Abstract
The reliable detection of airborne biological threat agents depends on several factors, including the performance criteria of the detector and its operational environment. One step in improving the detector's performance is to increase our knowledge of the biological aerosol background in potential operational environments. Subway stations are enclosed public environments, which may be regarded as potential targets for incidents involving biological threat agents. In this study, the airborne bacterial community at a subway station in Norway was characterized (concentration level, diversity, and virulence- and survival-associated properties). In addition, a SASS 3100 high-volume air sampler and a matrix-assisted laser desorption ionization–time of flight mass spectrometry-based isolate screening procedure was used for these studies. The daytime level of airborne bacteria at the station was higher than the nighttime and outdoor levels, and the relative bacterial spore number was higher in outdoor air than at the station. The bacterial content, particle concentration, and size distribution were stable within each environment throughout the study (May to September 2010). The majority of the airborne bacteria belonged to the genera Bacillus, Micrococcus, and Staphylococcus, but a total of 37 different genera were identified in the air. These results suggest that anthropogenic sources are major contributors to airborne bacteria at subway stations and that such airborne communities could harbor virulence- and survival-associated properties of potential relevance for biological detection and surveillance, as well as for public health. Our findings also contribute to the development of realistic testing and evaluation schemes for biological detection/surveillance systems by providing information that can be used to mimic real-life operational airborne environments in controlled aerosol test chambers.
INTRODUCTION
As of 2011, more than 120 cities worldwide have underground railway transportation systems (subways), which transport about 200 million people daily. Public places, and especially locations where people are confined in enclosed spaces, such as subway stations, may be regarded as potential targets for the dispersion of biological threat agents in air. Even though no successful deliberate dispersion of biological threat agents in subway environments has been previously reported, such incidents cannot be ruled out. The dispersion of sarin nerve gas (a chemical threat agent) by the Japanese religious cult, Aum Shinrikyo, killed 11 and injured over 5,000 persons in the Tokyo subway in 1995 (50). Aum Shinrikyo failed in causing anthrax infection when aerosolizing B. anthracis in Tokyo 2 years earlier, due to the use of a low-virulence vaccine strain (68).
Harmful concentrations of pathogenic microorganisms or their toxins in airborne environments could occur following a deliberate dispersion of biological threat agents but might also be a result of an unintentional release from natural sources. Most biological threat agents will not induce any immediate effects in humans even after fatal exposures, and in the absence of a reliable surveillance system, the public would most likely be unaware of an incident involving biological threat agents until exposed individuals seek medical assistance up to several days later. An early warning, detection, and response scheme could contribute to minimizing the consequences of such incidents (49). Both military and civilian societies/authorities have expressed an urgent need for detection/surveillance systems, and a lot of effort is being put into the development and “testing and evaluation” (T&E) of biological detectors. Biological detectors need to fulfill stringent requirements before they can be deployed for reliable surveillance purposes. The detectors' performance criteria, such as sensitivity and specificity, will be challenged during operation in complex airborne environments, which could lead to false-positive or -negative detection events reducing the detection equipment's credibility. Currently, few if any, available biological detectors have been able to meet the users' requirements regarding reliable sensitive and specific real-time monitoring of biological threat agents in different operational environments (49). This is partly due to the complex nature of microorganisms, and the natural occurrence of similar but nonpathogenic environmental relatives of the biological threat agents. T&E of biological detectors is in general performed in aerosol test chambers, where they are challenged with known amounts of live, attenuated, or killed biological threat agents, or their appropriate simulants, to determine important detector properties such as detection limits. However, it is also necessary to test biological detectors in realistic operational environments since there are several major differences between these environments and aerosol test chambers. In real-life environments, the biological detector will continuously be challenged with a complex and dynamic mixture of both biological and nonbiological airborne material, which should not interfere with its performance in detecting the biological threat agents of concern. Research has shown that several biological detectors do not perform optimally when tested in operational environments (20, 49). A lot of effort has been put into improving T&E methodologies for biological detectors by establishing aerosol test chamber systems that can mimic real-life environmental backgrounds (55, 59, 74), thus enabling operational testing of biological detectors by challenging them with biological threat agents in the presence of a more realistic background. The ability to mimic real-life conditions depends on the availability of detailed information about the environments that are to be mimicked. Such information is currently very limited, and further investigation of naturally occurring airborne microorganisms in potential target environments is therefore important. Information about naturally occurring airborne microorganisms in crowded public environments may also be important from a public health perspective, since elevated levels of airborne microorganisms are considered an important factor that affects indoor air quality and have been linked to adverse human health effects due to their potential toxigenic, allergenic, and infectious nature (19).
For the present study, we performed a detailed characterization of the cultivable airborne bacterial environment at the largest underground subway station in Oslo, Norway, from May to September of 2010. Daytime, nighttime, and outdoor reference samples were collected using a high-volume open-faced electret filter-based air sampler (SASS 3100) and analyzed by cultivation to enumerate total and spore-specific airborne bacterial concentrations. Microbiological, biochemical, and molecular methods were used to taxonomically classify the obtained bacterial isolates and to investigate virulence- and survival-associated properties, such as antibiotic resistance, hemolytic activity, and pigmentation. The results described here regarding the natural concentration level, composition, and variability of airborne bacteria in a subway station increase our knowledge about potential target airborne environments that biological detectors may be subjected to and provide information about the naturally occurring biological aerosols (bioaerosols) that biological detectors should be tested against during T&E in aerosol test chambers. Also, our work generates relevant baseline data that could be used when assessing human exposure to airborne bacteria in subway environments. In addition, we describe new methodologies for air quality analysis that might be of interest and applied in public health studies and safety assessments.
MATERIALS AND METHODS
Study location.
The study was conducted at the underground subway station Nationaltheatret, Oslo (ca. 600,000 inhabitants), Norway. On average one train departs every minute during the operating hours of the station, and the subway network yearly transports over 70 million people. The station is nonoperative between 1 and 5 a.m., except for maintenance activities. No heating, ventilation, and air conditioning system is installed at the station. Air samples were collected during a 5-month period, from May through September of 2010, at 2- to 3-week intervals (Table 1). Samples were collected at one location inside the subway station and at one outdoor location (reference). The indoor sampling was performed at the westbound concourse during the day (11 a.m. to 1 p.m.) and night (2 to 4 a.m.), and the outdoor reference sampling was performed during the day (9 to 11 a.m.) at a square adjacent to the stations entrance. Nighttime outdoor reference samples were not collected because of practical and security-related issues. Nighttime station samples and daytime outdoor samples were not collected on the first two and three sampling dates, respectively, since permissions to do such sampling were not available at these initial dates.
Table 1.
Overview of airborne bacterial concentrations and meteorological parameters
| Samplea | Cloud cover | Mean ± SD |
CFU m of air−3 ± SDb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Temp (°C) | Humidity (% RH) | TSA | R2A | TSA* | R2A* | TSA† | R2A† | ||
| 180510D | 17.5 ± 0.4 | 69.1 ± 3.8 | 480 ± 26 | 517 ± 22 | < | < | < | < | |
| 140610D | 18.3 ± 0.3 | 62.9 ± 6.4 | 461 ± 82 | 454 ± 74 | 11 ± 0 | 17 ± 10 | < | < | |
| 280610D | 19.6 ± 0.4 | 69.8 ± 3.3 | 352 ± 69 | 344 ± 20 | 15 ± 8 | 7 ± 8 | < | < | |
| 280610N | 18.9 ± 0.5 | 63.3 ± 2.0 | 13 ± 18 | 30 ± 13 | < | < | < | < | |
| 260710D | 19.4 ± 0.5 | 75.6 ± 0.5 | 222 ± 29 | 289 ± 67 | < | < | < | < | |
| 260710N | 20.4 ± 0.3 | 71.6 ± 1.5 | 17 ± 6 | 30 ± 14 | < | < | < | < | |
| 260710R | Overcast | 20.2 ± 1.2 | 65.2 ± 4.4 | 35 ± 17 | 76 ± 55 | 22 ± 24 | 76 ± 63 | < | 30 ± 14 |
| 160810D | 21.7 ± 0.5 | 69.5 ± 5.9 | 461 ± 212 | 276 ± 12 | < | < | < | < | |
| 160810N | 19.9 ± 0.2 | 74.7 ± 3.4 | 31 ± 3 | 30 ± 3 | < | < | < | < | |
| 160810R | Overcast | 19.0 ± 0.6 | 56.2 ± 3.3 | 181 ± 25 | 104 ± 33 | 20 ± 12 | 11 ± 10 | 6 ± 0 | < |
| 300810D | 19.3 ± 0.3 | 66.3 ± 6.9 | 441 ± 50 | 444 ± 112 | 11 ± 10 | 6 ± 6 | < | < | |
| 300810N | 18.8 ± 0.3 | 61.1 ± 1.4 | 43 ± 20 | 28 ± 17 | < | < | < | < | |
| 300810R | Sunny | 17.7 ± 1.1 | 46.0 ± 3.7 | 161 ± 19 | 228 ± 124 | 13 ± 3 | 6 ± 6 | < | < |
| 130910D | 19.3 ± 0.5 | 69.8 ± 5.0 | 469 ± 143 | 493 ± 43 | 65 ± 98 | 11 ± 6 | < | < | |
| 130910N | 18.3 ± 0.5 | 67.1 ± 1.1 | 6 ± 6 | 7 ± 3 | < | < | < | < | |
| 130910R | Sunny | 15.9 ± 1.7 | 67.4 ± 8.3 | 57 ± 26 | 137 ± 18 | 7 ± 8 | 11 ± 11 | < | < |
| 270910D | 16.6 ± 0.3 | 62.4 ± 7.4 | 341 ± 46 | 350 ± 47 | 26 ± 18 | 7 ± 3 | 22 ± 11 | 9 ± 6 | |
| 270910N | 14.9 ± 0.4 | 47.6 ± 0.8 | 7 ± 8 | < | < | < | < | < | |
| 270910R | Sunny | 12.6 ± 1.0 | 47.6 ± 3.2 | 70 ± 35 | 94 ± 15 | 7 ± 3 | 9 ± 12 | 9 ± 8 | < |
D, daytime station; N, nighttime station; R, daytime outdoor reference. N and R samples were not collected on the first sampling dates because the required permissions were not yet available.
Airborne bacterial concentrations are reported as the averages of the triplicate cultivation plates.
, Spore-specific cultivation (aerobic);
, spore-specific cultivation (anaerobic). <, below the limit of detection (5.5 CFU m−3).
Bioaerosol collection.
