Abstract
An α-glucosidase (HaG) with the following unique properties was isolated from Halomonas sp. strain H11: (i) high transglucosylation activity, (ii) activation by monovalent cations, and (iii) very narrow substrate specificity. The molecular mass of the purified HaG was estimated to be 58 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). HaG showed high hydrolytic activities toward maltose, sucrose, and p-nitrophenyl α-d-glucoside (pNPG) but to almost no other disaccharides or malto-oligosaccharides higher than trisaccharides. HaG showed optimum activity to maltose at 30°C and pH 6.5. Monovalent cations such as K+, Rb+, Cs+, and NH4+ increased the enzymatic activity to 2- to 9-fold of the original activity. These ions shifted the activity-pH profile to the alkaline side. The optimum temperature rose to 40°C in the presence of 10 mM NH4+, although temperature stability was not affected. The apparent Km and kcat values for maltose and pNPG were significantly improved by monovalent cations. Surprisingly, kcat/Km for pNPG increased 372- to 969-fold in their presence. HaG used some alcohols as acceptor substrates in transglucosylation and was useful for efficient synthesis of α-d-glucosylglycerol. The efficiency of the production level was superior to that of the previously reported enzyme Aspergillus niger α-glucosidase in terms of small amounts of by-products. Sequence analysis of HaG revealed that it was classified in glycoside hydrolase family 13. Its amino acid sequence showed high identities, 60%, 58%, 57%, and 56%, to Xanthomonas campestris WU-9701 α-glucosidase, Xanthomonas campestris pv. raphani 756C oligo-1,6-glucosidase, Pseudomonas stutzeri DSM 4166 oligo-1,6-glucosidase, and Agrobacterium tumefaciens F2 α-glucosidase, respectively.
INTRODUCTION
α-Glucosidase (EC 3.2.1.20, α-d-glucoside glucohydrolase) is a ubiquitous enzyme widely distributed in microorganisms, plants, and animals. It catalyzes liberation of α-glucose from the nonreducing ends of α-glucosides such as malto-oligosaccharides and sucrose. Its substrate specificity is significantly diverse, depending on the enzyme origins, and the enzymes are classified into three groups on the basis of their substrate specificities (10). Group I enzymes are more active toward heterogeneous substrates, represented by sucrose and synthetic glycosides such as p-nitrophenyl α-d-glucoside (pNPG) and phenyl α-d-glucoside, than toward homogeneous substrates such as maltose. Group II enzymes, known as maltases, prefer homogeneous substrates to heterogeneous substrates. Like group II enzymes, group III enzymes mainly hydrolyze homogeneous substrates but have high activity toward long-chain substrates like glycogen and soluble starch.
According to the sequence-based classification of glycoside hydrolase (GH), α-glucosidases are generally classified into two families, GH families 13 (subfamily 30) and 31 (9). Enzymes from bacteria, some yeast such as Saccharomyces cerevisiae, and insects are mainly classified into the former family, and those from plants, animals, some yeast such as Schizosaccharomyces pombe, and fungi are in the latter.
Enzymes of both families catalyze hydrolysis of α-glucosides, with net retention of the anomeric configuration, through a double displacement mechanism, where two acidic amino acid residues act as a catalytic nucleophile and a general acid/base catalyst (31). The general acid/base catalyst donates a proton to the glucosidic oxygen, and the catalytic nucleophile simultaneously attacks the anomeric carbon to form a β-glucosyl enzyme intermediate. Consequently, the general acid/base catalyst activates a water molecule as a general base catalyst to stimulate nucleophilic attack of the water molecule at the anomeric carbon of the glucosyl-enzyme intermediate. Transglucosylation can occur if a free hydroxy group of a sugar or alcohol, rather than water, attacks nucleophilically.
In the food and pharmaceutical industries, α-glucosidases play essential roles in the production of isomalto-oligosaccharides, nigero-oligosaccharides (48, 57), and some glucosides such as α-d-glucosyl vitamin E (33) and α-d-glucosylglycerol (αGG) (50). Glucosylation of nonsugar compounds is a useful technique for improving their physicochemical properties, such as stability and water solubility (26, 33, 47, 56).
αGG, a collective term for 2-O-α-d-glucosylglycerol (2-O-αGG), (2R)-1-O-α-d-glucosylglycerol [(2R)-1-O-αGG], and (2S)-1-O-α-d-glucosylglycerol [(2S-1-O-αGG], is a glucosylated compound observed in some osmotolerant bacteria and plants (18) and is also found in traditional Japanese liquors produced from rice (49). αGG has been reported to have some beneficial functions as a moisturizing agent in cosmetics (58) and potential as a health food material and therapeutic agent because it stimulates insulin-like growth factor behavior (17) and has antitumor effects (37).
αGG can be synthesized enzymatically from malto-oligosaccharides and glycerol with fungal α-glucosidase (50) or bacterial cyclodextrin glucanotransferase (EC. 2.4.1.19) (35). However, the production yield has been restricted because accumulation of by-products such as malto-oligosaccharides and di- or higher glucosylated glycerols was unavoidable as a result of the nature of the enzyme. In contrast, sucrose phosphorylase (EC. 2.4.1.7) has been reported to regioselectively produce 2-O-αGG from sucrose and glycerol in high yield (15). Although sucrose phosphorylase is an attractive biocatalyst for glucosides (29), not all types of glucosides can be synthesized using it, and research on its use for the production of various kinds of glucosides is still under development (1).
In this work, we focused on α-glucosidase as a biocatalyst for the production of glucosides because its substrate, malto-oligosaccharides, can be obtained by degradation of starch at low cost. We screened α-glucosidases showing high transglucosylation activity, and an enzyme with the desired properties was successfully obtained from a halophilic marine bacterium, Halomonas sp. strain H11, isolated from the surface of coral. In addition to its high transglucosylation activity, the enzyme has several unique properties: first, significant stimulation by monovalent cations such as NH4+, K+, Rb+, and Cs+, and second, very narrow specificity for substrate chain length and glucosidic bonds. In this article, the enzymatic properties and applications of α-glucosidase from Halomonas sp. H11 (HaG) are described.
MATERIALS AND METHODS
Isolation and identification of bacterial strains.
Bacterial strains from coral, collected in the sea near Okinawa, Japan, were propagated with a culture medium consisting of 1% (wt/vol) sodium arginate (Wako Pure Chemical Industries, Osaka, Japan), 0.5% (wt/vol) peptone (Becton Dickinson, Franklin Lakes, NJ), 0.5% (wt/vol) yeast extract (Becton Dickinson), 0.1% (wt/vol) KH2PO4 (Kanto Chemical, Tokyo, Japan), 0.02% (wt/vol) MgSO4 · 7H2O (Kanto Chemical), and 10% (wt/vol) NaCl; the pH was adjusted to 10 with separately sterilized 1% (wt/vol) Na2CO3 (Kanto Chemical). The culture medium was incubated at 30°C for 2 weeks, and a single colony was isolated from the resulting culture fluid on the same medium containing 2% (wt/vol) agar. Chromosomal DNA of the obtained bacterium was extracted using an Instragene matrix (Bio-Rad, Hercules, CA) and used as the template in PCR. Primestar HS DNA polymerase (Takara Bio, Shiga, Japan) and a primer set of 9F and 1510R (34) were used to amplify a region of 16S rRNA (see Table S1 in the supplemental material). A BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA) and primers 9F, 785F, 802R, and 1510R (34) were used in sequence analysis (see Table S1 in the supplemental material). An automatic DNA sequencer, the ABI Prism 3130xl genetic analyzer system (Applied Biosystems), and the software Chromaspro (version 1.4; Technelysium Pty., Tewantin, Australia) were used. Sequence similarity searches and phylogenetic classifications were performed using the software Aporon (version 2.0; Techno Suruga Laboratory, Shizuoka, Japan), the database Aporon DB-BA (version 6.0; Techno Suruga Laboratory), GenBank, the DNA Data Bank of Japan (DDBJ), and the EMBL Nucleotide Sequence Database. Cells grown on Marine Broth 2216 (Becton Dickinson) containing 2% (wt/vol) agar were used for the following tests. Morphological observations were carried out using an optical microscope, BX50F4 (Olympus, Tokyo, Japan). Cells were stained with Feyber G Nissui (Nissui Seiyaku, Tokyo, Japan) in a Gram-staining test. Other physiological tests were performed by the methods described by Barrow and Feltham (6) and using a kit, API 20NE (bioMérieux, Lyon, France).
