provide key and definite structural information about the architecture of GspER monomer and multimer.
5. GspFS, LY and MZ: the IMP stabilizers
GspFS is a polytopic integral membrane protein with a small periplasmic loop and two large cytoplasmic domains connected by three TM regions (figure 4) [90,91]. Two-hybrid studies have shown that the N-terminal domain of the E. chrysanthemi OutFS protein interacts both with OutER and OutLY [24] suggesting that OutFS could participate in the stability of the IMP [92]. Construction of a chimera between P. aeruginosa and P. putida XcpSF has shown that interaction with other T2SS components is mediated by the cytoplasmic domains [25].
GspLY is a bitopic IM protein organized in three domains, the C-terminal domain localized in the cytoplasmic compartment, the TM domain, and the periplasmic domain (figure 4) [22]. The structures of both cytoplasmic and periplasmic domains of EpsL in V. cholerae have been solved at 2.7 and 2.3 Å, respectively [93,94]. The cytoplasmic part is composed of subdomains I, II and III and was shown to interact with the N-terminal part of GspER through subdomains II and III [88].
GspMZ is a bitopic protein [22] with a short cytoplasmic domain, a TM domain and a periplasmic domain (figure 4) involved in homo-dimerization [24,95,96]. GspMZ was shown to be required for GspLY stability since the amount of the former is greatly dependent on the presence of the latter [23]. Studies on GspMZ variants in P. aeruginosa revealed that three periplasmic domains of the protein were found to be important for interaction with GspLY. Two distinct stabilizing domains were localized, respectively, at the beginning and at the end of the periplasmic part of the protein whereas the third one, localized next to the TM domain, also required the presence of the transperiplasmic protein GspCP to promote GspLY stabilization [97]. The influence of GspCP on the stability of the GspLY/GspMZ complex was also observed in X. campestris since GspLY dissociates faster from the GspLY/GspMZ complex than from the GspCP/LY/MZ one [98]. In addition, antibodies against GspMZ co-immunoprecipitated GspLY, GspCP, and GspER from detergent-solubilized cell extracts, confirming the existence of a complex containing these four proteins [99].
6. The pseudopilus: a central structure of the T2SS machine
Six of the 12 conserved gsp genes are dedicated to the formation of a periplasmic pilus-like structure called the pseudopilus (figure 1). Five of those genes, gspGT-KX encode the pseudopilins which are the constitutive elements of the pseudopilus, whereas gspOA encodes the prepilin peptidase involved in their maturation [26–28,100,101]. Like the closely related type IV pilins involved in type IV pilus formation, the five pseudopilins are synthesized as precursors with a short leader peptide of 6–7 mostly charged residues that is cleaved off by the prepilin peptidase PilD/GspOA. Mature pilins and pseudopilins are characterized by a highly conserved N-terminal hydrophobic domain of about 20 residues followed by a C-terminal extension specific for each pilin and pseudopilin [15]. Topology studies have shown that pilins and pseudopilins are bitopic IM proteins with a single N-terminal trans-membrane domain segment and a periplasmic C-terminal globular domain (figure 4) [27,101]. Similar to typical IM proteins, pseudopilins use the Sec/SRP pathway for their membrane targeting and insertion [90,102]. While it is an essential step, the significance and role of pseudopilin maturation by the peptidase is unknown. Nevertheless, the removal of cytoplasmic positive charges may facilitate extraction of the protein from the membrane. Interestingly, it was shown that pseudopilins co-fractionate with both IM and OM fractions [28,103], suggesting either a re-localization of these proteins to the OM after processing or more likely the formation of a supramolecular complex.
Among the five pseudopilins, GspGT is the most abundant and is therefore called the major pseudopilin [27], in contrast to GspHU, IV, JW and KX, which are named minor pseudopilins. Biochemical data obtained in X. campestris revealed the presence of the major pseudopilin, XpsGT, within a large complex of about 440 kDa [103]. This observation clearly favoured the formation of a pilus-like structure spanning the bacterial envelope. In agreement with this hypothesis, it was shown that when GspGT is overproduced it is able to assemble into an unusually long fibrillar structure protruding out of the cell, which closely resembles the type IV pilus [32,33,104,105]. Such a structure, also called a hyper pseudopilus (HPP), is only obtained upon overproduction of GspGT pseudopilins and probably represents an uncontrolled elongation of what could be a physiologically relevant pseudopilus. Based on crystallographic and electron microscopy data, an assembly model of GspGT into HPP has been generated by a molecular modelling approach. This pseudo-atomic model was experimentally validated and showed how protomers of GspGT interact with each other to form the pseudopilus [31,106,107]. The model proposes a right-handed helical organization of the T2SS pseudopilus, consistent with the type IV pilus structure [108].
The four minor pseudopilins GspHU–KX are unable to assemble into an HPP when overproduced [105], but their role in pseudopilus formation is undeniable. Results obtained with K. oxytoca or P. aeruginosa pseudopilins have shown that GspIV is essential for HPP formation and could play a major role during the initiation step of fibre formation [33,105]. It was also proposed that HPP elongation is controlled by GspKX [101] since its length varied depending on the number of GspKX subunits produced [104,105]. While none of the four minor pseudopilins have ever been detected in HPP structures [32,33,104], their presence in a native pseudopilus became clear in light of identified protein–protein interactions and resolved three-dimensional structures. The four minor pseudopilins share the typical αβ-fold commonly found in major pilins and pseudopilins with a long N-terminal α-helix involved in their helical oligomerization [29,107,109–118]. Three of the four minor pseudopilins (GspIV, JW and KX) have been co-crystallized and shown to form a ternary complex through their globular domains [29]. Recently, a quaternary complex containing the globular domains of the four minor pseudopilins was identified, indicating that the fourth minor pseudopilin, GspHU, also belongs to the minor pseudoplin complex [30]. Structural prediction using the entire amino acid sequence of the minor pseudopilins, i.e. including also the N-terminal hydrophobic domain, suggests a similar helical assembly as the one observed for GspGT. Since extra major pseudopilins could only be added beneath the minor pseudopilins complex [29], it was proposed that the tetrameric complex was localized at the tip of the GspGT pseudopilus. Given that GspHU was previously shown to interact with the major pseudopilin GspGT [103,119], GspHU could constitute the hinge between the tip and the core of the pseudopilus [30]. Taking into account the resemblance to the type IV piliation system, the phenotypic, structural and interaction network data lead to a reasonable model for pseudopilus architecture and biogenesis which consists of the prior assembly of the quaternary tip complex in the IM. The tip complex could then be driven to the secretin by the addition of major pseudopilin subunits underneath. Given that GspKX has a large globular domain and is located at the very end of the pseudopilus, its involvement in pseudopilus length control could consist of stopping pseudopilus growth when contacting the secretin. Therefore, when the physiological stoichiometry between GspKX and GspGT is respected, the length of the pseudopilus is likely to be restricted to the periplasmic space. Only when a large excess of GspGT is produced, may the pseudopilus grow without a tip and therefore pass through the secretin channel. This model, which requires experimental validation, is nevertheless supported by the direct interaction observed between minor pseudopilins and secretin [61,96].