Air samples were collected using a high-volume air sampler, SASS 3100 (Research International, Monroe, WA), using filter-based electret capture technology. The SASS 3100 instrument offers an user-adjustable airflow of between 50 and 360 liters per min (lpm), and the particle collection efficiencies are ca. 92% for particles in the 0.5- to 5.0-μm size range when sampling at 120 lpm and 78 to 79% for similar-sized particles at 320 lpm. The electret filter is composed of an injection-molded frame with an acoustically welded 44-mm diameter microfibrous capture disc, where each fiber has an electric field frozen into it. These fields will induce a charge in aerosols passing through the filter and provide an electret capture mechanism. The capture disc has a void volume of ca. 96% and an effective airflow velocity at the filter face of ∼3.5 m per s when sampling at 320 lpm. These properties translate into substantially lower particle impact speeds and pressure drops compared to most traditional dry filter collection methods, and the employed electret filter technology should therefore offer relatively benign capture conditions for delicate microorganisms even at high airflow rates (56). The instrument was mounted on a tripod with the filter at a height of 1.5 m above the ground, facing the tracks at a 45° downward angle from the horizontal position. The downward-facing angle was selected to avoid direct deposition of large particulates (>100 μm) that originated from sources immediately above or close to the sampler, since such particulates would not represent true aerosols due to their limited residence time in air (6). The tripod was positioned in the middle of the westbound concourse about 4 m from the train tracks. The airflow was 300 lpm, and the sampling period lasted for 2 h, corresponding to a total air sample of 36 m3. Sampled filters were placed back into their original sterile packaging, transported directly to the laboratory at room temperature, and processed within 2 h after sampling. The open-faced filter holder on the SASS 3100 instrument was disinfected with ethanol (70%) between samples to avoid cross-contamination. Field blanks were generated by mounting filters on the SASS 3100 instrument without drawing air on a few occasions and subjecting them to the same downstream procedures as the sampled filters. The collected particles were extracted from the filters into liquid using an extraction buffer (phosphate-buffered saline with 0.05% Triton X-100 [pH 7.4]) and the SASS 3010 extractor instrument (Research International) according to the manufacturer's standard instructions. The extractor instrument was disinfected with hydrogen peroxide (35%) or sodium hypochlorite solution (5,000 ppm free available chlorine), followed by multiple flushes with the extraction buffer, as per the manufacturer's recommendations, to avoid cross-contamination.
Particle and meteorological data collection.
Particle concentrations and size distribution data were measured with an optical particle counter (Aerotrak 8220; TSI, Shoreview, MN) mounted on a tripod, with the inlet pointing in the vertical direction at the same height as the SASS 3100. The instrument was equipped with an external temperature and humidity probe. Particle data were binned into size intervals corresponding to 0.5 to 1.0 μm, 1.0 to 2.0 μm, 2.0 to 3.0 μm, 3.0 to 4.0 μm, 4.0 to 5.0 μm, and >5.0 μm. Particle and meteorological data were collected simultaneously with the bioaerosol sampling and averaged over the entire 2-h sampling period.
Bacterial cultivation.
To enumerate total airborne cultivable bacteria, filter extracts (100 μl) were plated in triplicate using a standard spread plate method onto Trypticase soy agar (TSA; Merck, Darmstadt, Germany) and Reasoner's 2a (R2A) (Oxoid, Cambridge, United Kingdom) plates, supplemented with 100 μg of cycloheximide (Sigma-Aldrich, St. Louis, MO) ml−1 to avoid fungal growth (60). Incubation was performed at 30°C for 48 h before colony counting. To exclusively enumerate cultivable anaerobic and aerobic bacterial spores, filter extracts were heat shocked (75°C, 20 min) and then plated out and incubated as for vegetative bacteria (5). Anaerobic cultivation jars, AnaeroGen packs, and anaerobic indicators (Oxoid) were used to generate and verify anaerobic growth conditions. Airborne cultivable bacterial concentrations are presented as the average of the triplicate cultivation plates and expressed as CFU per cubic meter of air (CFU m−3) ± the standard deviation. The limit of detection (LOD) was calculated to be 5.5 CFU m−3 for the cultivation assay used, corresponding to the observation of at least one CFU on each of the triplicate plates. A representative selection of morphologically distinct colonies was isolated from the primary cultivation plates to obtain pure isolates for further characterization. Selected colonies were transferred to new TSA plates and incubated at 30°C for 48 h. The process was repeated until pure isolates were obtained. Long-time storage was done at −80°C in brain heart infusion broth (Oxoid) supplemented with 18% glycerol (Merck).
Partial 16S rRNA gene sequencing.
A single colony from each isolate was transferred to a polypropylene tube (1.5 ml; Axygen, Union City, CA) filled with PCR-grade H2O (100 μl; Ambion, Austin, TX) and submitted to five freeze-thaw cycles before centrifugation (10,000 × g, 2 min). Each cycle consisted of submersion in liquid nitrogen (1 min), followed by submersion in boiling water (1 min) and a brief vortex mixing. Real-time PCR was performed using the supernatant (2 μl) as a template in white 96-well PCR plates (Roche Diagnostics, Indianapolis, IN) on a LightCycler 480 instrument (Roche Diagnostics). Briefly, each reaction (30 μl) consisted of 2× SYBR green master mix (15 μl; Roche Diagnostics), PCR-grade H2O (9 μl), 10 μM forward and reverse primers (2 μl each), and sample (2 μl). Universal Bacteria 16S rRNA gene primers were used, including the forward primer 27F (5′-GAGTTTGATCMTGGCTCAG-3′) and the reverse primers 519R (5′-GWATTACCGCGGCKGCTG-3′) (40). The PCR program consisted of an initial denaturation (95°C, 5 min), 35 cycles of denaturation (95°C, 20 s), annealing (55°C, 10 s), and extension (72°C, 90 s), and finally a terminal extension (72°C, 5 min). Negative amplification controls were included on each PCR plate to verify the absence of contaminating DNA in the PCR reagents. Standard melting-curve and gel analyses were performed to verify amplification of specific PCR products. Purification and bidirectional sequencing using the primers 27F and 519R were performed at a commercial sequencing facility (Eurofins MWG Operon, Ebersberg, Germany). The sequence trace files were trimmed, aligned, and manually checked using BioNumerics 6.0 (Applied Maths, Sint-Martens-Latem, Belgium), and only sequences with read lengths greater than 400 bp and less than 1% ambiguous base calls were approved.
16S rRNA gene-based taxonomical classification.
Isolates were classified by submitting their 16S rRNA gene sequence to the Classifier and SeqMatch tools at the Ribosomal Database Project (RDP) website (http://rdp.cme.msu.edu, release 10, update 22) (75), yielding classification down to the genus level and best hits against the RDP database, respectively. In addition, a phylogenetic cluster analysis based on the 16S rRNA gene sequences was performed in BioNumerics 6.0 (Applied Maths) using the software's standard settings to allow selection of a single isolate to represent each observed environmental phylotype during further isolate characterization. An unweighted pair group method with arithmetic mean (UPGMA) algorithm was used for pairwise alignment, followed by construction and manual editing of a multiple alignment, and hierarchical clustering using a complete linkage algorithm. A 97% similarity cutoff was used to separate the isolates into operational taxonomic units (OTU) before selecting the isolate with the highest average similarity to the other isolates in the respective OTU.
MALDI-TOF MS-based taxonomical classification.
The representative isolate from each 16S rRNA gene-based OTU was classified using the Biotyper 2.0 microbial identification platform (Bruker Daltonics, Bremen, Germany) coupled to the MicroFlex matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) instrument (Bruker Daltonics). Pure bacterial colonies were prepared and analyzed according to the standard direct transfer method recommended by the manufacturer. Bacterial isolates were streaked for isolation on TSA plates and incubated at 30°C for 48 h before transferring a single colony onto a MSP 96 ground steel target (Bruker Daltonics) as triplicates. The α-cyano-4-hydroxycinnamic acid (HCCA) matrix (Bruker Daltonics) was prepared in accordance with the manufacturer's recommendations and overlaid each target spot (1 μl) immediately after the bacterial smear had dried. The target was loaded into the MicroFlex MALDI-TOF MS instrument immediately after the HCCA matrix had dried. The Biotyper 2.0 system was run in automatic classification mode, and the reference database used was the Bruker Taxonomy database (v3.1.1.0, containing 3,740 library entries).
Antibiotic resistance.
All isolates were assayed for resistance against various antibiotics by cultivation on TSA plates supplemented with nalidixic acid (NAL; 20 μg ml−1), ampicillin (AMP; 50 μg ml−1), tetracycline (TET; 10 μg ml−1), streptomycin (STR; 50 μg ml−1), or chloramphenicol (CHL; 25 μg ml−1). The antibiotics used were supplied by Sigma-Aldrich. Antibiotic resistance was assessed qualitatively by designating isolates as being resistant or sensitive based on the presence or absence of colony growth after incubation at 30°C for 48 h.
Hemolytic activity.
All isolates were initially screened for hemolytic activity by cultivation on Colombia agar plates supplemented with 5% sheep blood (Oxoid) at 30°C for 24 to 48 h. Isolates classified into the bacterial genus Bacillus that showed hemolytic activity were subsequently analyzed further to investigate virulence-associated factors. The hemolytic Bacillus spp. isolates were streaked out on Colombia agar plates supplemented with 5% bovine blood and incubated at 37°C. The plates were incubated until the clearing zone around the colonies was fully developed (24 to 72 h). The hemolytic activity was assessed as in the initial screening, but special attention was directed toward identifying hemolysis induced by small cyclic lipopeptide toxins (SCLPT), such as the surfactin-like peptides (surfactins, pumilacidins, and lichenysins), often characterized by an opaque and slowly growing clearing zone.
Nonribosomal peptide synthetases.
