Abstract
In longitudinal biomedical studies, there is often interest in the rate functions, which describe the functional rates of change of biomarker profiles. This paper proposes a semiparametric approach to model these functions as the realizations of stochastic processes defined by stochastic differential equations. These processes are dependent on the covariates of interest and vary around a specified parametric function. An efficient Markov chain Monte Carlo algorithm is developed for inference. The proposed method is compared with several existing methods in terms of goodness-of-fit and more importantly the ability to forecast future functional data in a simulation study. The proposed methodology is applied to prostate-specific antigen profiles for illustration. Supplementary materials for this paper are available online.
Keywords: Euler approximation, Functional data analysis, Gaussian process, Rate function, Stochastic differential equation, Semiparametric stochastic velocity model
1 Introduction
This paper focuses on semiparametric stochastic modeling of rate functions for functional data in a multi-subject setting, where the data consists of a set of subjects, and for each subject, the observations are discrete samples from a curve with additive measurement errors. The rate function describes the functional rate of change or slope with respect to time, and has been of recent interest in longitudinal biomedical studies (Mungas et al., 2005; Lloyd-Jones et al., 2007; Strasak et al., 2008; Kariyanna et al., 2010). For example, from subject-matter knowledge it may be the rate of change, rather than the level of some biomarker, that can explain and predict the disease outcomes. One challenge in this research is to model the rate function without making a strong parametric assumption. Further challenges include modeling the rate functions across the subjects and allowing it to depend on the covariates of interest.
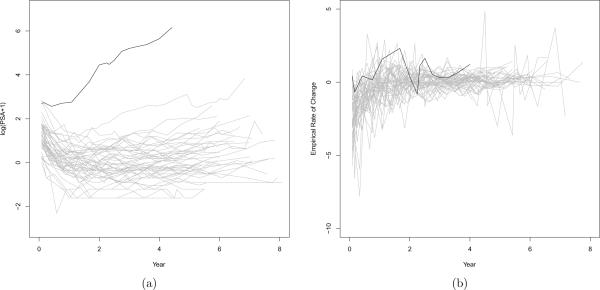
Our development has been largely motivated by a longitudinal study in prostate cancer patients (Proust-Lima et al., 2008), where prostate-specific antigen (PSA) profiles were collected for patients who received external beam radiation therapy (EBRT). PSA is roughly proportional to the prostate tumor size, and its rate of change has been shown to be associated with the recurrence of prostate cancer (Sartor et al., 1997). Figure 1(a) shows the log-transformed PSA level over time after EBRT treatment for 50 selected patients, and Figure 1(b) illustrates individual empirical rates of change, one for each subject. Figure 1(b) suggests that the individual rate of change of PSA roughly follows a common pattern. That is, it begins with a negative value caused by the EBRT, decreases over time in magnitude as the rate of tumor shrinkage gets lower, and eventually reaches a certain stable level. It is also apparent that rates of change vary considerably from this common pattern. For example, for the subject highlighted in black in Figure 1(b), his empirical rate of change fluctuates around zero and his PSA level appears very different from the others. Hence it is desirable to model the rate of change semiparametrically by incorporating empirical evidence or prior knowledge through a parametric function of time while accounting for deviation from the common pattern nonparametrically. Additionally, it is clear that for some subjects the long term stable rates of change are near zero, while for others they are positive. It is thus appealing not only to model a common stable rate of change across the subjects but also to let it follow a distribution, say a normal distribution with its mean depending on some baseline covariates. This flexibility will benefit the forecasts of future observations.
Figure 1.
PSA plots of (a) the raw data, (b) the empirical rate of change, which is defined as , for the give subject i with observation Yij at time tij. All profiles are plotted as the gray solid lines, except one profile highlighted in black color.
A number of methods have been used to study the rate of change in longitudinal studies. A popular approach is through a parametric linear mixed model (Laird and Ware, 1982; Diggle et al., 2002; Verbeke and Molenberghs, 2009), for example the random intercept and slope mixed model for disease progression (Zhang et al., 2008). This model assumes the subject's mean function follows a straight line with constant rate of change, which in turn is dependent on the covariates. In contrast to parametric models, the mean function have been modeled nonparametrically (Rice and Silverman, 1991; Wang and Taylor, 1995; Zeger and Diggle, 1994; Zhang et al., 1998; Verbyla et al., 1999). For these approaches, the rate function, as the first order derivative of the mean function, does not have any parametric form, and usually is not dependent on covariates. For other relevant literature that considers population dynamic models with multiple subjects see Wang et al. (2008), Paul et al. (2009) and Müller and Yao (2010). Additionally, in a time-varying coefficient model (Hastie and Tibshirani, 1993; Hoover et al., 1998) or functional mixed model (Guo, 2002; Morris and Carroll, 2006), the mean function Ui(t) of the ith subject is specified as . Hence, Ui(t) is a linear combination of several arbitrary smooth functions βk(t) with covariates Xik as the weights and depends on covariates linearly. Thus there seems to be a need for a model that allows flexible relationships between the rate function and covariates. Moreover, note that except for few approaches (Qin and Guo, 2006; Welham et al., 2006), nonparametric approaches seldom incorporate any prior knowledge from the subject-matter science, if available, in the modeling of the shape of the rate function.
Our goal is to develop a semiparametric stochastic model for the analysis of the rate function, which is called in this paper a semiparametric stochastic velocity model (SSVM). A key feature of SSVM is to utilize a stochastic process as a prior for the rate function, in a similar spirit to the work of Wahba (1978) and Zhu et al. (2011) for functional data in a single-subject setting. Formally, for each rate function for subject i ε N = {1, 2, …, n} and time , its prior is assumed to be a Gaussian process, conditional on xi = (xi0, xi1, …, xip)′, the vector of covariates for the ith subject. As an important special case of the proposed SSVM, we consider , where fxi(t) has a pre-specified parametric functional form dependent on covariates xi and is a scaled standard Wiener process. Hence, E{Vxi(t)} = fxi(t) implies that Vxi(t), the rate function of the ith subject, is expected to be centered about fxi(t), while the second term allows deviations from the parametric functional expectation fxi(t).
The remainder of the paper is organized as follows. Section 2 first presents the model and then is devoted to an important special case with the Ornstein-Uhlenbeck process as the prior for the rate function. Section 3 develops MCMC based methods for posterior inference and forecasting. Section 4 applies the methods to analyze the data of PSA profiles. Section 5 presents simulation results to evaluate and compare the performance of the proposed method with other existing methods. The paper concludes with a discussion in Section 6. Some supplementary materials related to the technical details of the proof of Theorem 1 are available online.