The pseudopilus assembly likely occurs as in the type IV piliation process with the energetic assistance of the GspER and PilB traffic ATPases, respectively. If the retraction process exists in type II secretion, it could be associated with a piston-like mechanism of the pseudopilus, but probably involves a different mechanism since there is no counterpart in the T2SS for the PilT ATPase that disassembles type IV pili [120,121]. Interestingly, the minor pseudopilin GspKX was found to interact with GspGT, and this interaction triggers a destabilization of GspGT [105]. An alternative retraction process can therefore be proposed for the T2SS pseudopilus, i.e. upon contact with the secretin pore, GspKX acquires, possibly upon conformational changes, the capacity to interact with GspGT, thus leading to pseudopilus collapse. An ATPase-free retraction event might thus be sufficient to support the disassembly of a short trans-periplasmic pseudopilus.
7. Substrate recognition and transport by the T2SS machine
The type II secretion apparatus is widespread in Gram-negative bacteria and a wide variety of enzymes and toxins use this pathway. We have already alluded to the species-specificity of this system; i.e. cognate exoproteins from one T2SS are not recognized by another machinery [75]. T2SS substrates are loaded on the nanomachine in the periplasm and translocated across the OM in a folded conformation [122–124]. Moreover, studies in E. chrysanthemi, K. oxytoca and P. aeruginosa have demonstrated that disulphide bridges are formed within exoproteins before secretion [125–128]. The high specificity demonstrated for T2SSs and their substrates, as well as their specific recognition in the periplasm among all other resident proteins, suggests the existence on folded substrates of a secretion motif that is required for T2SS recognition. Many studies have been carried out on T2SS-dependent exoenzymes in order to define this secretion motif, which is still a biological puzzle [129]. They all converge to the idea of a conformational signal gathering several motifs spread along the primary amino acid sequence of the protein. With the K. oxytoca pullulanase PulA, it was shown that two non-adjacent regions were together necessary to promote translocation of PulA-β-lactamase hybrid proteins across the OM [130]. Another study suggested that at least three regions of PulA might contain information that influences its secretion [131]. It was also suggested that P. aeruginosa exotoxin A (ToxA) contains two separate secretion signals [132], while alteration of another region also affects secretion efficiency [133]. Finally, the polygalacturonase PehA of E. carotovora was found to contain three separate domains involved in T2SS targeting [134,135]. As reported above, the secretion signal may be composed of residues from different locations in the linear polypeptide chain, which are brought together into a conformational patch during protein folding [136]. One alternative to a single structural motif is that successive specific interactions lead to the secretion of exoproteins. These interactions may involve different secretion signals that are not essential individually but are required simultaneously, or sequentially, for optimal secretion. Interestingly, secretion of the E. chrysanthemi cellulase Cel5 involves a transitory intramolecular interaction between the cellulase binding domain and a region close to the active site [137]. In this case, the exoprotein could adopt a secretion-competent conformation prior to secretion and another conformation once released in the extracellular medium [138]. This sequence of events is probably not the case with P. aeruginosa and V. cholerae T2SSs for which interactions found between substrates and secreton components were detected using purified secreted proteins [41,61,70]. Obviously the question of specific substrate recognition is far from being resolved. One may consider that co-evolution of T2SS machines together with their cognate substrate has resulted in a progressive adaptation to obtain an optimal fit.
Recently, several direct interactions have been identified between secreted substrates and periplasmic domains of secreton components. The previously identified interaction with the secretin [41] was confirmed by two independent studies which both used a highly sensitive technique, surface plasmon resonance or BIAcore [61,70]. By using this technique, Douzi et al. [70] have explored the P. aeruginosa T2SS periplasmic interaction network and, in addition to the secretin, two other substrate interactants, the transperiplasmic protein XcpPC and the pseudopilus tip. This set of interactions may suggest that the transperiplasmic element XcpPC might recruit the substrate and transfer it to the pseudopilus tip which then carries it towards the secretin through which it could be translocated (figure 5).
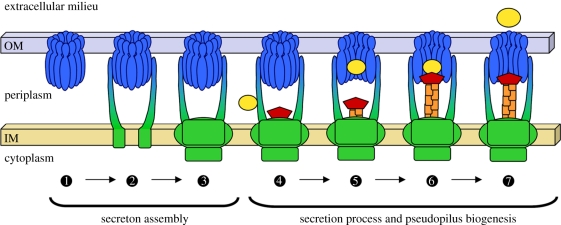
Figure 5.
Model of secreton assembly and operation. Schematic of secreton biogenesis as discussed in this review. The first part of the secreton to be assembled is proposed to be the secretin  . The next step could be the successive recruitment of the trans-periplasmic protein GspCP
. The next step could be the successive recruitment of the trans-periplasmic protein GspCP
 and of the inner membrane surface
and of the inner membrane surface  . The recognition of the substrate by the T2SS takes place in the periplasm and may involve a peripheral element of the secreton, GspCP
. The recognition of the substrate by the T2SS takes place in the periplasm and may involve a peripheral element of the secreton, GspCP
 . The substrate is then transferred to the secretin vestibule
. The substrate is then transferred to the secretin vestibule  in which it could contact the pseudopilus tip complex that is emerging from the inner membrane surface
in which it could contact the pseudopilus tip complex that is emerging from the inner membrane surface  . The exoprotein could then be released in the extracellular medium through the secretin pore
. The exoprotein could then be released in the extracellular medium through the secretin pore  . The secretin and periplasmic domain of GspCP are shown in blue, the components of the inner membrane surface are shown in green, the pseudopilus and the secreted proteins are shown in orange/red and yellow, respectively.