Purification of α-glucosidase from Halomonas sp. strain H11.
Halomonas sp. H11 was cultured aerobically at 37°C for 24 h with rotary shaking at 150 rpm in 2 liters of a medium consisting of 1% (wt/vol) soluble starch (Kanto Chemical), 0.5% (wt/vol) polypeptone (Wako Pure Chemical Industries), 0.5% (wt/vol) yeast extract, 3.5% (wt/vol) NaCl, 0.01% (wt/vol) KH2PO4, and 0.02% (wt/vol) MgSO4 · 7H2O; the pH was adjusted to 7.0 with 2 N NaOH. Bacterial cells were harvested by centrifugation and disrupted by sonication using an Astrason ultrasonic processor (XL; Misonix, Farmingdale, NY) in 50 ml of 20 mM HEPES-NaOH buffer (pH 7.0). The resulting cell debris was removed by centrifugation, and the supernatant was regarded as the crude extract. The pH of the crude extract was adjusted to 5.0 with 20 mM sodium acetate buffer (pH 5.0) and applied to a Toyopearl DEAE-650 M column (16 by 170 mm; Tosoh, Tokyo, Japan) equilibrated with 20 mM sodium acetate buffer (pH 5.0). After washing with the same buffer, the adsorbed protein was eluted using a linear gradient of NaCl (0 to 0.5 M; total volume, 60 ml; flow rate, 2 ml/min). The active fractions that eluted at about 0.1 M NaCl were collected, and ammonium sulfate was added to the solution to give a final concentration of 1.5 M. This was loaded on a Toyopearl Butyl-650 M column (16 by 200 mm; Tosoh) equilibrated with 20 mM HEPES-NaOH buffer containing 1.5 M ammonium sulfate (pH 7.0). The adsorbed protein was eluted using a descending linear gradient of ammonium sulfate (1.5 to 0 M; total volume, 120 ml; flow rate, 2 ml/min). The active fractions that eluted at about 1.0 M ammonium sulfate were collected and desalted by repeated dilution and concentration. The sample was concentrated by ultracentrifugation using an Amicon Ultra-15 centrifugal unit (nominal molecular weight limit, 10,000; Millipore, Billerica, MA), and the resulting concentrate was diluted with 20 mM HEPES-NaOH buffer (pH 7.0). Finally, the sample was concentrated to about 5 ml and applied to a Resource Q column (1 ml; GE Healthcare, Uppsala, Sweden) equilibrated with 20 mM HEPES-NaOH buffer (pH 7.0). After washing with 4 ml of the same buffer, the adsorbed protein was eluted using a linear gradient of NaCl (0 to 1.0 M; total volume, 30 ml; flow rate, 3 ml/min). The active fractions that eluted at about 0.1 M NaCl were collected and desalted as described above. To confirm the purity of the enzyme preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on a 12.5% polyacrylamide gel, as described by Laemmli (27). A molecular size marker, Mark12 unstained standard (Invitrogen, Carlsbad, CA), was used. The gel was stained with Rapid CBB Kanto (Kanto Chemical). Protein concentration was determined with a DC protein assay kit (Bio-Rad) in all procedures. Bovine serum albumin (Bio-Rad) was used as a standard protein. Since the enzyme activity was not affected by freeze-thawing, the enzyme preparation was frozen for long-term storage.
Enzyme activity assay.
For the standard assay, maltase activity was measured. A reaction mixture (0.1 ml) consisting of an appropriate concentration of enzyme, 0.2% (wt/vol) maltose (Nihon Shokuhin Kako, Tokyo, Japan), and 40 mM HEPES-NaOH buffer (pH 7.0) was incubated at 30°C for 10 min. The reaction was terminated by adding twice the volume of 2 M Tris-HCl buffer (pH 7.0), and the liberated glucose was measured using the glucose oxidase-peroxidase method (32) with a Glucose C II test kit (Wako Pure Chemical Industries). One unit of α-glucosidase activity was defined as the amount of enzyme that hydrolyzes 1 μmol of maltose per minute.
Effects of pH and temperature on activity and stability of HaG.
The optimum pH and optimum temperature were evaluated by measuring enzyme activities at given pH values and temperatures, respectively. The reaction pH was adjusted with 40 mM Britton-Robinson buffer (pH 3.0 to 12.0; composed of a mixture of 40 mM acetic acid, 40 mM phosphoric acid, and 40 mM glycine; the pH value was adjusted with NaOH) as the reaction buffer. The pH stability was determined on the basis of the residual activity after incubation of 0.36 μg/ml of enzyme solutions in 10 mM Britton-Robinson buffer at various pH values at 4°C for 24 h. The temperature stability was evaluated from the residual activity after incubating the enzyme in 20 mM HEPES-NaOH (pH 7.0) at given temperatures for 15 min. The activity before treatment was considered 100% of the residual activity.
Substrate specificity.
The initial hydrolytic rates for the following substrates were measured: maltose (Nihon Shokuhin Kako), maltotriose (Nihon Shokuhin Kako), maltotetraose (Seikagaku, Tokyo, Japan), maltopentaose (Seikagaku), maltohexaose (Seikagaku), isomaltose (Tokyo Chemical Industry, Tokyo, Japan), trehalose (Wako Pure Chemical Industries), nigerose (Wako Pure Chemical Industries), kojibiose (Wako Pure Chemical Industries), sucrose (Kanto Chemical), methyl α-d-glucoside (Taoka Chemical, Osaka, Japan), and soluble starch (Nacalai Tesque, Kyoto, Japan). A reaction mixture (0.1 ml) consisting of 7.2 μg/ml enzyme, 4 mM substrate (soluble starch, 0.2% [wt/vol]), and 40 mM HEPES-NaOH buffer (pH 7.0) was incubated at 30°C for 10 min. The liberated glucose was determined as described above. The enzyme reaction toward pNPG (Sigma, St. Louis, MO) was performed by incubating 0.1 ml of a mixture consisting of 4 mM substrate and 7.2 μg/ml enzyme in the same buffer. The reaction was stopped by adding twice the volume of 1 M Na2CO3. The absorbance at 405 nm was measured to determine the amount of p-nitrophenol (pNP) released. pNP (Wako Pure Chemical Industries) was used as a standard.
Effects of various salts on enzyme activity and stability.
Enzyme activities in the presence of 10 mM solutions of various salts (LiCl, NaCl, MgCl, KCl, CaCl2, MnCl2, FeCl2, CoCl2, CuCl, ZnCl2, AgNO3, CsCl, and NH4Cl) and 5 mM (NH4)2SO4 were assayed. The effects of ammonium ions on the temperature-activity profile were investigated by measuring the activity at various temperatures in the presence or absence of 10 mM NH4Cl. The thermal stability of HaG in the presence of 10 mM NH4Cl was evaluated as described above.
Kinetic parameters for maltose and pNPG in the presence of monovalent cations.
