Abstract
BACKGROUND
Xenotropic murine leukemia virus (MLV)-related virus (XMRV) and other related MLVs have been described with chronic fatigue syndrome (CFS) and certain types of prostate cancer. In addition, prevalence rates as high as 7% have been reported in blood donors, raising the risk of transfusion-related transmission. Several laboratories have utilized micro-neutralization assays as a surrogate marker for detection of anti-MLV serological responses – with up to 25% of prostate cancer patients reported to harbor neutralizing antibody responses.
STUDY DESIGN AND METHODS
We developed a high-throughput micro-neutralization assay for research studies on blood donors using retroviral vectors pseudotyped with XMRV-specific envelopes. Infection with these pseudotypes was neutralized by sera from both macaques and mice challenged with XMRV, but not pre-immune serum. 354 plasma samples from blood donors in the Reno/Tahoe area were screened for neutralization.
RESULTS
6.5% of donor samples gave moderate neutralization of XMRV, but not control pseudotypes. However, further testing by Western blot revealed no evidence of antibodies against MLVs in any of these samples. Furthermore, no evidence of infectious virus or viral nucleic acid was observed.
CONCLUSION
A micro-neutralization assay was developed for detection of XMRV, and can be applied in a high-throughput format for large scale studies. Although a proportion of blood donors demonstrated the ability to block XMRV envelope-mediated infection, we found no evidence that this inhibition was mediated by specific antibodies elicited by exposure to XMRV/MLV. It is likely that this moderate neutralization is mediated through another, non-specific mechanism.
Keywords: High-throughput micro-neutralization assay, XMRV, MLV, Pseudoviruses, Donor screening
INTRODUCTION
The short history of xenotropic murine leukemia virus (MLV)-related virus (XMRV) is one of controversy and discrepant results. Initial studies found XMRV nucleic acids and/or proteins in prostate cancers1,2 and even a low percentage of prostate tissues from individuals with no history of prostate cancer2. In contrast, several other studies have failed to detect XMRV in prostate cancer tissue3,4. Much of this controversy is likely explained by PCR and other nucleic acid contamination5,6. Despite XMRV originally being isolated from human prostate cancer samples1, it is in all likelihood a laboratory artifact, created by the passage of human prostate tissue through mice7. This resulted in infection with, and subsequent recombination between, at least two endogenous MLVs7. Cell lines created from this tissue, and harboring XMRV, were likely distributed to many laboratories working on prostate cancer.
The controversy surrounding the association between XMRV and chronic fatigue syndrome (CFS), is, if anything greater. It was reported by Lombardi and colleagues that two-thirds of CFS patients from the U.S. harbored XMRV compared to 4% of controls8. Importantly, this work was based on three separate lines of evidence: i) direct and indirect nucleic acid detection in PBMC, stimulated PBMC and plasma; ii) culture of replication-competent XMRV from plasma and PBMC by co-culture with human prostate cells and iii) serological evidence using a flow cytometry assay. In addition to the association with CFS, the presence of virus in plasma and blood cells, coupled with the relatively high prevalence observed in apparently healthy controls suggested that XMRV may be both blood transfusion-transmitted and a real threat to the safety of the U.S. blood supply. However, many other groups failed to detect XMRV in PBMC samples from CFS or healthy individuals9–13. At least two studies tried to fully replicate the initial study using PCR, culture and serology, without any convincing evidence of XMRV in either CFS patient or healthy controls14,15. Furthermore, more recent testing of specimens from the Lombardi et al study revealed some of the previously reported PCR-positive specimens were contaminated with XMRV-containing plasmid sequences leading to the partial retraction of these PCR results from the Lombardi et al. publication16. Additionally, a recent multi-laboratory blinded study using 15 previously reported XMRV/MLV-positive subjects as well as validated negative controls, demonstrated that virus culture assays used in Lombardi et al. were prone to cross-contamination17. Thus, this leaves only the serological results as possible evidence for the presence of XMRV or other MLVs in humans. In the same multi-laboratory study15 the assays used by Lombardi et al. detected a serological response in some specimens; however this reactivity was not consistent within replicates of the same plasma sample and no statistical association was observed in CFS patients compared to blood donors, while three other highly sensitive assays in the study failed to detect a serological response in any specimen17.