. The secretin and periplasmic domain of GspCP are shown in blue, the components of the inner membrane surface are shown in green, the pseudopilus and the secreted proteins are shown in orange/red and yellow, respectively.
8. A model for secreton assembly and mode of action
Type II protein secretion occurs in two steps. Secreted proteins are first exported across the IM and then released in the extracellular medium thanks to a sophisticated machine, the secreton. The secreton is composed of at least 12 different proteins embedded in the bacterial envelope and organized in a large multi-protein complex capable of secreting a wide range of folded exoproteins across the OM of Gram-negative bacteria. Based on structural data, protein–protein interactions, and phenotypic observations described in this review, it is possible to propose an innovative model for secreton biogenesis and functioning (figure 5). In this model, secreton biogenesis starts by the insertion of secretin in the OM, thus defining the secretion site [42]. In the second step, the transperiplasmic element GspCP binds the secretin, therefore allowing docking of IMP. Indeed, cellular localization experiments performed in V. cholerae have shown that, in contrast to secretin, GspCP needs GspDQ but not GspMZ which itself needs both GspCP and GspDQ for proper localization [42]. We propose that the transitory periplasmic T2SS substrates are first recruited by the peripheral element GspCP and then transferred to the secretin vestibule. The substrate could then be contacted by the pseudopilus tip, and could be pushed and expelled from the cell through the secretin by a growing pseudopilus. This tentative model is in good agreement with the majority of the data collected so far about the T2SSs, but as any working model, it should be challenged and used to design experimental approaches that will confirm or disprove the views presented in this review. For example: (i) Is there a dedicated location for the T2SS in the cell envelope? If yes, how is the first element of the system, say the secretin, targeted there? (ii) Even if we are getting closer to understand substrate recognition by the machinery, the identity of the secretion signal remains enigmatic. (iii) Does the binding of the substrate to XcpPC trigger the assembly of the whole system as previously shown for the type I secretion system [139] or does it only trigger pseudopilus elongation? (iv) Does the pseudopilus elongate upon contact of the substrate on GspCP or later when it is positioned in the secretin vestibule? (v) How is the substrate released from the secretin? Is it through a mechanical movement operated by the pseudopilus or following conformational changes within the secretin or both? (vi) Does the pseudopilus effectively retract? If yes, what are the molecular mechanism and energy source associated with this event?
Whereas all these questions remained to be addressed, it is remarkable to see the improvement of our understanding of the T2SS over the past few years. This is largely due to the ever increasing performance of structural and biochemical techniques that have generated a lot of new data and come to complement and back up all the original genetic data. Ideally, one would like to see the three-dimensional reconstruction of the whole T2SS machine and even better have this mega-structure in motion while transporting the exoprotein. Such level of achievement might not be so far away considering the advances that are currently being made in understanding motion in proteins using molecular dynamics computer simulations [31,140].
Acknowledgements
We should like to thank M. Tegoni, C. Cambillau, C. Bernard, S. Alphonse, K. T. Forest, L. Franz and D. Dyer for fruitful collaborations, as well as E. Durand, G. Ball, B. Ize, S. Bleves and G. Michel for their contribution to the research undertaken in our laboratory on type II secretion. Research on the bacterial type II secretion system in the laboratory of Romé Voulhoux is supported by the Centre National de la Recherche Scientifique (CNRS) and the ‘3D-Pilus’ young researcher ANR grant (ANR-JC 07-183230). Badreddine Douzi is supported by a BDI-PED PhD grant. Alain Filloux is supported by the Royal Society.
References
- 1.Filloux A., Bally M., Ball G., Akrim M., Tommassen J., Lazdunski A. 1990. Protein secretion in Gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 9, 4323–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filloux A., Michel G., Bally M. 1998. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22, 177–198 10.1111/j.1574-6976.1998.tb00366.x (doi:10.1111/j.1574-6976.1998.tb00366.x) [DOI] [PubMed] [Google Scholar]
- 3.Peabody C. R., Chung Y. J., Yen M. R., Vidal-Ingigliardi D., Pugsley A. P., Saier M. H., Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072 10.1099/mic.0.26364-0 (doi:10.1099/mic.0.26364-0) [DOI] [PubMed] [Google Scholar]
- 4.Cianciotto N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588 10.1016/j.tim.2005.09.005 (doi:10.1016/j.tim.2005.09.005) [DOI] [PubMed] [Google Scholar]
- 5.Pugsley A. P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57, 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.d'Enfert C., Ryter A., Pugsley A. P. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6, 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. 2010. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543 10.1016/j.ijmm.2010.08.005 (doi:10.1016/j.ijmm.2010.08.005) [DOI] [PubMed] [Google Scholar]
