Abstract
Listeria monocytogenes isolates from bovine hides and carcasses (n = 812) were mainly of serogroup 1/2a. All strains were positive for internalin genes. Several isolates were resistant to oxacillin (72.2%) or clindamycin (37.0%). These findings indicate that L. monocytogenes of beef origin can be considered a public health concern.
TEXT
Listeria monocytogenes may contaminate food of animal origin, including beef meat (7, 17). Although infection of humans due to L. monocytogenes has a low incidence, it is associated with a high mortality rate (6, 19). Serological and PCR techniques identified at least 13 L. monocytogenes serotypes; however, mainly isolates of 1/2a, 1/2b, 1/2c, and 4b serogroups are recovered from food and patients (4). Several putative virulence factors of L. monocytogenes have been described, and among them, inlA, inlC, and inlJ (encoding internalin-like proteins), lmo2672 (responsible for transcriptional regulator), and llsX (for listeriolysin S expression) seem to be the most important in the pathogenesis of human listeriosis (1, 3, 5, 9).
There is little information concerning L. monocytogenes antimicrobial resistance and especially little information about the isolates recovered from beef that has been determined by the microdilution method. It is known that Listeria strains are usually susceptible to most antibiotics; however, several resistant isolates have also been identified (3, 15, 16).
The aim of the present study was to determine the prevalence of L. monocytogenes in hides and carcasses of cattle slaughtered in Poland and to identify the virulence markers and antimicrobial resistance of the isolates.
A total of 406 cattle were used in the study. The samples from bovine hides and the corresponding carcasses (n = 812) were collected using a swab method, and L. monocytogenes was identified by the ISO 11290-1:1999 standard. Serotype determination was performed by multiplex PCR as described previously (4). The presence of virulence genes inlA, inlC, inlJ, lmo2672, and llsX was tested by PCR (Table 1) (10, 11). Antimicrobial susceptibility of the L. monocytogenes isolates was determined by using a Sensititre GPN3F plate according to the manufacturer's instructions (Trek Diagnostic Systems). The CLSI guidelines were used for the interpretation of the obtained MICs (2, 12).
Table 1.
PCR primers used for determination of L. monocytogenes virulence marker genes
| PCR test | Primer name | Sequence (5′→3′) | Primer final concn (μM) | Target gene | Size of PCR amplicon (bp) | Reference |
|---|---|---|---|---|---|---|
| mPCR1 | inlAF | ACGAGTAACGGGACAAATGC | 0.25 | inlA | 800 | 11 |
| inlAR | CCCGACAGTGGTGCTAGATT | 0.25 | ||||
| inlCF | AATTCCCACAGGACACAACC | 0.15 | inlC | 517 | ||
| inlCR | CGGGAATGCAATTTTTCACTA | 0.15 | ||||
| inlJF | TGTAACCCCGCTTACACACAGTT | 0.05 | inlJ | 238 | ||
| inlJR | AGCGGCTTGGCAGTCTAATA | 0.05 | ||||
| PCR2 | lmo2672F | CGGCACACTTGGATTCTCAT | 0.3 | lmo2672 | 481 | 10 |
| lmo2672R | AGGGCTAGTGACGGATGCTA | 0.3 | ||||
| PCR3 | llsXF | TTATTGCATCAATTGTTCTAGGG | 0.2 | llsX | 200 | 1 |
| llsXR | CCCCTATAAACATCATGCTAGTG | 0.2 |
It was found that 44 out of 406 hide samples (10.8%) were contaminated with L. monocytogenes, whereas 10 (2.5%) corresponding bovine carcasses were positive for this pathogen as shown by the presence of the 370-bp prs gene amplicon. Further PCR serotyping revealed that the majority of the isolates (47; 87.0%) were of the 1/2a serotype, and only 4 L. monocytogenes isolates were classified as 1/2c (Fig. 1). The remaining 3 strains belonged to serotype 1/2b (1 isolate) or 4b (2 isolates). All strains were positive for the inlA, inlC, inlJ, and lmo2672 virulence marker genes. Furthermore, 4 L. monocytogenes isolates from hides and belonging to serotype 4b (3 isolates) or 1/2a (1 strain) had another virulence marker gene, llsX. The antimicrobial resistance results for the 54 L. monocytogenes strains showed that all isolates were susceptible to ampicillin, gatifloxacin, levofloxacin, penicillin, rifampin, streptomycin, trimethoprim-sulfamethoxazole, and vancomycin (Table 2). On the other hand, many isolates were resistant to oxacillin (72.2% strains) and several were resistant to clindamycin (37.0%) or ceftriaxone (13.0%).
