Abstract
The 53-kb biosynthetic gene cluster for the novel anticholesterol natural product herboxidiene was identified in Streptomyces chromofuscus ATCC 49982 by genome sequencing and gene inactivation. In addition to herboxidiene, a biosynthetic intermediate, 18-deoxy-herboxidiene, was also isolated from the fermentation broth of S. chromofuscus ATCC 49982 as a minor metabolite.
TEXT
Heart disease is a major threat to human health and remains the world's top killer. Hypercholesterolemia is a primary risk factor for coronary heart disease. Lovastatin and other statins are widely used in clinics to lower cholesterol levels. However, some side effects of statins have been reported, such as muscle and liver problems (19). Thus, new anticholesterol agents are needed. Herboxidiene (compound 1, Fig. 1A) was isolated from Streptomyces chromofuscus A7847 (ATCC 49982) as a novel polyketide which selectively and effectively controls several annual weed species (10, 16). Later, compound 1 was found to effectively reduce plasma cholesterol and possess stronger low-density lipoprotein receptor upregulation activity than tricholstatin A and TMC-49A, representing a new type of anticholesterol molecule (13). Additionally, it also has strong cytotoxic activities and induces both G1 and G2/M arrest in human tumor cell line WI-38 (50% inhibitory concentrations range from 3.7 nM to 0.99 μM) (9, 23). Because of its promising biological activities, several chemical synthetic approaches toward compound 1 have recently been attempted (5, 7, 8, 26). One of the most recent chemical syntheses of compound 1 was accomplished from two lactate-derived chiral ketones in 14 steps with an 8% overall yield (21). However, these multistep chemical processes are not efficient and flexible enough for the synthesis of new herboxidiene analogs. Biosynthesis thus represents an attractive approach to generate structural diversity for more-active and less-toxic molecules.
Fig 1.
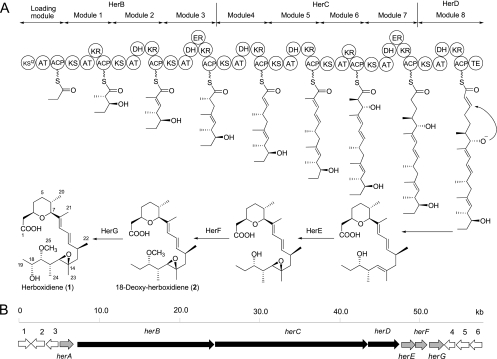
The proposed herboxidiene biosynthetic pathway and the sequenced biosynthetic gene cluster. (A) Proposed biosynthetic pathway of compound 1. (B) Organization of the herboxidiene biosynthetic gene cluster. ORFs involved in herboxidiene biosynthesis are shown as gray (regulatory and tailoring enzymes) and black (PKS) arrows.
For the bacterial strains and plasmids used in this study, see Table S1 in the supplemental material. To locate the herboxidiene biosynthetic gene cluster and better understand this pharmaceutically important strain, the genomic DNA of S. chromofuscus ATCC 49982 was extracted and sequenced using a 454 next-generation sequencing system, yielding approximately 9.6 Mb of sequence data (GC content of 71.25%). Analysis of the sequenced genome was performed with RAST (rapid annotation using subsystem technology) (2), which revealed 8,264 open reading frames (ORFs) encoding genes averaging 1,162 bp. Among the 8,264 putative proteins, 3,531 showed similarity to proteins with functional assignments. Only two contigs were found to contain a complete type I PKS gene cluster. Contig 013 (133,019 bp) contains a gene cluster that encodes a type I PKS consisting of a loading module and 11 extension modules. This PKS is proposed to synthesize a dodecaketide and thus is unlikely to be the herboxidiene synthase. Contig 008 (159,472 bp, GenBank accession number JN671974) was found to contain a gene cluster covering a 53-kb region (herA to -G, Fig. 1B) that encodes a nonaketide synthase and three tailoring enzymes, including an epoxidase, a methyltransferase, and a cytochrome P450 hydroxylase. The organization of the gene cluster is consistent with the structural characteristics of compound 1, thus representing a possible gene cluster involved in herboxidiene biosynthesis. The ORFs in the gene cluster and flanking regions are listed in Table 1.
Table 1.