- 8.Sikora A. E., Zielke R. A., Lawrence D. A., Andrews P. C., Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J. Biol. Chem. 286, 16 555–16 566 10.1074/jbc.M110.211078 (doi:10.1074/jbc.M110.211078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cianciotto N. P. 2009. Many substrates and functions of type II secretion: lessons learned from Legionella pneumophila. Future Microbiol. 4, 797–805 10.2217/fmb.09.53 (doi:10.2217/fmb.09.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandkvist M., Michel L. O., Hough L. P., Morales V. M., Bagdasarian M., Koomey M., DiRita V. J. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179, 6994–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H. M., Mizushima S., Lory S. 1993. A periplasmic intermediate in the extracellular secretion pathway of Pseudomonas aeruginosa exotoxin A. J. Bacteriol. 175, 7463–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zalewska-Piatek B., Bury K., Piatek R., Bruzdziak P., Kur J. 2008. Type II secretory pathway for surface secretion of DraD invasin from the uropathogenic Escherichia coli Dr+ strain. J. Bacteriol. 190, 5044–5056 10.1128/JB.00224-08 (doi:10.1128/JB.00224-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horstman A. L., Kuehn M. J. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277, 32 538–32 545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L., et al. 2008. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 190, 5512–5516 10.1128/JB.00514-08 (doi:10.1128/JB.00514-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filloux A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta. 1694, 163–179 10.1016/j.bbamcr.2004.05.003 (doi:10.1016/j.bbamcr.2004.05.003) [DOI] [PubMed] [Google Scholar]
- 16.Ball G., Durand E., Lazdunski A., Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43, 475–485 10.1046/j.1365-2958.2002.02759.x (doi:10.1046/j.1365-2958.2002.02759.x) [DOI] [PubMed] [Google Scholar]
- 17.Voulhoux R., Ball G., Ize B., Vasil M. L., Lazdunski A., Wu L. F., Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20, 6735–6741 10.1093/emboj/20.23.6735 (doi:10.1093/emboj/20.23.6735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker A. P., Vasil A. I., Filloux A., Ball G., Wilderman P. J., Vasil M. L. 2004. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol. Microbiol. 53, 1089–1098 10.1111/j.1365-2958.2004.04189.x (doi:10.1111/j.1365-2958.2004.04189.x) [DOI] [PubMed] [Google Scholar]
- 19.Filloux A., Bally M., Soscia C., Murgier M., Lazdunski A. 1988. Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol. Gen. Genet. 212, 510–513 10.1007/BF00330857 (doi:10.1007/BF00330857) [DOI] [PubMed] [Google Scholar]
- 20.Durand E., Alphonse S., Brochier-Armanet C., Ball G., Douzi B., Filloux A., Bernard C., Voulhoux R. 2011. The assembly mode of the pseudopilus: a hallmark to distinguish a novel secretion system subtype. J. Biol. Chem. 286, 24 407–24 416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrandez Y., Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol. Microbiol. 69, 1349–1357 10.1111/j.1365-2958.2008.06366.x (doi:10.1111/j.1365-2958.2008.06366.x) [DOI] [PubMed] [Google Scholar]
- 22.Bleves S., Lazdunski A., Filloux A. 1996. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J. Bacteriol. 178, 4297–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel G., Bleves S., Ball G., Lazdunski A., Filloux A. 1998. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology 144, 3379–3386 10.1099/00221287-144-12-3379 (doi:10.1099/00221287-144-12-3379) [DOI] [PubMed] [Google Scholar]
- 24.Py B., Loiseau L., Barras F. 2001. An inner membrane platform in the type II secretion machinery of gram-negative bacteria. EMBO Rep. 2, 244–248 10.1093/embo-reports/kve042 (doi:10.1093/embo-reports/kve042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arts J., de Groot A., Ball G., Durand E., El Khattabi M., Filloux A., Tommassen J., Koster M. 2007. Interaction domains in the Pseudomonas aeruginosa type II secretory apparatus component XcpS (GspF). Microbiology 153, 1582–1592 10.1099/mic.0.2006/002840-0 (doi:10.1099/mic.0.2006/002840-0) [DOI] [PubMed] [Google Scholar]
- 26.Nunn D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9, 402–408 10.1016/S0962-8924(99)01634-7 (doi:10.1016/S0962-8924(99)01634-7) [DOI] [PubMed] [Google Scholar]
- 27.Nunn D. N., Lory S. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175, 4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bally M., Filloux A., Akrim M., Ball G., Lazdunski A., Tommassen J. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6, 1121–1131 10.1111/j.1365-2958.1992.tb01550.x (doi:10.1111/j.1365-2958.1992.tb01550.x) [DOI] [PubMed] [Google Scholar]
- 29.Korotkov K. V., Hol W. G. 2008. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 15, 462–468 10.1038/nsmb.1426 (doi:10.1038/nsmb.1426) [DOI] [PubMed] [Google Scholar]
- 30.Douzi B., Durand E., Bernard C., Alphonse S., Cambillau C., Filloux A., Tegoni M., Voulhoux R. 2009. The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J. Biol. Chem. 284, 34 580–34 589 10.1074/jbc.M109.042366 (doi:10.1074/jbc.M109.042366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos M., Nilges M., Cisneros D. A., Francetic O. 2010. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc. Natl Acad. Sci. USA 107, 13 081–13 086 10.1073/pnas.1001703107 (doi:10.1073/pnas.1001703107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., Filloux A. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758 10.1128/JB.185.9.2749-2758.2003 (doi:10.1128/JB.185.9.2749-2758.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvonnet N., Vignon G., Pugsley A. P., Gounon P. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19, 2221–2228 10.1093/emboj/19.10.2221 (doi:10.1093/emboj/19.10.2221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brok R., Van Gelder P., Winterhalter M., Ziese U., Koster A. J., de Cock H., Koster M., Tommassen J., Bitter W. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294, 1169–1179 10.1006/jmbi.1999.3340 (doi:10.1006/jmbi.1999.3340) [DOI] [PubMed] [Google Scholar]
- 35.Nouwen N., Ranson N., Saibil H., Wolpensinger B., Engel A., Ghazi A., Pugsley A. P. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl Acad. Sci. USA 96, 8173–8177 10.1073/pnas.96.14.8173 (doi:10.1073/pnas.96.14.8173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Possot O. M., Letellier L., Pugsley A. P. 1997. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol. Microbiol. 24, 457–464 10.1046/j.1365-2958.1997.3451726.x (doi:10.1046/j.1365-2958.1997.3451726.x) [DOI] [PubMed] [Google Scholar]
- 37.Letellier L., Howard S. P., Buckley J. T. 1997. Studies on the energetics of proaerolysin secretion across the outer membrane of Aeromonas species. Evidence for a requirement for both the protonmotive force and ATP. J. Biol. Chem. 272, 11 109–11 113 [DOI] [PubMed] [Google Scholar]
