Abstract
Most genes for antibiotic resistance present in soil microbes remain unexplored because most environmental microbes cannot be cultured. Only recently has the identification of these genes become feasible through the use of culture-independent methods. We screened a soil metagenomic DNA library in an Escherichia coli host for genes that can confer resistance to kanamycin, gentamicin, rifampin, trimethoprim, chloramphenicol, or tetracycline. The screen revealed 41 genes that encode novel protein variants of eight protein families, including aminoglycoside acetyltransferases, rifampin ADP-ribosyltransferases, dihydrofolate reductases, and transporters. Several proteins of the same protein family deviate considerably from each other yet confer comparable resistance. For example, five dihydrofolate reductases sharing at most 44% amino acid sequence identity in pairwise comparisons were equivalent in conferring trimethoprim resistance. We identified variants of aminoglycoside acetyltransferases and transporters that differ in the specificity of the drugs for which they confer resistance. We also found wide variation in protein structure. Two forms of rifampin ADP-ribosyltransferases, one twice the size of the other, were similarly effective at conferring rifampin resistance, although the short form was expressed at a much lower level. Functional metagenomic screening provides insight into the large variability in antibiotic resistance protein sequences, revealing divergent variants that preserve protein function.
INTRODUCTION
Strains of bacteria that were resistant to antibiotics first appeared in hospitals in the 1930s and 1940s (18). Today, bacterial resistance affects all classes of antibiotics, endangering their ability to treat life-threatening infections (19). The mechanisms of this resistance include active efflux of the antibiotic from the microbial cell, destruction or modification of the antibiotic, and alteration of the intracellular target of the antibiotic, such as the ribosome (35). Antibiotic resistance is pervasive, with genes conferring resistance present in most bacterial genomes (6). In addition to their role as toxic agents, antibiotics may have many other physiological functions in bacterial communities, including a role in cell signaling and interspecies interaction (8, 11). Therefore, genes that did not evolve as resistance determinants can acquire the ability to protect microorganisms from antibiotics (21).
Much of our knowledge about antibiotic resistance mechanisms comes from bacteria pathogenic to humans. However, most antibiotics are produced by environmental microorganisms, suggesting that genes for antibiotic resistance emerged outside the clinic (5, 21). Studies of soil bacteria have shown that resistance is prevalent in this environment (27). Resistance mechanisms in environmental bacteria have the potential to become clinically relevant because the DNA within these bacteria can be shared among other diverse species through lateral gene transfer (19). An understanding of the evolution of resistance proteins could inform the design of new antibiotics capable of evading resistance.
Until recently, much of the genetic diversity found in the soil microenvironment has remained unexplored, because a large majority of bacteria cannot be cultured by standard laboratory techniques, with estimates that only 0.1 to 1% of species can be cultured. Therefore, the genomes of only a small proportion of bacteria have been accessible to researchers (15). Uncultured environmental bacteria potentially harbor many uncharacterized antibiotic resistance genes that could be transferred to clinically relevant bacteria (4). Several recent studies have uncovered new resistance genes from metagenomic samples. For example, a survey of antibiotic resistance in the human gut microbiome revealed many previously unsequenced resistance genes that are homologous to known genes (30). Functional screening of soil samples revealed many newly sequenced antibiotic resistance genes, even from pristine environments (1, 7, 16, 27, 33).
In this study, we used functional screening in an Escherichia coli host of a soil metagenomic library to find genes conferring resistance to one of six antibiotics. This strategy revealed examples of naturally occurring variants within several antibiotic resistance protein families, including rifampin ADP-ribosyltransferases (ARTs), dihydrofolate reductases (DHFRs), aminoglycoside acetyltransferases, and transporter proteins. We found variants within the dihydrofolate reductase protein family that differ remarkably at the sequence level but display similar efficacy as resistance proteins. We identified aminoglycoside acetyltransferases and transporter protein family variants that differ in the specificity of the drugs for which they confer resistance. We additionally identified two forms of rifampin ARTs, one that is twice the size of the other, and compared the efficacies of these two forms. Tapping into an uncultured resource also revealed proteins highly divergent from any known examples. This study increases the knowledge of resistance determinants in environmental bacteria, the diversity of possible sequences that can encode these determinants, and the functional consequences of this sequence variation.
MATERIALS AND METHODS
Soil description.
A soil sample was collected from an urban environment on the University of Washington campus in Seattle, Washington (47°39′N, 122°18′W). The sample was collected from the top 5 to 13 cm of earth and had a pH of 6.3 (1:1 ratio of soil to deionized water). The soil was passed through a sieve to remove any plant debris and roots, and DNA extraction was performed immediately without storage of the soil sample.
Bacterial strains and culture conditions.