2 Semiparametric Stochastic Velocity Model
2.1 Model Specification
Suppose that Yi(tij), j = 1, 2, …, mi, i = 1, 2, …, n, is the response of the ith subject at time tij and satisfies the following hierarchical model, SSVM:
| (1) |
| (2) |
| (3) |
where Uxi(t) is the mean function for the ith subject's outcome curve, Vxi(t) is the corresponding rate function and Wi(t) denotes the standard Wiener process. Note that in this specification, although the mean function is defined at continuous time , it is observed at discrete times only and is subject to measurement error. Equation (3) may be regarded as a prior for the rate function Vxi(t), in which the behavior of Vxi(t) is governed by a stochastic differential equation (SDE), with drift term a{Vxi(t); xi, ϕi} and diffusion term b{Vxi(t); xi, ϕi}, where xi and ϕi are the covariate vector and subject-specific parameter vector. We assume that the initial values with large value of variance to make it non-informative, and that the measurement error . Here Ik is the k×k identity matrix and denotes the k-dimensional normal distribution with mean vector m and covariance matrix Σ. Furthermore, [Uxi(t0), Vxi(t0)]′, εi(t) and Wi(t) are assumed mutually independent.
The SDE in equation (3) gives rise to a general class of Markovian Gaussian processes (Feller, 1970; Grimmett and Stirzaker, 2001). In our model, this stochastic process is considered as the prior for the rate function Vxi(t). According to the specific research interest or context of a given study, we can choose different forms for a{Vxi(t); xi, ϕi}, which measures the instantaneous mean or the expected conditional acceleration, and for b2{Vxi(t); xi, ϕi}, which reflects the instantaneous variance of the rate process. In particular, we have the SSVM-W, when a{Vxi(t); xi, ϕi} = 0 and b{Vxi(t); xi, ϕi} = σξ, and the prior for Vxi(t) is the Wiener process. The resulting mean function takes the form , which is the partially integrated Wiener process leading to a smoothing spline (Wahba, 1978; Wecker and Ansley, 1983; Ansley and Kohn, 1986) for a given subject. Note that this prior is independent of covariates.
For the PSA data analysis given in Section 4, we specify and b{Vxi(t); xi, ϕi} = σξ. This specification corresponds to an Ornstein-Uhlenbeck (OU) process for Vxi(t), and the resulting rate function is given by . More details and properties of the OU process can be found in Section 2.2 below. We refer to this specification as SSVM-OU. For the PSA data analysis, it is of interest to estimate the stable rate , since Vxi(t) will eventually stabilize and fluctuate around the level given by , which describes the long term rate of tumor growth after radiation treatment. In addition, to address the relationship between the long term tumor growth rate and the patients' baseline characteristics, we propose a linear model , where β = (β0, β1, …, βp)′ is the vector of fixed effect parameters and are random effects. This subject-specific SSVM-OU is very useful to understand the dynamics of tumor growth, to assess the effect of covariates, and to predict a patient's future PSA values using the baseline covariate information.
2.2 The OU and IOU Processes
The OU process was first proposed as a physical model for the velocity of a particle suspended in a fluid (Uhlenbeck and Ornstein, 1930). It describes a homeostasis system that fluctuates around some stable level and has been applied in biology (Trost et al., 2010), finance (Nicolato and Venardos, 2003) and engineering (Kulkarni and Rolski, 2009), among many others. In the statistics literature, Aalen and Gjessing (2004) studied the first-passage time of an OU process, and Taylor and Law (1998) modeled the serial correlation in a linear mixed model by an integrated OU (IOU) process with mean zero. The OU process is particularly suitable for the PSA profiles considered in this paper, where the rate function of tumor growth reaches a stable level that potentially depends on baseline covariates.
Now we present some properties for both the OU and IOU processes. For ease of exposition, we suppress the subject index i in the discussion. Let Uj := U(tj) and Vj := V(tj). The IOU and OU processes are given by, respectively,
| (4) |
| (5) |
Theorem 1 For IOU and OU processes at time tj, conditional on the values at time tj−1and parameters , ρ, σξ, the transition distribution is given by
δj = tj − tj−1, with conditional mean and covariance matrix given, respectively, by,
The proof is included in the supplementary materials.
Corollary 1 For δj → ∞ and fixed ρ > 0, such that exp(−ρδj) = o(1), then the conditional mean and variance in Theorem 1 can be approximated by,
where the errors in the approximation are and . The proof is straightforward by noting that ρδj → ∞ as δj satisfies exp (−ρδj) = o(1).
Corollary 2 For OU and IOU processes with ρ > 0 and δj = o(1), the approximate transition density denoted by is given by,
where is the normal density with mean and variance , and δ(·) is the Dirac Delta function.
This corollary can be proved by taking the component-wise first-order Taylor approximation of mj and Σj in Theorem 1 with respect to δj.
3 Inference and Forecasting
3.1 Posterior Distribution Approximation
In this section, we present Bayesian estimation for the mean function Uxi(t), the rate function Vxi(t) and parameters ϕi and σε for i = 1,2, –, n. Let [· | ·] denote the exact conditional density, the approximate conditional density and where . Similar notation is used for V, Y and x. For the model specified by equations (1), (2) and (3), we first consider the posterior density [ϕ | U, V, Y, x] for ϕ, where . The posterior distribution is given by
| (6) |
where [Uij, Vij | Ui;j−1M, Vi;j−1, ϕi, xi] is the exact transition density derived from the SDE in equation (3) and [U0, V0] and [ϕi] are non-informative prior densities. Unfortunately, except for a very few specific forms for the drift and diffusion terms in equation (3), [Uij, Vij | Ui;j−1, Vi;j−1, ϕi, xi] is usually analytically intractable. Even when the exact transition density does have a closed form, as is the case for the OU and IOU processes, for which the exact transition density is given in Theorem 1, the posterior density for ϕ still does not have an explicit form. Hence, we will use the Euler approximation to approximate the exact transition density, while applying the method of data augmentation (Tanner and Wong, 1987) to minimize the error in this approximation.
The strategy of combining data augmentation and Euler approximation to approximate the exact transition density has been discussed by Elerian et al. (2001), Eraker (2001), Roberts and Stramer (2001) and Durham and Gallant (2002), in the context of estimating parameters in the SDE for a single diffusion process observed at discrete times without measurement errors. Our approach is related to theirs, but with an important distinction that instead of being partially observed, both processes Vxi(t) and Uxi(t) are completely unobserved, and will be sampled as part of an MCMC algorithm. In this manner, we will estimate the processes Vxi(t), Uxi(t) and the parameters ϕ. It is worth pointing out that although augmentation only needs to take place for the latent process, augmenting the data themselves will facilitate the operation of the simulation smoother, as this algorithm requires observations (either observed or augmented) available at each corresponding time. In addition, the data augmentation allows us to create augmented longitudinal data with a common set of time points, and consequently this method enables us to handle longitudinal data with irregularly spaced times which may vary across the subjects.