- 38.Camberg J. L., Sandkvist M. 2005. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J. Bacteriol. 187, 249–256 10.1128/JB.187.1.249-256.2005 (doi:10.1128/JB.187.1.249-256.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patrick M., Korotkov K. V., Hol W. G., Sandkvist M. 2011. Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity. J. Biol. Chem. 286, 10 378–10 386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobbs M., Mattick J. S. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 10.1111/j.1365-2958.1993.tb01949.x (doi:10.1111/j.1365-2958.1993.tb01949.x) [DOI] [PubMed] [Google Scholar]
- 41.Shevchik V. E., Robert-Baudouy J., Condemine G. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16, 3007–3016 10.1093/emboj/16.11.3007 (doi:10.1093/emboj/16.11.3007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lybarger S. R., Johnson T. L., Gray M. D., Sikora A. E., Sandkvist M. 2009. Docking and assembly of the type II secretion complex of Vibrio cholerae. J. Bacteriol. 191, 3149–3161 10.1128/JB.01701-08 (doi:10.1128/JB.01701-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buddelmeijer N., Francetic O., Pugsley A. P. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secreton components PulL and PulM. J. Bacteriol. 188, 2928–2935 10.1128/JB.188.8.2928-2935.2006 (doi:10.1128/JB.188.8.2928-2935.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korotkov K. V., et al. 2011. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog. 7, e1002228. 10.1371/journal.ppat.1002228 (doi:10.1371/journal.ppat.1002228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senf F., Tommassen J., Koster M. 2008. Polar secretion of proteins via the Xcp type II secretion system in Pseudomonas aeruginosa. Microbiology 154, 3025–3032 10.1099/mic.0.2008/018069-0 (doi:10.1099/mic.0.2008/018069-0) [DOI] [PubMed] [Google Scholar]
- 46.Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., Cornelis G. R. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 10.1038/emboj.2010.84 (doi:10.1038/emboj.2010.84) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korotkov K. V., Gonen T., Hol W. G. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36, 433–443 10.1016/j.tibs.2011.04.002 (doi:10.1016/j.tibs.2011.04.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koster M., Bitter W., de Cock H., Allaoui A., Cornelis G. R., Tommassen J. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26, 789–797 10.1046/j.1365-2958.1997.6141981.x (doi:10.1046/j.1365-2958.1997.6141981.x) [DOI] [PubMed] [Google Scholar]
- 49.Collins R. F., Davidsen L., Derrick J. P., Ford R. C., Tonjum T. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183, 3825–3832 10.1128/JB.183.13.3825-3832.2001 (doi:10.1128/JB.183.13.3825-3832.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linderoth N. A., Model P., Russel M. 1996. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J. Bacteriol. 178, 1962–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kazmierczak B. I., Mielke D. L., Russel M., Model P. 1994. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 238, 187–198 10.1006/jmbi.1994.1280 (doi:10.1006/jmbi.1994.1280) [DOI] [PubMed] [Google Scholar]
- 52.Hardie K. R., Seydel A., Guilvout I., Pugsley A. P. 1996. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22, 967–976 10.1046/j.1365-2958.1996.01539.x (doi:10.1046/j.1365-2958.1996.01539.x) [DOI] [PubMed] [Google Scholar]
- 53.Guilvout I., Chami M., Engel A., Pugsley A. P., Bayan N. 2006. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 25, 5241–5249 10.1038/sj.emboj.7601402 (doi:10.1038/sj.emboj.7601402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo J., Brencic A., Darwin A. J. 2009. Analysis of secretin-induced stress in Pseudomonas aeruginosa suggests prevention rather than response and identifies a novel protein involved in secretin function. J. Bacteriol. 191, 898–908 10.1128/JB.01443-08 (doi:10.1128/JB.01443-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strozen T. G., Stanley H., Gu Y., Boyd J., Bagdasarian M., Sandkvist M., Howard S. P. 2011. Involvement of the GspAB complex in assembly of the type II secretion system secretin of Aeromonas and Vibrio species. J. Bacteriol. 193, 2322–2331 10.1128/JB.01413-10 (doi:10.1128/JB.01413-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viarre V., Cascales E., Ball G., Michel G. P., Filloux A., Voulhoux R. 2009. HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J. Biol. Chem. 284, 33 815–33 823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collin S., Guilvout I., Chami M., Pugsley A. P. 2007. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol. Microbiol. 64, 1350–1357 10.1111/j.1365-2958.2007.05743.x (doi:10.1111/j.1365-2958.2007.05743.x) [DOI] [PubMed] [Google Scholar]
- 58.Nickerson N. N., Tosi T., Dessen A., Baron B., Raynal B., England P., Pugsley A. P. 2011. Outer membrane targeting of secretin PulD protein relies on disordered domain recognition by a dedicated chaperone. J. Biol. Chem. 286, 38 833–38 843 10.1074/jbc.M111.279851 (doi:10.1074/jbc.M111.279851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 10.1126/science.1078973 (doi:10.1126/science.1078973) [DOI] [PubMed] [Google Scholar]
- 60.Genin S., Boucher C. A. 1994. A superfamily of proteins involved in different secretion pathways in Gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243, 112–118 10.1007/BF00283883 (doi:10.1007/BF00283883) [DOI] [PubMed] [Google Scholar]
- 61.Reichow S. L., Korotkov K. V., Hol W. G., Gonen T. 2010. Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232 10.1038/nsmb.1910 (doi:10.1038/nsmb.1910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korotkov K. V., Pardon E., Steyaert J., Hol W. G. 2009. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265 10.1016/j.str.2008.11.011 (doi:10.1016/j.str.2008.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wimley W. C. 2003. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 13, 404–411 10.1016/S0959-440X(03)00099-X (doi:10.1016/S0959-440X(03)00099-X) [DOI] [PubMed] [Google Scholar]
- 64.Bitter W., Koster M., Latijnhouwers M., de Cock H., Tommassen J. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27, 209–219 10.1046/j.1365-2958.1998.00677.x (doi:10.1046/j.1365-2958.1998.00677.x) [DOI] [PubMed] [Google Scholar]