To evaluate the effects of monovalent cations on the kinetic parameters for maltose and pNPG, the apparent kinetic parameters for these substrates in the presence of 10 mM concentration of activators were determined from the initial rates of hydrolysis at various substrate concentrations (2 to 20 mM for maltose and 0.001 to 8 mM for pNPG) by regressing to the Michaelis-Menten equation.
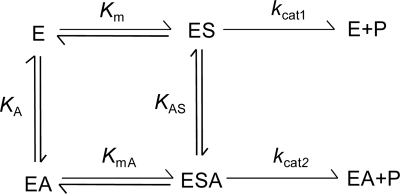
Activations for hydrolysis of maltose by monovalent cations were kinetically analyzed in detail, based on a nonessential activator model (12). The reaction scheme for a reaction activated by a nonessential activator is shown in Fig. 1. In this model, rate equations 1 and 2 below were given under the assumption of rapid equilibration:
| (1) |
where v is the initial hydrolysis rate, and A and S indicate the concentrations of the activator and substrate, respectively. The four dissociation constants in this equation are related as shown in Equation 2:
| (2) |
We determined the individual constants as follows. First, the initial hydrolytic rates were measured at various concentrations of maltose (2 to 20 mM), without an activator, to obtain kcat1 and Km. The values were calculated by fitting the experimental data to Equation 1 at A equal to 0 by nonlinear regression. Next, the hydrolytic rates were measured at a substrate concentration much higher than that for Km in the presence of various concentrations of activators (0.001 to 100 mM). The kcat2 and KAS values were calculated from the concentration in the activator-rate plot. Finally, the enzyme assay was carried out with maltose at the same concentration as that for Km in the presence of various concentrations of activators (0.001 to 100 mM). KA was determined from the concentration of the activator-hydrolytic rate plot as described above. Then KmA was obtained by substituting the other parameters for equation 2. To verify the calculated parameters, the experimental hydrolytic rates for 4.5 mM or 50 mM maltose in the presence of various concentrations of activators were compared with the theoretical values calculated from the obtained parameters.
Fig 1.
Kinetic model for the reaction activated by a nonessential activator. E, S, P, and A indicate enzyme, substrate, product, and activator, respectively. Km and KmA denote the dissociation constants for the ES complex and ESA ternary complex, respectively, to EA. KA and KAS are the dissociation constants for the EA complex and ESA ternary complex, respectively, to ES. kcat1 and kcat2 are rate constants in the absence and presence, respectively, of an activator.
Transglucosylation activity toward various alcohols.
The transglucosylation activities toward ethanol, glycerol, propylene glycol, 1-propanol, and 2-propanol (all reagents were purchased from Kanto Chemical) were qualitatively analyzed by thin-layer chromatography. Reaction mixtures (10 μl) containing 5% (vol/vol) alcohols, 10% (wt/vol) maltose, 50 mM HEPES-NaOH buffer (pH 7.0), 5 mM (NH4)2SO4, and 0.18 μg/μl of HaG were incubated at 37°C for 24 h. A 10-fold-diluted sample (1 μl) was analyzed by thin-layer chromatography (Silica Gel 60 F254; Merck, Darmstadt, Germany) with a solvent system consisting of 2-propanol–1-butanol–water at 2:2:1 (vol/vol/vol). Glucose and maltose (each 1% [wt/vol]) were used as standards. The chromatograms were visualized by spraying with a solvent consisting of 10% (vol/vol) sulfuric acid in methanol and heating.
Enzymatic synthesis of αGG.
The production efficiencies of αGG by HaG and Aspergillus niger α-glucosidase (AnG) were compared; AnG has been reported to produce αGG efficiently (50). A reaction mixture (1 ml) containing 20% (wt/vol) maltose, 20% (wt/vol) glycerol, 0.04 U/ml of purified HaG or AnG, and 40 mM reaction buffer (HEPES-NaOH buffer containing 5 mM ammonium sulfate, pH 7.0, for HaG and sodium acetate buffer, pH 5.0, for AnG) was incubated at 37°C. AnG was purchased from Amano Enzymes (Nagoya, Japan), and the activity was assayed using the same method as that used for HaG except for the reaction buffer, which was 40 mM sodium acetate buffer (pH 5.0). The total αGG was quantified by high-performance liquid chromatography (HPLC) under the following conditions: column, Ultron PS-80N.L (8.0 by 500 mm; Shinwa Chemical Industries, Kyoto, Japan); eluant, water; flow rate, 0.9 ml/min; column temperature, 50°C; detection, refractive index. To determine the ratio of stereoisomers of the product, αGG was purified with HPLC. αGG (5 mg) was trimethylsilylated using TMSI-C reagent (GL Sciences, Tokyo, Japan) and analyzed by gas chromatography (49).
N-terminal amino acid sequence analysis.
Purified α-glucosidase was transferred to a polyvinylidene difluoride membrane (Immobilon-PSQ, Millipore) using a semidry electroblotting apparatus (Bio-Rad). The protein band detected with Coomassie brilliant blue R-250 (Wako Pure Chemical Industries) was cut off and analyzed using a protein sequencer (Procise 492cLC; Applied Biosystems). To determine the internal amino acid sequence, purified HaG was digested by lysyl endopeptidase, and the resulting peptides were sequenced as follows. The purified enzyme (55 μg) was dissolved in 100 μl of 20 mM Tris-HCl (pH 8.0) containing 8 M urea. Then 3 μl of 0.02% (wt/vol) lysyl endopeptidase (Wako Pure Chemical Industries) was added, followed by incubation at 37°C overnight. The resultant peptides were separated by reverse-phase HPLC under the following conditions: column, ODS AS-303 (4.6 by 250 mm; YMC, Kyoto, Japan); flow rate, 1 ml/min; column temperature, 40°C; detection, UV 215 nm; elution, linear gradient of 0 to 100% acetonitrile in 0.1% trifluoroacetic acid for 60 min.
Cloning of HaG gene.
Chromosomal DNA of Halomonas sp. H11 was prepared using a previously described method (41) and digested with SalI or PstI (Takara Bio) to connect cassette DNA fragments supplied by an LA PCR in vitro cloning kit (Takara Bio). The resulting DNA fragments were used as a template in the PCR to amplify the HaG gene. All the primers used in this research are listed in Table S1 in the supplemental material. Ex Taq polymerase (Takara Bio) and a set of primers, HAG-N1, designed on the basis of the N-terminal amino acid sequence of HaG, and primer CS1, corresponding to the cassette sequence, were used. To amplify the desired region specifically, nested PCR was performed with primers HAG-N2 and CS2. The amplified DNA fragment was cloned into a pGEM-T Easy vector (Promega, Madison, WI) and sequenced with primers HAG-F′3, HAG-F′4, HAG-F′5, HAG-F′6, HAG-R′1, HAG-R′4, pGEM-up-F, and pGEM-dn-R. Sequence analyses were performed using a BigDye Terminator cycle sequencing kit (Beckman Coulter, Inc., Brea, CA) and a DNA sequencer, CEQ2000XL (Beckman Coulter). To obtain the remaining regions of the HaG gene, PCR was performed with two primer sets designed on the basis of the analyzed DNA sequence, HAG-I1 and CS1 and HAG-I2 and CSI. Nested PCR was followed by PCR with primers HAG-F′1 and CS2 and primers HAG-I3 and CS2, respectively. Amplified DNA fragments were cloned into the pGEM-T Easy vector and sequenced. A homology search was performed using the Basic Local Alignment Search Tool (BLAST) program (5).
Nucleotide sequence accession number.
The sequence data for HaG are available from DDBJ with accession number AB663683.
RESULTS
Identification of bacterial strain H11 isolated from coral.