Micro-neutralization assays have been used extensively as diagnostic and specificity tests for many viruses, including alphaviruses and influenza18,19,20. Indeed, neutralizing antibodies are typically formed as part of a highly specific response to conformational epitopes. Neutralization of XMRV in 11/40 (27.5%) of serum samples was observed in prostate cancer patients 21 suggesting that a micro-neutralization assay for XMRV would be feasible and useful. In this study, we generated a micro-neutralization assay for studies of blood donors looking for serological evidence of XMRV/MLV infection based on the dual envelope pseudovirus (DEP) assay system we recently developed22, which has been proven to be a rapid, sensitive, and specific high-throughput system for antiviral drug discovery targeting viral entry. This assay system is composed of two viruses. Entry of the target virus is driven by the XMRV envelope protein pseudotyped onto the core of a reporter retrovirus, while infection by a second, internal control pseudovirus is mediated by an unrelated envelope and is included to reduce the number of false positives. Using this assay, we screened 354 donors, and identified a small number with a neutralization signature warranting further testing.
MATERIALS AND METHODS
Sample collection
Anonymized plasma and whole blood aliquots were prepared using residual samples left over from pilot tubes collected for routine blood donation testing. The samples selected were from 354 different donations from the United Blood Services Reno facility. One or two EDTA plasma tube(s) were used for preparation of these aliquots depending on the unit collection type. From each EDTA tube two plasma aliquots were prepared, then the remaining sample was gently inverted to re-suspend, and three or four whole blood aliquots were prepared. All aliquots were frozen the day of preparation. Donor samples were coded to retain linkage only to the donor’s zip code of residence, age, gender, and race-ethnicity. Any linkage to personal donor information such as name, address, and telephone number was removed. All samples provided were anonymized prior to shipment to BSRI for subsequent testing. The Institutional Review Board of the University of California San Francisco approved the study protocol.
Cells and reagents
Human embryonic kidney 293T cells clone 17 (293T/17) and human prostate LNCaP cells were obtained from the ATCC and grown in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% FBS and Penicillin and Streptomycin (10 U/ml). LNCaP iGFP cells (DERSE, Detectors of Exogenous Retroviral Sequence Elements) were kindly provided by Vineet KewalRamani (NCI Frederick).
LacZ encoding polytropic MLV [termed LacZ(MCF13)] viruses were generated by first infecting NIH3T3 cells with replication-competent MCF13. The resulting cell line was then infected with replication-defective lacZ(A-MLV) pseudotype virus to introduce the lacZ gene23. CHO cells overexpressing murine ecotropic MLV receptor mCAT-1 (CERD9) have previously been described24,25.
Sera from wild mice experimentally or mock-infected infected with XMRV for 12 weeks26 were used as positive and negative controls. XMRV infected rhesus macaque (RII10 and RYH10) sera were kindly provided by John Hackett (Abbott)27.
Plasmids
XMRV envelope (env) was PCR amplified from 22Rv1 cells with 100% nucleic acid sequence identity to the XMRV 22Rv1/CWR-R1 env sequence (Genbank Accession Number: FN692043), and cloned into the pCAGGS vector with KpnI and NheI restriction sites. Plasmids encoding G protein of vesicular stomatitis virus (VSV-G), glycoprotein of Lassa virus (Lassa-GP), as well as the ecotropic MLV envelope, have been described previously28–32.
Pseudotyped viruses with HIV-based retroviral backbone were generated from two plasmids, one encoding env and the other encoding the HIV backbone with a reporter gene. pNL4-3 Luc-R−E− (pNL-luc), encodes a replication-incompetent variant of the HIV-1 molecular clone NL4-3, in which the nef gene has been replaced by a firefly luciferase (luc) reporter, and the env and vpr genes were inactivated, as previously described33. Similarly, pNL4-3 Ren-R−E− (pNL-ren) was constructed by swapping the firefly luciferase gene for Renilla luciferase22.