A universal PCR assay targeting the surfactin class of nonribosomal peptide synthetases (NRPS), developed by Tapi et al. (69), was performed to elucidate the nonribosomal peptide synthesis capabilities of the obtained hemolytic Bacillus spp. isolates. The PCR assay was performed according to the original publication and used the forward primer NRPS-F (5′-CGCGGMTACCGVATYGAGC-3′) and the reverse primer NRPS-R (5′-ATBCCTTTBTWDGAATGTCCGC-3′) that produced a PCR product between 419 and 431 bp for different types of NRPS within the surfactin class. The surfactin-producing strain Bacillus mojavensis B31 isolated from imported basil spices of unknown origin described by From et al. (24) was used as a positive control. A specific PCR assay for the cereulide synthase gene (ces), the NRPS that produces cereulide (B. cereus emetic toxin), developed by Fricker et al. (21), was also performed on the obtained hemolytic Bacillus spp. isolates. The PCR assay was performed according to the original publication and used the forward primer cesF (5′-GGTGACACATTATCATATAAGGTG-3′) and the reverse primer cesR (5′-ATBCCTTTBTWDGAATGTCCGC-3′) that produced a PCR product of 1,271 bp. DNA from a ces-positive strain (NVH 0137/09) and a ces-negative strain (NVH 1230/88), both isolated from commercial food products at the Norwegian School of Veterinary Science, was used as a positive and a negative control, respectively. Both PCR assays (NRPS and ces) were performed in a PTC-100 Peltier thermal cycler (MJ Research, Waltham, MA). Each reaction consisted of template DNA (1 μl), DyNAzyme II DNA polymerase/deoxynucleoside triphosphate mix (Finnzymes, Espoo, Finland), and a final primer concentration of 1 μM.
Phylogenetic clustering of hemolytic Bacillus spp. isolates.
The phylogenetic relationship between the hemolytic Bacillus spp. isolates obtained in the present study and closely related Bacillus spp., including species known to harbor NRPS-produced small cyclic peptides (SCPs), was investigated by hierarchical clustering based on their 16S rRNA gene sequences. Type strains of Bacillus spp. that were closely related to isolates obtained here were identified from the RDP SeqMatch results, and their 16S rRNA gene sequences were downloaded from the RDP database. To avoid large dendrograms with many closely related environmental isolates, Bionumerics 6.0 (Applied Maths) was used to cluster the hemolytic Bacillus spp. isolates obtained here into OTUs and select a single representative isolate from each OTU. The alignment and clustering methods were the same as those described for selecting OTU representatives from all of the obtained bacterial isolates, except that a 99% similarity cutoff was used. The partial 16S rRNA gene sequences from the OTU representative hemolytic Bacillus spp. isolates were then aligned with the RDP-derived full-length 16S rRNA gene sequences from closely related Bacillus spp. using the same alignment and clustering method, but without using any similarity cutoff. The RDP-derived 16S rRNA gene sequence from a type strain of B. megaterium (IAM 13418) and the partial 16S rRNA gene sequence from a closely related nonhemolytic isolate obtained here were included in the analysis. The RDP-derived 16S rRNA gene sequence from a type strain of E. coli (ATCC 11775T) was included in the analysis as an outgroup.
Cereulide and surfactin-like peptides.
A high-performance liquid chromatography-mass spectrometry assay targeting cereulide was performed on the hemolytic Bacillus spp. isolates that were positive for NRPS, as described elsewhere (31). Surfactin-like peptides, such as surfactins, pumilacidins, and lichenysins, were detected by a liquid chromatography mass spectrometry (LC-MS) assay, as described by From et al. (23), with minor modifications. Briefly, the hemolytic Bacillus spp. isolates positive for NRPS were grown on Colombia agar plates supplemented with 5% bovine blood at room temperature (∼22°C) for 72 h. Three colonies were collected and resuspended in ultrapure (18.2 mΩ cm1) H2O (500 μl), vortexed (10 s) and transferred to glass vials (10 ml). Acetone (3 ml) and chloroform (4 ml) were added, and the mixture was vigorously shaken (10 s) and centrifuged (1,600 × g, 3 min). The organic phase was transferred to another glass vial and evaporated to dryness at 60°C under a stream of air. The dry residue was dissolved in methanol (100 μl), followed by the addition of methanol-water (60:40, 300 μl), and mixed. The mixture was centrifuged (1,600 × g, 3 min), and the organic phase was recovered and centrifuged (5,600 × g, 2 min) through a Costar Spin-X centrifuge filter (0.22-μm-pore-size nylon; Corning, Corning, NY). Aliquots (25 μl) were injected into the LC-MS apparatus at intervals of 7 min. A Zorbax SB-C18 Rapid Resolution HT analytical column (2.1 mm by 5 mm 1.8 μm; Agilent Technologies, Santa Clara, CA) was used and operated at a constant temperature of 30°C. The mobile phase was a mixture of methanol-water (92:8) containing 0.1% formic acid. The pump was operated isocratically at a constant flow rate (300 μl min−1). The LC-MS instrumentation consisted of a Series 200 quaternary pump and autosampler (Perkin-Elmer, Foster City, CA) and an API 2000 MS system (Applied Biosystems, Foster City, CA) equipped with a Turbo-Ion-Spray source operated in ESI-positive mode. The turbo probe vaporizer temperature of the interface was fixed at 400°C. Surfactin-like peptides were elucidated in the m/z range from 1,008 through 1,076.
Pigmentation.
All OTU representative isolates showing visible colony pigmentation were selected for further pigment characterization by LC-MS. Isolates were cultured on TSA plates and incubated at 30°C for 48 h. Using a standard laboratory scale, ∼200 mg of biomass was scraped using a loop into aluminum foil-wrapped polypropylene tubes (1.5 ml; Axygen). The biomass was washed (1 ml) and resuspended (500 μl) in phosphate-buffered saline before the addition of Ready-Lyse lysozyme (EpiCentre, Madison, WI) to a final concentration of 10 U μl−1, followed by incubation at room temperature for 10 min. The biomass was pelleted (10,000 × g, 5 min) and lyophilized to facilitate extraction in organic solvents. Organic extraction of pigments was performed with methanol (200 μl) on a shaker plate at room temperature for 1 h. After centrifugation (10,000 × g, 5 min), the supernatant was transferred into amber glass vials and stored at −80°C. LC-MS analysis was performed on an Agilent TOF mass spectrometer coupled to an Agilent 1100 series LC system equipped with a diode array detector recording the UV/VIS spectra between 200 and 650 nm. Mobile phases were methanol-water (50:50) in channel A and dichloromethane-methanol-water (45:50:5) in channel B. A Zorbax RP C8 4.6-by-150-mm column was used, the flow was kept at 800 μl min−1, and 20 μl of the extract was injected for each run. It was assumed that most of the pigments belong to the chemical group of carotenoids and, hence, the chromatographic conditions were optimized using astaxanthin (Sigma-Aldrich) and β-carotene (Sigma-Aldrich) as representative external standards. The hydrophilic initial elution conditions should also retain more hydrophilic pigments, if present in the extract. The following gradient was used: 0% B for 0 to 2 min, linear gradient to 100% B after 10 min, and then 100% B for additional 5 min. The data acquisition was also performed during the 6 min re-equilibration period. Analytes were ionized using atmospheric pressure chemical ionization with following settings: 325°C dry temperature, 350°C vaporizer temperature, 50-lb/in2 nebulizer pressure, and 5.0 liters min−1 dry gas. Reference solution for correction of mass axes on the mass spectrometer was continuously added after column chromatography to the mobile phase using a T-fitting. Tentative identification of pigments was performed by inspection of the diode array chromatograms and corresponding total ion chromatograms. The UV/VIS peaks were assigned peak maximum values, and the corresponding mass spectra were evaluated for correlating m/z peaks (i.e., extracted ion chromatogram with the same profile as the extracted wavelength chromatogram). Determined accurate mass and absorption peak maximum values were used to search the Dictionary of Natural Products (version 19.2) database (http://dnp.chemnetbase.com).
Statistical analysis.
Airborne cultivable bacterial concentrations and particle data were found to be far from well modeled by a normal distribution based on a normality plot and the Lillefors test (43). The nonparametric Mann-Whitney U test (47) was therefore used to test for differences in the concentration of airborne bacteria between the environments and the cultivation media and also to test for differences in the total particle concentrations between environments. Particle size distribution data were analyzed using the Pearson product-moment correlation coefficient (52) to determine the correlation among the various particle size bins within and between the environments. Pearson product-moment correlation was also used to investigate the correlation between the total particle concentrations and total airborne bacterial concentrations independent of sampling time and location. Differences in the observed bacterial diversity between environments and cultivation media were tested using the Pearson chi-square test (53). The significance level was set at P < 0.05 for all statistical tests.
RESULTS
Airborne bacterial concentration.
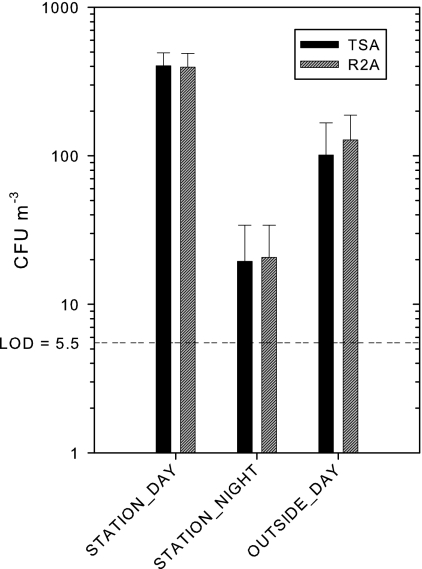
Total and spore-specific airborne bacterial concentrations were determined on TSA and R2A growth media for each individual sample (Table 1). The average concentrations on TSA were 403 ± 91, 19 ± 15, and 101 ± 66 CFU m−3 for daytime and nighttime sampling at the station and daytime outdoor reference sampling, respectively (Fig. 1). The average concentrations on R2A were 396 ± 93, 21 ± 13, and 128 ± 60 CFU m−3 for daytime and nighttime sampling at the station and daytime outdoor reference sampling, respectively (Fig. 1). The average aerobic spore concentrations on TSA were 17 ± 21 and 14 ± 7 CFU m−3, for daytime sampling at the station and daytime outdoor reference sampling, respectively. All average anaerobic spore concentrations and aerobic spore concentrations for the nighttime sampling at the station were below the LOD (5.5 CFU m−3) on TSA and are therefore not reported. The average aerobic spore concentrations on R2A were 7 ± 5 and 23 ± 30 CFU m−3 for daytime sampling at the station and daytime outdoor reference sampling, respectively. Except for the average daytime outdoor concentration of 6 ± 13 CFU m−3, all average anaerobic spore concentrations and the aerobic spore concentration for the nighttime station samples were below the LOD (5.5 CFU m−3) on R2A and are therefore not reported. The obtained results showed that daytime concentrations at the subway station were 20- and 3.5-fold higher (P < 0.05) than the nighttime concentrations at the station and the daytime outdoor reference concentrations, respectively. The aerobic spore fraction (spore specific to total cultivation ratio) was 4.7-fold higher (P < 0.05) at the outdoor reference compared to the subway station, accounting for ca. 16% of the total concentration of cultivable airborne bacteria at the outdoor location and ca. 3.4% at the station. The two growth media (TSA and R2A) did not show significant differences (P > 0.586) in total cultivable bacterial concentrations. No bacterial growth was observed in any of the field blanks during this study.