To carry out the data augmentation and the Euler approximation, we first specify time points at which data would be augmented. Let denotes the set of augmentation times for the ith subject. Consequently, the time interval τij between adjacent data points, either observed or augmented, is less than τc. In addition, let denote the set of all possible time points of the observed and augmented data across subjects. With further data augmentation at times , each subject would have either observed or augmented data at the common time set . The Euler approximation to equations (2) and (3) for leads to the following difference equations:
| (7) |
| (8) |
where and j = 1, 2, …, m. Thus, the conditional posterior density for ϕ is approximated by,
| (9) |
where with and similarly for and . Note that the approximate transition density ≺ Uij, Vij | Ui,j−1, Vi,j−1, ϕi, xi ≻ in equation (9) is given by,
| (10) |
which is derived from equations (7) and (8). This implies that it is feasible to directly sample from the posterior distribution of ϕ, if the conjugate priors for ϕ are chosen.
With regard to the posterior samples of Uxi(t) and Vxi(t) for , we follow equations (7) and (8) to come up with their approximations, denoted by (t) and (t), with linear interpolation for t between tj−1 and tj for j = 1, 2, …, m. Bouleau and Lepingle (1992) showed that under some regularity conditions, with constant Ci, the Lp-norm of the approximation error for Vxi(t) is bounded at the rate of ; that is,
where ∥f∥p = {∫Ω |f(z)|pdμ(z)}1/p for a real function f on the space (Ω, ) with measure μ on random variable z. This indicates that if m is sufficiently large, then (t) will approach to its continuous counterpart Vxi(t) with arbitrary precision. Similar arguments hold for (t). Note that we will sample m instead of mi data points for (t) and (t) with possibly m ≫ mi. Hence, the benefit of introducing augmented data is two fold: (i) it reduces the error of approximation, when (t) or (t), instead of (t) or (t), is used to replace Uxi(t) or Vxi(t); (ii) it gives a more accurate approximation to the exact transition density, as shown by Pedersen (1995), which benefits estimation of model parameters ϕ. Under the assumption that m is large enough such that the approximation error is small, for the ease of exposition, we still use Vxi(t) instead of (t) throughout the rest of the paper. Uxi(t) is treated similarly.
In the MCMC algorithm to update the values of Uxi(t) and Vxi(t) for , we draw samples from
| (11) |
where , ≺ Uij, Vij | Ui,j−1, ϕi, xi ≻ is given in equation (10) and [U0, V0] is a non-informative prior. Equivalently, the posterior density (11) may be derived from a state space model representation (Durbin and Koopman, 2001), which is a useful reformulation of the SSVM in equations (1), (2) and (3) when it is discretized using the Euler approximation and data augmentation.
Consider an example where Vxi(t) follows the OU process and ≺ Uij, Vij | Ui,j−1, ϕi, xi ≻ is given in Corollary 2. Let denote the observed or augmented data for n subjects at time tj, and let be the latent states with . The corresponding SSVM can be expressed as a state space model, given as follows:
where Fj = InI ⊗ Fij, Gj = In ⊗ Gij, Fij = [1, 0, 0]′ with ⊗ denoting Kronecker product,
Likewise, when Vxi(t) follows a Wiener process, the corresponding reformulation as a state space model can be obtained in a similar manner.
In this paper we have adopted the MCMC method for Bayesian inference. In the literature other likelihood-based or sampling-based methods have also been developed for nonlinear and/or non-Gaussian state space models, including Kitagawa's (1987) numeric algorithm using piecewise linear approximation, Durbin and Koopman's (1997) simulated maximum likelihood estimation, Jørgensen et al.'s (1999) Kalman estimating equations and some recent work on sequential Monte Carlo methods using particulate filtering (Gordon et al., 1993; Pitt and Shephard, 1999; Liu, 2008; Andrieu et al., 2010), among others.
3.2 MCMC Algorithm
Under the state space model formulation, Gibbs sampler was first developed to sample one latent state θj at a time, this was later improved by various algorithms that use simultaneous block-based sampling schemes (e.g. Frühwirth-Schnatter 1994; Carter and Kohn 1994). The simulation smoother proposed first by de Jong and Shephard (1995) and later improved by Durbin and Koopman (1997) provides a remarkably efficient sampling tool. It draws samples of θj through sampling independent innovations ξj, rather than realizations of a Markov process, so the entire sampling is based on very low dimensional distributions and free of autocorrelation. Thus, the rate of mixing and moreover burn-in can be achieved quickly. We will use the simulation smoother in our implementation.
The proposed MCMC algorithm iterates through the following steps.
Draw augmented data according to at times for the ith subject, i = 1, 2, …, n.
Update latent states Uxi(t) and Vxi(t) for from the posterior density (11) by using the simulation smoother.
- Update ϕ by sampling from the posterior density (9). In particular, when Vxi(t) follows an OU process and is discretized through the Euler approximation, the collection of equations (8) can be equivalently reformulated as a linear mixed model,
where , , with Vj = [V1j, V2j, …, Vnj and . Further, β* = [ρβ′, −ρ]′, b* = ρν, ν = [ν1, ν2, …, νn]′, , . As a result, the set of model parameters is , can be sampled straightforwardly by using the standard Gibbs sampler in the linear mixed model (Ruppert et al., 2003, Chap. 16) with non-informative conjugate priors, , , and . Here denotes the inverse gamma distribution with shape parameter a and scale parameter b. - Update by sampling from the following posterior distribution
where the prior distribution for is .
3.3 Bayesian Posterior Forecasting
The proposed model is useful to forecast processes of interest, including Uxi(t), Vxi(t) and Yi(t), for . With the availability of posterior samples for Uxi(t), Vxi(t), ϕi and σε with i = 1, 2, …, n and , it is straightforward to derive Bayesian posterior forecasting. Note that the posterior forecasting distributions are,
and
Thus, we draw , and from and for r = 1,2, …, where , , and are the rth posterior samples from the MCMC algorithm. If [Uxi(t), Vxi(t) | Uxi(tm), Vxi(tm), ϕi, x] does not have a closed form, the approximate transition density ≺ Uxi(t), Vxi(t) | Uxi(tm), Vxi(tm), ϕi, x ≻ could be used instead along with data augmentation.