A bacterial strain, H11, isolated from coral was selected as the enzyme source because it appeared to produce an enzyme harboring strong transglucosylation activity toward glycerol when it was cultivated with the medium containing soluble starch. A phylogenetic tree constructed with its 16S rRNA sequence showed that the H11 strain is closely related to Halomonas aquamarina, Halomonas meridiana, and Halomonas axialensis and is likely to be categorized in the Halomonas genus (see Fig. S1 in the supplemental material). Its 16S rRNA sequence showed 99.5% identities to the 16S rRNA sequences of both H. aquamarina DMS 30161 and H. meridiana DMS 5425. Physiological tests revealed that the H11 strain is rod shaped (0.7 to 0.8 by 1.2 to 2.0 μm), pale yellow, smooth surfaced, Gram negative, mobile, and capable of growing at 4 to 45°C. It showed catalase, oxidase, urease, and cytochrome oxidase activities. Otherwise, all of the following were negative: nitrate reduction; production of indole; glucose oxidation; arginine dihydrolase and β-galactosidase activities; lipase activity for Tween 80; hydrolysis of esculin, gelatin, starch, and casein; and use of glucose, l-arabinose, d-mannose, d-mannitol, N-acetylglucosamine, maltose, potassium gluconate, n-capric acid, adipic acid, dl-malic acid, sodium citrate, and phenyl acetate. These features were not totally identical to those of any Halomonas species reported so far (23, 53). Therefore, the H11 strain was named Halomonas sp. H11 and deposited at the National Biological Resource Center (Chiba, Japan) as NBRC 108813.
Purification of HaG.
HaG was purified from a cell extract to homogeneity by several types of column chromatography (see Table S2 in the supplemental material). After hydrophobic column chromatography with Toyopearl Butyl-650 M, the total enzyme activity significantly increased because of activation by NH4+, described in detail later. The purified enzyme showed a single band in SDS-PAGE (see Fig. S2 in the supplemental material), and its molecular mass was estimated to be 58 kDa. It showed 50.9-fold higher specific activity than that of the crude extract.
Effects of pH and temperature.
The effects of pH and temperature on enzyme activity and stability were investigated, and these are summarized in Fig. S3 in the supplemental material. The maximum activity was observed at pH 6.5 when the buffer was 20 mM Britton-Robinson buffer. In 40 mM HEPES-NaOH (pH 7.0), the optimum temperature for HaG was 30°C, and interestingly, it exhibited more than 50% of the maximum activity even at 4°C. As will be mentioned in detail later, the monovalent cation in the buffer used was important for both pH activity and temperature activity. HaG was stable in the range from medium pH to the alkaline side at 4°C for 24 h. After treatment at 4 to 60°C for 15 min, the original activity was retained below 45°C but abruptly decreased above 50°C.
Substrate specificity.
The hydrolytic rates for various substrates were measured (Table 1). HaG showed high activity for maltose, pNPG, and sucrose, in this order. The hydrolysis rates for isomaltose, trehalose, nigerose, and maltotriose were negligible, and kojibiose was not hydrolyzed at all. Malto-oligosaccharides longer than maltotriose, starch, and methyl α-d-glucoside were rarely hydrolyzed.
Table 1.
Hydrolytic rates for various substrates
| Substrate | Hydrolytic rate (s−1) | Relative hydrolysis activitya (%) |
|---|---|---|
| Maltose | 0.763 | 100 |
| Isomaltose | 0.016 | 1.1 |
| Trehalose | 0.024 | 2.6 |
| Nigerose | 0.032 | 4.2 |
| Kojibiose | NDb | ND |
| Maltotriose | 0.040 | 5.3 |
| Maltotetraose | ND | ND |
| Maltopentaose | ND | ND |
| Maltohexaose | ND | ND |
| Sucrose | 0.253 | 33 |
| Methyl-α-d-glucoside | ND | ND |
| Soluble starch | ND | ND |
| pNPG | 0.439 | 57.5 |
Reaction rates on 4 mM each substrate (expect soluble starch was 0.2% [wt/vol]) are shown, where the activity to maltose is 100 for calculating relative activity.
ND, not detectable.
Effects of various salts on HaG activity.
The effects of various salts on HaG activity were investigated. The relative activities in the presence of a 10 mM salt concentration, where the activity with no salt was regarded as 100%, were as follows: LiCl, 151%; NaCl, 116%; KCl, 601%; RbCl, 579%; CsCl, 332%; NH4Cl, 548%; MgCl2, 30%; CaCl2, 20%; CoCl2, 1%; and ZnCl2, MnCl2, FeCl2, CuCl, and AgNO3, not detectable. (NH4)2SO4 also caused an increase in HaG activity to 545% when the molar concentration of NH4+ was equal to that of NH4Cl. The results indicate that HaG activity was obviously activated by monovalent cations. On the other hand, HaG activity was inhibited by divalent cations such as Mg2+ and Ca2+.
Kinetic analysis for activation of HaG by monovalent cations.
The results described above showed that monovalent cations, particularly K+, Rb+, NH4+, and Cs+, were strong activators for HaG. To clarify the effects of these monovalent cations, the apparent kinetic parameters for maltose and pNPG in the absence or presence of a 10 mM concentration of activators were measured (Table 2). The apparent Km (Kmapp) and apparent kcat (kcatapp) values for maltose in the absence of monovalent cations were 4.50 mM and 0.80 s−1, respectively. These values were most improved to 1.45 mM and 7.43 s−1 by NH4+ and K+, respectively. The kcatapp/Kmapp value was increased 22.4-fold by NH4+ relative to that without any monovalent cations. For pNPG, Kmapp and kcatapp were 4.64 mM and 1.00 s−1, respectively, in the absence of an activator. They were almost the same as those for maltose. On the other hand, Kmapp was significantly improved to about 0.02 mM in the presence of all the monovalent cations tested. Although the changes were less, kcatapp values were also improved, to 4.36 s−1, 3.29 s−1, 1.55 s−1, and 3.59 s−1 by K+, Rb+, Cs+, and NH4+, respectively. The kcatapp/Kmapp values were increased 969-fold, 893-fold, 372-fold, and 929-fold by K+, Rb+, Cs+, and NH4+, respectively.
Table 2.
Apparent kinetic parameters for maltose and pNPG in the absence or presence of monovalent cationsa
| Activator | Maltose |
pNPG |
||||||
|---|---|---|---|---|---|---|---|---|
| kcatapp (s−1) | Kmspp (mM) | kcatapp/Kmapp (s−1 mM−1) | kcatapp/Kmapp (fold) | kcatapp (s−1) | Kmspp (mM) | kcatapp/Kmapp (s−1 mM−1) | kcatapp/Kmapp (fold) | |
| None | 0.80 | 4.50 | 0.18 | 1 | 1.00 | 4.64 | 0.22 | 1 |
| K+ | 7.43 | 2.06 | 3.61 | 20.3 | 4.36 | 0.021 | 209 | 969 |
| Rb+ | 6.73 | 1.92 | 3.51 | 19.7 | 3.29 | 0.017 | 193 | 893 |
| Cs+ | 5.23 | 2.88 | 1.82 | 10.2 | 1.55 | 0.019 | 80.2 | 372 |
| NH4+ | 5.77 | 1.45 | 3.98 | 22.4 | 3.59 | 0.018 | 200 | 929 |
The hydrolysis rate was assayed in the presence of 10 mM activator (cation) at various substrate concentrations. Parameters were calculated by regressing the experimental data to the Michaelis-Menten equation.