Fig 1.
Average total cultivable airborne bacterial concentration levels on TSA and R2A from the three environments studied: daytime station (n = 8), nighttime station (n = 6), and daytime outdoor reference (n = 5). Standard deviations are indicated by error bars.
Particle concentration and size distribution.
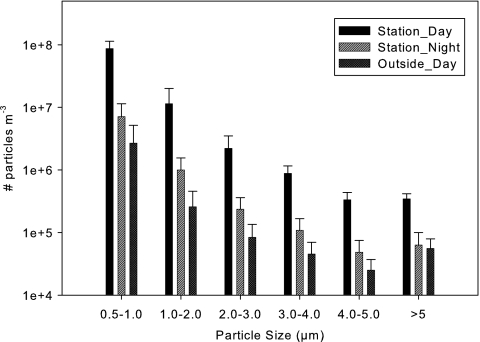
Particle concentrations and size distributions were stable within each environment throughout the study, and the relative differences between the sampled environments were consistently observed (Fig. 2). The average cumulative particle concentrations over the entire measured size range showed that daytime station samples had 11.9-fold higher (P < 0.05) particle level than the nighttime station samples and that the nighttime station samples had 2.8-fold higher (P < 0.05) particle level than the daytime outdoor reference samples. The particle size distribution profiles showed a decrease in particle concentrations related to increased particle size, and this observation was conserved between all samples and environments (r = 0.995, P < 0.05), with the smallest size bin (0.5 to 1.0 μm) accounting for >80% of the total particle counts. The average count median diameters were similar between the environments, 0.86 ± 0.01, 0.88 ± 0.03, and 0.88 ± 0.02 μm, while the average mass median diameters varied, 3.79 ± 0.37, 4.71 ± 0.50, and 5.80 ± 0.21 μm, for the daytime station, nighttime station, and daytime outdoor samples, respectively. The total particle concentrations and total cultivable airborne bacterial concentrations showed a significant correlation when calculated independent of time and location (r = 0.896, P < 0.05), indicating that the level of airborne bacteria was related to the total particle concentration.
Fig 2.
Average airborne particle concentrations and size distributions from the three environments studied: daytime station (n = 8), nighttime station (n = 6), and daytime outdoor reference (n = 5). Standard deviations are indicated by error bars.
Meteorological data.
Temperature and relative humidity (RH) are reported as an average of a 2-h sampling period for each individual sample (Table 1). Average temperatures of 19.3 ± 0.4, 19.3 ± 0.4, and 18.2 ± 1.2°C were observed for the daytime station, nighttime station, and the daytime outdoor samples, respectively. The average RH values were 69.9% ± 4.9%, 67.6% ± 1.9%, and 58.7% ± 4.9% for the daytime station, nighttime station, and daytime outdoor samples, respectively.
Airborne bacterial diversity.
A total of 429 bacterial colonies were isolated from TSA and R2A primary cultivation plates. High-quality partial 16S rRNA gene sequences were obtained from 94% (291/308) and 92% (111/121) of the isolates from total and spore-specific cultivations, respectively. Thus, 402 bacterial isolates were successfully characterized, while the remaining 27 were discarded due to no growth after freeze storage or inability to yield 16S rRNA gene sequence data with >400 acceptable base calls and <1% ambiguities. Isolates were distributed between three major phyla, Actinobacteria (48%), Firmicutes (43%), and Proteobacteria (9%) from total cultivation and Actinobacteria (3%), Firmicutes (96%), and Proteobacteria (1%) from spore-specific cultivation, respectively. A total of 37 bacterial genera were observed (Table 2), with the majority belonging to the genera Micrococcus (32%), Staphylococcus (20%), Bacillus (18%), Pseudomonas (4%), Microbacterium (3%), and Streptomyces (3%) from total cultivation and Bacillus (68%), Staphylococcus (15%), and Paenibacillus (9%) from spore-specific cultivation. More than 80% of the isolates originating from spore-specific cultivations were classified into the bacterial genus Bacillus or the closely related genera Paenibacillus, Viridibacillus, Tumebacillus, Brevibacillus, and Lysinibacillus, consistent with their spore-forming capabilities, but also into the genus Staphylococcus (15%), which is not consistent with spore-forming capabilities. Although there was no significant difference in the distribution of bacterial genera at the subway station and the outside reference environment (P = 0.19), Bacillus spp. were more frequently observed at the outside reference location than at the station, accounting for 28 and 17% of the total cultivation isolates, respectively. The opposite was observed for Micrococcus spp. and Streptomyces spp., which accounted for 36 and 3% of the total cultivation isolates at the station and 23 and 0% at the outside reference location, respectively. There was a significant difference between the daytime and nighttime distributions of bacterial genera at the station (P < 0.05). Bacillus and Micrococcus spp. were observed more frequently during the daytime, where they accounted for 17 and 36% of the isolates from daytime samples and 5 and 27% from nighttime samples, respectively, whereas Staphylococcus spp. were found to account for 25% of the isolates at night and 19% during the day. Several bacterial genera were uniquely present, although in low abundance, at nighttime compared to daytime, such as the genera Corynebacterium, Erwinia, Gordonia, Rothia, and Serratia. A significant difference in the genus distribution was observed between the two culture media that we used (P < 0.05). Micrococcus, Pseudomonas, and Staphylococcus spp. were recovered more frequently from TSA than R2A, accounting for 40, 6, and 23% of the isolates on TSA and 24, 2, and 16% on R2A, respectively. Dermacoccus and Rhodococcus spp. were recovered only from R2A, where each genus accounted for 4% of the isolates.
Table 2.
Genus distribution of the obtained bacterial isolates
| Genus | Total cultivation |
Spore-specific cultivationa |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Station |
Outside (day) |
Total |
Station |
Outside (day) |
Total |
|||||||||||
| Day |
Night |
Day |
Night |
|||||||||||||
| nb | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| Arthrobacter | 1 | <1 | 2 | 5 | 2 | 4 | 5 | 2 | ||||||||
| Bacillus | 33 | 17 | 2 | 5 | 16 | 28 | 51 | 18 | 44 | 73 | 1 | 50 | 30 | 61 | 75 | 68 |
| Brevibacillus | 1 | 2 | 1 | <1 | ||||||||||||
| Brevundimonas | 2 | 1 | 2 | <1 | ||||||||||||
| Cellulosimicrobium | 1 | 2 | 1 | <1 | ||||||||||||
| Comamonas | 1 | <1 | 1 | <1 | ||||||||||||
| Corynebacterium | 2 | 5 | 2 | <1 | ||||||||||||
| Curtobacterium | 1 | 2 | 1 | <1 | ||||||||||||
| Dermacoccus | 5 | 3 | 1 | 2 | 6 | 2 | ||||||||||
| Dietzia | 1 | <1 | 1 | <1 | ||||||||||||
| Enhydrobacter | 1 | <1 | 1 | <1 | ||||||||||||
| Erwinia | 1 | 2 | 1 | <1 | ||||||||||||
| Exiguobacterium | 1 | <1 | 1 | <1 | ||||||||||||
| Gordonia | 1 | 2 | 1 | <1 | ||||||||||||
| Janibacter | 1 | <1 | 1 | <1 | ||||||||||||
| Kocuria | 3 | 2 | 1 | 2 | 4 | 1 | 1 | 2 | 1 | <1 | ||||||
| Lysinibacillus | 1 | <1 | 1 | 2 | 2 | <1 | 1 | 2 | 1 | <1 | ||||||
| Microbacterium | 6 | 3 | 2 | 5 | 1 | 2 | 9 | 3 | ||||||||
| Micrococcus | 69 | 36 | 12 | 27 | 13 | 23 | 94 | 32 | ||||||||
| Paenibacillus | 3 | 2 | 1 | 2 | 3 | 5 | 7 | 2 | 4 | 7 | 6 | 12 | 10 | 9 | ||
| Paenisporosarcina | 1 | <1 | 1 | <1 | 1 | 2 | 1 | <1 | ||||||||
| Pantoea | 1 | <1 | 1 | 2 | 2 | <1 | ||||||||||
| Paracoccus | 3 | 2 | 3 | 1 | ||||||||||||
| Planococcus | 2 | 1 | 2 | <1 | ||||||||||||
| Plantibacter | 1 | 2 | 1 | <1 | ||||||||||||
| Pseudomonas | 9 | 5 | 1 | 2 | 3 | 5 | 13 | 4 | ||||||||
| Rhodococcus | 1 | <1 | 3 | 7 | 1 | 2 | 5 | 2 | ||||||||
| Roseomonas | 1 | <1 | 1 | 2 | 2 | <1 | ||||||||||
| Rothia | 1 | 2 | 1 | <1 | 1 | 2 | 1 | 2 | 2 | 2 | ||||||
| Serratia | 1 | 2 | 1 | <1 | ||||||||||||
| Sphingomonas | 1 | 2 | 1 | <1 | ||||||||||||
| Sporosarcina | 1 | 2 | 1 | <1 | ||||||||||||
| Staphylococcus | 37 | 19 | 11 | 25 | 10 | 18 | 58 | 20 | 6 | 10 | 1 | 50 | 10 | 20 | 17 | 15 |
| Streptomyces | 6 | 3 | 2 | 5 | 8 | 3 | ||||||||||
| Tumebacillus | 1 | 2 | 1 | <1 | ||||||||||||
| Viridibacillus | 1 | 2 | 1 | <1 | 1 | 2 | 1 | <1 | ||||||||
| Weissella | 1 | <1 | 1 | <1 | ||||||||||||
| Sum | 190 | 44 | 57 | 291 | 60 | 2 | 49 | 111 | ||||||||
Some isolates obtained from the spore-specific cultivations were from bacterial genera not known to harbor spore-forming members, such as Staphylococcus.
n, number of isolates.
Biotyper 2.0 taxonomical classification.
Using the MALDI-TOF MS-based Biotyper 2.0 microbial identification system to classify the OTU representative isolates (n = 84) resulted in 40% species-consistent and 60% genus-consistent results compared to the 16S rRNA gene-based classifications (see Table S1 in the supplemental material). Only one isolate failed to generate an approvable mass spectrum, whereas 36% of the isolates yielded only low-scoring classification results, i.e., score values below 1.7. When considering the 64% (n = 54) of the isolates with score values above 1.7, 61% of the isolates were species-consistently classified and a total of 94% were genus-consistently classified, respectively. Of the 6% of isolates (n = 3) with score values above 1.7 that did not show genus-consistent classification results, the bacterial genera (n = 2) or species (n = 1) given by the 16S rRNA gene-based classification were missing from the Bruker Taxonomy database. Of the total number of isolates that were analyzed (n = 84), seven genera and 24 species given by the 16S rRNA gene-based classification were missing from the Bruker Taxonomy database.