4 Application to the PSA Data
We apply the proposed SSVM-OU to analyze the PSA data discussed in Section 1. The prior of the rate function Vxi(t) is assumed to be the OU process with and b{Vxi(t); xi, ϕi} = σξ in equation (3). A total of 739 observations are obtained for 50 subjects. The number of observations for each subject varies from 13 to 24. The initial observation for all subjects is at one month (0.083 years) after EBRT treatment, and the time for the last observation ranges from 3.833 to 8.083 years, with the average of 6.050 years. To reduce the approximation error discussed in Section 3.1, we further augment the data to let the time interval between adjacent data points, either observed or augmented, be less than 0.0208 years. The appropriateness of this choice of time interval is confirmed using the simulation studies in Section 5. We investigate the association of the pretreatment covariates (i.e. baseline PSA, Gleason score and T-stage) with the stable PSA rate via the model , where is a random effect; XPi denotes the log-transformed baseline PSA for the ith subject, centered around the mean of 2.3; XGi is equal to 1 if Gleason score is above or equal to level 7, and is 0 otherwise; XTi takes the value of 1 if T-stage is at level 2 or higher, and is 0 otherwise. We leave out the last observation for each subject as well as the observations after year 5 as validation data to assess the forecasting ability of the model.
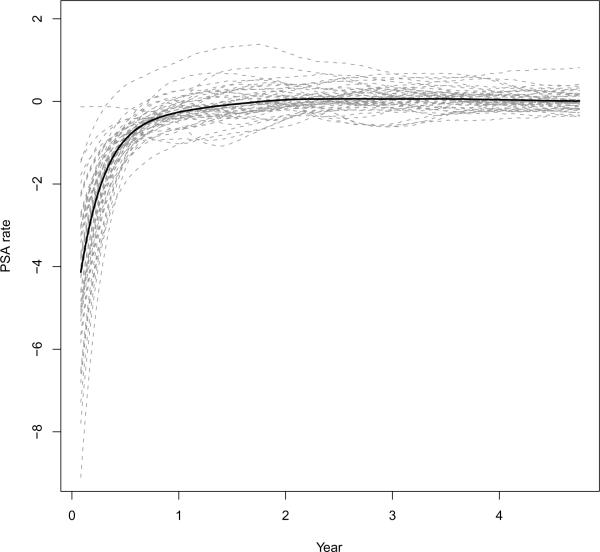
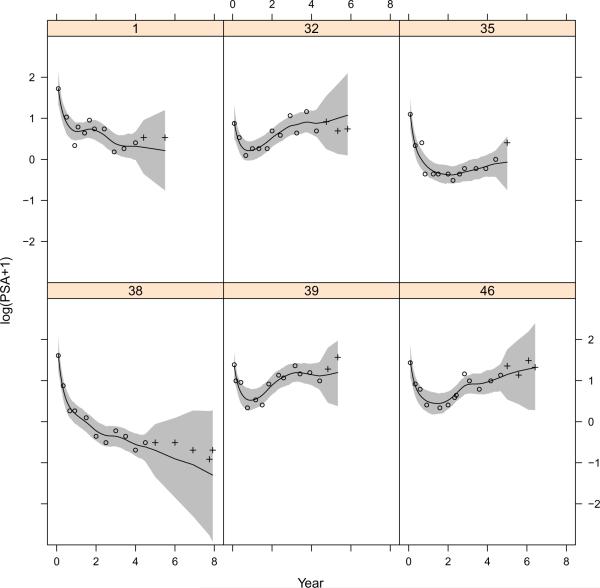
The posterior draws are obtained from the proposed MCMC algorithm with 20,000 iterations, discarding the first 10,000 as the burn-in stage and subsequently saving every 10th draws. The trace plots suggest the algorithm converges fast and mixes well. Table 1 presents the posterior summary statistics for the parameters. Baseline PSA and T-stage are found to have significant effect on the PSA stable rate. This result suggests that Baseline PSA and T-stage are predictive of the long term rate of change for PSA, which is in agreement with the finding by Lieberfarb et al. (2002). Figure 2 displays E[Vxi(t) | Y], the posterior means of the rate function for each subject (shown as dashed lines), and E[V (t) | Y] = E[E[Vxi(t) | Y]], the posterior mean of the rate function in the population (shown as a solid line). It is clear that although the rate function for the population is smooth and may be specified by a parametric form, the individual rate functions are much more wiggly, vary significantly across subjects and would be difficult to model parametrically. Figure 3 shows the posterior means and credible intervals of Uxi(t) for six randomly selected subjects, including the forecasted Uxi(t) after year 5. Note that the width of the forecasted credible intervals is comparable to the theoretical results given in Corollary 1.
Table 1.
PSA data: Posterior means and quantiles of parameters for the SSVM-OU and LMM.
| Model | Parameter | Mean | SD | 2.5% | 50% | 97.5% |
|---|---|---|---|---|---|---|
| SSVM-OU | 0.044 | 0.004 | 0.037 | 0.044 | 0.053 | |
| 1.365 | 0.297 | 0.921 | 1.320 | 2.108 | ||
| ρ | 3.721 | 0.360 | 3.101 | 3.690 | 4.464 | |
| 0.054 | 0.015 | 0.031 | 0.051 | 0.089 | ||
| β 0 | −0.171 | 0.085 | −0.335 | −0.169 | −0.004 | |
| β 1 | 0.139 | 0.072 | 0.001 | 0.139 | 0.277 | |
| β 2 | 0.242 | 0.095 | 0.060 | 0.237 | 0.438 | |
| β 3 | 0.061 | 0.103 | −0.157 | 0.064 | 0.269 | |
|
|
||||||
| LMM | β 20 | 0.061 | 0.066 | −0.072 | 0.058 | 0.200 |
| β 21 | 0.116 | 0.056 | 0.008 | 0.117 | 0.225 | |
| β 22 | 0.260 | 0.076 | 0.116 | 0.260 | 0.411 | |
| β 23 | 0.046 | 0.078 | −0.105 | 0.046 | 0.193 | |
Figure 2.
Posterior means of Vxi(t) for each subject as gray dashed lines and the population-level rate function V (t) as black solid line
Figure 3.
Plots of training data points (ο), validation data points (+), posterior means (−) and 95% credible intervals (gray shades) of Uxi(t) for six randomly selected subjects.