Pseudotyped viruses with MLV-based retroviral backbone were generated from three plasmids: XMRV env, MLV-based firefly luciferase reporter (MRP-luc)34 and MLV gag/pol expression plasmid pHIT6035.
Virion production
HIV-based pseudovirions were produced essentially as previously described30 by transfecting 293T/17 cells with 10 μg of the corresponding HIV construct (pNL-luc or pNL-ren vector) and 30 μg of plasmid encoding the viral envelope per 10-cm dish using the calcium phosphate transfection method. Similarly, MLV-based pseudovirions were produced by transfecting 5 μg of each of the three plasmid constructs per 10-cm dish. The next day, expression was induced with sodium butyrate (10 mM) for 6 h before washing the cells once with PBS then replacing the medium. Forty hours after transfection, the supernatant was filtered through a 0.45 μm pore size filter and frozen at −80°C. If required, virions were concentrated by ultracentrifuge concentration at 28,000 rpm in a SW28 rotor (Beckman) through a 20% sucrose cushion for 1.5 h at 4°C. The pellets were resuspended in HBSS buffer and aliquoted for storage at −80°C. Resulting reporter viruses were classified according to retroviral backbone, reporter system and viral envelope, for example MLV-luc(XMRV Env) or HIV-ren(Lassa GP).
LacZ encoding polytropic MLV was harvested from 3T3LacZMCF13 cells, filtered through a 0.45 μm pore size filter and frozen at −80°C.
Micro-neutralization assay
Neutralization assays were performed in 96-well white tissue culture plates (Nunc). Donor serum samples were prepared from plasma by adding Thrombin (King Pharmaceuticals) in 0.5M MgCl2/CaCl2 solution and then removing fibrin clots. The serum supernatant was transferred to a new tube and heat inactivated at 56°C for 30 min. A volume of 10 μL of serially diluted test sera or medium alone were transferred to assay wells, followed by 30 μL of either a single or a two reporter virus mixture depending on the purpose of the assay, and incubated for 1hr at room temperature before addition of 40μL of 293T/17 or LNCaP cells (500,000 cells/ml) to all wells. Plates were incubated for two days at 37°C and 5% CO2 and firefly and Renilla luciferase reporter expression was determined sequentially as described in22. For the initial high-throughput micro-neutralization assays, sera samples with final dilutions of 80 and 240-fold were tested and each experiment repeated twice.
Neutralization dose response
For generation of neutralization dose response curves with selected donor sera, samples were serially diluted starting from 40- or 80-fold initial dilutions. Assays were performed in triplicate. Infection of pseudoviruses MLV-luc(XMRV Env) and MLV-luc(VSV G) in 293T/17 cells and infection of MLV-luc(MLV-E Env) and MLV-luc(VSV G) in CERD9 cells were detected using the Bright-Glo™ Luciferase Assay System (Promega). Infection of LacZ encoding polytropic MLV in 293T/17 cells was detected using the Galacto-Light Plus System for chemiluminescent reporter detection of β-Galactosidase (Applied Biosystems). Additionally, the percent of cells infected with LacZ encoding polytropic MLV was measured with cell fixation and visualization of blue color development under a microscope using a β-gal staining kit (Invitrogen).
Western blot
Western blot (WB) analysis was performed to detect anti-XMRV/MLV antibodies in selected donor sera and healthy controls as previously described12,36. Briefly, XMRV-infected DU145 prostate cells (C7) were grown in complete HuMEC serum-free medium supplemented with 1% HuMEC and 50ug/ml bovine pituitary extract (Invitrogen). Tissue culture supernatants were clarified by centrifugation and by passage through a 0.45 μm filter. XMRV was purified from 150 ml C7 supernatant using the ViraTrap Retrovirus Maxiprep Kit (Bioland Scientific LLC) following the manufacturer’s protocol. A volume of 150 μl of purified XMRV was denatured with SDS-PAGE sample buffer at 95°C for 10 min and viral proteins were separated by gel electrophoresis in a NuPAGE 4-12% Bis-Tris gel (Invitrogen) for WB testing as previously described but modified by using horseradish peroxidase conjugated protein G instead of protein A/G36,37. Seroreactivity was defined by reactivity to viral envelope and/or gag proteins of the expected size as seen in the positive control anti-sera. This WB test accurately detected XMRV antibodies in three experimentally infected macaques equivalent to detection using recombinant proteins in recently described immunoassays27.
