Abstract
Targeted drug delivery offers an opportunity for the development of safer and more effective therapies for the treatment of cancer. In this study, we sought to identify short, cell-internalizing peptide ligands that could serve as directive agents for specific drug delivery in hematologic malignancies. By screening of human leukemia cells with a combinatorial phage display peptide library, we isolated a peptide motif, sequence Phe-Phe/Tyr-Any-Leu-Arg-Ser (FF/YXLRS), which bound to different leukemia cell lines and to patient-derived bone marrow samples. The motif was internalized through a receptor-mediated pathway, and we next identified the corresponding receptor as the transmembrane glycoprotein neuropilin-1 (NRP-1). Moreover, we observed a potent anti-leukemia cell effect when the targeting motif was synthesized in tandem to the pro-apoptotic sequence D(KLAKLAK)2. Finally, our results confirmed increased expression of NRP-1 in representative human leukemia and lymphoma cell lines and in a panel of bone marrow specimens obtained from patients with acute lymphoblastic leukemia or acute myelogenous leukemia compared with normal bone marrow. These results indicate that NRP-1 could potentially be used as a target for ligand-directed therapy in human leukemias and lymphomas and that the prototype CGFYWLRSC-GG-D(KLAKLAK)2 is a promising drug candidate in this setting.
Introduction
The development of targeted drug-delivery strategies for safer and more effective therapy in human hematologic malignancies has been a long-standing goal for clinical and basic investigators. We reasoned that profiling of human leukemia- and lymphoma-derived cell lines with combinatorial libraries might yield ligand peptide sequences that bind to specific internalizing receptors on cell surfaces and may potentially lead to the discovery of new or unrecognized therapeutic targets. Such targeting motifs could also serve as vehicles for the preferential delivery of cytotoxic agents to leukemia and lymphoma cells.
Several cell surface-binding peptides recognizing receptors in the membranes of lymphoma and leukemia cell lines have been reported.1–5 The selected peptide ligands are readily internalized by cells and may therefore be potentially useful in ligand-directed drug delivery. Recently, we described an arginine-rich motif that is internalized into leukemia and lymphoma cells through the macropinocytotic pathway; however, the precise cell surface receptor has yet to be identified.6 In effect, there is currently a relative lack of well-defined ligand-receptor systems for targeting human leukemia or lymphoma cells. The identification and validation of ligand-receptor pairs for these hematologic cancer cells relative to normal leukocytes would potentially represent a differential strategy and perhaps even improve disease outcomes.
In this study, we used a combinatorial phage display–based subtractive selection7–9 to identify ligand motifs that bind to specific cell surface receptors on human leukemia and lymphoma cell lines, as well as to primary human acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL) bone marrow specimens obtained from patients. We assessed the capacity of the newly selected peptides to be internalized by leukemia cells and used this criterion to select ligands that could serve as carriers for ligand-directed drug delivery. We describe a cell-internalizing motif, Phe-Phe/Tyr-Any-Leu-Arg-Ser (FF/YXLRS), and characterize its corresponding receptor as neuropilin-1 (NRP-1). Moreover, the functional relevance of this internalizing receptor was evaluated in the context of disease progression via the targeted delivery of a pro-apoptotic peptidomimetic to leukemia and lymphoma cells. We also show that the expression of NRP-1 was elevated in a panel of human leukemia and lymphoma cell lines relative to the controls. Finally, our results show increased levels of NRP-1 in bone marrow specimens from AML and ALL patients compared with normal human bone marrow specimens. Because neuropilins are also expressed in many human solid tumors,10 the FF/YXLRS motif may have several targeting applications. Our results define a new ligand CGFYWLRSC peptide/NRP-1 receptor system that offers a potential drug-delivery approach for therapies against human leukemias and lymphomas.
Methods
Leukemia and lymphoma cell lines
The human cell lines MOLT-4 (T-cell ALL), CCRF-CEM (T-cell ALL), HL-60 (acute promyelocytic leukemia), OCI-AML3 (AML), K562 (chronic myelogenous leukemia), RPMI-8226 (myeloma), SR-786 (anaplastic large T-cell lymphoma), and U937 (monocytic lymphoma) were all obtained from cryopreserved samples at The University of Texas M. D. Anderson Cancer Center. Cells were cultured and maintained in 5% carbon dioxide in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin as described in previous studies.4,6,9
Leukemia patient and control tissue samples
Primary leukemia and lymphoma samples from patients who had signed an informed consent in accordance with the Declaration of Helsinki were obtained as cryopreserved leukemia specimens from the Leukemia Cell Bank at the University of Texas M. D. Anderson Cancer Center, and the use of patient samples was approved by the institutional review board. Normal bone marrow samples were commercially obtained as cryopreserved specimens (Lonza).
Peptide solid-phase synthesis and purification
All peptides were synthesized, cyclized, and purified to our specifications by AnaSpec. Purification was by reverse-phase high-performance liquid chromatography to > 95% purity. Identification of the peptides was carried out by analysis with matrix-assisted laser desorption time-of-flight mass spectrometry. Peptides containing 2 cysteine residues derived from the CX7C library were cyclized by air oxidation before purification. The glycinylglycine (Gly-Gly) linker was added as a spacer to prevent steric hindrance.