E. coli strain DH10B (Invitrogen) was used for the metagenomic library construction. E. coli strain TOP10F′ (Invitrogen) was used for cloning of FLAG-tagged DHFRs and rifampin ARTs. E. coli cells were routinely cultured in LB medium at 37°C. Plating media contained 1.5% Bacto agar. Antibiotics were purchased from Sigma-Aldrich.
Construction of a soil metagenomic plasmid library.
DNA was extracted from 10 g of the soil sample using the PowerMax soil DNA isolation kit (Mo Bio). Extracted DNA was sheared by sonication, and fragments between 1 and 3 kb were purified by gel extraction. The recovered DNA was end repaired and ligated into the pUC18 vector that was linearized with the blunt cutting enzyme SmaI. We ligated soil DNA with pUC18 plasmid and transformed the library into E. coli DH10B cells. The library was plated onto LB medium containing ampicillin (50 μg/ml), and colonies were allowed to grow overnight at 37°C. The colonies were then washed from the plates and combined into an amplified library stock.
Selection of antibiotic-resistant clones.
The metagenomic library was plated to over 100 times coverage onto media containing inhibitory concentrations of chloramphenicol (5 μg/ml), kanamycin (20 μg/ml), gentamicin (10 μg/ml), tetracycline (10 μg/ml), rifampin (20 μg/ml), or trimethoprim (20 μg/ml). Plates were incubated at 37°C for 24 h and then at room temperature for an additional 24 to 48 h if there were no colonies after the original incubation time. Plasmids were isolated from individual clones and transformed into new E. coli DH10B cells to confirm that the plasmid was responsible for the resistance phenotype.
Identification of antibiotic resistance genes.
Metagenomic insert DNA was Sanger sequenced using primers specific for the pUC18 plasmid and additional primers designed as necessary to sequence full inserts. Open reading frames were identified using the blastx program from the Basic Local Alignment Search Tool (2).
Phylogenetic analyses.
Multiple-sequence alignments were conducted using the ClustalW online tool (17). Alignments were then visualized using Jalview (34). Phylograms were constructed using the neighbor-joining method with the MEGA5 software package (31).
Cloning DHFRs and rifampin ADP-ribosyltransferases.
Primers were designed to amplify individual DHFR and rifampin ART genes from the original metagenomic clones. PCR products were then ligated into the pGEM-T vector (Promega). Plasmids containing the gene of interest in the correct orientation were then digested with SpeI and NdeI, and DNA containing the FLAG tag sequence flanked by these cut site ends was inserted into the construct 5′ to the cloned gene. The constructs were transformed into E. coli TOP10F′ cells.
Spot assays.
E. coli DH10B cultures were grown to an optical density at 600 nm (OD600) of 0.5 in LB containing 50 μg/ml ampicillin while shaking at 37°C. E. coli TOP10F′ cultures were grown to an OD600 of 0.5 in LB containing 50 μg/ml ampicillin while shaking at 37°C and were allowed to grow for an additional hour with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce expression of the cloned gene product. Ten microliters of 10-fold serial dilutions of each bacterial culture was spotted onto LB medium containing antibiotic. DH10B culture dilutions ranged from undiluted to 1:105. TOP10F′ culture dilutions ranged from 1:10 to 1:106. IPTG was added to plates for TOP10F′ cultures to induce expression of the cloned gene. All cultures were spotted onto LB medium containing ampicillin (50 μg/ml) as a control.
Western blot analysis.
E. coli from 2 ml of log-phase cultures at equal optical density readings were pelleted and resuspended in SUTEB buffer (0.01% bromophenol blue, 10 mM ethylenediaminetetraacetic acid, 1% SDS, 10 mM Tris-HCl [pH 6.8], 8 M urea). The samples were heated to 100°C for 10 min and then sonicated for 10 s, and debris was pelleted by centrifugation. Equal volumes of protein extract were loaded onto a 4 to 12% bis-Tris gel and run in morpholineethanesulfonic acid (MES) running buffer (Invitrogen). Proteins were immobilized onto a nitrocellulose membrane and probed with an anti-FLAG antibody (Sigma-Aldrich).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this study are available in GenBank under accession numbers JF924866 to JF924904.
RESULTS
Antibiotic resistance genes identified in a soil metagenomic library.
Using microbial DNA extracted from a soil sample, we constructed in an E. coli host a metagenomic plasmid library containing a total of 1.4 million cloned genes. The average insert size was 2 kb, providing a total of 2.8 × 109 base pairs of DNA in the library. The library was screened for cloned genes that conferred on E. coli the ability to grow in the presence of chloramphenicol, kanamycin, gentamicin, rifampin, trimethoprim, or tetracycline. The chosen antibiotics represent both naturally occurring and synthetic drugs that target different pathways within the microbial cell, including synthesis of RNA (rifampin), proteins (chloramphenicol, kanamycin, gentamicin, and tetracycline), and folate (trimethoprim).