Antibiotic resistance.
About 27% of the total cultivation isolates (n = 291) showed antibiotic resistance against at least two of the antibiotics tested, whereas 10% did not show resistance against any of the antibiotics tested. Resistance against three, four, and all of the antibiotics used was seen in 12, 1, and 1 of the isolates, respectively. The distribution of resistance in the total cultivation isolates were as follows: 27% AMP, 9% STR, 75% NAL, 4% TET, and 5% CHL. About 32% of the spore-specific cultivation isolates (n = 111) showed antibiotic resistance against at least two of the antibiotics tested, whereas 22% did not show resistance against any of the antibiotics tested. Resistance against 3, 4, and all of the antibiotics used was seen in 3, 1, and none of the isolates, respectively. The distribution of resistance in the spore-specific cultivation isolates was as follows: 44% AMP, 19% STR, 37% NAL, 2% TET, and 14% CHL, showing that these isolates had less frequent resistance to NAL and TET and more frequent resistance against AMP, STR, and CHL than total cultivation isolates. The majority of isolates from the dominant genera had frequent resistance against NAL (Table 3), except isolates of Bacillus (14%) and Paenibacillus (0%) spp. Arthrobacter and Kocuria spp. showed no resistance, while Micrococcus spp. had a very low frequency of resistance (1 to 6%), against all of the antibiotics that were tested except NAL. Paenibacillus spp. isolates were the only isolates to show frequent (57%) resistance against STR. Paenibacillus spp. isolates also had a very low frequency of resistance against all of the other antibiotics that were tested. Very few isolates displayed resistance against TET and CHL, except isolates of Pseudomonas, Microbacterium, and Rhodococcus spp., which still had a relatively low frequency of resistance (10 to 20%). No differences in the distribution of antibiotic resistance were observed between the environments, except that isolates derived from spore-specific cultivation had a higher frequency of CHL resistance at the station (17%) compared to the outdoor reference location (8%).
Table 3.
Distribution of antibiotic resistance in the dominant bacterial genera
| Genus | Total cultivation |
Spore-specific cultivationa |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL |
AMP |
STR |
CHL |
TET |
NAL |
AMP |
STR |
CHL |
TET |
|||||||||||
| % | nb | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | |
| Arthrobacter | 100 | 5/5 | 0 | 0/5 | 0 | 0/5 | 0 | 0/5 | 0 | 0/5 | ||||||||||
| Bacillus | 14 | 7/51 | 61 | 31/51 | 16 | 8/51 | 8 | 4/51 | 2 | 1/51 | 20 | 15/75 | 40 | 30/75 | 13 | 10/75 | 17 | 13/75 | 3 | 2/75 |
| Dermacoccus | 100 | 6/6 | 0 | 0/6 | 17 | 1/6 | 0 | 0/6 | 0 | 0/6 | ||||||||||
| Kocuria | 100 | 4/4 | 0 | 0/4 | 0 | 0/4 | 0 | 0/4 | 0 | 0/4 | ||||||||||
| Microbacterium | 100 | 9/9 | 0 | 0/9 | 0 | 0/9 | 11 | 1/9 | 11 | 1/9 | ||||||||||
| Micrococcus | 98 | 92/94 | 4 | 4/94 | 6 | 6/94 | 2 | 2/94 | 1 | 1/94 | ||||||||||
| Paenibacillus | 0 | 0/7 | 0 | 0/7 | 57 | 4/7 | 0 | 0/7 | 0 | 0/7 | 20 | 2/10 | 20 | 2/10 | 90 | 9/10 | 20 | 2/10 | 0 | 0/10 |
| Pseudomonas | 54 | 7/13 | 31 | 4/13 | 8 | 1/13 | 23 | 3/13 | 15 | 2/13 | ||||||||||
| Rhodococcus | 100 | 5/5 | 40 | 2/5 | 0 | 0/5 | 0 | 0/5 | 20 | 1/5 | ||||||||||
| Staphylococcus | 93 | 54/58 | 31 | 18/58 | 10 | 6/58 | 5 | 3/58 | 2 | 1/58 | 100 | 17/17 | 88 | 15/17 | 6 | 1/17 | 0 | 0/17 | 0 | 0/17 |
| Streptomyces | 100 | 8/8 | 38 | 3/8 | 0 | 0/8 | 0 | 0/8 | 0 | 0/8 | ||||||||||
Some isolates obtained from the spore-specific cultivations were from bacterial genera not known to harbor spore-forming members, such as Staphylococcus.
n, number of resistant isolates/total number of isolates.
Hemolytic activity.
About 22% (87/402) of the bacterial isolates showed hemolysis in the initial screening on sheep blood agar plates. These were distributed between the bacterial genera Bacillus (87%), Staphylococcus (10%), Erwinia (1%), and Pseudomonas (1%). Of the total number of Bacillus spp. isolates obtained (n = 125), 61% were shown to be hemolytic in the initial screening on sheep blood agar plates. All isolates showed beta-hemolysis, except two that showed alpha-hemolysis. All of the hemolytic Bacillus isolates (n = 76) were further investigated to elucidate hemolytic and virulence-associated properties (see Table S2 in the supplemental material). In the subsequent analysis performed on bovine blood agar plates, 87% of the isolates from the initial screen on sheep blood were hemolytic. The two isolates that induced alpha-hemolysis on sheep blood were nonhemolytic on bovine blood. Phylogenetic clustering of the hemolytic Bacillus spp. isolates and type strains of closely related Bacillus spp. showed that the obtained isolates clustered into at least three distinct groups (see Fig. S1 in the supplemental material). Each group harbored known producers of SCPs, including B. cereus (cereulide), B. pumilus (pumilacidin), and B. subtilis (surfactin). A total of 30% of the isolates showing hemolysis on bovine blood displayed putative SCLPT-induced hemolysis, characterized by an opaque and slow-growing clearing zone. These isolates were limited to the B. pumilus and B. subtilis groups, except for one isolate that was closely related to B. mycoides and B. weihenstephanensis in the B. cereus group. A total of 35% of all of the Bacillus spp. isolates obtained and 60% of the Bacillus spp. isolates that were hemolytic on bovine blood,were positive for NRPS. NRPS-positive isolates were distributed between all three identified Bacillus groups but, interestingly, all hemolytic isolates showing high similarity to B. pumilus (n = 5) were NRPS positive. The presence of cereulide synthase (ces) or cereulide were not detected in any of the hemolytic Bacillus spp. isolates analyzed here. The LC-MS analysis performed on all NRPS- positive isolates, except for one isolate that was discarded due to slow growth, revealed that 77% of the isolates produced SCLPTs. This corresponded to 50% of the Bacillus isolates that were hemolytic on bovine blood and 26% of the total number of Bacillus isolates obtained in the present study. Isolates positive for SCLPTs were limited to the B. pumilus and B. subtilis groups, except for three isolates that were closely related to B. thuringiensis (n = 2) and B. cereus (n = 1) in the B. cereus group. More than 60% of the SCLPT-positive isolates were classified as B. altitudinis and B. stratosphericus in the B. pumilus group.
Pigmentation.
About 48% (141/291) of the total cultivation isolates displayed visible colony pigmentation, while 8% (9/111) of the spore-specific cultivation isolates displayed visible colony pigmentation. The observed pigmentation included many shades of yellow, orange, pink, and red, with the majority being yellow. No major differences in colony pigmentation were observed between the environments. Bacterial isolates from the station showed a 50% frequency of pigmentation, while the outdoor isolates showed ca. 40%, which could probably be attributed to the presence of more Micrococcus spp. and fewer Bacillus spp., typically yellow pigmented and nonpigmented, respectively, at the station compared to the outdoor reference. A total of 28% (24/84) of the OTU representative isolates displayed visible colony pigmentation and were further characterized by a LC-MS method optimized for detection of carotenoids (see Table S3 in the supplemental material). From these, 54% of the isolates yielded data that led to tentative pigment identification. About 29% of the isolates led to appropriate UV/VIS spectra but no corresponding ion masses that enabled determination of accurate mass, while 17% of the isolates yielded no appropriate UV/VIS spectra. No further attempts were made to identify colored compounds in these 11 extracts. Several carotenoids were observed, and although only tentatively identified, triophaxanthin, alloxanthin, amarouciaxanthin, sarcinaxanthin, cycloviolaxanthin, a diglycosylated variant of zeaxanthin, a glycosylated variant of OH-chlorobactene, and a derivative of staphyloxanthin (8-apo-caroten-8-oic acid) were observed. Sarcinaxanthin was observed in six different isolates from the bacterial phylum Actinobacteria, distributed between the bacterial families Microbacteriaceae (n = 5) and Micrococcaceae (n = 1).
DISCUSSION
We evaluated here the airborne bacterial community at a subway station in Norway. The concentration level, composition, and variability of airborne bacteria were investigated, along with virulence- and survival-associated bacterial properties, such as hemolytic activity, antibiotic resistance, pigmentation, and spore fraction. A methodology scheme for investigating airborne bacteria based on the SASS 3100 high-volume electret filter-based air sampler and for rapid classification of bacterial isolates using the Biotyper 2.0 MALDI-TOF MS-based microbial identification system was also demonstrated. The observed bacterial content, particle size distribution, and concentration level were stable, except for some sporadic deviations, within each environment (daytime station, nighttime station, and daytime outdoor reference) despite the seasonal variation (spring-summer-fall) throughout the study (May to September 2010). In contrast to this, several differences were seen between the sampled airborne environments, such as a higher daytime concentration level of airborne bacteria at the subway station compared to the outdoor location and a higher concentration level of airborne bacteria at the subway station during the day than at night. These results, combined with observed differences in the bacterial diversity between the environments, suggested that anthropogenic sources, such as passengers and train traffic, were major contributors to airborne bacteria at the Norwegian subway station.