Phage display random peptide library screening
A phage display random peptide library based on the fUSE5 vector displaying an insert with the general arrangement CX7C (C, cysteine; X, any residue) was designed and constructed with diversity between 108 to 109 unique sequences. We used biopanning and rapid analysis of selective interactive ligands (BRASIL) methodology to select cell-binding phage clones.7–9 In brief, human leukemia and lymphoma cells were collected, washed in phosphate-buffered saline (PBS) containing 2.5mM EDTA (ethylenediaminetetraacetic acid), and resuspended in RPMI 1640 medium containing 1% bovine serum albumin (BSA; BRASIL binding buffer) at 104 cells/mL. The cell suspension (200 μL) was incubated with 109 transducing units (TUs) of the CX7C phage library at 4°C. Phage selection was performed with an excess of the competing Arg-Gly-Asp (RGD) synthetic integrin-binding peptide motif to minimize and eliminate the recovery of RGD-containing ligands.8,9 After 2 hours, the phage/cell mixture (aqueous phase) was gently transferred to the top of a nonmiscible organic phase (200 μL/tube) consisting of dibutyl phthalate:cyclohexane 9:1 (vol/vol), and was centrifuged at 10 000g for 10 minutes at 4°C. The tube was quickly frozen in liquid nitrogen, and the cell-phage pellet was isolated. Cell-bound phage were recovered by infection of a host bacteria strain (Escherichia coli K91/kan). After 4 subsequent rounds of selection, phage-infected bacteria were grown as individual colonies, and inserts from randomly picked single clones were sequenced as described in previous studies.6–9
Phage-binding assay to cell surfaces
Phage-binding assays were performed as described in previous studies.7–9 A total of 106 cells were incubated in BRASIL binding buffer with 107 TU of positive control phage,6 various test phage, or insertless phage (negative control) at 4°C for 2 hours. Cell-bound phage were separated from unbound phage using BRASIL selection7–9 and recovered by bacterial infection of the host (E coli K91/kan), and then bacterial colonies in serially diluted plates were counted.
Phage internalization assay into leukemia cells
To permit potential receptor-mediated internalization, K562 cells (105) were incubated with 109 TU of test phage or insertless phage at 37°C overnight. After washes with serum-free RPMI 1640 containing 0.1% (wt/vol) trypsin to remove cell surface-bound, noninternalized phage, cells were adhered to glass chamber slides precoated with poly-D-lysine. Subsequently, cells were fixed and rendered permeable with precooled methanol, blocked with 5% normal goat serum, and stained with either a rabbit anti-fd bacteriophage antibody (Sigma-Aldrich) or with a nonimmune rabbit immunoglobulin G (IgG; Dako) diluted 1:1000 (vol/vol) in PBS containing 1% BSA at room temperature for 2 hours. Cells were washed with PBS and incubated with a fluorescein isothiocyanate (FITC)–conjugated goat anti–rabbit antibody (Jackson ImmunoResearch Laboratories) diluted 1:200 (vol/vol) in PBS containing 1% BSA at room temperature for 1 hour. Samples were mounted with Vectashield containing DAPI (Vector Laboratories).
Phage-binding assay to recombinant proteins
Phage overlay assays on leukemia and lymphoma samples were used to characterize ligand-receptor binding. Commercially available recombinant proteins (R&D Systems) were immobilized in 96-well plates (1 μg/well) at 4°C overnight. Unbound proteins were removed by washing, followed by a blocking step with PBS containing 1% BSA. Individual phage clones (at 109 TU each) were added to their respective wells and incubated at room temperature for 2 hours. Unbound phage were removed by several wash steps, followed by recovery of bound phage by host E coli K91/kan bacterial infection. Phage binding was quantified by standard bacterial colony counting.6–9
Flow cytometric analysis
Human leukemia and lymphoma cells were rinsed with PBS, blocked with 1 μg of human IgG (Sigma-Aldrich) per 105 cells for 15 minutes at room temperature. Cells were incubated in labeling buffer (PBS containing 0.2% BSA, 0.1% NaN3, and 5% heat-inactivated donkey serum) with 5 μL of 200 μg/mL of rabbit anti–human NRP-1 antibody (Santa Cruz Biotechnology) on ice for 1 hour, washed with PBS containing 0.2% BSA and 0.1% NaN3, and incubated with a PerCP-conjugated donkey anti–rabbit IgG (Santa Cruz Biotechnology) diluted to 1:70 (vol/vol) in labeling buffer for 30 minutes. After several rinses in PBS containing 0.2% BSA and 0.1% NaN3, cells were resuspended in PBS containing 0.2% BSA and analyzed by flow cytometry with an FACSCanto II (BD Bioscience).
RNA interference
Targeted short interfering RNA (siRNA) against NRP-1 and target controls were commercially obtained (Santa Cruz Biotechnology). siRNAs were transfected into cells with the siRNA Reagent System (Santa Cruz Biotechnology).
Targeting the pro-apoptotic peptidomimetic to leukemia and lymphoma cells
We assessed the capacity of the lead ligand peptide CGFYWLRSC to deliver the pro-apoptotic moiety D(KLAKLAK)211 into leukemia and lymphoma cells by cytotoxicity assays in vitro. Leukemia and lymphoma cells (2 × 104 cells/well) were plated in 96-well dishes containing RPMI 1640 supplemented with 10% FBS (vol/vol), penicillin, and streptomycin. The cells were incubated with increasing concentrations of the CGFYWLRSC-GG-D(KLAKLAK)2 peptide or with control peptides consisting of an admixture of CGFYWLRSC + D(KLAKLAK)2 at equimolar concentrations for 20 hours at 37°C. Cell viability was measured using the DHL cell proliferation and viability assay (AnaSpec).
Leukemia cell death assay
OCI-AML3 cells (2 × 105 cells/well in 1 mL) were plated in 6-well plates containing RPMI 1640 supplemented with 10% FBS (vol/vol), penicillin, and streptomycin. The cells were incubated with 20μM CGFYWLRSC-GG-D(KLAKLAK)2 or with 20μM CGFYWLRSC + D(KLAKLAK)2 control peptide for 20 hours at 37°C. Untreated cells served as a control to measure background levels of cell death in these conditions. After the incubation, the cells were stained with FITC-conjugated annexin V antibody and propidium iodine using the annexin V–FITC apoptosis detection kit (Sigma-Aldrich). The cells were subsequently analyzed by flow cytometry.