- 65.Dong C., Beis K., Nesper J., Brunkan-Lamontagne A. L., Clarke B. R., Whitfield C., Naismith J. H. 2006. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–269 10.1038/nature05267 (doi:10.1038/nature05267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandran V., Fronzes R., Duquerroy S., Cronin N., Navaza J., Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015 10.1038/nature08588 (doi:10.1038/nature08588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kowalska K., Soscia C., Combe H., Vasseur P., Voulhoux R., Filloux A. 2010. The C-terminal amphipathic alpha-helix of Pseudomonas aeruginosa PelC outer membrane protein is required for its function. Biochimie 92, 33–40 10.1016/j.biochi.2009.10.004 (doi:10.1016/j.biochi.2009.10.004) [DOI] [PubMed] [Google Scholar]
- 68.Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. 2003. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426, 862–866 10.1038/nature02208 (doi:10.1038/nature02208) [DOI] [PubMed] [Google Scholar]
- 69.Reichow S. L., Korotkov K. V., Gonen M., Sun J., Delarosa J. R., Hol W. G., Gonen T. 2011. The binding of cholera toxin to the periplasmic vestibule of the type II secretion channel. Channels 5, 215–218 10.4161/chan.5.3.15268 (doi:10.4161/chan.5.3.15268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Douzi B., Ball G., Cambillau C., Tegoni M., Voulhoux R. 2011. Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J. Biol. Chem. 286, 40 792–40 801 (doi:10.1074/jbc.M111.294843 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerard-Vincent M., Robert V., Ball G., Bleves S., Michel G. P., Lazdunski A., Filloux A. 2002. Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 44, 1651–1665 10.1046/j.1365-2958.2002.02991.x (doi:10.1046/j.1365-2958.2002.02991.x) [DOI] [PubMed] [Google Scholar]
- 72.Login F. H., Shevchik V. E. 2006. The single transmembrane segment drives self-assembly of OutC and the formation of a functional type II secretion system in Erwinia chrysanthemi. J. Biol. Chem. 281, 33 152–33 162 (doi:10.1074/jbc.M606245200 ) [DOI] [PubMed] [Google Scholar]
- 73.Korotkov K. V., Krumm B., Bagdasarian M., Hol W. G. 2006. Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae. J. Mol. Biol. 363, 311–321 10.1016/j.jmb.2006.08.037 (doi:10.1016/j.jmb.2006.08.037) [DOI] [PubMed] [Google Scholar]
- 74.Login F. H., Fries M., Wang X., Pickersgill R. W., Shevchik V. E. 2010. A 20-residue peptide of the inner membrane protein OutC mediates interaction with two distinct sites of the outer membrane secretin OutD and is essential for the functional type II secretion system in Erwinia chrysanthemi. Mol. Microbiol. 76, 944–955 10.1111/j.1365-2958.2010.07149.x (doi:10.1111/j.1365-2958.2010.07149.x) [DOI] [PubMed] [Google Scholar]
- 75.Lindeberg M., Salmond G. P., Collmer A. 1996. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol. 20, 175–190 10.1111/j.1365-2958.1996.tb02499.x (doi:10.1111/j.1365-2958.1996.tb02499.x) [DOI] [PubMed] [Google Scholar]
- 76.de Groot A., Koster M., Gerard-Vincent M., Gerritse G., Lazdunski A., Tommassen J., Filloux A. 2001. Exchange of Xcp (Gsp) secretion machineries between Pseudomonas aeruginosa and Pseudomonas alcaligenes: species specificity unrelated to substrate recognition. J. Bacteriol. 183, 959–967 10.1128/JB.183.3.959-967.2001 (doi:10.1128/JB.183.3.959-967.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savvides S. N., et al. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22, 1969–80 10.1093/emboj/cdg223 (doi:10.1093/emboj/cdg223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Misic A. M., Satyshur K. A., Forest K. T. 2010. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 400, 1011–1021 10.1016/j.jmb.2010.05.066 (doi:10.1016/j.jmb.2010.05.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Planet P. J., Kachlany S. C., DeSalle R., Figurski D. H. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl Acad. Sci. USA 98, 2503–2508 10.1073/pnas.051436598 (doi:10.1073/pnas.051436598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sandkvist M., Bagdasarian M., Howard S. P., Dirita V. J. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14, 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turner L. R., Lara J. C., Nunn D. N., Lory S. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175, 4962–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Py B., Loiseau L., Barras F. 1999. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289, 659–670 10.1006/jmbi.1999.2803 (doi:10.1006/jmbi.1999.2803) [DOI] [PubMed] [Google Scholar]
- 83.Possot O., Pugsley A. P. 1994. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol. 12, 287–299 10.1111/j.1365-2958.1994.tb01017.x (doi:10.1111/j.1365-2958.1994.tb01017.x) [DOI] [PubMed] [Google Scholar]
- 84.Ball G., Chapon-Herve V., Bleves S., Michel G., Bally M. 1999. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Camberg J. L., Johnson T. L., Patrick M., Abendroth J., Hol W. G., Sandkvist M. 2007. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 26, 19–27 10.1038/sj.emboj.7601481 (doi:10.1038/sj.emboj.7601481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shiue S. J., Kao K. M., Leu W. M., Chen L. Y., Chan N. L., Hu N. T. 2006. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 25, 1426–1435 10.1038/sj.emboj.7601036 (doi:10.1038/sj.emboj.7601036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robien M. A., Krumm B. E., Sandkvist M., Hol W. G. 2003. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333, 657–674 10.1016/j.jmb.2003.07.015 (doi:10.1016/j.jmb.2003.07.015) [DOI] [PubMed] [Google Scholar]
- 88.Abendroth J., Murphy P., Sandkvist M., Bagdasarian M., Hol W. G. 2005. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J. Mol. Biol. 348, 845–855 10.1016/j.jmb.2005.02.061 (doi:10.1016/j.jmb.2005.02.061) [DOI] [PubMed] [Google Scholar]
- 89.Chen Y., Shiue S. J., Huang C. W., Chang J. L., Chien Y. L., Hu N. T., Chan N. L. 2005. Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J. Biol. Chem. 280, 42 356–42 363 [DOI] [PubMed] [Google Scholar]
- 90.Arts J., van Boxtel R., Filloux A., Tommassen J., Koster M. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076 10.1128/JB.01236-06 (doi:10.1128/JB.01236-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas J. D., Reeves P. J., Salmond G. P. 1997. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF . Microbiology 143, 713–720 10.1099/00221287-143-3-713 (doi:10.1099/00221287-143-3-713) [DOI] [PubMed] [Google Scholar]