This study used a SASS 3100 high-volume electret filter-based air sampler, and comparable airborne bacterial concentration levels to previous studies in subway environments were obtained (34, 38, 62), suggesting that this instrument may be suitable for sampling of airborne bacteria for cultivation analysis. Preliminary results from another sampling campaign performed by the Norwegian Defense Research Establishment at the same subway station (unpublished data) show that the SASS 3100 provides airborne bacterial concentration level estimates comparable to those obtained with the MAS-100 (high-volume impactor) and Andersen sampler (six-stage cascade impactor). Future harmonization or standardization of the sampling, processing, and analysis methodologies (e.g., air sampling equipment and cultivation conditions) used in bioaerosol characterization studies of indoor/outdoor environments is needed to facilitate improved interpretation and comparison of the obtained results.
The results reported here may aid in improving the development of biological detection/surveillance equipment and enable T&E schemes that can be used to evaluate biological detectors' operational performance by simulating complex real-life environments in controlled aerosol test chambers during challenge tests with biological threat agents. Such T&E schemes could improve the performance criteria of biological detection/surveillance equipment to meet the users' requirements to such equipment with respect to reliable sensitive and specific real-time monitoring of biological threat agents in different operational environments. Furthermore, the present report may provide public health authorities with baseline data for airborne bacteria, which can be used when assessing human exposure limits or evaluating other aspects of air quality, in subways or other enclosed environments where people may be confined.
Airborne bacterial concentration.
Bioaerosol surveys investigating airborne bacteria have been performed at subway stations in various countries, including the United States (7), Japan (35, 62), Korea (34, 38, 41), China (18), Russia (8), Egypt (4), the United Kingdom (27), and Hungary (66, 67), but have not previously been carried out in Norway. The airborne bacterial concentrations reported in these studies ranged from not detected to 104 CFU m−3, and, in general, the content of airborne bacteria was higher in subway stations compared to adjacent outdoor air and also higher at stations located deeper underground than in more shallow stations (34). Due to the lack of standardized equipment, analytical methods, and variable goals of the different studies, several different air sampling instruments and cultivation conditions have been used in the published studies. Thus, the comparison of the results is challenging since the various methodologies have been shown to provide different results even when subjected to the same bioaerosol challenge (3, 42, 64, 76). The daytime concentration of airborne cultivable bacteria (∼400 CFU m−3) found at the subway station in Oslo was within the range of previous reports from Korea and Japan (34, 38, 62) but 10- to 100-fold lower than that found in China and Egypt (4, 18). This discrepancy could be due to the use of different air sampling and cultivation methods, geographical differences in the atmospheric concentration of airborne bacteria, and physical differences between the subway stations, such as their size, layout, type of ventilation system, and not least the number of passengers. The daytime concentration of airborne bacteria at the station was higher (3.5-fold) than in adjacent outdoor air, a finding consistent with previous reports (34). The increased daytime concentration observed at the station compared to outdoor air could suggest that anthropogenic sources are major contributors to airborne bacteria in the subway station since these sources were more abundant at the station compared to the outdoor location. The observation that the nighttime airborne bacterial concentration at the station was lower than the station (20-fold) and outdoor (5.7-fold) daytime concentrations further strengthens this hypothesis since anthropogenic sources were nearly absent from the station during the night. Bacterial spores are considered the most resistant form of bacteria, and spores of B. anthracis (the causative agent of anthrax) are on the Centers for Disease Control and Prevention's Category A list of biological threat agents (12). However, little information is available about the amount of airborne bacterial spores in the subway environment. To our knowledge, the study by Awad (4) in Egypt is the only previous report addressing the airborne bacterial spore content in the subway environment. Our results showed that the bacterial spore fraction was higher (4.7-fold) in outdoor air than at the subway station, which is consistent with the previous report from Egypt that also found a higher spore fraction in outdoor air (4). These observations are probably due to exposure to solar/UV-radiation in the outdoor environment that primarily inactivates vegetative bacteria or to differences in the composition of bacterial sources between the subway station and the outside environment. The daytime concentration of airborne bacterial spores that we observed at the subway station, about 12 CFU m−3, is 100-fold lower than that noted in the study from Egypt (4), thus indicating that only low airborne concentrations of bacterial spores were present at the Norwegian subway station. The LOD for the cultivation assay used here was 5.5 CFU m−3, and many of the spore-specific cultivations fell below this threshold.
Airborne bacterial diversity.
The presence and activities of anthropogenic sources might be linked to increased concentrations of airborne bacteria in subway stations, which is consistent with the observation that these environments frequently are dominated by members of the bacterial genera Micrococcus, Bacillus and Staphylococcus, all of which contain several species representatives associated with an anthropogenic origin (4, 62). The dominant bacterial genera observed in all sampled environments in the present study were Micrococcus, Bacillus, and Staphylococcus, which is consistent with previous cultivation studies in the subway environment (38). Micrococcus and Bacillus spp., which are regularly found on human skin (39), were observed at higher frequencies at the subway station during the day than at night, and Micrococcus spp. were observed at higher frequencies at the subway station than in outdoor air, thus further strengthening the hypothesis that anthropogenic sources are a major contributor to airborne bacteria in subway environments. Bacillus spp. were observed more frequently in outdoor air than at the station, suggesting local source differences or specific inactivation of non-spore-forming bacteria in the outdoor atmosphere. Several bacterial genera, such as Corynebacterium, Erwinia, Gordonia, Rothia, and Serratia, were exclusively recovered from the station at nighttime but might still be present at the station during daytime even if they were not recovered. A plausible explanation for this might be that the higher cultivable airborne bacterial concentration observed at the station during daytime and also the higher frequency of Bacillus spp. and Micrococcus spp. could bias the isolation process into missing genera present only in very low abundances. Our results also showed differences in the airborne bacterial diversity obtained with the two cultivation media (TSA and R2A), thus highlighting the importance of using multiple growth media to reveal a more complete diversity estimate and the challenges associated with comparing results obtained with different growth media. Our spore-specific cultivation assay used a heat-shock procedure to inactivate vegetative bacterial cells and activate bacterial spores, which have been commonly applied to determine the content of bacterial spores in food products and soil (28). As expected, most isolates originating from spore-specific cultivations were classified into the bacterial genus Bacillus or the closely related genera such as Paenibacillus, Viridibacillus, Tumebacillus, Brevibacillus, and Lysinibacillus, consistent with their spore-forming capabilities, but also into the genus Staphylococcus (15%), which is not consistent with spore-forming capabilities. This discrepancy might indicate that the obtained Staphylococcus spp. isolates were heat resistant. Even though an extensive literature survey was performed by the authors, no reports were found that have previously described the finding of Staphylococcus spp. that were heat resistant to the heat-shocking procedure used here (75°C, 20 min). However, ongoing work at the Norwegian Defense Research Establishment has provided similar observations of heat resistance in Staphylococcus spp. found in wastewater treatment plants (unpublished data). Alternatively, the bacterial cells might have been protected from the treatment if they were imbedded in larger airborne particles, such as cell aggregates, biofilms, skin flakes, or other composite matrices, as have been shown for other injuring stresses such as solar/UV radiation and desiccation (46, 71). It is generally acknowledged that spore-forming bacteria, such as Bacillus spp., are present mainly as spores in the environment (65). This is supported by the high amount of Bacillus spp. that were derived from spore-specific cultivations in the present study, although we cannot exclude the possibility of Bacillus spp. being present as both spores and vegetative cells in the airborne environment. Direct comparison of the number of Bacillus spp. isolates derived from the total and spore-specific cultivations, which could have been used to elucidate whether Bacillus spp. were exclusively present as spores, was not appropriate on the basis of some important cultivation and isolation differences between the total and spore-specific cultivations. All colonies from spore-specific cultivation plates were isolated, whereas only a representative selection was isolated from total cultivation plates. In addition, the germination efficiencies could be different for bacterial spores that were activated by heat shocking prior to cultivation and those that were not (16, 37).
Particle concentration and size distribution.
The particle concentration and size distribution obtained at the subway station were comparable to those reported by Birenzvige et al. from the Washington, DC, subway (7). Over the entire measured size range, increased particle concentrations were seen at the station during daytime compared to nighttime, and this is probably related to anthropogenic daytime sources that were nearly absent at night, such as passengers and train traffic. The total particle concentrations and total cultivable airborne bacterial concentrations showed significant correlation (r = 0.896). This indicated that the level of airborne bacteria was related to the total particle concentration, but the ratio (total particles to cultivable bacteria) was as high as 106 airborne particles per CFU detected when considering the entire particle size range measured. This is in agreement with the study from Washington, DC, which showed that fluorescent airborne particles accounted for <1% of the total particle counts (7), which would translate into a total particle to total cultivable bacteria ratio much higher than 102 airborne particles per CFU detected.
Biotyper 2.0 MALDI-TOF MS-based classification.
We focused here on rapid screening of bacterial isolates obtained from airborne bacteria at a subway station using the Biotyper 2.0 microbial identification system, and thus the standard direct transfer method recommended by the manufacturer for screening purposes was exclusively used. The obtained results showed correlation, but also some discrepancies, between the time-consuming and costly 16S rRNA gene-based classification, and the rapid MALDI-TOF MS-based Biotyper 2.0 system. About 64% of the isolates analyzed with the Biotyper 2.0 yielded a score value above 1.7, and hence 36% were assigned with no reliable identification since 1.7 is the lower cutoff score value used by the Biotyper 2.0 system. Still, one and six of these low-scoring isolates yielded species- and genus-consistent classification results compared to the 16S rRNA gene-based classification, respectively, although this could not have been appreciated without the a priori knowledge from the 16S rRNA gene-based classification. When only considering the isolates that yielded a score value above 1.7, 61% were species-consistently classified and 94% were genus-consistently classified compared to the 16S rRNA gene-based classification. Of the 84 isolates that were analyzed (Biotyper 2.0 system), seven genera and 24 species given by the 16S rRNA gene-based classification were missing from the Bruker Taxonomy database. Adding these into the database could probably increase the consistency between the two classification methods. The coverage of environmental bacteria in the Bruker Taxonomy database and other taxonomic reference databases in general are gradually increasing, but there is still a bias toward clinically relevant and highly studied bacteria, and the coverage is still low for the myriad of newly identified environmental bacteria (11). The Biotyper 2.0 system allows users to construct their own libraries, thus enabling them to increase the database coverage for organisms of special interest in their research. Taken together, the Biotyper 2.0 microbial identification platform is a rapid and powerful tool for low-cost screening of environmental bacterial isolates, but an increased coverage of such bacteria is urgently needed in the reference database before the full potential of this screening method can be appreciated. Care must also be taken to obtain high-quality spectra, balancing between the rapidness of the direct transfer method and the more laborious protein extraction method that is recommended by the manufacturer to increase spectrum quality.