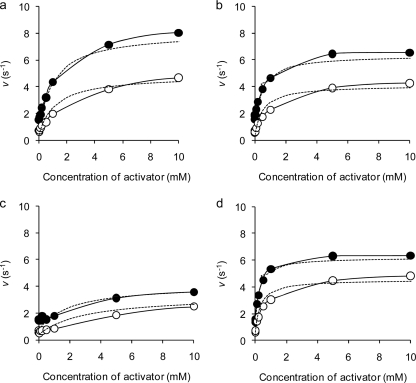
Next, the kinetic parameters for maltose hydrolysis in a nonessential activation model were measured to evaluate the activation by K+, Rb+, NH4+, and Cs+ (Table 3). The measured reaction rates for 4.5 mM or 50 mM maltose with various concentrations of activators fitted well to the theoretical lines, indicating that activation by monovalent cations was explained well by a nonessential activation model (Fig. 2). The activation degree reached almost maximum at 10 mM each activator and even over the range of 10 to 100 mM. The actual hydrolytic rates for 4.5 mM and 50 mM maltose in the absence of an activator were 0.71 s−1 and 1.53 s−1, respectively, whereas those for 4.5 mM maltose increased 8.3-fold, 5.9-fold, 5.0-fold, and 7.1-fold in the presence of 50 mM K+, Rb+, Cs+, and NH4+, respectively. Similarly, the rates for 50 mM maltose were increased 5.7-fold, 4.0-fold, 2.7-fold, and 4.3-fold by the same concentration of the respective monovalent cations.
Table 3.
Kinetic parameters for hydrolysis of maltose in nonessential activator model
| Activator | kcat1 (s−1) | kcat2 (s−1) | Km (mM) | KmA (mM) | KA (mM) | KAS (mM) |
|---|---|---|---|---|---|---|
| K+ | 0.8 | 8.6 | 4.5 | 3.52 | 1.35 | 1.06 |
| Rb+ | 0.8 | 6.7 | 4.5 | 2.88 | 0.66 | 0.42 |
| Cs+ | 0.8 | 4.2 | 4.5 | 1.19 | 7.14 | 1.88 |
| NH4+ | 0.8 | 6.4 | 4.5 | 1.86 | 0.54 | 0.22 |
Fig 2.
Activator-reaction rate plots for hydrolysis of maltose. Reaction rates for 4.5 mM (open circles) and 50 mM (solid circles) maltose were measured in the presence of various concentrations of monovalent cations. The averages of at least two measurements less than 5% standard deviation are shown. Theoretical lines (dotted lines) were drawn on the basis of Equation 1 and measured kinetic parameters. (a) K+; (b) Rb+; (c) Cs+; (d) NH4+.
The KA and KAS values for NH4+, which mirror the affinity of the activator with the enzyme and the enzyme-substrate complex, respectively, were the lowest among the monovalent cations tested, followed by Rb+, K+, and Cs+, in this order. The KmA values were as follows: Cs+, 1.19 mM; NH4+, 1.86 mM; Rb+, 2.88 mM; and K+, 3.52 mM. The kcat2 value for the hydrolysis of maltose in the presence of K+ was the best of those for the monovalent cations tested, and those with Rb+, NH4+, and Cs+ followed, in this order.
Effects of monovalent cations on pH-activity and temperature-activity.
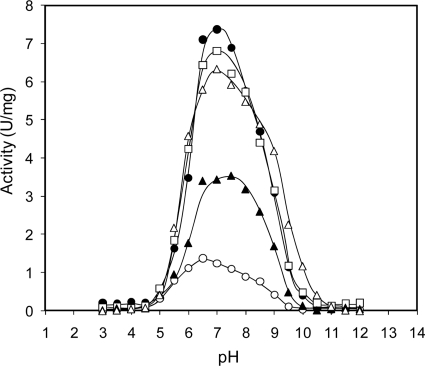
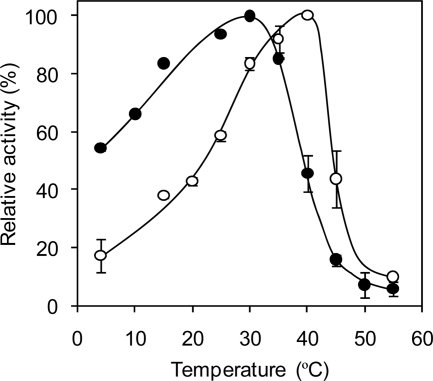
The effects of NH4+ on enzyme activity at various pH values and temperatures were evaluated. As shown in Fig. 3, the activities at all pH values were raised by monovalent cations in the following order: K+, Rb+, NH4+, and Cs+. These ions shifted the bell-shaped pH-activity curve to the alkaline side. The optimum pH in the absence of monovalent cations was 6.5, that in the presence of 10 mM K+, Rb+, and NH4+ was 7.0, and that in the presence of Cs+ was 7.5. NH4+ was most effective in increasing the activity around the alkaline side. Interestingly, the optimum temperature for HaG activity was shifted from 30°C to 40°C in the presence of 10 mM NH4+ (Fig. 4), although stability was not affected (data not shown). The actual activity in the presence of NH4+ at 40°C was 11.6-fold higher than that in its absence.
Fig 3.
pH-activity profiles of HaG in the presence of monovalent cations. Enzyme activity was assayed at various pH values. The averages of at least two measurements less than 5% standard deviation are shown. Open circles, no monovalent cation; solid circles, K+; open triangles, Rb+; solid triangles, Cs+; open squares, NH4+.
Fig 4.
NH4+ effect on temperature-activity curve of HaG. Enzyme activity was measured at various temperatures in the absence or presence of 10 mM NH4+. The activities at 30°C and 40°C in the presence and absence, respectively, of NH4+ were regarded as 100%. The averages on at least two measurements are shown. Solid and open circles, HaG activity measured in the presence of 0 and 10 mM NH4+, respectively.
Transglucosylation toward various alcohols and enzymatic synthesis of α-d-glucosylglycerol.
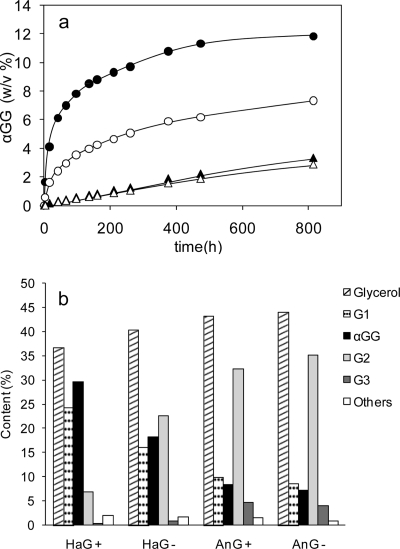
The transglucosylation activities toward various alcohols, i.e., ethanol, glycerol, propylene glycol, 1-propanol, and 2-propanol, were investigated. Transglucosylation products were detected in all the reactions tested (data not shown). As expected from the results on substrate specificity, no oligosaccharides were produced by transglucosylation. The efficiencies of the synthesis of αGG by HaG and AnG were compared in the absence and presence of 10 mM NH4+. The time course of αGG production is shown in Fig. 5a. The production rate and yield of αGG by HaG in the presence of NH4+ were the highest among the four cases tested. The carbohydrate contents (percent, wt/wt) of the reaction mixture at 816 h are summarized in Fig. 5b. In the reaction of HaG in the presence of NH4+, the content of αGG represented 29.4% (wt/wt) of the total carbohydrates (the molar yield of αGG from converted maltose was 92.7%), and the αGG content for the reaction of AnG was about 8% (wt/wt), independent of NH4+. The accumulations of glucose and trisaccharides (G3) were considerably less in the reaction with HaG than in that with AnG. The compositions of the stereoisomers of αGG at 816 h were analyzed by gas chromatography, revealing that the ratios of the constituent isomers 2-O-αGG/(2R)-1-O-αGG/(2S)-1-O-αGG were 10:52:38 for HaG and 7:73:20 for AnG.
Fig 5.