Patient sample immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections of bone marrow aspirate clot or biopsy specimens. Sections were deparaffinized, rehydrated, and submitted to heat-induced epitope retrieval as described in Pileri et al.12 Samples were incubated with an anti-NRP-1 (A-12) mouse monoclonal antibody (Santa Cruz Biotechnology) or a negative control mouse IgG at room temperature in a dark humidified chamber for 3 hours. Detection of the primary antibody was achieved with the EnVision+ system (DakoCytomation) containing secondary antibodies conjugated to a horseradish peroxidase complex. Slides were incubated at room temperature in a dark, humidified chamber for 30 minutes, and developed with the chromogen 3,3′-diaminobenzidine/H2O2 (DakoCytomation). Slides were counterstained with hematoxylin, dehydrated, mounted, and coverslipped.
Statistical analysis
All data are reported as the average mean ± SEM. Unpaired Student t tests were used to determine statistical significance (n = 6, unless otherwise specified). P values less than .05 were considered statistically significant.
Results
Identification of a new leukemia-targeting motif: selection, binding, and internalization into leukemia and lymphoma cells
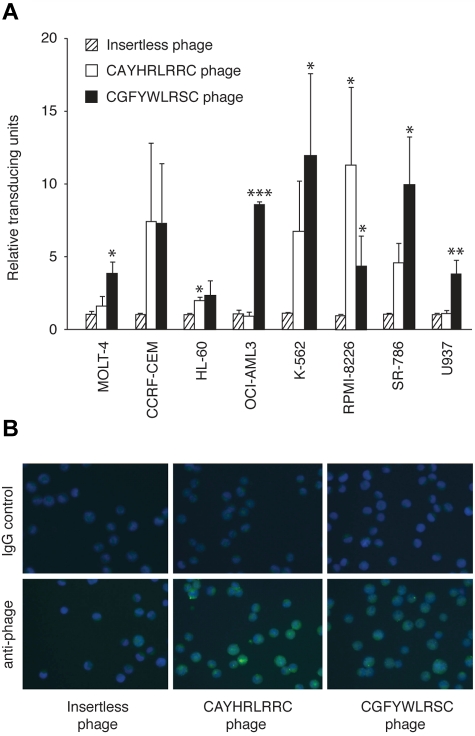
We screened for ligand peptides that internalized into leukemia and lymphoma cells, affording the potential of intracellular drug delivery. First, a cyclic phage display peptide library (CX7C) was screened on MOLT-4 cells to isolate peptide motifs.7–9 After 4 rounds of serial selection, binding phage clones (n = 96) were recovered from MOLT-4 cells and individually sequenced. This approach has identified the leukemia-binding peptide CAYHRLRRC (57% of the clones) containing the motif Arg-Leu-Arg-Arg (RLRR), which has been reported previously by Nishimura et al.6 However, a newly identified consensus motif, FF/YXLRS, accounted for 22% of the selected peptide sequences; specifically, 2 similar sequences, CGFYWLRSC (5%) and CSFFYLRSC (17%), were found. Together, these 2 distinct leukemia-targeting motifs (RLRR and FF/YXLRS) exhibited the highest frequency (79%) of all of the selected clones. First, we studied the binding characteristics of the novel peptide-displaying phage to leukemia cell lines. A representative phage clone displaying the motif FF/YXLRS bound to all (n = 8) leukemia/lymphoma cell lines studied relative to the negative control (Figure 1A), and the differential binding pattern of RLRR and FF/YXLRS suggested that the motifs likely target different receptors. Subsequently, the isolated peptide-displaying phage were studied for their internalization capabilities into leukemia cells. The CGFYWLRSC peptide internalized into leukemia cells nearly in a comparable level to our previously identified macropinocytosis marker6 (Figure 1B). The CSFFYLRSC peptide, on the contrary, did not mediate as efficient phage internalization (data not shown). Therefore, we elected to pursue functional studies with the CGFYWLRSC peptide as the lead FF/YXLRS-containing ligand for leukemia targeting, and the CSFFYLRSC peptide was not pursued further here despite its higher selection (17% versus 5%). In effect, the higher selection could also be paradoxical due to preferential coding utilization in bacteria.
Figure 1.
Identification of a new leukemia-targeting motif. (A) Binding of CGFYWLRSC-, CAYHRLRRC-, or insertless phage to ALL (T-lymphoblastic) cell lines MOLT-4 and CCRF-CEM, AML cell lines HL-60 (promyelocytic) and OCI-AML3 (myelocytic), CML cell line K562, multiple myeloma cell line RPMI-8226, and lymphoma cell lines SR-786 (T-cell lymphoma) and U937 (monocyte lymphoma), respectively. The data are averages of triplicates per assay from 2 independent experiments. *P < .05; **P < .01; and ***P < .001. (B) Comparison of internalization of phage-displaying peptides into leukemia cells. The FITC-labeled anti-phage stain is shown relative to that of the IgG isotype control. The blue color indicates the DAPI nuclear stain. Magnification, 500×.