For comparison, we also analyze the PSA data using smoothing splines and a parametric linear mixed-effects model(LMM). The model fits are evaluated by the Deviance Information Criterion (DIC, Speigelhalter et al., 2003). Note that , where the posterior mean deviance measures the goodness of fit and the “effective number of parameters” pD measures the model complexity. According to Speigelhalter et al. (2003), DIC may be regarded asymptotically as a generalization of the Akaike information criterion (AIC). Similar to AIC, a smaller value of DIC indicates a better trade-off between the fit to the data and the complexity of model. We further compare the forecasting ability of these three models on the validation data points. For the smoothing spline approach, we obtain the estimates of Vxi(t) from the SSVM-W with a Wiener process as the prior for Vxi(t), where a{Vxi(t); xi,ϕi} = 0 and b{Vxi(t); xi,ϕi} = σξ in equation (3). As mentioned in Section 2.1, the estimation of Vxi(t) from this model, is equivalent to estimation by a smoothing spline with a common smoothing parameter . The exact transition density in this SSVM-W, is given by Wecker and Ansley (1983) as
with
and is used in the proposed MCMC algorithm. The forecasting of future observations is outlined in Section 3.3 for the SSVM-OU and SSVM-Ws. The parametric linear mixed model is similar to the one given by Proust-Lima et al. (2008),
| (12) |
where the mean function Uxi(t) consists of three parts: (i) post-therapy level , (ii) short-term evolution , and (iii) long-term evolution . In addition, f1(t) = (1 + t)−1.5 − 1 and f2(t) = t; the fixed effects a non-informative prior with large value of ; the random effects where Σν,lmm is a diagonal matrix with its main diagonal entries ; measurement error . We further assume noninformative prior distributions with small values of a and b for , , , , and respectively. The MCMC algorithm for the linear mixed model (Ruppert et al., 2003, Chap. 16) is applied to draw the posterior samples with the same burn-in stage and thinning scheme as for the MCMC algorithm for the SSVM-OU. Table 1 presents the posterior summary of the parameters β20, β21, β22, and β23, which are involved in the long-term evolution in equation (12). Note that these parameters in the LMM are designed to measure the association between the long term stable level and the covariates of the interest, similar to the parameter β0, β1,β2, and β3 in the SSVM-OU. Given the rth samples , and , the forecasts of PSA at time t for the ith subject can be drawn from , where .
The values of DIC for SSVM-OU and SSVM-W are 71.809 and 119.400 respectively, both of which are significantly lower than that of LMM (151.048). Thus, SSVM-OU fits the data best among these three models. This implies that the parametric LMM is less able to capture longitudinal dynamics of subject's trajectories than the other two SSVMs. Next, to compare the prediction capability among these three models, we predict the 164 validation data points and evaluate their posterior predictive ability. Table 2 presents relative bias and mean squared error (MSE) of the point forecast based on the posterior mean, as well as corresponding coverage rate and averaged length of credible interval. For the 69 validation data points within 1 year distance from the last training data points, the SSVM-OU performs best, with the smallest MSE. For the remaining validation data points at later times, the SSVM-W outperforms the other two in terms of relative bias and MSE. However, for the coverage rate, the SSVM-OU intervals are closest to the nominal 95% level, whereas those from the SSVM-W are too wide to be clinically useful. This may be due to the nonstationary variance of the latent process of SSVM-W.
Table 2.
PSA data: Posterior forecasting of the validation data points. The relative bias is defined as for the posterior mean of validation data point Y.
| Method | Type | Relative Bias | MSE | Coverage Rate | Interval Length |
|---|---|---|---|---|---|
| SSVM-OU | ≤ 1 year | −0.143 | 0.076 | 1 | 1.403 |
| > 1 year | −0.913 | 0.581 | 0.966 | 2.356 | |
| All | −0.644 | 0.404 | 0.912 | 2.023 | |
| SSVM-W | ≤ 1 year | −0.047 | 0.098 | 1 | 2.379 |
| > 1 year | 0.031 | 0.403 | 1 | 8.329 | |
| All | 0.040 | 0.296 | 1 | 6.250 | |
| LMM | ≤ 1 year | 0.205 | 0.108 | 0.899 | 1.226 |
| > 1 year | 0.387 | 0.568 | 0.672 | 1.610 | |
| All | 0.323 | 0.407 | 0.748 | 1.476 |
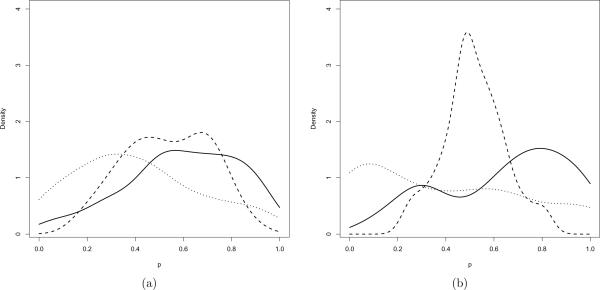
Besides evaluation of the point forecasts and the corresponding credible intervals, we further use the probability integral transform (PIT, Dawid, 1984; Gneiting et al., 2007) value to assess the predictive performance of the probabilistic forecasts. This forecast can be expressed as the posterior predictive cumulative distribution functions (CDFs) Fij(Y), where Y is the forecasted validation data point at time tij for the ith subject and is assumed to be generated from the true unknown CDF Gij(Y). For the observed validation data point Yij, the PIT value pij = Fij(Yij) should have a uniform distribution, if Fij(Y) = Gij(Y) for every i and j. We estimate Fij(t) by the empirical CDF , which is based on the Bayesian posterior forecasting draws for the three models. The corresponding smoothed density plots of are displayed in Figure 4. The density of for the SSVM-OU is left skewed, indicating the forecasts are slightly under predicted, while the density for the linear mixed model is right skewed and the forecasts are slightly over predicted. The density for the SSVM-W is hump-shaped, implying the posterior predictive distribution is over dispersed and the credible intervals are too wide on average. While none of the models gives the ideal PIT plots, the plots of SSVM-OU and the LMM are reasonably close to a uniform density..
Figure 4.
PIT density plots for (a) tij ≤ 1 year, (b) tij > 1 year of SSVM-OU (−), SSVM-W (- - -), LMM (⋯)
5 A Simulation Study
We carry out a simulation study to (i) assess the performance of the proposed MCMC algorithm in estimating the model parameters and stable rates and (ii) compare the performance of the proposed SSVM-OU with the other two methods for forecasting future observations. We generate 100 replicated datasets from the SSVM-OU with the model parameter set close to those estimated from the analysis of the PSA data. Each dataset includes 20 subjects each with 13 observations and three validation data points per subject. The observations are equidistantly spaced with time interval 0.416, equal to the median of time intervals in the PSA data. The three validation data points are at 0.08, 0.5 and 1 years after the last observation, respectively. To investigate the influence of data augmentation on the estimation of the model parameters, we analyze the same dataset using the proposed MCMC algorithm without data augmentation, and with 9 and 19 augmented data points between the consecutive observed data points. The corresponding time interval between the adjacent data points, either observed or augmented, decreases from 0.416 in the original datasets to 0.0416 and 0.0208 for the MCMC algorithm with 9 and 19 augmented data points between neighboring observations.