Enzymatic synthesis of αGG. (a) Time course of production of αGG. αGG (percent, wt/vol) produced in the reaction mixture with 20% (wt/vol) maltose, 20% (wt/vol) glycerol, and 0.04 U/ml enzymes. Open and solid circles, reaction with purified HaG in the absence and presence of 10 mM NH4+, respectively; open and solid triangles, reaction with purified HaG in the absence and presence of α-glucosidase from Aspergillus niger, respectively. (b) Content (percent, wt/wt) of carbohydrates at a reaction time of 816 h. Plus and minus signs, presence and absence of NH4+, respectively. The carbohydrate content is equivalent to the area percentage of the detector, i.e., the refractive index response, in HPLC analysis. G1, glucose; αGG, α-glucosylglycerol; G2, disaccharides; G3, trisaccharides.
Sequence analysis of HaG.
The N-terminal amino acid sequence of purified HaG was MQDNMMWWRGGVIYQIYPRS. Primers were designed on the basis of this sequence, and the HaG gene was obtained by the cassette PCR method (22). The HaG gene consisted of 1,617 bases and encoded 538 amino acids (Fig. 6). The theoretical molecular mass and pI were 61,139 Da and 4.75, respectively. The partial internal peptide sequence TLGAPEANPY was found in this sequence. Four regions, I, II, III, and IV, conserved in the GH family 13 enzymes, were found in the amino acid sequence of HaG, namely, DQVISH, GFRLDTVNF, EIGD, and ATSNHD. HaG was therefore obviously classified in GH family 13. The amino acid sequence of HaG exhibited high identities, of the order of 87%, 79%, 78%, 69%, and 68%, to those of putative α-amylases (provably α-glucosidase) from Halomonas sp. strain TD01 (GenBank accession number EGP19311), Chromohalobacter salexigens DSM 3043 (GenBank accession number ABE57826), Halomonas elongata DSM 2581 (GenBank accession number CBV43561), Marinomonas posidonica IVIA-Po-181 (GenBank accession number AEF56441), and Marinomonas sp. strain MED121 (GenBank accession number EAQ64921), respectively. Targeting validated proteins, HaG showed 60%, 58%, 57%, and 56% identities to α-glucosidase from Xanthomonas campestris WU-9701 (GenBank accession number BAC87873), oligo-1,6-glucosidase from X. campestris pv. raphani 756C (GenBank accession number AEL07660), oligo-1,6-glucosidase from Pseudomonas stutzeri DSM 4166 (GenBank accession number AEA85435), and α-glucosidase from Agrobacterium tumefaciens F2 (GenBank accession number EGP58908), respectively. Identities to α-glucosidase from Geobacillus sp. strain HTA-462 (Protein Data Bank [PDB] accession number 2ZE0) (43), trehalulose synthase from Pseudomonas mesoacidophia (PDB accession number 2PWH), oligo-1,6-glucosidase from Bacillus cereus (PDB accession number 1UOK) (54), and dextran glucosidase from Streptococcus mutans (PDB accession number 2ZIC) (20), all of whose crystal structures have been solved, were 36%, 35%, 33%, and 31%, respectively.
Fig 6.
Multiple-sequence alignment of amino acid sequences of HaG and some α-glucosidases. Amino acid sequences of HaG and α-glucosidases from Xanthomonas campestris WU-9701 (GenBank accession number BAC87873), Agrobacterium tumefaciens F2 (GenBank accession number EGP58908), and Geobacillus sp. HTA-462 (PDB accession number 2ZE0), oligo-1,6-glucosidase from Bacillus cereus (PDB accession number 1UOK), and dextran glucosidase from Streptococcus mutans (PDB accession number 2ZIC) were aligned using the Clustal Omega program (16, 45). Asterisks, colons, and periods indicate completely conserved, strongly similar, and weakly similar residues, respectively, as mentioned on the website (http://www.ebi.ac.uk/Tools/msa/clustalo/help/faq.html#23). α and β above the sequence indicate the predicted α helices and β strand, respectively. Four conserved regions, I, II, III, and IV, common in GH family 13 are shaded. N-terminal and partial internal amino acid sequences analyzed by Edman degradation are underlined.
DISCUSSION
In this work, we describe a unique α-glucosidase which exhibits strong transglucosylation activity toward alcohols and glycerol and has additional remarkable features, namely, stimulation by monovalent cations and narrow substrate specificity. The enzyme is probably psychrophilic and prefers a moderate pH (Fig. 3; see Fig. S3 in the supplemental material). These conditions are well suited to the marine environment where Halomonas sp. H11 was isolated. Halomonas sp. H11 was negative for the hydrolysis of starch and utilization of maltose or glucose under the conditions of the physiological tests. However, it could utilize starch when soluble starch was contained in the culture medium and produced HaG. This suggests that expression of enzymes that are related to glycoside hydrolysis is probably induced by derivatives of starch.
HaG was found to have specific activity toward maltose, sucrose, and pNPG but rarely had activity toward longer malto-oligosaccharides or disaccharides other than maltose (Table 1). Such a narrow specificity for the substrate chain length by HaG is remarkable because bacterial α-glucosidases generally hydrolyze not only maltose but also longer malto-oligosaccharides (21, 24, 36).
On the basis of multiple-sequence alignment, HaG was predicted to have a longer β → α loop 4, which connects β-strand 4 and α-helix 4, than other enzymes (Fig. 6). The length of β → α loop 4 is the determinant of the specificity for substrate chain length in GH family 13 α-glucosidases and related enzymes, such as oligo-1,6-glucosidase and dextran glucosidase (40). The shorter loop of dextran glucosidase, which forms open substrate binding sites to accommodate long-chain substrates, has been shown to be associated with a high preference for long-chain substrates. Compared with α-glucosidases and oligo-1,6-glucosidase, HaG has an even longer β → α loop 4. The length of the putative β → α loop 4 of α-glucosidase from X. campestris WU-9701 (GenBank accession number BAC87873), which hydrolyzes almost only maltose among the malto-oligosaccharides, as does HaG (52), is the same as that of HaG. We expect that such an unusually long β → α loop 4 might contribute to the narrow specificity for substrate chain length in HaG.
HaG was greatly stimulated by monovalent cations, namely, K+, Rb+, NH4+, and Cs+. Although numerous enzymes have been documented to be activated by monovalent cations in the plant and animal world (44), most types of known α-glucosidases in GH family 13 are independent of monovalent cations. There have been only two reports documenting stimulation of α-glucosidase activity by monovalent cations. Crude α-glucosidases from fast-growing rhizobia and A. tumefaciens were reported to be stimulated by K+, NH4+, and Rb+ (19), and purified α-glucosidase from Rhizobium sp. strain USDA 4280 was reported to be stimulated to 141% and 207% by addition of 10 mM K+ and NH4+, respectively (7). However, such α-glucosidases have as yet rarely been investigated in detail. HaG exhibited high sequence identity (56%) to α-glucosidase from A. tumefaciens F2 (GenBank accession number EGP58908), implying that these enzymes share a monovalent cation binding site structure and activation mechanism.
We determined the activation kinetics and enzymological properties of HaG. The monovalent cation activators K+, Rb+, NH4+, and Cs+ for HaG have similar ionic diameters. In particular, the ionic diameters of Rb+ and NH4+ are almost the same (Rb+, 1.48 Å; NH4+, 1.43 Å). These ions showed almost the same KA and KAS values (Table 3). In addition, the kcat2 values for maltose hydrolysis in the presence of these ions were also similar. The KA and KAS values for Cs+, which has an ionic diameter of 1.70 Å, were higher than those for the other ions. The kcat2 value was highest with the smallest ion, K+, whose diameter is 1.33 Å. These results indicate that the activator binding site of HaG is suitable for binding to monovalent ions with ionic diameters of about 1.4 Å.