Deduced functions of ORFs in the herboxidiene biosynthetic gene cluster
| Gene | No. of amino acids | Protein homologue (accession no.) | % Identity/similarity | Proposed function(s) |
|---|---|---|---|---|
| orf1 | 269 | HypB of Rhodococcus jostii RHA1 (ABG96388) | 74/88 | Hydrogenase nickel incorporation protein HypB |
| orf2 | 340 | MviM of Streptomyces bingchenggensis BCW-1 (ADI12327) | 82/90 | Oxidoreductase domain protein |
| orf3 | 261 | Hfi of Streptomyces violaceusniger Tü 4113 (AEM84847) | 86/90 | Xylose isomerase domain protein |
| herA | 341 | PurR of S. violaceusniger Tü 4113 (YP_004815128) | 77/83 | Transcriptional regulator, LacI family |
| herB | 6394 | PldAI of Streptomyces platensis Mer-11107 (BAH02268) | 56/67 | Type I polyketide synthase |
| Loading module | KS, AT, ACP | |||
| Module 1 | KS, AT, KR, ACP | |||
| Module 2 | KS, AT, DH, KR, ACP | |||
| Module 3 | KS, AT, DH, ER, KR, ACP | |||
| herC | 7138 | PldAII of S. platensis Mer-11107 (BAH02269) | 56/66 | Type I polyketide synthase |
| Module 4 | KS, AT, DH, KR, ACP | |||
| Module 5 | KS, AT, DH, KR, ACP | |||
| Module 6 | KS, AT, KR, ACP | |||
| Module 7 | KS, AT, DH, ER, KR, ACP | |||
| herD | 2030 | ORF17 of Streptomyces aizunensis NRRL B-11277 (AAX98192) | 55/65 | Type I polyketide synthase |
| Module 8 | KS, AT, DH, KR, ACP, TE | |||
| herE | 477 | PldD of S. platensis Mer-11107 (BAH02274) | 57/70 | Epoxidase |
| herF | 297 | MitM of Streptomyces lavendulae NRRL 2564 (AF127374_14) | 48/61 | O-Methyltransferase |
| herG | 422 | P450 of S. platensis (CBX53644) | 50/63 | Cytochrome P450 monooxygenase |
| orf4 | 177 | NAT_SF of Streptomyces griseoaurantiacus M045(ZP_08289959) | 66/74 | GCN5-like N-acetyltransferase |
| orf5 | 422 | SSFG-07305 of Streptomyces ghanaensis ATCC 14672 (ZP_06581609) | 65/78 | Hypothetical protein |
| orf6 | 278 | SSFG_07304 of S. ghanaensis ATCC 14672 (ZP_06581608) | 62/76 | Abhydrolase |
Among the seven herboxidiene biosynthetic genes, herB, -C, and -D encode a multimodular type I PKS comprising a loading module and eight extension modules. The organization of the PKS and proposed herboxidiene biosynthetic pathway are shown in Fig. 1A. Ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) domains are essential for chain elongation and exist in all of the extension modules. Ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains were found occasionally between AT and ACP for reductive modifications of the β-keto group formed from each extension step. The loading module in HerB contains a KSQ domain whose active-site Cys is replaced by a Glu, while the KS domains in the eight extension modules harbor the typical active-site sequence DTACS (17). KSQ lacks condensation activity but retains the ability to decarboxylate dicarboxylic acid starters (4, 17). In this herboxidiene biosynthetic pathway, the KSQ domain is proposed to decarboxylate methylmalonyl-ACP to yield propionyl-ACP, which is subsequently used as the starter unit to initiate elongation. AT domains are responsible for selecting and transferring the starter and extender units for polyketide chain synthesis. In keeping with the structure of the resulting molecule, the ATs in modules 1, 2, 3, 5, and 6, as well as AT0 in the loading module, are presumed to be specific for methylmalonyl coenzyme A (CoA), while the other ATs are proposed to take malonyl-CoA (8, 25). Further alignment analysis revealed that all of the AT domains contain the conserved motifs which correlate with the malonyl-CoA and methylmalonyl-CoA specificity, as shown in Fig. 2A (11, 22). All nine ACP domains exhibit the GxDS motif in which the conserved Ser residue is required for posttranslational attachment of the 4′-phosphopantetheinyl cofactor (6, 20). Sequence analysis (Fig. 2B) showed that KR3, -4, -5, -6, -7, and -8 all contain the same conserved motifs to produce an “R” β-hydroxyl group, thus belonging to type B1. KR1 is predicted to be a type A1 KR that forms an “S” hydroxyl group (11). However, the conserved motifs of KR2 are obscure and its type remains unclear. Six DH and two ER domains were found in the herboxidiene PKS. Alignment of the DH domains revealed the conserved HxxxGxxxxP motif, which contains the proposed active-site His (1, 6). Both ER3 and ER7 contain the highly conserved sequence LxHxxxGGVG, which is the proposed NADPH-binding site (6). All of these catalytic domains work together to yield a linear 19-carbon nonaketide, which is released from the PKS by a C-terminal thioesterase (TE) domain that contains the conserved GxSxG and PGDH motifs (27). A tetrahydropyran ring is subsequently formed between C-3 and C-7, likely through a spontaneous process, as shown in Fig. 1A.