NRP-1 as the corresponding cell surface receptor in leukemia cells
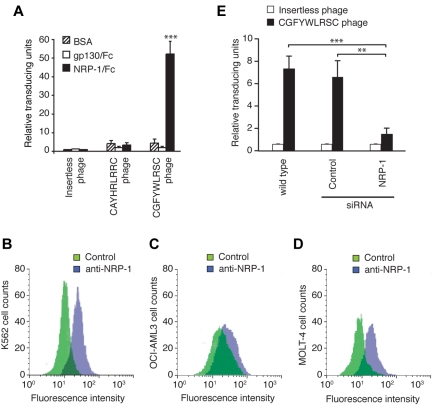
To determine the internalization mechanism of the lead CGFYWLRSC peptide, we searched online protein databases for natural proteins that this motif might mimic. The peptide sequence demonstrated partial alignments with many unrelated proteins, and we thus queried whether any homology would exist for a common interaction pattern. We noted that several weak, nonlinear short matches were identified with proteins putatively forming a multiprotein complex, including VEGF-165 (vascular endothelial growth factor-165), NRP-2, plexin A1, and plexin B1 (data not shown); interestingly, these proteins are all known to interact with NRP-1. We therefore carried out a series of microtiter binding assays to assess whether FF/YXLRS-containing peptides indeed recognize NRP-1. Notably, the CGFYWLRSC phage selectively bound to NRP-1 but not to the GP130-like monocyte receptor (Figure 2A), which had appeared as another candidate receptor for the consensus motif in the search of the protein database, thus suggesting specificity. Moreover, the CGFYWLRSC phage did not show any significant binding to NRP-2, semaphorin 3A, or semaphorin 6A, which are known to form complexes with NRP-1, nor did it bind to unrelated recombinant proteins TrkB and NGFR (data not shown). Further supporting the specificity of the CGFYWLRSC phage to NRP-1, we observed that the insertless phage or the phage displaying the unrelated sequence CAYHRLRRC did not bind to NRP-1 (Figure 2A). Subsequently, we confirmed the expression of NRP-1 in the leukemia cell lines by flow cytometry analysis. K562 and OCI-AML3 cells showed the highest levels of NRP-1, followed by MOLT-4 cells (Figure 2B-D), results generally correlating with the CGFYWLRSC phage binding (Figure 1A). Finally, we performed RNA interference to diminish NRP-1 mRNA in target leukemia cells, and found that the CGFYWLRSC phage binding was reduced in NRP-1 siRNA-treated leukemia cells, but not in leukemia cells treated with control siRNA (Figure 2E). These data suggest that NRP-1 is a receptor for the lead ligand peptide CGFYWLRSC.
Figure 2.
Validation of NRP-1 as a receptor for the CGFYWLRSC peptide. (A) Phage binding to recombinant rat NRP-1/Fc chimera (rrNRP-1/Fc) or to control GP130-Fc protein was assessed. Results shown are averages of 3 independent assays. ***P < .001. (B-D) Flow cytometry analysis of cell-surface expression of NRP-1 on K562 (B), OCI-AML3 (C), and MOLT-4 (D) leukemia cell lines. Cells were stained as follows: control rabbit IgG and FITC-goat anti–rabbit IgG (green), rabbit anti–human NRP-1 and FITC-goat anti–rabbit IgG (violet). (E) Binding of the CGFYWLRSC phage to OCI-AML3 cells transfected with NRP-1-targeting siRNA or with the nontargeting control siRNA. Untransfected wild-type cells and insertless phage were used as controls. **P < .01; ***P < .001.
Ligand-directed delivery of a targeted drug candidate to NRP-1
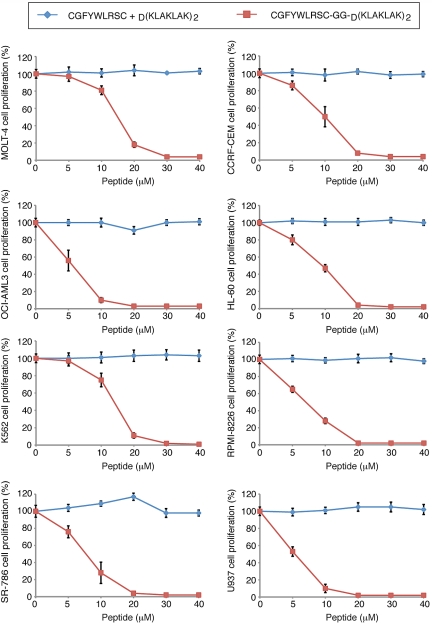
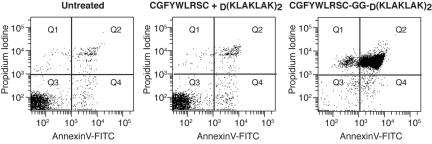
Given that NRP-1 is overexpressed in leukemia cell lines and bone marrow samples, and that NRP-1 mediates phage internalization, we next asked whether NRP-1 would act as a suitable target for ligand-directed drug delivery. We studied the targeted delivery of the pro-apoptotic peptidomimetic D(KLAKLAK)2 linked to the CGFYWLRSC peptide as an anti-leukemia therapy. D(KLAKLAK)2, upon internalization by target cells, causes apoptosis by physical interference and disruption of mitochondrial membranes.11 CGFYWLRSC-GG-D(KLAKLAK)2 decreased cell viability at relatively low concentrations (5-30μM range) in human leukemia and lymphoma cell lines, and in most of the cases the maximum inhibition was reached at ≤ 20μM (Figure 3). The control CGFYWLRSC + D(KLAKLAK)2 peptide had no effect at equimolar concentrations. These results suggest a potential therapeutic window for our new drug candidate. The cells were further analyzed by annexin V–FITC/propidium iodide staining to assess the extent of cell death induced by the CGFYWLRSC-GG-D(KLAKLAK)2 peptide (Figure 4). The OCI-AML3 cells treated with 20μM CGFYWLRSC-GG-D(KLAKLAK)2 peptide for 20 hours had undergone a nearly complete apoptosis induction (> 95% of the cells; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), while the cells treated with the control CGFYWLRSC + D(KLAKLAK)2 peptide had only background levels of cell death, which was similar to those observed in untreated control cells (4%-6% of the cells; supplemental Table 1).
Figure 3.
Assessment of the cytotoxic effect of CGFYWLRSC-GG-D(KLAKLAK)2 peptide on leukemia cells in vitro. The effects of increasing doses of CGFYWLRSC + D(KLAKLAK)2 peptides or the CGFYWLRSC-GG-D(KLAKLAK)2 peptide are shown. Cell viability was measured via the DHL cell proliferation and viability assay (AnaSpec). The cell lines were (A) MOLT-4, (B) CCRF-CEM, (C) OCI-AML3, (D) HL-60, (E) K562, (F) RPMI-8226, (G) SR-786, and (H) U937. Results shown represent the average of 3 independent experiments.
Figure 4.