Screens of the library revealed antibiotic-resistant transformants carrying 39 unique cloned genes (Table 1). All clones but one code for genes that are related to known antibiotic resistance genes. However, their nucleotide sequences and encoded proteins differ from any known genes, with several of the proteins sharing less than 40% amino acid sequence identity with their closest match in GenBank (Table 1). We used PhyloPythia in an attempt to determine the source organisms of the DNA in each clone (23). This analysis assigned 19 clones to the phylum Proteobacteria, 8 clones to Actinobacteria, and 2 clones to Firmicutes; for the remaining 10 clones, the phylum could not be identified (see Table S1 in the supplemental material). Six of the clones code for two possible antibiotic resistance proteins (see Fig. S1A in the supplemental material). To determine whether both resistance genes were functional, we individually subcloned both potential resistance genes from each clone and tested for the ability of each individual gene to confer antibiotic resistance to an E. coli host. For all but two of the clones, only a single resistance determinant was found (Fig. S1B to F). For clone tet4, two distinct functional resistance genes were identified (Fig. S1B). For clone chlor2, neither of the two genes tested conferred resistance to E. coli, suggesting that the resistance determinant is housed elsewhere in the metagenomic insert DNA.
Table 1.
Summary of antibiotic-resistant clones recovered from functionally screening a soil metagenomic library
| Clone | GenBank accession no. | Resistance gene annotation | Most similar protein GenBank accession no./organism | % amino acid identity |
|---|---|---|---|---|
| kan1 | JF924866 | 6′-N-Acetyltransferase | NP_105746/Mesorhizobium loti MAFF303099 | 71.1 |
| kan2 | JF924867 | 6′-N-Acetyltransferase | ZP_04608289/Micromonospora sp. strain ATCC 39149 | 55.9 |
| kan3 | JF924868 | 6′-N-Acetyltransferase | ZP_06823301/Streptomyces sp. strain SPB74 | 38.0 |
| kan4 | JF924869 | 6′-N-Acetyltransferase | ZP_06823301/Streptomyces sp. SPB74 | 39.3 |
| gent1 | JF924870 | 3′-N-Acetyltransferase | CBI71176/uncultured bacterium | 61.2 |
| chlor1 | JF924871 | Undetermined | ||
| chlor2 | JF924872 | Undetermined | ||
| chlor3 | JF924873 | MFS transporter | ABK64017/Janthinobacterium lividum | 53.5 |
| rif1 | JF924874 | Rifampin ADP-ribosyltransferase | ZP_07113423/Oscillatoria sp. strain PCC 6506 | 69.2 |
| rif2 | JF924875 | Rifampin ADP-ribosyltransferase | ZP_07113423/Oscillatoria sp. PCC 6506 | 69.6 |
| rif3 | JF924876 | Rifampin ADP-ribosyltransferase | ZP_07113423/Oscillatoria sp. PCC 6506 | 76.9 |
| rif4 | JF924877 | Rifampin ADP-ribosyltransferase | ZP_07113423/Oscillatoria sp. PCC 6506 | 75.3 |
| rif5 | JF924878 | Rifampin ADP-ribosyltransferase | ZP_07113423/Oscillatoria sp. PCC 6506 | 77.7 |
| rif6 | JF924879 | Rifampin ADP-ribosyltransferase | YP_948552/Arthrobacter aurescens TC1 | 72.8 |
| rif7 | JF924880 | Rifampin monooxygenase | YP_002281165/Rhizobium leguminosarum bv. Trifolii WSM2304 | 72.4 |
| tri1 | JF924881 | Dihydrofolate reductase | YP_004145638/Pseudoxanthomonas suwonensis 11-1 | 66.5 |
| tri2 | JF924882 | Dihydrofolate reductase | ZP_03629733/bacterium Ellin514 | 64.7 |
| tri3 | JF924883 | Dihydrofolate reductase | YP_857866/Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 37.6 |
| tri4 | JF924884 | Dihydrofolate reductase | YP_001021245/Methylibium petroleiphilum PM1 | 57.6 |
| tri5 | JF924885 | Dihydrofolate reductase | YP_003383770/Kribbella flavida DSM 17836 | 52.8 |
| tri6 | JF924886 | Dihydrofolate reductase | XP_001980195/Drosophila erecta | 33.9 |
| tri7 | JF924887 | Dihydrofolate reductase | YP_001685012/Caulobacter sp. strain K31 | 66.7 |
| tri8 | JF924888 | Dihydrofolate reductase | ZP_08196415/Nocardioidaceae bacterium Broad-1 | 60.5 |
| tri9 | JF924889 | Dihydrofolate reductase | YP_001364161/Kineococcus radiotolerans SRS30216 | 59.1 |