- 92.Robert V., Filloux A., Michel G. P. 2005. Role of XcpP in the functionality of the Pseudomonas aeruginosa secreton. Res. Microbiol. 156, 880–86 10.1016/j.resmic.2005.04.002 (doi:10.1016/j.resmic.2005.04.002) [DOI] [PubMed] [Google Scholar]
- 93.Abendroth J., Bagdasarian M., Sandkvist M., Hol W. G. 2004. The structure of the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae: an unusual member of the actin-like ATPase superfamily. J. Mol. Biol. 344, 619–633 10.1016/j.jmb.2004.09.062 (doi:10.1016/j.jmb.2004.09.062) [DOI] [PubMed] [Google Scholar]
- 94.Abendroth J., Kreger A. C., Hol W. G. 2009. The dimer formed by the periplasmic domain of EpsL from the type 2 secretion system of Vibrio parahaemolyticus. J. Struct. Biol. 168, 313–322 10.1016/j.jsb.2009.07.022 (doi:10.1016/j.jsb.2009.07.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sandkvist M., Hough L. P., Bagdasarian M. M., Bagdasarian M. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol. 181, 3129–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Douet V., Loiseau L., Barras F., Py B. 2004. Systematic analysis, by the yeast two-hybrid, of protein interaction between components of the type II secretory machinery of Erwinia chrysanthemi. Res. Microbiol. 155, 71–75 10.1016/j.resmic.2003.10.001 (doi:10.1016/j.resmic.2003.10.001) [DOI] [PubMed] [Google Scholar]
- 97.Robert V., Hayes F., Lazdunski A., Michel G. P. 2002. Identification of XcpZ domains required for assembly of the secreton of Pseudomonas aeruginosa. J. Bacteriol. 184, 1779–1782 10.1128/JB.184.6.1779-1782.2002 (doi:10.1128/JB.184.6.1779-1782.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsai R. T., Leu W. M., Chen L. Y., Hu N. T. 2002. A reversibly dissociable ternary complex formed by XpsL, XpsM and XpsN of the Xanthomonas campestris pv. campestris type II secretion apparatus. Biochem. J. 367, 865–871 10.1042/BJ20020909 (doi:10.1042/BJ20020909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Possot O. M., Vignon G., Bomchil N., Ebel F., Pugsley A. P. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182, 2142–2152 10.1128/JB.182.8.2142-2152.2000 (doi:10.1128/JB.182.8.2142-2152.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nunn D. N., Lory S. 1992. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc. Natl Acad. Sci. USA 89, 47–51 10.1073/pnas.89.1.47 (doi:10.1073/pnas.89.1.47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bleves S., Voulhoux R., Michel G., Lazdunski A., Tommassen J., Filloux A. 1998. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol. 27, 31–40 10.1046/j.1365-2958.1998.00653.x (doi:10.1046/j.1365-2958.1998.00653.x) [DOI] [PubMed] [Google Scholar]
- 102.Francetic O., Buddelmeijer N., Lewenza S., Kumamoto C. A., Pugsley A. P. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189, 1783–1793 10.1128/JB.01230-06 (doi:10.1128/JB.01230-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu N. T., Leu W. M., Lee M. S., Chen A., Chen S. C., Song Y. L., Chen L. Y. 2002. XpsG, the major pseudopilin in Xanthomonas campestris pv. campestris, forms a pilus-like structure between cytoplasmic and outer membranes. Biochem. J. 365, 205–211 10.1042/BJ20020194 (doi:10.1042/BJ20020194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vignon G., Kohler R., Larquet E., Giroux S., Prevost M. C., Roux P., Pugsley A. P. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185, 3416–3428 10.1128/JB.185.11.3416-3428.2003 (doi:10.1128/JB.185.11.3416-3428.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Durand E., Michel G., Voulhoux R., Kurner J., Bernadac A., Filloux A. 2005. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J. Biol. Chem. 280, 31 378–31 389 [DOI] [PubMed] [Google Scholar]
- 106.Campos M., Francetic O., Nilges M. 2010. Modeling pilus structures from sparse data. J. Struct. Biol. 173, 436–444 10.1016/j.jsb.2010.11.015 (doi:10.1016/j.jsb.2010.11.015) [DOI] [PubMed] [Google Scholar]
- 107.Kohler R., et al. 2004. Structure and assembly of the pseudopilin PulG. Mol. Microbiol. 54, 647–664 10.1111/j.1365-2958.2004.04307.x (doi:10.1111/j.1365-2958.2004.04307.x) [DOI] [PubMed] [Google Scholar]
- 108.Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 10.1016/j.molcel.2006.07.004 (doi:10.1016/j.molcel.2006.07.004) [DOI] [PubMed] [Google Scholar]
- 109.Parge H. E., Forest K. T., Hickey M. J., Christensen D. A., Getzoff E. D., Tainer J. A. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378, 32–38 10.1038/378032a0 (doi:10.1038/378032a0) [DOI] [PubMed] [Google Scholar]
- 110.Craig L., et al. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 10.1016/S1097-2765(03)00170-9 (doi:10.1016/S1097-2765(03)00170-9) [DOI] [PubMed] [Google Scholar]
- 111.Forest K. T., Tainer J. A. 1997. Type-4 pilus-structure: outside to inside and top to bottom—a minireview. Gene 192, 165–169 10.1016/S0378-1119(97)00008-5 (doi:10.1016/S0378-1119(97)00008-5) [DOI] [PubMed] [Google Scholar]
- 112.Craig L., Pique M. E., Tainer J. A. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 10.1038/nrmicro885 (doi:10.1038/nrmicro885) [DOI] [PubMed] [Google Scholar]
- 113.Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. 2008. Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae. J. Mol. Biol. 377, 91–103 10.1016/j.jmb.2007.08.041 (doi:10.1016/j.jmb.2007.08.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. 2008. The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus. J. Mol. Biol. 375, 471–486 10.1016/j.jmb.2007.10.035 (doi:10.1016/j.jmb.2007.10.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lam A. Y., Pardon E., Korotkov K. V., Hol W. G., Steyaert J. 2009. Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J. Struct. Biol. 166, 8–15 10.1016/j.jsb.2008.11.008 (doi:10.1016/j.jsb.2008.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Franz L. P., Douzi B., Durand E., Dyer D. H., Voulhoux R., Forest K. T. 2011. Structure of the minor pseudopilin XcpW from the Pseudomonas aeruginosa type II secretion system. Acta Crystallogr. D Biol. Crystallogr. 67, 124–130 10.1107/S0907444910051954 (doi:10.1107/S0907444910051954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alphonse S., Durand E., Douzi B., Waegele B., Darbon H., Filloux A., Voulhoux R., Bernard C. 2009. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J. Struct. Biol. 169, 75–80 10.1016/j.jsb.2009.09.003 (doi:10.1016/j.jsb.2009.09.003) [DOI] [PubMed] [Google Scholar]