Fig 2.
Alignment of divergent motifs in the AT and KR domains of the herboxidiene PKS. (A) Methylmalonyl-CoA-specific and malonyl-CoA-specific ATs. The underlined residues are associated with malonyl-CoA and methylmalonyl-CoA specificity. (B) Alignment of the conserved motifs in KRs. KR1 belongs to A1-type KRs (1, no LDD; 2, W; 3, no H). KR3, -4, -5, -6, -7, and -8 fall into the B1 type (1, LDD; 4, no P). The conserved motifs of KR2 are obscure, and the type of this domain remains unclear.
Three genes (herE, herF, and herG) encoding post-PKS tailoring enzymes were found downstream of the PKS genes. HerE shows 58% identity to the epoxidase PldD for pladienolide biosynthesis (15) and is considered responsible for the epoxidation of the C-14–C-15 double bond. HerF is a putative O-methyltransferase, which likely methylates the 17-OH. HerG shows 50% identity with a cytochrome P450 monooxygenase in the biotransformation of terfenadine (14) and is predicted to catalyze the C-18 hydroxylation.
A putative regulatory gene, herA, is located upstream of the six biosynthetic genes, which is proposed to be involved in herboxidiene biosynthesis as a pathway-specific regulator. HerA belongs to the LacI transcriptional regulator family and contains a ligand binding domain (18, 24). No transporter genes are present in the gene cluster and flanking regions. Since compound 1 was found to be a major extracellular metabolite in the fermentation broth of S. chromofuscus ATCC 49982, the excretion of this compound may occur through diffusion or actions of certain general transporters in the cells.
To confirm that the discovered gene cluster is really involved in herboxidiene biosynthesis, AT4 in HerC was disrupted by means of single-crossover homologous recombination (3, 12). Briefly, a 1.18-kb HindIII/EcoRI fragment (see Fig. S1 in the supplemental material) was amplified from the genomic DNA with primers A and B (5′-AAAGAATTCCTGAAGTCGAACATCGGGCACA-3′ and 5′-AAAAAGCTTCATCAGCGGGGAGTGGAAGG-3′) and inserted into the HindIII/EcoRI site of pKC1139 to yield pLS-6 for insertional inactivation of herC. The plasmid was introduced into S. chromofuscus by intergeneric conjugation from Escherichia coli ET12567 (3, 12). The apramycin-resistant colonies appearing at 37°C were identified as the integrating mutants, in which a single-crossover homologous recombination event took place (see Fig. S2A in the supplemental material). The genotypes of the resulting strains were further confirmed by PCR amplification with both genome- and vector-specific primers including Pcheck1 (5′-GCGTGACGGACCCAGTAGGC-3′), Pcheck2 (5′-GAGCGGATAACAATTTCACACAGG-3′) (RV-M), Pcheck3 (5′-GTACGGGCAGAACCGGGACC-3′), Pcheck4 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) (M13-47), Pcheck5 (5′-GCGACGTACGGGCAGAACC-3′), and Pcheck6 (5′-CGTGACGGACCCAGTAGGC-3′). The 1.35- and 1.26 -kb fragments were, respectively, amplified from the mutant mHER1001 using Pcheck1/Pcheck2 and Pcheck3/Pcheck4 (see Fig. S2B in the supplemental material), indicating that pLS-6 was inserted into the genome. A PCR that spanned the disrupted region was also conducted using Pcheck5/Pcheck6. In contrast to the wild type, the 1.37-kb gene fragment in herC could not be amplified from the genomic DNA of mHER1001 (see Fig. S2C in the supplemental material), which eliminated the possibility of the additional herC copy in the mutant. All of the PCR products were ligated into pJET1.2 and confirmed by sequencing. Thus, we confirmed the disruption of AT4 in HerC in mHER1001.