| tri10 | JF924890 | Dihydrofolate reductase | YP_003674069/Methylotenera versatilis 301 | 44.6 |
| tri11 | JF924891 | Dihydrofolate reductase | YP_923504/Nocardioides sp. strain JS614 | 69.4 |
| tri12 | JF924892 | Dihydrofolate reductase | YP_001193819/Flavobacterium johnsoniae UW101 | 51.3 |
| tri13 | JF924893 | Dihydrofolate reductase | YP_004039327/Methylovorus sp. strain MP688 | 43.0 |
| tri14 | JF924894 | Dihydrofolate reductase | ABB72057/Salmonella enterica subsp. enterica serovar Infantis | 24.4 |
| tri15 | JF924895 | Dihydrofolate reductase | YP_923504/Nocardioides sp. JS614 | 71.0 |
| tri16 | JF924896 | Dihydrofolate reductase | ZP_03629733/bacterium Ellin514 | 70.1 |
| tri17 | JF924897 | Dihydrofolate reductase | YP_003126057/Chitinophaga pinensis DSM 2588 | 46.7 |
| tri18 | JF924898 | Dihydrofolate reductase | YP_003766287/Amycolatopsis mediterranei U32 | 37.7 |
| tri19 | JF924899 | Dihydrofolate reductase | YP_001757175/Methylobacterium radiotolerans JCM 2831 | 56.6 |
| tri20 | JF924900 | MFS transporter | YP_823123/Candidatus Solibacter usitatus Ellin6076 | 51.3 |
| tet1 | JF924901 | MFS transporter | CCA59776/Streptomyces venezuelae ATCC 10712 | 68.1 |
| tet2 | JF924902 | MFS transporter | ZP_06970071/Ktedonobacter racemifer DSM 44963 | 37.4 |
| tet3 | JF924903 | MFS transporter | ZP_07088554/Chryseobacterium gleum ATCC 35910 | 77.0 |
| tet4 | JF924904 | ABC transporter | ZP_07389559/Paenibacillus curdlanolyticus YK9 | 83.1 |
| ABC transporter | ZP_07389558/Paenibacillus curdlanolyticus YK9 | 84.8 |
The antibiotic resistance genes fall into eight distinct protein families (Table 1). Cloned genes conferring resistance to the aminoglycoside antibiotics kanamycin and gentamicin code for aminoglycoside acetyltransferases. Six other clones code for transporters of two different protein families, six others for rifampin ADP-ribosyltransferases, and 19 others for dihydrofolate reductases (DHFRs). The remainder include one glutathione S-transferase domain protein, one rifampin monooxygenase, and one clone, chlor1, whose resistance mechanism is uncharacterized. Since the phenotype of E. coli carrying chlor1 was weak compared to that of the other clones (requiring 5 days of growth compared to 1 or 2 days), we did not pursue determination of the resistance mechanism.
Related genes vary in their resistance profiles.
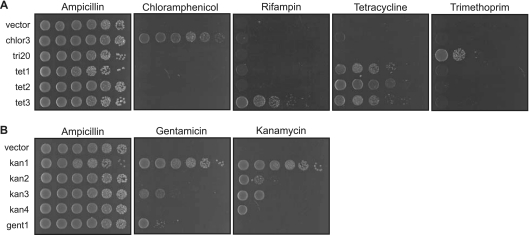
Efflux of antibiotics is a common mechanism of resistance found for many different types of antibiotics (26). We recovered six cloned genes coding for potential drug transporters, which fall into two different superfamilies. Five clones code for major facilitator (MFS) transporters, while the remaining clone contains two ATP-binding cassette (ABC) transporters. We assessed the resistance profiles of E. coli carrying these clones by a spot dilution assay. Although antibiotic resistance is traditionally assayed by determining the MIC that kills bacteria, we felt that spot assays were more appropriate to assess the metagenomic clones. In this study, the resistance genes are located randomly within the insert sequences of a high-copy-number plasmid, and the encoded resistance proteins are expressed at different levels. As a result, determination of MICs in this setting would not accurately represent the resistance determinant's efficacy in its native genomic setting. Spot assays allow simple and clear visualization of whether a metagenomic clone allows its E. coli host to survive antibiotic challenge. We tested the ability of the E. coli carrying the MFS transporter-containing clones to grow in the presence of each of the six antibiotics used in this study, with growth on ampicillin (based on the vector-expressed β-lactamase gene) as a loading control. All but one clone were specific for the single antibiotic for which they could confer resistance (Fig. 1A). A single clone, tet3, conferred growth in the presence of two antibiotics, tetracycline and rifampin.
Fig 1.