Table 3 presents simulation results for the estimation of model parameters, assessed by relative bias, MSE of posterior means, coverage rate and average length of credible interval. All results indicate clearly that the data augmentation is critical to obtain proper estimates of the second moment parameters, , , and ρ. Their relative biases and MSEs decrease significantly even by adding 9 data points between adjacent observations. For example, the relative bias of ρ reduces from 0.47 to 0.052 and the MSE drops from 2.704 to 0.0360. Augmentation with 19 data points can further improve the relative bias in the estimation of parameters and ρ, and no additional improvement results from more aggressive augmentation (the results not shown here). The data augmentation, however, has little effect on the relative bias for the estimation of parameters of interest, β1, β2 and β3, implying that the consistent estimation for these parameters may be obtained using observed data. Yet, the data augmentation has noticeable effects on the coverage rates, because it affects the variance of posterior distributions.
Table 3.
Simulation results on the estimation of SSVM-OU parameters. The relative bias is defined as for the posterior mean of the parameter ϕ.
| Data Augmented | Parameter | Truth | Relative Bias | MSE | Coverage Rate | Interval Length |
|---|---|---|---|---|---|---|
| 0 | 0.05 | −0.202 | 1.294e−04 | 0.678 | 0.026 | |
| 1.00 | −0.408 | 1.689e−01 | 0.044 | 0.461 | ||
| ρ | 3.50 | −0.470 | 2.704e+00 | 0.000 | 0.101 | |
| 0.05 | 2.101 | 1.123e−02 | 0.211 | 0.299 | ||
| β 0 | −0.15 | 0.03 | 1.992e−02 | 1.000 | 1.150 | |
| β 1 | 0.15 | 0.139 | 3.140e−02 | 1.000 | 1.307 | |
| β 2 | 0.25 | −0.006 | 1.693e−02 | 1.000 | 1.120 | |
| β 3 | 0.10 | −0.105 | 1.995e−02 | 1.000 | 1.166 | |
|
|
||||||
| 9 | 0.05 | 0.012 | 2.952e−05 | 0.967 | 0.024 | |
| 1.00 | −0.041 | 2.253e−02 | 0.967 | 0.800 | ||
| ρ | 3.50 | −0.052 | 3.596e−02 | 0.256 | 0.273 | |
| 0.05 | 0.548 | 9.410e−04 | 0.989 | 0.133 | ||
| β 0 | −0.15 | 0.035 | 2.033e−02 | 0.978 | 0.647 | |
| β 1 | 0.15 | 0.135 | 3.133e−02 | 0.956 | 0.739 | |
| β 2 | 0.25 | 0.005 | 1.673e−02 | 0.989 | 0.634 | |
| β 3 | 0.10 | −0.099 | 2.006e−02 | 0.956 | 0.660 | |
|
|
||||||
| 19 | 0.05 | 0.013 | 3.921e−05 | 0.956 | 0.024 | |
| 1.00 | 0.007 | 2.497e−02 | 0.967 | 0.802 | ||
| ρ | 3.50 | −0.018 | 8.104e−03 | 0.911 | 0.288 | |
| 0.05 | 0.494 | 7.960e−04 | 0.989 | 0.126 | ||
| β 0 | −0.15 | 0.022 | 2.013e−02 | 0.967 | 0.625 | |
| β 1 | 0.15 | 0.138 | 3.135e−02 | 0.944 | 0.713 | |
| β 2 | 0.25 | 0.003 | 1.686e−02 | 0.978 | 0.612 | |
| β 3 | 0.10 | −0.113 | 1.980e−02 | 0.978 | 0.635 | |
For the data simulated from the SSVM-OU, we further forecast the validation data points by the SSVM-OU, SSVM-W and LMM (12). Table 4 compares the forecasting ability of the posterior mean and credible intervals for the three models, evaluated by the relative bias, MSE, coverage rate and interval length. As we expected, the relative biases of posterior means of the forecasting draws from the SSVM-OU are smaller than those from the other models and the corresponding interval lengths are narrower. Furthermore, it is of interest to study the sensitivity of the forecasting ability of SSVM-OU. We simulate another 100 datasets from the LMM specified as equation (12) in which the parameters are the same as those obtained from the PSA data analysis. In addition, the number of subjects, and the number of observations and the validation data points, are set identical to those used to generate datasets from the above SSVM-OU. The forecasting results are given in the second part of Table 4. We find that SSVM-OU has comparable performance to the LMM (for the short-term forecast at time 0.08), with smaller relative bias but slightly larger MSE and wider interval length. For the long-term forecast at time 0.5 or 1, SSVM-OU performs worse than the LMM but is better than SSVM-W.
Table 4.
Simulation results on forecasting by three models
| Simulation Model | Fitted Model | Year Distance | Relative Bias | MSE | Coverage Rate | Interval Length |
|---|---|---|---|---|---|---|
| SSVM-OU | SSVM-OU | 0.08 | 0.010 | 0.162 | 0.949 | 1.116 |
| 0.5 | 0.019 | 0.232 | 0.951 | 1.336 | ||
| 1 | 0.011 | 0.343 | 0.951 | 1.639 | ||
| SSVM-W | 0.08 | 0.008 | 0.270 | 0.957 | 1.455 | |
| 0.5 | −0.144 | 6.700 | 1 | 9.640 | ||
| 1 | −0.024 | 39.430 | 1 | 23.974 | ||
| LMM | 0.08 | −0.165 | 1.508 | 0.994 | 3.465 | |
| 0.5 | −0.061 | 2.206 | 0.936 | 3.547 | ||
| 1 | −0.303 | 3.442 | 0.742 | 3.661 | ||
|
| ||||||
| LMM | SSVM-OU | 0.08 | 0.001 | 0.034 | 0.915 | 0.483 |
| 0.5 | 0.077 | 0.069 | 0.974 | 0.781 | ||
| 1 | 0.092 | 0.155 | 0.996 | 1.249 | ||
| SSVM-W | 0.08 | −0.021 | 0.046 | 0.920 | 0.565 | |
| 0.5 | 0.075 | 0.384 | 1.000 | 2.152 | ||
| 1 | −0.040 | 1.905 | 1.000 | 5.085 | ||
| LMM | 0.08 | −0.007 | 0.028 | 0.943 | 0.456 | |
| 0.5 | 0.026 | 0.030 | 0.943 | 0.478 | ||
| 1 | −0.006 | 0.034 | 0.952 | 0.510 | ||
6 Discussion
This paper considers modeling and inference for the rate functions in longitudinal studies with an application in the analysis of PSA biomarker profiles. For a given subject, the rate of change is described by a rate function whose prior is assumed to follow a Gaussian process conditional on the covariates. A key feature of this approach is that the Gaussian process is specified by an SDE and is expected to be centered on a pre-specified parametric function, while allowing significant deviations from this functional expectation nonparametrically. We have focused on the case where the rate function follows an OU process, motivated by analyzing PSA profiles. The same modeling strategy and inference method should be widely useful in the setting when we aim to model the rate function semiparametrically.