In order to isolate the standard of compound 1 for analysis of its production by the mutant, the wild-type strain was fermented in 4 liters of YM medium (0.4% glucose, 0.4% yeast extract, 1% malt extract, pH 7.3) at 28°C and 240 rpm for 8 days. The fermentation broth was extracted three times with 3.5 liters of ethyl acetate. Fractionation of the concentrated extract on a Diaion HP-20 column and further purification by high-performance liquid chromatography (HPLC) yielded 17 mg of compound 1 and 1.3 mg of a minor metabolite, compound 2. For the spectral data obtained for compounds 1 and 2, see the supplemental material. Electrospray ionization-mass spectrometry of compound 2 showed a quasimolecular ion [M − H]− at m/z 421, indicating that the molecular mass of this compound is 422 Da, which is 16 Da less than the molecular mass of compound 1. This suggested that compound 2 could be the 18-deoxy precursor of compound 1. The 1H nuclear magnetic resonance spectrum of compound 2 was very similar to that of compound 1 (see the supplemental material), except that the signals at δ 3.77 (dq, 6.5, 6.2 Hz, H-18) and 1.10 (d, 6.5 Hz, H3-19) in compound 1 were replaced by those assignable to a methylene group at δ 1.55 and 1.62 and a methyl group at δ 0.86 (t, 7.6 Hz) in compound 2, respectively. The 1H-1H correlation spectroscopy spectrum of compound 2 showed that the methylene group (δ 1.55 and 1.62) correlated with the methyl group (δ 0.86) and H-17 (δ 3.10). Therefore, the structure of compound 2 was determined as 18-deoxy-herboxidiene (Fig. 1A), which is likely to be an intermediate in herboxidiene biosynthesis.
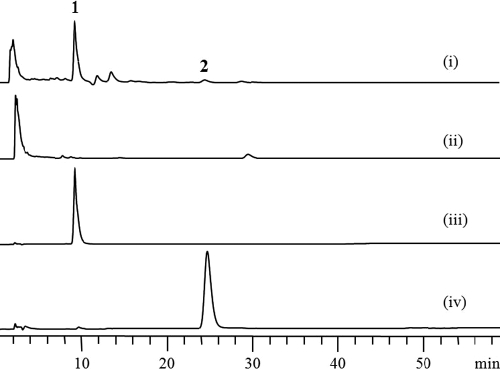
To examine the effect of the disruption of herC on herboxidiene biosynthesis, both S. chromofuscus ATCC 49982 and mHER1001 were grown in 100 ml of YM medium and extracted as described above. The extracts were analyzed using an Agilent ZORBAX SB-C18 column (5 μm, 4.6 by 250 mm) on an Agilent 1200 HPLC apparatus, eluted with 75% methanol-water (containing 0.1% trifluoroacetic acid) at 1 ml/min for 60 min. HPLC trace ii in Fig. 3 clearly shows that the production of both compounds 1 and 2 was abolished in mHER1001, indicating that herC is essential for herboxidiene biosynthesis. Successful inactivation of this gene cluster not only revealed its involvement in herboxidiene biosynthesis but also confirmed the genetic manipulability of the herboxidiene-producing strain, which makes it feasible to further conduct genetic and metabolic engineering of herboxidiene biosynthesis for novel analogs.
Fig 3.