Variation in resistance profiles of related protein family members. (A) Spot assay of E. coli expressing metagenomic clones coding for MFS drug transporters. Tenfold serial dilutions of log-phase cultures were spotted onto LB containing chloramphenicol (10 μg/ml), trimethoprim (20 μg/ml), tetracycline (5 μg/ml), or rifampin (20 μg/ml). The E. coli cultures were also spotted on plates containing ampicillin (50 μg/ml) as a positive control. None of the clones conferred growth on media containing kanamycin (20 μg/ml) or gentamicin (10 μg/ml). (B) Spot assay of E. coli expressing metagenomic clones coding for aminoglycoside acetyltransferases. Tenfold serial dilutions of log-phase cultures were spotted onto LB containing kanamycin (20 μg/ml) or gentamicin (10 μg/ml). The E. coli cultures were also spotted on plates containing ampicillin (50 μg/ml) as a positive control for growth.
Kanamycin and gentamicin are aminoglycoside antibiotics produced by soil microorganisms that act by inhibiting protein synthesis. Resistance to aminoglycosides is often due to enzymes that modify the antibiotic, rendering it ineffective (14, 35). The five clones that conferred resistance to kanamycin and gentamicin code for aminoglycoside acetyltransferases. However, E. coli carrying these five clones exhibited different specificity profiles. With two clones (kan1 and kan3), the E. coli cells carrying the clone were able to grow in the presence of either aminoglycoside, while with the remaining three clones (kan2, kan4, and gent1), the E. coli cells could grow on either kanamycin or gentamicin (Fig. 1B). While there is high amino acid sequence identity between one acetyltransferase (labeled kan3b_aac) from clone kan3 and the protein encoded by clone kan4 (92% identity [see Fig. S2 in the supplemental material]), there is low sequence conservation among the other aminoglycoside acetyltransferases, ranging between 12% and 31% sequence identity in pairwise alignments (Fig. S2).
A highly divergent type II dihydrofolate reductase.
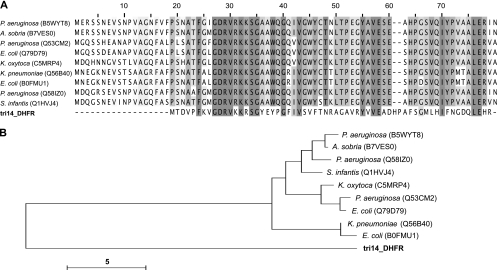
Trimethoprim inhibits bacterial dihydrofolate reductases. One mechanism of resistance is by exogenous expression of another DHFR that is not susceptible to the drug (12, 13). Two types of genetically and structurally unrelated bacterial DHFR proteins have been characterized (12). Type I DHFRs, ∼180 amino acids in length, are often chromosomally encoded and function as monomers. Type II DHFRs, ∼80 amino acids in length, are plasmid encoded and function as homotetramers. Screening on trimethoprim led to the identification of 19 clones coding for DHFRs. On the basis of the top BLAST hits, only one of these clones (referred to as tri14_DHFR) appears to code for a type II DHFR. This protein is only 61 amino acids long and shares 24% amino acid identity with its closest match in GenBank (Table 1). Sequence alignment of tri14_DHFR with other plasmid gene-encoded sequences highlights many amino acid residues that are conserved in all type II DHFRs except for the metagenomic tri14_DHFR protein (Fig. 2A). Pairwise alignments of tri14_DHFR with other type II DHFRs show 22% to 24% amino acid identity (see Fig. S3 in the supplemental material). Phylogenetic clustering highlights that tri14_DHFR is evolutionarily distant from any other plasmid gene-encoded DHFRs (Fig. 2B).
Fig 2.
A highly divergent type II dihydrofolate reductase. (A) Multiple-sequence alignment of tri14_DHFR and 9 unique type II DHFR sequences from the Pfam database. Sequences from Pseudomonas aeruginosa, Aeromonas sobria, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, and Salmonella enterica serovar Infantis are shown. Several sequences in the database were identical, and in these cases, only one representative sequence was included in the alignment. The degree of shading indicates the degree of amino acid conservation at each site. Gaps introduced to maximize sequence alignment are indicated by the hyphens. (B) Unrooted neighbor-joining tree of aligned amino acid sequences from type II DHFRs from the Pfam database and our study. Identifiers in parentheses are UniProt accession numbers. Bar = 5 changes/site.
Trimethoprim resistance is conferred by dihydrofolate reductases with large sequence variation.