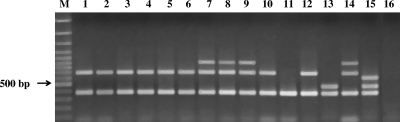
Fig 1.
Agarose gel electrophoresis of examples of DNA amplicons generated by multiplex PCR for identification of L. monocytogenes serotypes. Lanes 1 to 6, L. monocytogenes 1/2a (hide samples); lanes 7 to 9, L. monocytogenes 1/2c (hide and carcass samples); lane 10, L. monocytogenes 1/2a (carcass sample); lane 11, L. ivanovii ATCC 19119; lane 12, L. monocytogenes 05CEB424LM (1/2a); lane 13, L. monocytogenes 06CEB406LM (1/2b); lane 14, L. monocytogenes 06CEB405LM (1/2c); lane 15, L. monocytogenes 06CEB422LM (4b); lane 16, negative control.
Table 2.
Antimicrobial resistance of L. monocytogenes isolates tested in the study
| Antimicrobial group | Antimicrobial | Antimicrobial class according to WHOa | No. (%) of isolates |
||
|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | |||
| Aminoglycosides | Gentamicin | CI | 53 (98.1) | 0 | 1 (1.9) |
| Streptomycin | CI | 54 (100) | 0 | 0 | |
| Ansamycin | Rifampin | CI | 54 (100) | 0 | 0 |
| Cephalosporins | Ceftriaxone | CI | 32 (59.2) | 15 (27.8) | 7 (13.0) |
| Glycopeptides | Vancomycin | CI | 54 (100) | 0 | 0 |
| Lincosamides | Clindamycin | I | 5 (9.3) | 29 (53.7) | 20 (37.0) |
| Macrolides | Erythromycin | CI | 53 (98.1) | 1 (1.9) | 0 |
| Oxazolidinones | Linezolid | CI | 51 (94.4) | 3 (5.6) | 0 |
| Penicillins | Ampicillin | CI | 54 (100) | 0 | 0 |
| Penicillin | CI | 54 (100) | 0 | 0 | |
| Oxacillin | HI | 15 (27.8) | 0 | 39 (72.2) | |
| Fluoroquinolones | Ciprofloxacin | CI | 43 (79.6) | 11 (20.4) | 0 |
| Gatifloxacin | CI | 54 (100) | 0 | 0 | |
| Levofloxacin | CI | 54 (100) | 0 | 0 | |
| Streptogramins | Quinupristin-dalfopristin | CI | 52 (96.2) | 1 (1.9) | 1 (1.9) |
| Potentiated sulfonamide | Trimethoprim-sulfamethoxazole | HI | 54 (100) | 0 | 0 |
| Tetracyclines | Tetracycline | HI | 53 (98.1) | 0 | 1 (1.9) |
CI, critically important; I, important; HI, highly important (2).
Our results indicate that L. monocytogenes may contaminate beef carcasses during the slaughter process. Molecular serotyping revealed that serotype 1/2a was predominant, irrespective of the sample origin. Very few isolates were of the 1/2c, 4b, or 1/2b serogroup.
Worldwide, several studies were conducted to determine the prevalence of L. monocytogenes in bovine samples. This pathogen was identified in cattle hides (13.3%) and carcasses (2.8%) at processing plants in the United States; the most prevalent serotype, as in the present study, was 1/2a, and only few strains were of serogroup 1/2b or 4b (8). Another study performed by Rivera-Betancourt et al. (18) indicated that the bacteria were found on 9.9% of 1,033 examined hide samples, whereas bacteria were found in the carcasses (n = 522) to a much smaller extent, i.e., 1.1%.
Many putative virulence markers in L. monocytogenes have been identified, and the surface-associated internalins are claimed to play a role in the pathogenesis of human listeriosis (14). In the present study, three main internalin genes (inlA, inlC, and inlJ) as well as the lmo2672 marker were detected in all 54 L. monocytogenes isolates, which suggests that these strains may be potentially virulent for consumers. Similar results were obtained by Mammina et al. (13) in a study of 54 humans. The L. monocytogenes isolates belonged to three main serotypes (1/2a, 1/2b, and 4b).