Antibiotic resistance.
To our knowledge, antibiotic resistance profiling of the airborne bacterial community in a subway station has not previously been addressed, but several studies have addressed antibiotic resistance of airborne bacteria in other specific environments (14, 17, 25, 30, 58), such as cattle, swine, or poultry farms (1, 26, 57). The results obtained in the present study show that most dominant bacterial genera have a very high frequency of resistance to NAL, except for Bacillus and Paenibacillus spp., which had 14 and 0% frequencies of resistance, respectively. Bacillus was the only genus to show a high (>50%) frequency of resistance against AMP, while Paenibacillus was the only genus to show a high (>50%) frequency of resistance against STR. The airborne environment harbored bacteria with antibiotic resistance that covered all of the antibiotics tested, and more than 27% of the obtained isolates were resistant against at least two of them. This shows that airborne bacteria in the subway environment contain a pool of antibiotic resistance determinants against a broad range of antibiotics. If these determinants are mobile, or mobilized, they could be transferred to pathogenic or opportunistic pathogenic bacteria, either in the environment or after human inhalation. This could possibly have an impact on public health, and further elucidation of this pool of antibiotic resistance determinants and their presence on mobile genetic elements (e.g., plasmids, integrons, and transposons) is warranted.
Hemolytic activity and small cyclic peptides.
The hemolytic properties of airborne microorganisms in the subway environment have only been briefly studied previously (8, 62). About 22% of the obtained bacterial isolates in the present study were hemolytic. This is less than what was found in a study from Korea (62), which reported that 34% of the isolates were hemolytic. These researchers reported 27% beta-hemolysis and 7% alpha-hemolysis, while we found ca. 21% beta-hemolysis and less than 1% alpha-hemolysis in our initial screen on sheep blood and ca. 19% beta-hemolysis and no alpha-hemolysis in the subsequent screen on bovine blood. The majority of our hemolytic isolates (87%) were members of the bacterial genus Bacillus. More than 50% of the total number of Bacillus isolates obtained here were hemolytic on both sheep and bovine blood. The hemolytic Bacillus isolates clustered into at least three distinct phylogenetic groups, corresponding to known producers of NRPS-produced SCPs, including B. cereus (cereulide), B. pumilus (pumilacidin), and B. subtilis (surfactin). Cereulide (B. cereus emetic toxin) causes food poisoning (22, 65). We investigated here the ecology of cereulide-producing strains in airborne bacteria and in a subway environment. In our study, the presence of cereulide or the NRPS gene (ces) was not detected in any of the hemolytic Bacillus isolates, while in foods usually ca. 5% of strains are ces positive (22). Surfactin-like hemolytic peptides, such as surfactins, pumilacidins, and lichenycins, are a family of structurally similar SCLPTs that are produced by several B. subtilis group species, such as B. subtilis, B. pumilus, and B. licheniformis. In the present study, we investigated the distribution of SCLPTs in airborne Bacillus spp. from a subway environment, showing that 50% of the hemolytic Bacillus spp. isolates and 26% of the total number of Bacillus spp. isolates produced SCLPTs. Surprisingly, SCLPT-positive isolates were limited to the B. pumilus and B. subtilis group, except for three isolates that were closely related to B. thuringiensis (n = 2) and B. cereus (n = 1) in the B. cereus group. The percentage of SCLPT-producing isolates is very high compared to what has been reported by From et al., who found that only 8 of 333 isolates from food and water produced SCLPTs (24). We currently have no explanation for the finding of such a high number of SCLPT-positive airborne isolates compared to that found in foods and water. To our knowledge, there are no available surveys on the percentage of SCLPT-containing Bacillus spp. in the airborne environment.
Pigmentation.
The atmosphere is generally considered a hostile environment for microorganisms due to stress, such as desiccation, radiation, oxygen toxicity, and pollutants (13). Solar/UV radiation has been shown to influence the airborne survival of bacteria, and pigmentation is a mechanism adopted by several types of bacteria to protect them against photo-oxidative damage (72). Our study presented an in-depth characterization of pigmentation in airborne bacteria at a subway station. About 48% of the total cultivation isolates were pigmented, while only 8% of the spore-specific cultivation isolates were pigmented. One of the main protective effects of pigmentation is protection against oxidative damage due to solar/UV radiation, and since bacterial spores are highly resistant to UV/solar radiation (63), the finding of a substantially lower percentage of pigmented spore-forming bacteria is not surprising but still noteworthy. A high percentage of pigmented bacteria has been found in ambient outdoor atmosphere, and positive selection toward pigmented bacteria has been demonstrated (73). Carotenoids, with more than 700 different representatives isolated from natural sources, represent an abundant group of natural pigments in both the prokaryote and eukaryote kingdoms, exhibiting colors from dark red to bright yellow (15). Yellow, orange, and red pigments are abundantly found in members of the bacterial genera Micrococcus, Corynebacterium, Mycobacterium, and Nocardia, all of which contain carotenoids (61). More than 50% of the pigmented isolates analyzed by LC-MS in the present study led to tentative pigment identification, spanning a broad range of different carotenoids. Although the pigments were only tentatively identified, the finding of sarcinaxanthin in an isolate classified as M. luteus further strengthens our identifications since this pigment was originally isolated from S. lutea (the former name of M. luteus) (45). Carotenoids have more recently also been implicated as potential bacterial virulence factors, since they may quench destructive effects of oxygen radicals, a killing mechanism used by immune cells during the innate immune response to bacterial infections (29, 44, 54). A derivative of staphyloxanthin, 8-apo-caroten-8-oic acid, was found in an isolate that was classified as S. warneri, and this is of interest since staphyloxanthin is the predominant pigment in another bacterial species within the same genus, namely, S. aureus (48). Staphyloxanthin has recently been implicated as a potential virulence factor involved in protecting the bacteria against oxygen radical killing used by neutrophils during the innate immune response to bacterial infections (44).
Future directions.
We have addressed the bacterial fraction of the bioaerosol community and its characteristics at a subway station. Our aim was to investigate the presence of naturally occurring airborne bacteria in an environment regarded as a potential target for incidents involving the deliberate use of biological threat agents and where biological detectors might be used. Information about the bioaerosol background of the detectors operational location is important for its performance criteria. However, other types of airborne microorganisms (e.g., fungi and viruses) and their products (e.g., toxins) could also be important for biological detection/surveillance, as well as for public health. Further investigations of the subway environment, or other operational environments, should therefore seek to also address airborne fungi, viruses, and toxins.
The temporal variability of the bioaerosol background might influence the operational performance of biological detectors, and while the present study revealed differences between the daytime and nighttime bioaerosol environment at a subway station, an apparent limitation is the temporal resolution of the cultivation data since only two samples were collected each day. In an attempt to more specifically address the diurnal variation of the bioaerosol background at the subway station, we are currently conducting follow-up studies involving a multiday continuous sampling campaign using the high-volume impactor MAS-100. This approach will hopefully yield cultivation data with a much higher temporal resolution than what was obtained here and thereby allow a more in-depth investigation of the diurnal variability of the bioaerosol background.
Cultivation has been the traditional method used to determine airborne bacterial concentrations, but since generally <1% of environmental bacteria are cultivable by standard laboratory methods (2), cultivation-independent analyses have become widespread. In the present study, the obtained bacterial isolates were distributed among only three phyla, Actinobacteria, Proteobacteria, and Firmicutes, and the proportion of Gram-positive bacteria was >90%. The predominance of these phyla and the high content of Gram-positive bacteria are common for cultivation-dependent studies of airborne bacteria (36) but likely represent only a fraction of the complete diversity (32, 33, 51). To further elucidate the airborne bacterial diversity at the Norwegian subway station, we are currently conducting a culture-independent diversity study using 16S rRNA gene-based PhyloChip microarrays (9, 10).
The regional deposition of bioaerosols inside the human respiratory tract is related to particle size, which is an important property of airborne bacteria-containing particles when assessing their potential health hazard (70). To our knowledge, only one study (available in Korean) has previously addressed the size distribution of cultivable bacterium-containing particles in a subway environment (41). We are currently conducting further studies at the Nationaltheatret subway station using the Andersen sampler (six-stage cascade impactor) to investigate the size distribution of airborne bacterium-containing particles.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Norwegian Defense Research Establishment.