Leukemia cell death assay. Untreated OCI-AML3 cells served as a control to determine the background levels of cell death under the experimental conditions used. Cells were incubated with 20μM CGFYWLRSC + D(KLAKLAK)2 control peptide or with 20μM CGFYWLRSC-GG-D(KLAKLAK)2 for 20 hours at 37°C. annexin V–FITC/propidium iodide staining was used to detect the level of cell death via flow cytometry.
Differential expression of NRP-1 in leukemia versus normal bone marrow
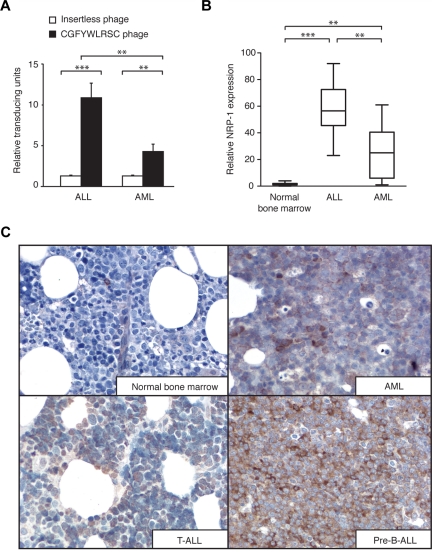
Given the expression of NRP-1 in the leukemia cells, we next asked whether NRP-1 is present in the bone marrow of leukemia patients relative to normal human bone marrow control and whether its expression varies within different leukemia types. First, we analyzed a patient-derived panel of AML (n = 5) and ALL (n = 7) bone marrow specimens for CGFYWLRSC-phage binding. All of the samples tested showed enhanced binding relative to the insertless phage (Figure 5A). Immunohistochemistry confirmed the expression of NRP-1 in the bone marrow specimens (Figure 5B), and also showed a weak expression of NRP-1 in 6 of 7 bone marrow specimens from healthy donors (Figure 5B), suggesting a basal medullary expression of NRP-1. In comparison, leukemia bone marrow specimens showed high levels of NRP-1 on the cell surface of all ALL samples (n = 6) studied; specifically, 5 precursor B-cell ALL and one T-cell ALL. Four human precursor B-cell ALL samples exhibited the most robust staining for NRP-1 among the panel (n = 37 samples) bone marrow specimens analyzed here (Figure 5B). Compared with ALL, AML bone marrow specimens (n = 24) exhibited moderate levels of NRP-1, generally correlating with the CGFYWLRSC phage binding to leukemia cells (Figure 5A). However, in two-thirds of the cases (16 of 24 cases), the level of NRP-1 was well above the baseline observed in normal bone marrow (Figure 5B, and data not shown). Representative samples are shown compared with normal human bone marrow (Figure 5C). To further study NRP-1 expression, we used flow cytometry to compare the median expression level of NRP-1 antibody relative to that of control IgG on bone marrow specimens (n = 9) from different patient and normal donors. AML (n = 3) and ALL (n = 3) bone marrow cells analyzed showed significantly higher levels of NRP-1 expression (P < .05 in AML and P < .01 in ALL) compared with normal bone marrow cells (n = 3) (Supplemental Figure 1A). ALL bone marrow cells had the highest median expression of NRP-1 relative to the control IgG, whereas AML cells had a moderate expression, results consistent with the immunohistochemistry staining. To analyze whether NRP-1 expression varied among certain cell types, we gated bone marrow cell populations (Supplemental Figure 1B) and evaluated NRP-1 expression in these populations (Supplemental Figure 1C). These results showed elevated NRP-1 expression in several cell populations, indicating that the NRP-1 is not specifically expressed by any particular leukocyte, but rather is generally overexpressed in leukemia bone marrow. Future studies with a larger cohort of samples will shed light on NRP-1 expression in human leukemia.
Figure 5.
NRP-1 expression in leukemia bone marrow samples. (A) Binding of peptide-displaying phage or insertless control phage to bone marrow specimens of AML and ALL patients. **P < .01; ***P < .001. (B) Immunohistochemical staining for NRP-1 on ALL, AML, and normal bone marrow specimens. The amount of positively stained cells in the specimen represents the approximate levels of NRP-1 expression. *P < .05; **P < .01; ***P < .001. (C) Representative examples of immunohistochemical staining of NRP-1 (brown) on AML, T-cell ALL, and pre-B–cell ALL and normal bone marrow specimens are shown. Magnification, 500×.
Discussion
Ligand peptides that enable targeted drug delivery to tumor cells offer new horizons for therapy. We introduce a new ligand motif, FF/YXLRS, which is capable of targeting leukemia and lymphoma cells. Our lead sequence, CGFYWLRSC, mediated phage binding and internalization of a magnitude generally comparable with the macropinocytosis marker peptide CAYHRLRRC,6 but the receptor and the internalizing mechanism were apparently different. Interestingly, our FF/YXLRS motif contains a tripeptide sequence, Leu-Arg-Ser (LRS) that has been commonly reported in the literature. We and others have previously reported several different LRS-sequence-containing peptides in different settings,13–16 indicating that this tripeptide sequence is likely to be less specific or not sufficient to differentiate binding to tumor versus normal cells. In the present study, we show a significant binding of CGFYWLRSC-displaying phage to NRP-1. Our findings suggest that the CGFYWLRSC peptide binds to NRP-1 and/or to a protein complex containing NRP-1. Moreover, RNA interference with NRP-1 inhibited phage binding, indicating that NRP-1 might be a singular receptor for CGFYWLRSC and that the internalization of CGFYWLRSC could be mediated by cell surface NRP-1. It has recently been reported that NRP-1 mediates internalization of peptides with a C-terminally exposed arginine (or rarely lysine) residue with a R/KXXR/K motif.17,18 However, no such motif is present in the NRP-1-binding ligand peptide described here. In addition, our peptide does not contain another known polybasic NRP-1-binding motif, RRXR, also identified by phage display,19 and differs from the other described phage-display–derived peptides that bind to NRP-1.7,20,21
To evaluate the drug-delivery potential of the novel NRP-1–binding sequence CGFYWLRSC, we synthesized the lead peptide in tandem with an apoptosis-inducing moiety, D(KLAKLAK)2, an amphipathic peptidomimetic that disrupts mitochondrial membranes,11 and studied the cytotoxic effect on malignant cells in vitro. We observed differential killing of leukemia and lymphoma cells by the CGFYWLRSC-targeted pro-apoptotic drug candidate. D(KLAKLAK)2 itself was not internalized by cells and required an appropriate intracellular drug delivery system to induce apoptosis. Moreover, contrary to previously reported NRP-1–binding peptides that have antitumor activity,22 CGFYWLRSC alone did not have a detectable effect on leukemia cell proliferation or viability. Our results demonstrating apoptosis in cells treated with the CGFYWLRSC peptide-targeted D(KLAKLAK)2 indicate that CGFYWLRSC mediated delivery into the cells and subsequently targeted their mitochondrial membrane via the D(KLAKLAK)2 motif. The ligand CGFYWLRSC peptide therefore appears to be a promising candidate for cancer-targeted drug-delivery systems, and our results indicate that CGFYWLRSC-GG-DKLAKLAK2 might serve as an anti–leukemia/lymphoma agent.