One can extend our model to discrete outcomes and to include the covariates in equation (1). Moreover, a similar modeling and inference approach can be applied to analyze the acceleration function, which is the second-order derivative of the mean function. In addition, for simplicity, we assume the stable rates depend on the covariates through a parametric distribution, which could potentially be replaced by a nonparametric distribution with a stick-breaking process as its prior.
The MCMC algorithm is currently programmed in R (R Development Core Team, 2008). For the PSA application with 50 subjects and 225 observed or augmented data points per subject, it took about 4 hours per 1000 MCMC iterations on a PC with 2.93GHz Intel(R)Core(TM)2Duo CPU. In contrast, it took about 15 minutes per 1000 MCMC iterations if the model was fit with only observed data. One way to speed up computation is to develop a C or C++ program for the proposed method, which is one of our future research tasks. Our computation-related experiences have suggested that the computation time is approximately linearly proportional to the number of subjects. Hence, we anticipate that with fast computation software this algorithm can be applied to handle studies with relatively large sample sizes.
Supplementary Material
Acknowledgments
This research of the second author was partially supported by National Cancer Institute grants CA110518 and CA69568, and the research of the third author was partially supported by National Science Foundation (DMS0904177). The authors are thankful to the first author's dissertation committee members Dr. Naisysin Wang and Dr. Brisa Sanchez for the helpful discussions, and to the editor, associate editor and two anonymous reviewers for valuable comments and suggestions.
Footnotes
Supplementary Materials Supplementary Materials are available at the JASA website http://pubs.amstat.org/loi/jasa.
References
- Aalen OO, Gjessing HK. Survival models based on the Ornstein-Uhlenbeck process. Lifetime Data Analysis. 2004;10:407–423. doi: 10.1007/s10985-004-4775-9. [DOI] [PubMed] [Google Scholar]
- Andrieu C, Doucet A, Holenstein R. Particle Markov chain Monte Carlo methods. Journal of the Royal Statistical Society: Series B (Statistical Methodology. 2010;72:269–342. [Google Scholar]
- Ansley CF, Kohn R. On the Equivalence of Two Stochastic Approaches to Spline Smoothing. Journal of Applied Probability. 1986;23:391–405. [Google Scholar]
- Bouleau N, Lepingle D. Numerical Methods for Stochastic Process. Wiley; New York: 1992. [Google Scholar]
- Carter CK, Kohn R. On Gibbs sampling for state space models. Biometrika. 1994;81:541–553. [Google Scholar]
- Dawid AP. Present position and potential developments: Some personal views: Statistical theory: The prequential approach. Journal of the Royal Statistical Society: Series A (Statistics in Society) 1984;147:278–292. [Google Scholar]
- de Jong P, Shephard N. The simulation smoother for time series models. Biometrika. 1995;82:339–350. [Google Scholar]
- Diggle PJ, Heagerty P, Liang KY, Zeger S. Analysis of longitudinal data. Oxford University Press; Oxford: 2002. [Google Scholar]
- Durbin J, Koopman SJ. Monte Carlo maximum likelihood estimation for non-Gaussian state space models. Biometrika. 1997;84:669–684. [Google Scholar]
- Durbin J, Koopman SJ. Time Series Analysis by State Space Methods. Oxford University Press; Oxford: 2001. [Google Scholar]
- Durham GB, Gallant AR. Numerical techniques for maximum likelihood estimation of continuous-time diffusion processes. Journal of Business & Economic Statistics. 2002;20:297–338. [Google Scholar]
- Elerian O, Chib S, Shephard N. Likelihood inference for discretely observed nonlinear diffusions. Econometrica. 2001;69:959–993. [Google Scholar]
- Eraker B. MCMC Analysis of Diffusion Models With Application to Finance. Journal of Business & Economic Statistics. 2001;19:177–191. [Google Scholar]
- Feller W. An Introduction to Probability Theory and Its Application. Springer Verlag; New York: 1970. [Google Scholar]
- Frühwirth-Schnatter S. Data augmentation and dynamic linear models. Journal of Time Series Analysis. 1994;15:183–202. [Google Scholar]
- Gneiting T, Balabdaoui F, Raftery AE. Probabilistic forecasts, calibration and sharpness. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2007;69:243–268. [Google Scholar]
- Gordon NJ, Salmond DJ, Smith AFM. Novel approach to nonlinear/non-Gaussian Bayesian state estimation. Radar and Signal Processing, IEE Proceedings F. 1993;140:107–113. [Google Scholar]
- Grimmett G, Stirzaker D. Probability and Random Processes. Oxford University Press; Oxford: 2001. [Google Scholar]
- Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R. Varying-coefficient models. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1993;55:757–796. [Google Scholar]
- Hoover DR, Rice JA, Wu CO, Yang LP. Nonparametric smoothing estimates of time-varying coefficient models with longitudinal data. Biometrika. 1998;85:809–822. [Google Scholar]
- Jørgensen B, Lundbye-Christensen S, Song PX-K, Sun L. A state space model for multivariate longitudinal count data. Biometrika. 1999;86:169–181. [Google Scholar]
- Kariyanna SS, Light RP, Agarwal R. A longitudinal study of kidney structure and function in adults. Nephrology Dialysis Transplantation. 2010;25:1120–1226. doi: 10.1093/ndt/gfp654. [DOI] [PubMed] [Google Scholar]
- Kitagawa G. Non-Gaussian state-space modeling of nonstationary time series. Journal of the American Statistical Association. 1987;82:1032–1041. [Google Scholar]
- Kulkarni V, Rolski T. Fluid model driven by an Ornstein-Uhlenbeck process. Probability in the Engineering and Informational Sciences. 2009;8:403–417. [Google Scholar]
- Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Lieberfarb ME, Schultz D, Whittington R, Malkowicz B, Tomaszewski JE, Weinstein M, Wein A, Richie JP, D'Amico AV. Using PSA, biopsy Gleason score, clinical stage, and the percentage of positive biopsies to identify optimal candidates for prostate-only radiation therapy. International Journal of Radiation Oncology Biology Physics. 2002;53:898–903. doi: 10.1016/s0360-3016(02)02812-2. [DOI] [PubMed] [Google Scholar]
- Liu JS. Monte Carlo strategies in scientific computing. Springer Verlag; New York: 2008. [Google Scholar]