A wide spectrum of antibiotics, of 11 different groups, were used in this study to assess the resistance of the L. monocytogenes isolates (Table 2). The results revealed that the strains were sensitive to most antibiotics tested, except oxacillin, to which 72.2% of the isolates were resistant; some strains were also resistant to clindamycin (37.0%). A similar set of antimicrobials was used by Lyon et al. (12), who found that as many as 90% of L. monocytogenes isolates were resistant to oxacillin. Furthermore, the results for ceftriaxone, ciprofloxacin, and clindamycin susceptibility revealed that several strains displayed intermediate resistance, i.e., 38%, 34%, and 27% of isolates, respectively. Similar results for those antibiotics were also found in our studies (27.8%, 20.4%, and 53.7% of the strains tested, respectively).
The resistance of L. monocytogenes to antimicrobials currently used in human therapy was also investigated by other authors (3, 15, 16). Most of the strains isolated from raw meat, food, or food-processing environments were susceptible to antimicrobials except oxacillin; some isolates were also resistant to ampicillin, clindamycin, gentamicin, tetracyclines, and penicillin. These results are in agreement with the antimicrobial resistance pattern obtained in the present study. On the other hand, a recent study on L. monocytogenes isolates from food and the environment (n = 202) performed by Granier et al. (7) showed the resistance of 4 strains (2.0%) only; 2 of them were resistant to tetracycline (MIC > 8 μg/ml), and another 2 were resistant to erythromycin (MIC > 2 μg/ml). As shown in the present study, only 1 out of 54 isolates tested was resistant to tetracycline. All these results indicate the need for further investigation of the antimicrobial resistance profile of L. monocytogenes, especially the strains isolated from the food chain.
In summary, our results regarding the presence of virulence markers and antimicrobial resistance of L. monocytogenes indicate that contamination of beef may be a public health concern.
ACKNOWLEDGMENT
This work was financially supported by the EU FP6 ProSafeBeef Project, grant no. FOOD-CT-2006-36241.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Clayton EM, Hill C, Cotter PD, Ross RP. 2011. Real-time PCR assay to differentiate listeriolysin S-positive and-negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 77:163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49:132–141 [DOI] [PubMed] [Google Scholar]
- 3. Conter M, et al. 2009. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 128:497–500 [DOI] [PubMed] [Google Scholar]
- 4. Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doumith M, et al. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Granier SA, et al. 2011. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl. Environ. Microbiol. 77:2788–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guerini MN, et al. 2007. Listeria prevalence and Listeria monocytogenes serovar diversity at cull cow and bull processing plants in the United States. J. Food Prot. 70:2578–2582 [DOI] [PubMed] [Google Scholar]
- 9. Kreft J, Vazquez-Boland JA. 2001. Regulation of virulence genes in Listeria. Int. J. Microbiol. 291:145–157 [DOI] [PubMed] [Google Scholar]
- 10. Liu D, Ainsworth AJ, Austin FW, Lawrence ML. 2003. Characterization of virulent and avirulent Listeria monocytogenes strains by PCR amplification of putative transcriptional regulator and internalin genes. J. Med. Microbiol. 52:1065–1070 [DOI] [PubMed] [Google Scholar]
- 11. Liu D, Lawrence ML, Austin FW, Ainsworth AJ. 2007b. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 71:133–140 [DOI] [PubMed] [Google Scholar]
- 12. Lyon SA, Berrang ME, Fedorka-Cray PJ, Fletcher DL, Meinersmann RJ. 2008. Antimicrobial resistance of Listeria monocytogenes isolated from a poultry further processing plant. Foodborne Pathog. Dis. 5:253–259 [DOI] [PubMed] [Google Scholar]
- 13. Mammina C, et al. 2009. Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 47:2925–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGann P, Raengpradub S, Ivanek R, Wiedmann M, Boor KJ. 2008. Differential regulation of Listeria monocytogenes internalin and internalin-like genes by σB and PrfA as revealed by subgenomic microarray analyses. Foodborne Pathog. Dis. 4:417–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morvan A, et al. 2010. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 54:2728–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pesavento G, Ducci B, Nieri D, Comodo N, Lo Nostro A. 2010. Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 21:708–713 [Google Scholar]
- 17. Rhoades JR, Duffy G, Koutsoumanis K. 2009. Prevalence and concentration of verocytotoxigenic Escherichia coli, Salmonella enterica and Listeria monocytogenes in the beef production chain: a review. Food Microbiol. 26:357–376 [DOI] [PubMed] [Google Scholar]
- 18. Rivera-Betancourt M, et al. 2004. Prevalence of Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella in two geographically distant commercial beef processing plants in the United States. J. Food Prot. 67:295–302 [DOI] [PubMed] [Google Scholar]
- 19. Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microb. Infect. 9:1236–1243 [DOI] [PubMed] [Google Scholar]