We have also done pilot studies in the severe combined immune-deficient (SCID) leukemia mouse model, in which some early therapeutic benefit was observed. In retrospect, however, the SCID leukemia mouse model of AML used was likely too aggressive; because the overall performance status of the SCID mice deteriorated from widespread tumor burden, we were unable to clearly establish a survival benefit. Furthermore, the kidney toxicity, presumably associated with the renal clearance of D residues from the D(KLAKLAK)2 motif, made it challenging to evaluate later time points and survival. Other models, doses, and treatment schedules will be performed in the future under Good Manufacturing Practice conditions. In an independent ongoing preclinical program (also under Good Manufacturing Practice conditions), we have now determined that the dose-limiting toxicity of the D(KLAKLAK)2 motif is indeed renal and that this toxicity is unrelated to the specific ligand peptide used (W.A., R.P., unpublished data, November 2010). We are currently starting to address the issue of nephrotoxicity. However, our primary purpose here is to report a new ligand-receptor interaction that could potentially be used as a drug-delivery system in human leukemia. Ultimately, detailed future studies will be required to address the capacity of CGFYWLRSC to mediate targeted drug delivery in vivo.
The major sites of expression of NRP-1 are neurons and endothelial cells, and NRP-1 has an essential role in normal neuronal and vascular development.23–25 NRP-1 is known to bind to 2 unrelated natural ligands, class 3 semaphorins, a family of secreted polypeptides with key roles in axonal guidance,26,27 and various members of the VEGF family.28 NRP-1 acts in conjunction with membrane-associated signal transducers, such as VEGF receptor tyrosine kinases (eg, VEGFR2) and plexins, the transmembrane receptors of the semaphorin family.29,30 Recent studies have shown that NRP-1 also interacts with several heparin-binding growth factors.31 In addition to its function as an essential coreceptor in neuronal guidance and angiogenesis,23–28 NRP-1 has also been implicated as a novel mediator of the primary immune response.32,33 Moreover, the expression of NRP-1 can be stimulated in response to tissue injury or hypoxic conditions,34,35 and it is highly expressed in diverse solid tumors, such as prostate, breast, pancreatic, lung, ovarian, and gastrointestinal carcinomas.10,28,36,37 Increased expression of NRP-1 has been correlated with tumor growth and vascularization in vivo and with invasiveness in human cancer.38–41 Furthermore, new data indicate that angiogenesis is an important requirement for the development and progression of human hematologic malignancies such as leukemias and lymphomas. Accordingly, increased angiogenesis in the bone marrow of patients with AML has been reported, as well as the expression of NRP-1 in CML and AML.42–45 The increased cell-surface expression of NRP-1 is also correlated with poor survival of AML patients46 and with the percentage of blasts in the peripheral blood and bone marrow of AML patients47; these results indicate that levels of NRP-1 are correlated with the disease severity and biologic progression.
In a systemic leukemia mouse model, AML progression was induced by VEGF, and a soluble form of NRP-1 significantly inhibited AML progression.48 In the present study, we found a nearly linear correlation between cell-surface levels of NRP-1 and phage binding to leukemia cell lines. We observed that in several bone marrow specimens, which were obtained from variety of leukemia subtypes, NRP-1 expression was elevated in ALL patients compared with AML patients, a pilot result suggesting a slightly higher median expression level of NRP-1 in the bone marrow derived from ALL versus the AML sample. Whether human NRP-1 overexpression varies among certain types of leukemia will need to be further studied with larger patient cohorts. Nevertheless, these studies show that NRP-1 expression is increased in the bone marrow of ALL and AML patients compared with normal bone marrow. In preliminary studies, we have also observed that NRP-1 expression is apparently increased in several cell populations from bone marrow of leukemia patients. These data indicate that NRP-1 is not markedly expressed by any one particular type of leukocyte, but rather may be generally overexpressed in leukemic bone marrow, perhaps as a response to a hypoxic microenvironment.49 Therefore, we reasoned that the NRP-1-binding ligands should exhibit an increased homing specificity toward leukemic bone marrow, and that NRP-1 could serve as an effective target for receptor-mediated drug delivery.
Our results support the conclusion that NRP-1 can be considered as a potential target in leukemia and lymphoma therapies, and that it can serve in the capacity of ligand-directed drug delivery. We conclude that the NRP-1-binding motif FF/YXLRS is a promising candidate for targeted drug development and lead optimization.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH and DOD, the Specialized Program in Research Excellence (SPORE) in Leukemia from The University of Texas M. D. Anderson Cancer Center, the Gillson-Longenbaugh Foundation (to R.P. and W.A.), and The Kimberly Patterson Fellowship for Leukemia Research (to D.E.J.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.K., D.E.J., C.E.B.-R., A.J.Z, A.K., B.L.E., and F.C.M. performed research; K.K., D.E.J, C.E.B.-R., A.J.Z., A.K., F.C.M., E.K., R.P., and W.A. designed and analyzed the research; K.K., D.E.J, C.E.B.-R., A.J.Z., H.M.K., J.E.C., S.O., E.K., R.P., and W.A. wrote and/or edited the article; and B.L. provided reagents.