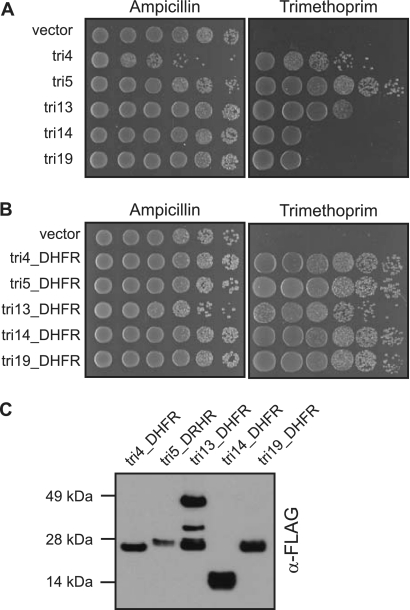
The amino acid sequences of the 19 DHFRs are diverse (see Fig. S4A in the supplemental material). A spot assay of E. coli carrying five of these cloned genes showed a broad range of resistance, ranging over more than 3 orders of magnitude (Fig. 3A). Pairwise alignments reveal that the DHFRs range between 7% and 44% amino acid sequence identity (Fig. S4B). The different levels of resistance could be due to intrinsic differences in the proteins' sensitivity to trimethoprim or to differences in protein expression, since the DHFRs are found in random positions within the insert sequences and expressed using transcriptional and translational signals from a variety of bacterial species. To test their sensitivity, we cloned FLAG-tagged versions of the five DHFRs that appeared to confer different levels of resistance to trimethoprim and expressed these DHFRs from the same promoter in E. coli. Despite their sequence diversity and the resistance range of the original isolates, when the DHFRs were cloned and expressed off the same promoter, they all conferred similar levels of resistance to E. coli (Fig. 3B). A Western blot indicated that the tagged DHFRs were expressed at roughly comparable levels, with only a fewfold range between the more poorly expressed tri5_DHFR and the better expressed tri14-DHFR (Fig. 3C). Thus, the differences in resistance conferred by the original metagenomic clones are likely due primarily to different levels of expression as a result of their genomic context.
Fig 3.
Similar trimethoprim resistance profiles conferred by diverse dihydrofolate reductases. (A) Spot assay of E. coli expressing metagenomic clones conferring trimethoprim resistance. Tenfold serial dilutions of log-phase cultures were spotted onto LB containing trimethoprim (20 μg/ml). The E. coli cultures were also spotted on plates containing ampicillin (50 μg/ml) as a positive control for growth. (B) Spot assay of E. coli expressing FLAG-tagged DHFRs. Tenfold serial dilutions of log-phase cultures were spotted onto LB containing trimethoprim (80 μg/ml) or ampicillin (50 μg/ml) as a positive control for growth. (C) Western blot of FLAG-tagged DHFRs. An equal amount of protein extract from E. coli cells expressing FLAG-tagged DHFRs was loaded in each lane, and the Western blot was probed with anti-FLAG antibody (α-FLAG).
While most of the DHFRs appear as only a single band on a Western blot, for tri13_DHFR, two strong bands, as well as a third weaker band, were present (Fig. 3C). One band migrated at the estimated molecular mass of 25 kDa, while a second band migrated at twice this molecular mass, suggesting that this protein may form a dimer. Type I DHFRs are generally thought to exist as monomers within the cell, with only very few examples of dimerized forms of these enzymes (12, 20, 33).
Characterization of two forms of rifampin ADP-ribosyltransferases.
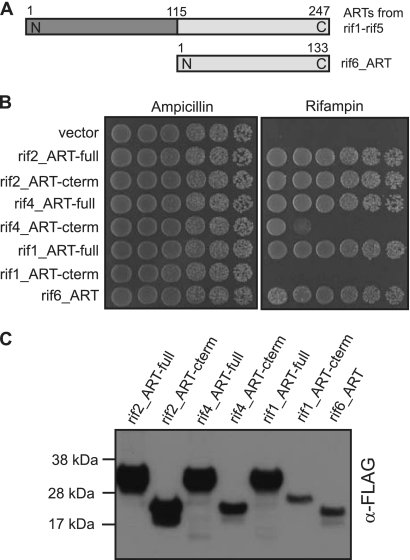
Rifampin inhibits bacterial DNA-dependent RNA polymerases. A common mode of resistance involves modification of the antibiotic, either by oxidation or ribosylation (36). Screening against rifampin identified seven cloned genes. One clone encodes a rifampin monooxygenase, while the other six encode rifampin ADP-ribosyltransferases. The rifampin ARTs fall into two groups. Five of the proteins are ∼250 amino acids long, while rif6_ART is only 133 amino acids long. rif6_ART aligns with the C termini of the longer rifampin ARTs (Fig. 4A). While short rifampin ARTs have been characterized (3), there are few examples of long rifampin ARTs that have been sequenced. None of these have been functionally evaluated, leaving the functional relevance of the additional 115 N-terminal amino acids unknown.
Fig 4.