We thank Abdelghani Ahmed and Victor Hormazabal at the Norwegian School of Veterinary Science for technical assistance during the characterization of bacterial hemolytic activity and LC-MS studies on cereulide and surfactin-like peptides. We thank Anja Valen and Vilde Sørvik Eggen, Norwegian Defense Research Establishment, who contributed to the project with technical and statistical assistance, respectively.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alvarado CS, et al. 2009. Seasonal changes in airborne fungi and bacteria at a dairy cattle concentrated animal feeding operation in the southwest United States. J. Environ. Health 71:40–44 [PubMed] [Google Scholar]
- 2. Amann R, Ludwig W, Schleifer K. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. An HR, Mainelis G, Yao M. 2004. Evaluation of a high-volume portable bioaerosol sampler in laboratory and field environments. Indoor Air 14:385–393 [DOI] [PubMed] [Google Scholar]
- 4. Awad AHA. 2002. Environmental study in subway metro stations in Cairo, Egypt. J. Occup. Health 44:112–118 [Google Scholar]
- 5. Barbeau B, et al. 1997. A modified method for the enumeration of aerobic spore-forming bacteria. Can. J. Microbiol. 43:976–980 [Google Scholar]
- 6. Baron P, Willeke K, Kulkarni P. (ed). 2011. Aerosol measurement: principles, techniques, and applications, 3rd ed John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 7. Birenzvige A, et al. 2003. Aerosol characteristics in a subway environment. Aerosol Sci. Technol. 37:210–220 [Google Scholar]
- 8. Bogomolova E, Kirtsideli I. 2009. Airborne fungi in four stations of the St. Petersburg underground railway system. Int. Biodeterior. Biodegrad. 63:156–160 [Google Scholar]
- 9. Brodie EL, et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brodie EL, et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cayrou C, Raoult D, Drancourt M. 2010. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of environmental organisms: the Planctomycetes paradigm. Environ. Microbiol. Rep. 2:752–760 [DOI] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention 2011. Category A list. Centers for Disease Control and Prevention, Atlanta, GA: [Online.] [Google Scholar]
- 13. Cox CS, Wathes CM. 1995. Bioaerosols handbook. Lewis Publishers, Boca Raton, FL [Google Scholar]
- 14. Decraene V, Ready D, Pratten J, Wilson M. 2008. Air-borne microbial contamination of surfaces in a UK dental clinic. J. Gen. Appl. Microbiol. 54:195–203 [DOI] [PubMed] [Google Scholar]
- 15. Dembitsky V. 2005. Astonishing diversity of natural surfactants. III. Carotenoid glycosides and isoprenoid glycolipids. Lipids 40:535–557 [DOI] [PubMed] [Google Scholar]
- 16. Desrosier NW, Heiligman F. 1956. Heat activation of bacterial spores. J. Food Sci. 21:54–62 [Google Scholar]
- 17. Donderski W. 2005. Microbiological contamination of air within the city of Torun. Pol. J. Environ. Studies 14:223–230 [Google Scholar]
- 18. Dong S, Yao M. 2010. Exposure assessment in Beijing, China: biological agents, ultrafine particles, and lead. Environ. Monitoring Assess. 170:331–343 [DOI] [PubMed] [Google Scholar]
- 19. Douwes J, Thorne P, Pearce N, Heederik D. 2003. Bioaerosol health effects and exposure assessment: progress and prospects. Ann. Occup. Hyg. 47:187–200 [DOI] [PubMed] [Google Scholar]
- 20. Emanuel P, Caples M. 2011. Chemical, biological, radiological technology survey, 2011. U.S. Army Edgewood Chemical Biological Center, APG, MD [Google Scholar]
- 21. Fricker M, Messelhausser U, Busch U, Scherer S, Ehling-Schulz M. 2007. Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 73:1892–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. From C, Hormazabal V, Granum PE. 2007. Food poisoning associated with pumilacidin-producing Bacillus pumilus in rice. Int. J. Food Microbiol. 115:319–324 [DOI] [PubMed] [Google Scholar]
- 23. From C, Hormazabal V, Hardy SP, Granum PE. 2007. Cytotoxicity in Bacillus mojavensis is abolished following loss of surfactin synthesis: implications for assessment of toxicity and food poisoning potential. International J. Food Microbiol. 117:43–49 [DOI] [PubMed] [Google Scholar]
- 24. From C, Pukall R, Schumann P, Hormazabal V, Granum PE. 2005. Toxin-producing ability among Bacillus spp. outside the Bacillus cereus group. Appl. Environ. Microbiol. 71:1178–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gandara A, et al. 2006. Isolation of Staphylococcus aureus and antibiotic-resistant Staphylococcus aureus from residential indoor bioaerosols. Environ. Health Perspect. 114:1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibbs SG, et al. 2006. Isolation of antibiotic-resistant bacteria from the air plume downwind of a swine confined or concentrated animal feeding operation. Environ. Health Perspect. 114:1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilleberg SB, Faull JL, Graeme-Cook KA. 1998. A preliminary survey of aerial biocontaminants at six London underground stations. Int. Biodeterior. Biodegrad. 41:149–152 [Google Scholar]
- 28. Gould GW. (ed). 1971. Methods for studying bacterial spores, vol 6A Academic Press, London, England [Google Scholar]
- 29. Hassett D, Cohen M. 1989. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 3:2574–2582 [DOI] [PubMed] [Google Scholar]
- 30. Heo Y, et al. 2010. Size-resolved culturable airborne bacteria sampled in rice field, sanitary landfill, and waste incineration sites. J. Environ. Monit. 12:1619–1624 [DOI] [PubMed] [Google Scholar]
- 31. Hormazábal V, Østensvik Ø, O'Sullivan K, Granum PE. 2004. Quantification of Bacillus cereus emetic toxin (cereulide) in figs using LC/MS. J. Liquid Chromatogr. Relat. Technol. 27:2531–2538 [Google Scholar]
- 32. Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hugenholtz P, Pace NR. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:190–197 [DOI] [PubMed] [Google Scholar]
- 34. Hwang SH, Yoon CS, Ryu KN, Paik SY, Cho JH. 2010. Assessment of airborne environmental bacteria and related factors in 25 underground railway stations in Seoul, Korea. Atmos. Environ. 44:1658–1662 [Google Scholar]
- 35. Kawasaki T, et al. 2010. Distribution and identification of airborne fungi in railway stations in Tokyo, Japan. J. Occup. Health 52:186–193 [DOI] [PubMed] [Google Scholar]
- 36. Kellogg CA, Griffin DW. 2006. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 21:638–644 [DOI] [PubMed] [Google Scholar]
- 37. Keynan A, Evenchik Z, Halvorson HO, Hastings JW. 1964. Activation of bacterial endospores. J. Bacteriol. 88:313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim KY, Kim YS, Kim D, Kim HT. 2011. Exposure level and distribution characteristics of airborne bacteria and fungi in Seoul metropolitan subway stations. Ind. Health 49:242–248 [DOI] [PubMed] [Google Scholar]
- 39. Kloos WE, Musselwhite MS. 1975. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Environ. Microbiol. 30:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt EGM. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 41. Lee CM. 2004. Characterization of airborne bioaerosol concentration in public facilities (Korean). J. Environ. Sci. 13:215–222 [Google Scholar]
- 42. Li C-S. 1999. Evaluation of microbial samplers for bacterial microorganisms. Aerosol Sci. Technol. 30:100–108 [Google Scholar]
- 43. Liaaen-Jensen S, Weeks OB, Strang RH, Thirkell D. 1967. Identity of the C-50-carotenoid dehydrogenans-P439 and sarcinaxanthin. Nature 214:379–380 [DOI] [PubMed] [Google Scholar]
- 44. Lighthart B, Shaffer BT. 1997. Increased airborne bacterial survival as a function of particle content and size. Aerosol Sci. Technol. 27:439–446 [Google Scholar]
- 45. Lilliefors H. 1967. On the Kolmogorov-Smirnov test for normality with mean and variance unknown. J. Am. Stat. Assoc. 62:399–402 [Google Scholar]
- 46. Liu GY, et al. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202:209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Mathematical Stat. 18:50–60 [Google Scholar]
- 48. Marshall JH, Wilmoth GJ. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147:900–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. National Research Council 2005. 2005 Sensor systems for biological agent attacks: protecting buildings and military bases. NRC Committee on Materials and Manufacturing Processes for Advanced Sensors. National Academies Press, Washington, DC [Google Scholar]
- 50. Okumura T, et al. 1996. Report on 640 victims of the Tokyo subway sarin attack. Ann. Emerg. Med. 28:129–135 [DOI] [PubMed] [Google Scholar]
- 51. Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740 [DOI] [PubMed] [Google Scholar]
- 52. Pearson K. 1895. Note on regression and inheritance in the case of two parents. Proc. R. Soc. 58:241 [Google Scholar]
- 53. Pearson K. 1900. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos. Magazine 50:157–175 [Google Scholar]
- 54. Provvedi R, et al. 2008. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 190:7859–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ratnesar-Shumate S, et al. 2011. Improved method for the evaluation of real-time biological aerosol detection technologies. Aerosol Sci. Technol. 45:635–644 [Google Scholar]
- 56. Research International 2011, posting date Evaporation rates for microorganisms captured by Research International's electret filters. Research International, Seattle, WA: [Online.] [Google Scholar]
- 57. Rule AM, Evans SL, Silbergeld EK. 2008. Food animal transport: a potential source of community exposures to health hazards from industrial farming (CAFOs). J. Infect. Public Health 1:33–39 [DOI] [PubMed] [Google Scholar]
- 58. Safatov AS, et al. 2008. To what extent can viable bacteria in atmospheric aerosols be dangerous for humans? Clean Soil Air Water 36:564–571 [Google Scholar]
- 59. Samuels AC, et al. (ed). 2006. Test methodology development for biological agent detection systems. Proc. SPIE 6378:637802–1 [Google Scholar]
- 60. Sánchez-Monedero MA, Aguilar MI, Fenoll R, Roig A. 2008. Effect of the aeration system on the levels of airborne microorganisms generated at wastewater treatment plants. Water Res. 42:3739–3744 [DOI] [PubMed] [Google Scholar]
- 61. Schlegel H, Jannasch H. 2006. Prokaryotes and their habitats, p 137–184 In Dworkin M, et al. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 62. Seino K, Takano T, Nakamura K, Watanabe M. 2005. An evidential example of airborne bacteria in a crowded, underground public concourse in Tokyo. Atmos. Environ. 39:337–341 [Google Scholar]
- 63. Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525 [DOI] [PubMed] [Google Scholar]
- 64. Shahamat M, et al. 1997. Evaluation of media for recovery of aerosolized bacteria. Aerobiologia 13:219–226 [Google Scholar]
- 65. Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579–606 [DOI] [PubMed] [Google Scholar]
- 66. Szam L, Nikodemusz I, Csatai L, Vedres I, Dakay M. 1980. Airborne microflora found in some stations of the metro in the Hungarian capital of Budapest (author's transl). Zentralbl. Bakteriol. B 170:199–208 [PubMed] [Google Scholar]
- 67. Szam L, Vedres I, Csatai L, Nikodemusz I. 1983. Further microbiological studies of the air in a newly built (under the pavement) section of the underground railway in Budapest (author's transl.). Zentralbl. Bakteriol. Mikrobiol. Hyg. B 177:312–318 [PubMed] [Google Scholar]
- 68. Takahashi H, et al. 2004. Bacillus anthracis incident, Kameido, Tokyo, 1993. Emerg. Infect. Dis. 10:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tapi A, Chollet-Imbert M, Scherens B, Jacques P. 2010. New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl. Microbiol. Biotechnol. 85:1521–1531 [DOI] [PubMed] [Google Scholar]
- 70. Thomas RJ, et al. 2008. Characterization and deposition of respirable large- and small-particle bioaerosols. Appl. Environ. Microbiol. 74:6437–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tong Y, Lighthart B. 1998. Effect of simulated solar radiation on mixed outdoor atmospheric bacterial populations. FEMS Microbiol. Ecol. 26:311–316 [Google Scholar]
- 72. Tong Y, Lighthart B. 1997. Solar radiation has a lethal effect on natural populations of culturable outdoor atmospheric bacteria. Atmos. Environ. 31:897–900 [Google Scholar]
- 73. Tong Y, Lighthart B. 1997. Solar radiation is shown to select for pigmented bacteria in the ambient outdoor atmosphere. Photochem. Photobiol. 65:103–106 [Google Scholar]
- 74. Valdes JJ, et al. 2011. Background discrimination of bioaerosols. ITEA J. 32:181–190 [Google Scholar]
- 75. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yao M, Mainelis G. 2007. Analysis of portable impactor performance for enumeration of viable bioaerosols. J. Occup. Environ. Hyg. 4:514–524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.