Conflict-of-interest disclosure: The University of Texas and some of its researchers (W.A. and R.P.) have equity in Mercator Therapeutics, which is subject to certain restrictions under university policy; the university manages and monitors the terms of these arrangements in accordance to its conflict-of-interest policies. The authors declare no other competing financial interests.
Correspondence: Wadih Arap, David H. Koch Center, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: warap@mdanderson.org; or Renata Pasqualini, David H. Koch Center, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: rpasqual@mdanderson.org.
References
- 1.Jäger S, Jahnke A, Wilmes T, et al. Leukemia targeting ligands isolated from phage display peptide libraries. Leukemia. 2007;21(3):411–420. doi: 10.1038/sj.leu.2404548. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi S, Mok H, Parrott MB, et al. Selection of chronic lymphoblastic leukemia binding peptides. Cancer Res. 2003;63(17):5213–5217. [PubMed] [Google Scholar]
- 3.McGuire MJ, SamLi KN, Chang YC, Brown KC. Novel ligands for cancer diagnosis: Selection of peptide ligands for identification and isolation of B-cell lymphomas. Exp Hematol. 2006;34(4):443–452. doi: 10.1016/j.exphem.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Koivunen E, Ranta TM, Annila A, et al. Inhibition of beta(2) integrin-mediated leukocyte cell adhesion by leucine-leucine-glycine motif-containing peptides. J Cell Biol. 2001;153(5):905–916. doi: 10.1083/jcb.153.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanidakis M, Karjalainen K, Jaalouk DE, et al. Role of leukemia cell invadosome in extramedullary infiltration. Blood. 2009;114(14):3008–3017. doi: 10.1182/blood-2008-04-148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura S, Takahaski S, Kamikatahira H, et al. Combinatorial targeting of the macropinocytotic pathway in leukemia and lymphoma cells. J Biol Chem. 2008;283(17):11752–11762. doi: 10.1074/jbc.M708849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giordano RJ, Cardó-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat Med. 2001;7(11):1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- 8.Christianson DR, Ozawa MG, Pasqualini R, Arap W. Techniques to decipher molecular diversity by phage display. Methods Mol Biol. 2007;357:385–406. doi: 10.1385/1-59745-214-9:385. [DOI] [PubMed] [Google Scholar]
- 9.Kolonin MG, Bover L, Sun J, et al. Ligand-directed surface profiling of human cancer cells with combinatorial peptide libraries. Cancer Res. 2006;66(1):34–40. doi: 10.1158/0008-5472.CAN-05-2748. [DOI] [PubMed] [Google Scholar]
- 10.Bielenberg DR, Pettaway CA, Takashima S, Klagsbrun M. Neuropilin in neoplasms: Expression, regulation, and function. Exp Cell Res. 2006;312(5):584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Ellerby HM, Arap W, Ellerby LM, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med. 1999;5(9):1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 12.Pileri SA, Ascani S, Milani M, et al. Acute leukaemia immunophenotyping in bone-marrow routine sections. Br J Hematol. 1999;105(2):394–401. [PubMed] [Google Scholar]
- 13.Pulli T, Koivunen E, Hyypiä T. Cell-surface interaction of Echovirus 22*. J Biol Chem. 1997;272(34):21176–21180. doi: 10.1074/jbc.272.34.21176. [DOI] [PubMed] [Google Scholar]
- 14.Koivunen E, Arap W, Valtanen H, et al. Tumor targeting with a selective gelatinase inhibitor. Nat Biotechnol. 1999;17(8):768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 15.Berntzen G, Brekke OH, Mousavi SA, et al. Characterization of an FcγRI-binding peptide selected by phage display. Protein Eng Des Sel. 2006;19(3):121–128. doi: 10.1093/protein/gzj011. [DOI] [PubMed] [Google Scholar]
- 16.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380(6572):364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 17.Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci U S A. 2009;106(38):16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugahara KN, Teesalu T, Karmali PP, et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328(5981):1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong TM, Chen YL, Wu YY, et al. Targeting neuropilin 1 as an antitumor strategy in lung cancer. Clin Cancer Res. 2007;13(16):4759–4768. doi: 10.1158/1078-0432.CCR-07-0001. [DOI] [PubMed] [Google Scholar]
- 20.Giordano RJ, Anobom CD, Cardó-Vila M, et al. Structural basis for the interaction of a vascular endothelial growth factor mimic peptide motif and its corresponding receptors. Chem Biol. 2005;12(10):1075–1083. doi: 10.1016/j.chembiol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Giordano RJ, Cardó-Vila M, Salameh A, et al. From combinatorial peptide selection to drug prototype (I): Targeting the vascular endothelial growth factor receptor pathway. Proc Natl Acad Sci U S A. 2010;107(11):5112–5117. doi: 10.1073/pnas.0915141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr MP, Byrne AM, Condron CM, et al. A peptide corresponding to the neuropilin-1-binding site on VEGF165 induces apoptosis of neuropilin-1-expressing breast tumor cells. B J Cancer. 2005;92(2):328–333. doi: 10.1038/sj.bjc.6602308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami A, Kitsukawa T, Takagi S, Fujisawa H. Developmentally regulated expression of a cell surface protein, neuropilin, in the mouse nervous system. J Neurobiol. 1996;29(1):1–17. doi: 10.1002/(SICI)1097-4695(199601)29:1<1::AID-NEU1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Herzog Y, Kalcheim C, Kahane N, Reshef R, Neufeld G. Differential expression of neuropilin-1 and neuropilin-2 in arteries and veins. Mech Dev. 2001;109(1):115–119. doi: 10.1016/s0925-4773(01)00518-4. [DOI] [PubMed] [Google Scholar]