- 118.Korotkov K. V., Gray M. D., Kreger A., Turley S., Sandkvist M., Hol W. G. 2009. Calcium is essential for the major pseudopilin in the type 2 secretion system. J. Biol. Chem. 284, 25 466–25 470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuo W. W., Kuo H. W., Cheng C. C., Lai H. L., Chen L. Y. 2005. Roles of the minor pseudopilins, XpsH, XpsI and XpsJ, in the formation of XpsG-containing pseudopilus in Xanthomonas campestris pv campestris. J. Biomed. Sci. 12, 587–599 10.1007/s11373-005-7372-3 (doi:10.1007/s11373-005-7372-3) [DOI] [PubMed] [Google Scholar]
- 120.Satyshur K. A., Worzalla G. A., Meyer L. S., Heiniger E. K., Aukema K. G., Misic A. M., Forest K. T. 2007. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15, 363–376 10.1016/j.str.2007.01.018 (doi:10.1016/j.str.2007.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Misic A. M., Satyshur K. A., Forest K. T. 2011. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 400, 1011–1021 10.1016/j.jmb.2010.05.066 (doi:10.1016/j.jmb.2010.05.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hirst T. R., Holmgren J. 1987. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc. Natl Acad. Sci. USA 84, 7418–7422 10.1073/pnas.84.21.7418 (doi:10.1073/pnas.84.21.7418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hirst T. R., Holmgren J. 1987. Transient entry of enterotoxin subunits into the periplasm occurs during their secretion from Vibrio cholerae. J. Bacteriol. 169, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hardie K. R., Schulze A., Parker M. W., Buckley J. T. 1995. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol. Microbiol. 17, 1035–1044 10.1111/j.1365-2958.1995.mmi_17061035.x (doi:10.1111/j.1365-2958.1995.mmi_17061035.x) [DOI] [PubMed] [Google Scholar]
- 125.Bortoli-German I., Brun E., Py B., Chippaux M., Barras F. 1994. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol. 11, 545–553 10.1111/j.1365-2958.1994.tb00335.x (doi:10.1111/j.1365-2958.1994.tb00335.x) [DOI] [PubMed] [Google Scholar]
- 126.Pugsley A. P. 1992. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc. Natl Acad. Sci. USA 89, 12 058–12 062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Braun P., Ockhuijsen C., Eppens E., Koster M., Bitter W., Tommassen J. 2001. Maturation of Pseudomonas aeruginosa elastase. Formation of the disulfide bonds. J. Biol. Chem. 276, 26 030–26 035 [DOI] [PubMed] [Google Scholar]
- 128.Braun P., Tommassen J., Filloux A. 1996. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol. Microbiol. 19, 297–306 10.1046/j.1365-2958.1996.381908.x (doi:10.1046/j.1365-2958.1996.381908.x) [DOI] [PubMed] [Google Scholar]
- 129.Filloux A. 2010. Secretion signal and protein targeting in bacteria: a biological puzzle. J. Bacteriol. 192, 3847–3849 10.1128/JB.00565-10 (doi:10.1128/JB.00565-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sauvonnet N., Pugsley A. P. 1996. Identification of two regions of Klebsiella oxytoca pullulanase that together are capable of promoting beta-lactamase secretion by the general secretory pathway. Mol. Microbiol. 22, 1–7 10.1111/j.1365-2958.1996.tb02650.x (doi:10.1111/j.1365-2958.1996.tb02650.x) [DOI] [PubMed] [Google Scholar]
- 131.Francetic O., Pugsley A. P. 2005. Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J. Bacteriol. 187, 7045–7055 10.1128/JB.187.20.7045-7055.2005 (doi:10.1128/JB.187.20.7045-7055.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McVay C. S., Hamood A. N. 1995. Toxin A secretion in Pseudomonas aeruginosa: the role of the first 30 amino acids of the mature toxin. Mol. Gen. Genet. 249, 515–525 10.1007/BF00290577 (doi:10.1007/BF00290577) [DOI] [PubMed] [Google Scholar]
- 133.Voulhoux R., Taupiac M. P., Czjzek M., Beaumelle B., Filloux A. 2000. Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa. J. Bacteriol. 182, 4051–4058 10.1128/JB.182.14.4051-4058.2000 (doi:10.1128/JB.182.14.4051-4058.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palomaki T., Pickersgill R., Riekki R., Romantschuk M., Saarilahti H. T. 2002. A putative three-dimensional targeting motif of polygalacturonase (PehA), a protein secreted through the type II (GSP) pathway in Erwinia carotovora. Mol. Microbiol. 43, 585–596 10.1046/j.1365-2958.2002.02793.x (doi:10.1046/j.1365-2958.2002.02793.x) [DOI] [PubMed] [Google Scholar]
- 135.Palomaki T., Saarilahti H. T. 1995. The extreme C-terminus is required for secretion of both the native polygalacturonase (PehA) and PehA-Bla hybrid proteins in Erwinia carotovora subsp carotovora. Mol. Microbiol. 17, 449–559 10.1111/j.1365-2958.1995.mmi_17030449.x (doi:10.1111/j.1365-2958.1995.mmi_17030449.x) [DOI] [PubMed] [Google Scholar]
- 136.Sandkvist M. 2001. Biology of type II secretion. Mol. Microbiol. 40, 271–283 10.1046/j.1365-2958.2001.02403.x (doi:10.1046/j.1365-2958.2001.02403.x) [DOI] [PubMed] [Google Scholar]
- 137.Py B., Chippaux M., Barras F. 1993. Mutagenesis of cellulase EGZ for studying the general protein secretory pathway in Erwinia chrysanthemi. Mol. Microbiol. 7, 785–793 10.1111/j.1365-2958.1993.tb01169.x (doi:10.1111/j.1365-2958.1993.tb01169.x) [DOI] [PubMed] [Google Scholar]
- 138.Chapon V., Simpson H. D., Morelli X., Brun E., Barras F. 2000. Alteration of a single tryptophan residue of the cellulose-binding domain blocks secretion of the Erwinia chrysanthemi Cel5 cellulase (ex-EGZ) via the type II system. J. Mol. Biol. 303, 117–123 10.1006/jmbi.2000.4103 (doi:10.1006/jmbi.2000.4103) [DOI] [PubMed] [Google Scholar]
- 139.Thanabalu T., Koronakis E., Hughes C., Koronakis V. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17, 6487–6496 10.1093/emboj/17.22.6487 (doi:10.1093/emboj/17.22.6487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Karplus M. 2002. Molecular dynamics simulations of biomolecules. Acc Chem Res. 35, 321–323 10.1021/ar020082r (doi:10.1021/ar020082r) [DOI] [PubMed] [Google Scholar]