The Kmapp and kcatapp values for maltose and pNPG were improved by the addition of K+, Rb+, Cs+, and NH4+ (Table 2). In particular, the Kmapp values for pNPG were significantly improved, regardless of the type of monovalent cation. Enzyme-pNPG complexes might be stabilized by interaction between the negative charge of the nitrate group of pNP and a monovalent cation bound to the substrate binding site of the enzyme. Monovalent cation activators may therefore be involved in the formation of the substrate binding sites of HaG.
Unlike the minority of α-glucosidases, which are stimulated by monovalent cations, one group of α-amylases classified in the same GH family 13, i.e., α-amylases from mammals and Pseudoalteromonas haloplanktis, are allosterically activated by chloride ions (11, 30). In all chloride-dependent α-amylases whose crystal structures have been solved (2, 3, 8, 28, 38, 39, 46), Cl− binds to a common site, close to the center of the catalytic barrel in the vicinity of the active site. Some of these amylases have been further investigated by structural and kinetic analyses with mutant enzymes (4, 30); Cl− may play a role by polarizing the catalytic water molecule and interacting with the general acid/base catalyst to elevate the acid dissociation constant (pKa) value of the Glu residue (14). In the case of HaG, the enzyme activity at the alkaline side was enhanced by addition of monovalent cations (Fig. 3). This indicated that the pKa value of the acid/base catalyst was increased, as observed in chloride-dependent α-amylases. Monovalent cations might interact with the second catalytic Asp, which is situated at the conserved region IV and forms hydrogen-bonding interactions with the general acid/base catalyst, thereby increasing the pKa value of the acid/base (30).
It was notable that monovalent cations increased the optimum temperature of HaG (Fig. 4), but as yet we have no explanation for this phenomenon. The temperature-activity profiles in the absence and presence of NH4+ were similar to those of psychrophilic and mesophilic α-amylases (13). We expect that a study of the interactions of monovalent cations and HaG will provide some insights to aid understanding of the environmental adaptation of the enzyme.
HaG efficiently transferred glucosyl residues to some alcohols. The precise structures of these products were not analyzed, but based on the properties of HaG, they were thought to be α-glucosides linked to the OH groups of alcohols. The glucosylation efficiency toward glycerol to produce αGG was superior to that of AnG when their hydrolysis activities for maltose were compared (Fig. 5), implying that HaG has stronger transglucosylation activity than does AnG. The synthesis of αGG was more efficient in the presence of NH4+, suggesting that a monovalent cation stimulated not only hydrolysis but also the transglucosylation activity of HaG. As deduced from the substrate specificity (Table 1), smaller amounts of trisaccharides were produced in the reaction with HaG than in that with AnG. Such an efficient production rate and the yield of αGG, with few by-products, are very attractive properties of HaG. The transglucosylation reaction for glycerol was carried out with the enzymes in a considerably smaller amount (0.04 U/ml) to distinguish the effects of activator and the differences between HaG and AnG. It should be noted that the length of the reaction time can be shortened if the amount of enzymes is increased (data not shown).
A sequence similarity search by BLAST showed that putative proteins with high similarities to HaG are distributed in moderate halophilic, marine (Halomonas spp. and Marinomonas spp.), and plant pathogen-related (Agrobacterium spp. and Xanthomonas spp.) bacteria. It seems that these bacteria have in common an adaptation to saline or nitrate-rich environments. Some kinds of Halomonas spp. have been reported to exist at extreme ocean depths (23, 42, 51), and Halomonas elongata DMS 3043 is known to accumulate compatible solutes such as ectoine and betaine to balance high osmotic pressure (55). αGG is also known to be accumulated as a compatible solute in cyanobacteria and some osmotolerant bacteria to adopt a high osmotic pressure. In cyanobacteria, the synthesis of αGG is carried out by two enzymes, GG-phosphate synthase (EC 2.4.1.213) and GG-phosphate phosphatase (EC. 3.1.3.69), with glycerol 3-phosphate and ADP-glucose (25). As there is no report that α-glucosidases might be involved in the formation of αGG in any living organisms and the general task of α-glucosidase is producing glucose from the α-glucoside for a carbon source, we do not yet have an idea why HaG hydrolyzes maltose specifically and exhibits strong transglucosylation activity toward glycerol and some other alcohols. α-Glucosidases similar to HaG in marine bacteria are expected to have properties similar to those of HaG. Further investigation of the relations between enzymes and their environment is required.
Supplementary Material
Footnotes
Published ahead of print 6 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aerts D, et al. 2011. Transglucosylation potential of six sucrose phosphorylases toward different classes of acceptors. Carbohydr. Res. 346:1860–1867 [DOI] [PubMed] [Google Scholar]
- 2. Aghajari N, Feller G, Gerday C, Haser R. 1998. Crystal structures of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7:564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aghajari N, Feller G, Gerday C, Haser R. 1998. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 6:1503–1516 [DOI] [PubMed] [Google Scholar]
- 4. Aghajari N, Feller G, Gerday C, Haser R. 2002. Structural basis of α-amylase activation by chloride. Protein Sci. 11:1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 6. Barrow GI, Feltham RKA. 1993. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed Cambridge University Press, New York, NY [Google Scholar]
- 7. Berthelot K, Delmotte FM. 1999. Purification and characterization of an α-glucosidase from Rhizobium sp. (Robinia pseudoacacia L.) strain USDA 4280. Appl. Environ. Microbiol. 65:2907–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brayer GD, Luo Y, Withers SG. 1995. The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci. 4:1730–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cantarel BL, et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiba S. 1988. α-Glucosidases, p 104–116 In Amylase Research Society of Japan (ed), Handbook of amylases and related enzymes, 1st ed Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 11. D'Amico S, Gerbay C, Feller G. 2000. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene 253:95–105 [DOI] [PubMed] [Google Scholar]
- 12. Dixon M, Webb EC. 1979. Enzyme inhibition and activation, p 389–391 In Dixon M, Webb EC. (ed), Enzymes, 3rd ed Academic Press, New York, NY [Google Scholar]
- 13. Feller G, et al. 1994. Stability and structural analysis of α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 222:441–447 [DOI] [PubMed] [Google Scholar]
- 14. Feller G, Bussy O, Houssier C, Gerday C. 1996. Structural and functional aspects of chloride binding to Alteromonas haloplanctis α-amylase. J. Biol. Chem. 271:23836–23841 [DOI] [PubMed] [Google Scholar]
- 15. Goedl C, Sawangwan T, Mueller M, Schwarz A, Nidetzky B. 2008. A high-yielding biocatalytic process for the production of 2-O-(α-d-glucopyranosyl)-sn-glycerol, a natural osmolyte and useful moisturizing ingredient. Angew. Chem. Int. Ed. Engl. 47:10086–10089 [DOI] [PubMed] [Google Scholar]