- Lloyd-Jones DM, Liu K, Colangelo LA, Yan LL, Klein L, Loria CM, Lewis CE, Savage P. Consistently stable or decreased body mass index in young adulthood and longitudinal changes in metabolic syndrome components: the Coronary Artery Risk Development in Young Adults Study. Circulation. 2007;115:1004–1011. doi: 10.1161/CIRCULATIONAHA.106.648642. [DOI] [PubMed] [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-based functional mixed models. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HG, Yao F. Empirical dynamics for longitudinal data. The Annals of Statistics. 2010;38:3458–3486. [Google Scholar]
- Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, Mack WJ, Kramer JH, Weiner MW, Schuff N, et al. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65:565–571. doi: 10.1212/01.wnl.0000172913.88973.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolato E, Venardos E. Option pricing in stochastic volatility models of the Ornstein-Uhlenbeck type. Mathematical Finance. 2003;13:445–466. [Google Scholar]
- Paul D, Peng J, Burman P. Semiparametric modeling of autonomous nonlinear dynamical systems with applications. 2009. submitted. [Google Scholar]
- Pedersen AR. A new approach to maximum likelihood estimation for stochastic differential equations based on discrete observations. Scandinavian Journal of Statistics. 1995;22:55–71. [Google Scholar]
- Pitt MK, Shephard N. Filtering Via Simulation: Auxiliary Particle Filters. Journal of the American Statistical Association. 1999;94:590–591. [Google Scholar]
- Proust-Lima C, Taylor JMG, Williams S, Ankerst D, Liu N, Kestin L, Bae K, Sandler H. Determinants of change in prostate-specific antigen over time and its association with recurrence after external beam radiation therapy for prostate cancer in five large cohorts. International Journal of Radiation Oncology Biology Physics. 2008;72:782–791. doi: 10.1016/j.ijrobp.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Guo W. Functional mixed-effects model for periodic data. Biostatistics. 2006;7:225–234. doi: 10.1093/biostatistics/kxj003. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. ISBN 3-900051-07-0. [Google Scholar]
- Rice JA, Silverman BW. Estimating the mean and covariance structure nonparametrically when the data are curves. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1991;53:233–243. [Google Scholar]
- Roberts GO, Stramer O. On inference for partially observed nonlinear diffusion models using the Metropolis-Hastings algorithm. Biometrika. 2001;88:603–621. [Google Scholar]
- Ruppert D, Wand MP, Carroll RJ. Semiparametric Regression. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- Sartor CI, Strawderman MH, Lin XH, Kish KE, McLaughlin PW, Sandler HM. Rate of PSA rise predicts metastatic versus local recurrence after definitive radiotherapy. International Journal of Radiation Oncology Biology Physics. 1997;38:941–947. doi: 10.1016/s0360-3016(97)00082-5. [DOI] [PubMed] [Google Scholar]
- Speigelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) Journal of the Royal Statistical Society B: (Statistical Methodology) 2003;64:583–616. [Google Scholar]
- Strasak AM, Kelleher CC, Klenk J, Brant LJ, Ruttmann E, Rapp K, Concin H, Diem G, Pfeiffer KP, Ulmer H. Longitudinal change in serum gamma-glutamyltransferase and cardiovascular disease mortality: a prospective population-based study in 76 113 Austrian adults. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28:1857–1865. doi: 10.1161/ATVBAHA.108.170597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner MA, Wong WH. The calculation of posterior distributions by data augmentation. Journal of the American Statistical Association. 1987;82:528–540. [Google Scholar]
- Taylor JMG, Law N. Does the covariance structure matter in longitudinal modelling for the prediction of future CD4 counts? Statistics in Medicine. 1998;17:2381–2394. doi: 10.1002/(sici)1097-0258(19981030)17:20<2381::aid-sim926>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Trost DC, Overman EA, II, Ostroff JH, Xiong W, March P. A model for liver homeostasis using modified mean-reverting Ornstein-Uhlenbeck process. Computational and Mathematical Methods in Medicine. 2010;11:27–47. [Google Scholar]
- Uhlenbeck GE, Ornstein LS. On the Theory of the Brownian Motion. Physical Review. 1930;36:823–841. [Google Scholar]
- Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. Springer Verlag; New York: 2009. [Google Scholar]
- Verbyla AP, Cullis BR, Kenward MG, Welham SJ. The analysis of designed experiments and longitudinal data by using smoothing splines. Journal of the Royal Statistical Society: Series C (Applied Statistics) 1999;48:269–311. [Google Scholar]
- Wahba G. Improper Priors, Spline Smoothing and the Problem of Guarding Against Model Errors in Regression. Journal of the Royal Statistical Society B: (Statistical Methodology) 1978;40:364–372. [Google Scholar]
- Wang S, Jank W, Shmueli G, Smith P. Modeling price dynamics in eBay auctions using differential equations. Journal of the American Statistical Association. 2008;103:1100–1118. [Google Scholar]
- Wang Y, Taylor JMG. Inference for smooth curves in longitudinal data with application to an AIDS clinical trial. Statistics in Medicine. 1995;14:1205–1205. doi: 10.1002/sim.4780141106. [DOI] [PubMed] [Google Scholar]
- Wecker WE, Ansley CF. The Signal Extraction Approach to Nonlinear Regression and Spline Smoothing. Journal of the American Statistical Association. 1983;78:81–89. [Google Scholar]
- Welham SJ, Cullis BR, Kenward MG, Thompson R. The analysis of longitudinal data using mixed model L-splines. Biometrics. 2006;62:392–401. doi: 10.1111/j.1541-0420.2005.00500.x. [DOI] [PubMed] [Google Scholar]
- Zeger SL, Diggle PJ. Semiparametric models for longitudinal data with application to CD4 cell numbers in HIV seroconverters. Biometrics. 1994;50:689–699. [PubMed] [Google Scholar]
- Zhang D, Lin XH, Raz J, Sowers M. Semiparametric Stochastic Mixed Models for Longitudinal Data. Journal of the American Statistical Association. 1998;93:710–719. [Google Scholar]
- Zhang P, Song PX-K, Qu A, Greene T. Effcient estimation for patient-specific rates of disease progression using nonnormal linear mixed models. Biometrics. 2008;64:29–38. doi: 10.1111/j.1541-0420.2007.00824.x. [DOI] [PubMed] [Google Scholar]
- Zhu B, Song PX-K, Taylor JMG. Biometrics. 2011. Stochastic Functional Data Analysis: A Diffusion Model-Based Approach. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.