Characterization of two forms of rifampin ADP-ribosyltransferases. (A) Schematic of long and short rifampin ADP-ribosyltransferases. The rifampin ARTs from the rif1, rif2, rif3, rif4, and rif5 clones are between 147 and 150 amino acids long. The rifampin ART from clone rif6 is 133 amino acids long and aligns with the C termini of the longer rifampin ARTs. (B) Spot assay of E. coli expressing FLAG-tagged full-length proteins and C termini of rifampin ADP-ribosyltransferases. Tenfold serial dilutions of log-phase cultures were spotted onto plates containing rifampin (20 μg/ml) or ampicillin (50 μg/ml) as a positive control for growth. A full-length rifampin ADP-ribosyltransferase (-full) or the C terminus of the ART protein (-cterm) is indicated at the end of the ART designation. (C) Western blot of FLAG-tagged rifampin ARTs. An equal amount of protein extract from E. coli cells expressing FLAG-tagged rifampin ARTs was loaded in each lane, and the Western blot was probed with anti-FLAG antibody.
There is high sequence conservation in the N-terminal region of the five long rifampin ARTs we identified (see Fig. S5 in the supplemental material), suggesting that this region contains residues important for the protein's function. We investigated three of the long rifampin ARTs (rif1_ART, rif2_ART, and rif4_ART) and compared them to the single short rifampin ART we obtained (rif6_ART) to determine whether the N-terminal region of the long protein is necessary for conferring rifampin resistance. A multiple-sequence alignment of these four proteins shows that there is high sequence conservation among them (Fig. S6). We expressed both the full-length protein and the C-terminal fragment (which aligns with the short ART) of the long rifampin ARTs in E. coli to determine whether the C-terminal region alone could confer rifampin resistance. Comparable levels of protein expression and rifampin resistance were observed for all full-length ARTs (Fig. 4B and C). A similar level of protein expression and rifampin resistance was also observed for the C terminus of rif2_ART. However, the C-terminal regions of rif4_ART and rif1_ART were expressed at lower levels in E. coli and were less effective at conferring rifampin resistance than the full-length versions of these variants (Fig. 4B and C). These results suggest that only the C-terminal region of the long ARTs is necessary to confer rifampin resistance and that the level of rifampin resistance correlated with the level of protein expression in E. coli (Fig. 4B and C). In contrast, rif6_ART was expressed at a low level in E. coli, comparable to the expression of the rif1_ART C terminus, yet it conferred rifampin resistance more than 5 orders of magnitude greater than the rif1_ART C terminus (Fig. 4B and C).
DISCUSSION
We used functional screening of a soil metagenomic DNA library to identify naturally occurring variants of antibiotic resistance genes, which allows recovery of genes independent of their sequence. By selecting only for activity—in contrast to a random mutagenesis strategy which generates many detrimental mutations—functional screening directly indicates significantly divergent amino acid sequences that can be tolerated in a protein. None of the genes we identified exactly matches any in GenBank, and in many cases these genes are highly divergent from previously characterized family members. Other examples of antibiotic resistance protein variants have been identified by functional screening, including variants of β-lactamases, aminoglycoside acetyltransferases, chloramphenicol acetyltransferases, and efflux pumps (1, 7, 16, 27, 30, 33).
The sequence data generated from functional metagenomic studies could be useful for assessing protein structure and function, particularly for protein families for which there are few known structures. For example, because membrane proteins are difficult to crystallize, only limited structural data exist for transporter proteins, with no structural tetracycline efflux pump data available (9, 32). Additionally, there are only four structures of aminoglycoside acetyltransferases, and no common catalytic residues are known (14). The amino acid diversity identified by metagenomic sequencing can provide information about mutational tolerance and potentially identify key residues.
The extent of sequence deviation in these resistance proteins compared to related proteins in GenBank is striking. For example, we identified a type II DHFR that shares only 24% sequence identity with its closest match in GenBank. Pairwise comparisons of tri14_DHFR with other proteins in the same family show only 22% to 24% sequence identity. We show that protein variants with highly diverse amino acid sequences are able to function similarly in conferring antibiotic resistance, as is the case for DHFRs which confer resistance to trimethoprim. This class of proteins may be a particular challenge for designing effective drugs, given the huge array of possible sequence variants in existence. A recent metagenomic screen identified a new type of reductase that confers trimethoprim resistance, further highlighting this challenge (33).
We found diversity not only in amino acid sequence but also in the overall structure of the proteins. We isolated several long-form rifampin ARTs, although the majority of known rifampin ARTs are of the short variety. This is the first study to examine the functional implications of deleting the N-terminal regions of these long ARTs, which suggests that these additional amino acids are not required for function. However, our results suggest that the short rifampin ART may be much more effective at conferring resistance than the C termini of the long rifampin ARTs. While the level of rifampin resistance correlated with the level of protein expression for the C-terminal regions of the long rifampin ARTs, the short rifampin ART conferred a high level of resistance even though it was only weakly expressed in E. coli.