- 16. Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harada N, Zhao J, Kurihara H, Nakagata N, Okajima K. 2010. Effects of topical application of α-d-glucosylglycerol on dermal levels of insulin-like growth factor-I in mice and facial skin elasticity in humans. Biosci. Biotechnol. Biochem. 74:759–765 [DOI] [PubMed] [Google Scholar]
- 18. Hincha DK, Hagemann M. 2004. Stabilization of model membranes during drying by compatible solutes involved in the stress tolerance of plants and microorganisms. Biochem. J. 383:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoelzle I, Streeter JG. 1989. Stimulation of α-glucosidases from fast-growing rhizobia and Agrobacterium tumefaciens by K+, NH4+, and Rb+. Can. J. Microbiol. 36:223–227 [Google Scholar]
- 20. Hondoh H, et al. 2008. Substrate recognition mechanism of α-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans. J. Mol. Biol. 378:911–920 [DOI] [PubMed] [Google Scholar]
- 21. Hung VS, et al. 2005. α-Glucosidase from a strain of deep-sea Geobacillus: a potential enzyme for the biosynthesis of complex carbohydrates. Appl. Microbiol. Biotechnol. 68:757–765 [DOI] [PubMed] [Google Scholar]
- 22. Isegawa Y, et al. 1992. Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): application to sequencing 6.5 kb genome segment of hantavirus strain B-1. Mol. Cell. Probes 6:467–475 [DOI] [PubMed] [Google Scholar]
- 23. Kaye JZ, Márquez MC, Ventosa A, Baross JA. 2004. Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54:499–511 [DOI] [PubMed] [Google Scholar]
- 24. Kelly CT, Fogarty W. 1983. Microbial α-glucosidases. Process Biochem. 18:6–12 [Google Scholar]
- 25. Klähn S, Hagemann M. 2011. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 13:551–562 [DOI] [PubMed] [Google Scholar]
- 26. Kurosu J, et al. 2002. Enzymatic synthesis of α-arbutin by α-anomer-selective glucosylation of hydroquinone using lyophilized cells of Xanthomonas campestris WU-9701. J. Biosci. Bioeng. 93:328–330 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 28. Larson SB, Greenwood A, Cascio D, Day J, McPherson A. 1994. Refined molecular structure of pig pancreatic α-amylase at 2.1 Å resolution. J. Mol. Biol. 235:1560–1584 [DOI] [PubMed] [Google Scholar]
- 29. Luley-Goedl C, Nidetzky B. 2010. Carbohydrate synthesis by disaccharide phosphorylases: reactions, catalytic mechanisms and application in the glycosciences. Biotechnol. J. 5:1324–1338 [DOI] [PubMed] [Google Scholar]
- 30. Maurus R, et al. 2005. Structural and mechanistic studies of chloride induced activation of human pancreatic α-amylase. Protein Sci. 14:743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCarter JD, Withers SG. 1996. Unequivocal identification of Asp-214 as the catalytic nucleophile of Saccharomyces cerevisiae α-glucosidase using 5-fluoro glycosyl fluorides. J. Biol. Chem. 271:6889–6894 [DOI] [PubMed] [Google Scholar]
- 32. Miwa I, Okuda J, Maeda K, Okuda G. 1972. Mutarotase effect on colorimetric determination of blood glucose with β-d-glucose oxidase. Clin. Chim. Acta 37:538–540 [DOI] [PubMed] [Google Scholar]
- 33. Murase H, Yamauchi R, Kato K, Kunieda T, Terao J. 1997. Synthesis of a novel vitamin E derivative, 2-(α-d-glucopyranosyl) methyl-2,5,7,8-tetramethylchroman-6-ol, by α-glucosidase-catalyzed transglycosylation. Lipids 32:73–78 [DOI] [PubMed] [Google Scholar]
- 34. Nakagawa K, Kawasaki H. 2001. DNA sequence analysis, p 83–117 In Society for Actinomycetes Japan (ed), Identification manual of Actinomycetes. Business Center for Academic Societies Japan, Tokyo, Japan [Google Scholar]
- 35. Nakano H, et al. 2003. Synthesis of glycosyl glycerol by cyclodextrin glucanotransferases. J. Biosci. Bioeng. 95:583–588 [PubMed] [Google Scholar]
- 36. Nakao M, et al. 1994. Structure and expression of gene coding for thermostable α-glucosidase with broad substrate specificity from Bacillus. sp. SAM1606. Eur. J. Biochem. 220:293–300 [DOI] [PubMed] [Google Scholar]
- 37. Nitta A, Takenaka F, Iki M, Matsumura E, Sakaguchi M. October 2007. Antitumor agents containing a-d-glucosylglycerols and food and cosmetics containing them. JP patent 2007-262023
- 38. Qian M, Haser R, Payan F. 1993. Structure and molecular model refinement of pig pancreatic α-amylase at 2.1 Å resolution. J. Mol. Biol. 231:785–799 [DOI] [PubMed] [Google Scholar]
- 39. Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. 1996. Structure of human salivary α-amylase at 1.6 Å resolution: implication for its role in the oral cavity. Acta Crystallogr. D Biol. Crystallogr. 52:435–446 [DOI] [PubMed] [Google Scholar]
- 40. Saburi W, Mori H, Saito S, Okuyama M, Kimura A. 2006. Structural elements in dextran glucosidase responsible for high specificity to long chain substrate. Biochim. Biophys. Acta 1764:688–698 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Sánchez-Porro C, Kaur B, Mann H, Ventosa A. 2010. Halomonas titanicae sp. nov., a halophilic bacterium isolated from the RMS Titanic. Int. J. Syst. Evol. Microbiol. 60:2768–2774 [DOI] [PubMed] [Google Scholar]
- 43. Shirai T, Hung VS, Morinaka K, Kobayashi T, Ito S. 2008. Crystal structure of GH13 α-glucosidase GSJ from one of the deepest sea bacteria. Proteins 73:126–133 [DOI] [PubMed] [Google Scholar]
- 44. Shulter CH. 1970. Enzymes activated by monovalent cations. Science 168:789–795 [DOI] [PubMed] [Google Scholar]
- 45. Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strobl S, et al. 1998. Crystal structure of yellow meal worm α-amylase at 1.64 Å resolution. J. Mol. Biol. 278:617–628 [DOI] [PubMed] [Google Scholar]
- 47. Suzuki Y, Suzuki K. 1991. Enzymatic formation of 4G-α-d-glucopyranosyl-rutin. Agric. Biol. Chem. 55:181–187 [PubMed] [Google Scholar]
- 48. Takaku H. 1988. Manufacture of oligosaccharides, p 212–222 In Amylase Research Society of Japan (ed), Handbook of amylases and related enzymes, 1st ed Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 49. Takenaka F, Uchiyama H, Imamura T. 2000. Identification of α-d-glucosylglycerol in sake. Biosci. Biotechnol. Biochem. 64:378–385 [DOI] [PubMed] [Google Scholar]
- 50. Takenaka F, Uchiyama H. 2000. Synthesis of α-d-glucosyl glycerol by α-glucosidase and some its characteristics. Biosci. Biotechnol. Biochem. 64:1821–1826 [DOI] [PubMed] [Google Scholar]
- 51. Tamegai H, et al. 2006. Halomonas sp. strain DT-W, a halophile from the 11,000 m depth of the Mariana Trench. Extremophiles 5:27–33 [Google Scholar]
- 52. Usami S, et al. February 2001. Production of glycoside with α-glucosidase and new α-glucosidase and its production. JP patent 2001-46096
- 53. Vreeland RH. 2005. Genus I. Halomonas, p 300–313 In Garrity GM, et al. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2, part B Springer, New York, NY [Google Scholar]
- 54. Watanabe K, Hata Y, Kizaki H, Katsube Y, Suzuki Y. 1997. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269:142–153 [DOI] [PubMed] [Google Scholar]
- 55. Wohlfarth A, Severin J, Galinski EA. 1990. The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J. Gen. Microbiol. 135:705–712 [Google Scholar]
- 56. Yamamoto I, Muto N. 1992. Bioavailability and biological activity of l-ascorbic acid 2-O-alpha-glucoside. J. Nutr. Sci. Vitaminol. Spec. No. 1992:161–164 [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto T, Unno T, Sugawara M, Goda T. 1999. Properties of a nigeose and nigerosylmaltooligosaccharides-suppplemented syrup. J. Appl. Glycosci. 46:475–482 [Google Scholar]
- 58. Yoshida K, Takenaka F, Nitta A, Iki M. June 2007. α-d-Glucopyranosyl glycerol derivatives as antiallergic agents, health foods, and cosmetics. JP patent 2007-137862
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.