- 25.Gu C, Rodriguez ER, Reimert DV, et al. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5(1):45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90(4):739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 27.Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90(4):753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 28.Klagsbrun M, Takashima S, MamLuk R. The role of neuropilin in vascular and tumor biology. Adv Exp Med Biol. 2002;515:33–48. doi: 10.1007/978-1-4615-0119-0_3. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi T, Fournier A, Nakamura F, et al. Plexin-neuropilin 1 complexes form functional semaphorin-3A receptors. Cell. 1999;99(1):59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld G, Kessler O, Herzog Y. The interaction of neuropilin-1 and neuropilin-2 with tyrosine-kinase receptors for VEGF. Adv Exp Med Biol. 2002;515:81–90. doi: 10.1007/978-1-4615-0119-0_7. [DOI] [PubMed] [Google Scholar]
- 31.West DC, Rees CG, Duchesne L, et al. Interaction of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J Biol Chem. 2005;280(14):13457–13464. doi: 10.1074/jbc.M410924200. [DOI] [PubMed] [Google Scholar]
- 32.Wülfing C, Rupp F. Neuropilin-1: another neuronal molecule in the “immunological synapse.“. Nat Immunol. 2002;3(5):418–419. doi: 10.1038/ni0502-418. [DOI] [PubMed] [Google Scholar]
- 33.Tordjman R, Lepelletier Y, Lemarchandel V, et al. A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat Immunol. 2002;3(5):477–482. doi: 10.1038/ni789. [DOI] [PubMed] [Google Scholar]
- 34.Brusselmans K, Bono F, Collen D, Herbert JM, Carmeliet P, Dewerchin M. A novel role for vasculature endothelial growth factor as an autocrine survival factor for embryonic stem cells during hypoxia. J Biol Chem. 2005;280(5):3493–3499. doi: 10.1074/jbc.M406613200. [DOI] [PubMed] [Google Scholar]
- 35.Matthies AM, Low QEH, Lingen MW, DiPietro LA. Neuropilin-1 participates in wound angiogenesis. Am J Pathol. 2002;160(1):289–296. doi: 10.1016/S0002-9440(10)64372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 37.Guttmann-Raviv N, Kessler O, Shraga-Heled N, Lange T, Herzog Y, Neufeld G. The neuropilins and their role in tumorigenesis and tumor progression. Cancer Lett. 2006;231(1):1–11. doi: 10.1016/j.canlet.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 38.Pan Q, Chanthery Y, Liang WC, et al. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell. 2007;11(1):53–67. doi: 10.1016/j.ccr.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Hansel DE, Wilentz RE, Yeo CJ, Schulick RD, Montgomery E, Maitra A. Expression of neuropilin-1 in high-grade dysplasia, invasive cancer, and metastasis of the human gastrointestinal tract. Am J Surg Pathol. 2004;28(3):347–356. doi: 10.1097/00000478-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Latil A, Bieche I, Pesche S, et al. VEGF overexpression in clinically localized prostate tumors and neuropilin-1 overexpression in metastatic forms. Int J Cancer. 2000;89(2):167–171. doi: 10.1002/(sici)1097-0215(20000320)89:2<167::aid-ijc11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 41.Bachelder RE, Crago A, Chung JC, et al. Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res. 2001;61(15):5736–5740. [PubMed] [Google Scholar]
- 42.Padró T, Ruiz S, Bieker R, Büet, et al. Increased angiogenesis in the bone marrow of patients with acute myeloid leukemia. Blood. 2000;95(8):2637–2644. [PubMed] [Google Scholar]
- 43.Hussong JW, Rodgers GM, Shami PJ. Evidence of increased angiogenesis in patients with acute myeloid leukemia. Blood. 2000;95(1):309–313. [PubMed] [Google Scholar]
- 44.Vales A, Kondo R, Aichberger KJ, et al. Myeloid leukemias express a broad spectrum of VEGF receptors including neuropilin-1 (NRP-1) and NRP-2. Leuk Lymphoma. 2007;48(10):1997–2007. doi: 10.1080/10428190701534424. [DOI] [PubMed] [Google Scholar]
- 45.Yamada Y, Oike Y, Ogawa H, et al. Neuropilin-1 on hematopoietic cells as a source of vascular development. Blood. 2003;101(5):1801–1809. doi: 10.1182/blood-2002-01-0119. [DOI] [PubMed] [Google Scholar]
- 46.Kreuter M, Woelke K, Bieker R, et al. Correlation of neuropilin-1 overexpression to survival in acute myeloid leukemia. Leukemia. 2006;20(11):1950–1954. doi: 10.1038/sj.leu.2404384. [DOI] [PubMed] [Google Scholar]
- 47.Lu L, Zhang L, Xiao ZJ, Lu SH, Yang RC, Han ZC. Neuropilin-1 in acute myeloid leukemia: Expression and role in proliferation and migration of leukemia. Leuk Lymphoma. 2008;49(2):331–338. doi: 10.1080/10428190701809149. [DOI] [PubMed] [Google Scholar]
- 48.Schuch G, Machluf M, Bartsch G, et al. In vivo administration of vascular endothelial growth factor (VEGF) and its antagonist, soluble neuropilin 1, predicts a role of VEGF in the progression of acute myeloid leukemia in vivo. Blood. 2002;100(13):4622–4628. doi: 10.1182/blood.V100.13.4622. [DOI] [PubMed] [Google Scholar]
- 49.Konopleva M, Tabe Y, Zeng Z, Andreef M. Therapeutic targeting of microenvironmental interactions in leukemia: mechanisms and approaches. Drug Resist Updat. 2009;12(4–5):103–113. doi: 10.1016/j.drup.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.