Further characterization of environmental antibiotic resistance protein variants will benefit studies of human health as well as studies of ecological questions. Knowledge of the sequence variation of resistance proteins could direct the design of new antibiotics that are less susceptible to resistance. Additionally, while antibiotic resistance is important in a clinical setting, these genes evolved in a nonclinical setting (22). Proteins that function as antibiotic resistance determinants often have other functions in their natural setting (11). For example, the chromosomally encoded 2′-N-acetyltransferase in Providencia stuartii can function as a resistance protein by acetylating aminoglycosides, but it also modifies peptidoglycan in the cell wall and is necessary to maintain normal cell morphology and division (24, 25). It is therefore beneficial to relate these protein variants to their functions in natural environments. Identification of variants through functional screening is not limited to antibiotic resistance proteins. A body of functional screening data is emerging for diverse protein families, including DNA polymerases, carbohydrate-active enzymes, and a variety of biocatalysts (10, 28, 29).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Thomas, Doug Fowler, Lea Starita, and Carlos Araya for advice and discussions and Colin Manoil, Maitreya Dunham, and Sam Miller for critical review of the manuscript.
This work was partially supported by grant P41 RR11823 from the NIH. K.M.M. was supported by training grant T32 HG000035 from the NIH. S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3:243–251 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baysarowich J, et al. 2008. Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc. Natl. Acad. Sci. U. S. A. 105:4886–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Costa VM, Griffiths E, Wright GD. 2007. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10:481–489 [DOI] [PubMed] [Google Scholar]
- 5. D'Costa VM, et al. 2011. Antibiotic resistance is ancient. Nature 477:457–461 [DOI] [PubMed] [Google Scholar]
- 6. D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377 [DOI] [PubMed] [Google Scholar]
- 7. Donato JJ, et al. 2010. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 76:4396–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fajardo A, Martìnez JL. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11:161–167 [DOI] [PubMed] [Google Scholar]
- 9. Fluman N, Bibi E. 2009. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta 1794:738–747 [DOI] [PubMed] [Google Scholar]
- 10. Hess M, et al. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467 [DOI] [PubMed] [Google Scholar]
- 11. Hoffman L, D'Argenio D, Bader M, Miller S. 2007. Microbial recognition of antibiotics: ecological, physiological, and therapeutic implications. Microbe 2:175–182 [Google Scholar]
- 12. Howell EE. 2005. Searching sequence space: two different approaches to dihydrofolate reductase catalysis. ChemBioChem 6:590–600 [DOI] [PubMed] [Google Scholar]
- 13. Huovinen P, Sundstrom L, Swedberg G, Skold O. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jana S, Deb JK. 2006. Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 70:140–150 [DOI] [PubMed] [Google Scholar]
- 15. Kellenberger E. 2001. Exploring the unknown. The silent revolution of microbiology. EMBO Rep. 2:5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lang KS, et al. 2010. Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil by using functional metagenomics. Appl. Environ. Microbiol. 76:5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 18. Levy S. 1982. Microbial resistance to antibiotics: an evolving and persistent problem. Lancet 320:83–88 [DOI] [PubMed] [Google Scholar]
- 19. Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 20. Loveridge EJ, Rodriguez RJ, Swanwick RS, Allemann RK. 2009. Effect of dimerization on the stability and catalytic activity of dihydrofolate reductase from the hyperthermophile Thermotoga maritima. Biochemistry 48:5922–5933 [DOI] [PubMed] [Google Scholar]
- 21. Martinez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367 [DOI] [PubMed] [Google Scholar]
- 22. Martinez JL. 2009. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 276:2521–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. 2007. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods 4:63–72 [DOI] [PubMed] [Google Scholar]
- 24. Payie K, Clarke A. 1997. Characterization of gentamicin 2′-N-acetyltransferase from Providencia stuartii: its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J. Bacteriol. 179:4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Payie K, Rather P, Clarke A. 1995. Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptidoglycan in Providencia stuartii. J. Bacteriol. 177:4303–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poole K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56:20–51 [DOI] [PubMed] [Google Scholar]
- 27. Riesenfeld CS, Goodman RM, Handelsman J. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981–989 [DOI] [PubMed] [Google Scholar]
- 28. Simon C, Daniel R. 2009. Achievements and new knowledge unraveled by metagenomic approaches. Appl. Microbiol. Biotechnol. 85:265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simon C, Herath J, Rockstroh S, Daniel R. 2009. Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl. Environ. Microbiol. 75:2964–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sommer MOA, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thaker M, Spanogiannopoulos P, Wright G. 2010. The tetracycline resistome. Cell. Mol. Life Sci. 67:419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Torres-Cortés G, et al. 2011. Characterization of novel antibiotic resistance genes identified by functional metagenomics on soil samples. Environ. Microbiol. 4:1101–1114 [DOI] [PubMed] [Google Scholar]
- 34. Waterhouse AM, Procter JB, Martin DMA, Clamp MI, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wright GD. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175–186 [DOI] [PubMed] [Google Scholar]
- 36. Wright GD. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57:1451–1470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.