HPLC analysis of herboxidiene biosynthesis in S. chromofuscus ATCC 49982 and mHER1001 at 238 nm. Traces: i, products of S. chromofuscus ATCC 49982; ii, products of S. chromofuscus mHER1001; iii, purified standard of compound 1; iv, purified standard of compound 2.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a Scientist Development Grant (09SDG2060080) from the American Heart Association.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akey DL, et al. 2010. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure 18:94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bierman M, et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 4. Bisang C, et al. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502–505 [DOI] [PubMed] [Google Scholar]
- 5. Blakemore PR, Kocieński PJ, Morley A, Muir K. 1999. A synthesis of herboxidiene. J. Chem. Soc. Perkin 1 1999:955–968 [Google Scholar]
- 6. Donadio S, Katz L. 1992. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51–60 [DOI] [PubMed] [Google Scholar]
- 7. Edmunds AJ, Arnold G, Hagmann L, Schaffner R, Furlenmeier H. 2000. Synthesis of simplified herboxidiene aromatic hybrids. Bioorg. Med. Chem. Lett. 10:1365–1368 [DOI] [PubMed] [Google Scholar]
- 8. Ghosh AK, Li J. 2011. A stereoselective synthesis of (+)-herboxidiene/GEX1A. Org. Lett. 13:66–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasegawa M, et al. 2011. Identification of SAP155 as the target of GEX1A (herboxidiene), an antitumor natural product. ACS Chem. Biol. 6:229–233 [DOI] [PubMed] [Google Scholar]
- 10. Isaac BG, Ayer SW, Elliott RC, Stonard RJ. 1992. Herboxidiene: a potent phytotoxic polyketide from Streptomyces sp. A7847. J. Org. Chem. 57:7220–7226 [Google Scholar]
- 11. Keatinge-Clay AT. 2007. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 14:898–908 [DOI] [PubMed] [Google Scholar]
- 12. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 13. Koguchi Y, et al. 1997. Trichostatin A and herboxidiene up-regulate the gene expression of low density lipoprotein receptor. J. Antibiot. 50:970–971 [DOI] [PubMed] [Google Scholar]
- 14. Lombard M, Salard I, Sari MA, Mansuy D, Buisson D. 2011. A new cytochrome P450 belonging to the 107L subfamily is responsible for the efficient hydroxylation of the drug terfenadine by Streptomyces platensis. Arch. Biochem. Biophys. 508:54–63 [DOI] [PubMed] [Google Scholar]
- 15. Machida K, et al. 2008. Organization of the biosynthetic gene cluster for the polyketide antitumor macrolide, pladienolide, in Streptomyces platensis Mer-11107. Biosci. Biotechnol. Biochem. 72:2946–2952 [DOI] [PubMed] [Google Scholar]
- 16. Miller-Wideman M, et al. 1992. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847—taxonomy, fermentation, isolation, physicochemical and biological properties. J. Antibiot. 45:914–921 [DOI] [PubMed] [Google Scholar]
- 17. Moffitt MC, Neilan BA. 2003. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J. Mol. Evol. 56:446–457 [DOI] [PubMed] [Google Scholar]
- 18. Nguyen CC, Saier MH., Jr 1995. Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors. FEBS Lett. 377:98–102 [DOI] [PubMed] [Google Scholar]
- 19. Noël B. 2004. Autoimmune disease and other potential side-effects of statins. Lancet 363:2000. [DOI] [PubMed] [Google Scholar]
- 20. Paitan Y, Orr E, Ron EZ, Rosenberg E. 1999. Genetic and functional analysis of genes required for the post-modification of the polyketide antibiotic TA of Myxococcus xanthus. Microbiology 145:3059–3067 [DOI] [PubMed] [Google Scholar]
- 21. Pellicena M, Kramer K, Romea P, Urpi F. 2011. Total synthesis of (+)-herboxidiene from two chiral lactate-derived ketones. Org. Lett. 13:5350–5353 [DOI] [PubMed] [Google Scholar]
- 22. Reeves CD, et al. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 40:15464–15470 [DOI] [PubMed] [Google Scholar]
- 23. Sakai Y, et al. 2002. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. II. The effects on cell cycle progression and gene expression. J. Antibiot. 55:863–872 [DOI] [PubMed] [Google Scholar]
- 24. Weickert MJ, Adhya S. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869–15874 [PubMed] [Google Scholar]
- 25. Wu Y, Kang Q, Shen Y, Su W, Bai L. 2011. Cloning and functional analysis of the naphthomycin biosynthetic gene cluster in Streptomyces sp. CS. Mol. Biosyst. 7:2459–2469 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Panek JS. 2007. Total synthesis of herboxidiene/GEX 1A. Org. Lett. 9:3141–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zirkle R, Black TA, Gorlach J, Ligon JM, Molnar I. 2004. Analysis of a 108-kb region of the Saccharopolyspora spinosa genome covering the obscurin polyketide synthase locus. DNA Seq. 15:123–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.