qRT-PCR
RNA was extracted from 100 μL of selected donor whole blood samples using Qiagen Viral RNA Mini kit. The isolated RNA was subjected to reverse transcription by MLV reverse transcriptase (RT; Roche). The resulting cDNA was amplified in a real-time PCR reaction and quantified in a Roche LightCycler 480. qRT-PCR was performed with FastStart Taq polymerase (Roche) in 45 amplification cycles of 95°C and 60°C for 30sec each. Two primer pairs were used, integrase [F2 (5′-AACCTGATGGCAGATCAAGC-3′), R2 (5′-CCCAGTTCCCGTAGTCTTTTGAG-3′), and XMRV probe (5′-FAM-AGTTCTAGAAACCTCTACACTC-BHQ1-3′)]13 or gag [Q445F (5′-GGACTTTTTGGAGTGGCTTTGTT-3′), Q528R (5′-GCGTAAAACCGAAAGCAAAAAT-3′), and XMRV probe F480PRO-BHQ (5′-FAM-ACAGAGACACTTCCCGCCCCCG-BHQ1-3′)]38. A cut-off of 40 CTs was used as evidence for the presence of XMRV/MLV sequences in a specimen. Positive controls represented recombinant plasmid spiked into whole blood samples in a dilution series from 106 to 104 copies/ml.
Nested RT-PCR amplification of XMRV sequences
Nested RT-PCR was performed as described39. Briefly, RNA was extracted from 0.5 ml donor plasma using the QIAamp Ultrasens Virus kit (Qiagen) and subjected to reverse transcription employing the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). Culture supernatant of the XMRV-producing prostate cancer cell line 22Rv1 was used at a 10−5 dilution as a positive control for RNA isolation. For amplification of XMRV gag sequences, 5 μl of the transcribed cDNA were used for the first round of 40-cycle amplification with primers 419F (5′-ATCAGTTAACCTACCCGAGTCGGAC-3′) and 1154R (5′-GCCGCCTCTTCTTCATTGTTCTC-3′)8 and HotStart-IT FideliTaq Master Mix (USB). Nested PCR was performed for 45-cycle amplification with 5 μl of the first round PCR product and two different primer pairs, Gag-I-F (5′-TCTCGAGATCATGGGACAGA-3′) and Gag-I-R (5′-AGAGGGTAAGGGCAGGGTAA-3′) or NP116 (5′-CATGGGACAGACCGTAACTACC-3′) and NP117 (5′-GCAGATCGGGACGGAGGTTG-3′)40. To monitor assay sensitivity, plasmid DNA containing a cloned fragment of XMRV gag12 was included in each PCR run at concentrations from 1 to 100 copies/μl. PCR and RT-PCR of GAPDH controls with primer pairs, Forward (5′-CATGTTCCAATATGATTCAC-3′) and Reverse (5′-CCTGGAAGATGGTGATG-3′), were performed to ensure similar levels of DNA and RNA input in each round of amplification.
Propagation of infectious XMRV in indicator cells
DERSE (Detectors of Exogenous Retroviral Sequence Elements) indicator cells were developed at the National Cancer Institute by stable transfection of pBabe.iGFP-puro into LNCaP cells. The intron interrupted GFP gene from pBabe.iGFP-puro is only expressed after mobilization by an infecting gammaretrovirus for a second round of infection41. To test for the presence of infectious XMRV in selected donor plasma, DERSE.Li-G cells were inoculated with donor plasma or control plasma and spin infection, as described in39. GFP expression was monitored every 3 to 4 days for a total period of 3 weeks. As a positive control, culture supernatant of the XMRV-producing prostate cancer cell line 22Rv1 (containing roughly 109 copies/ml) was used as an inoculum at 10−2, 10−4, and 10−6 dilution.
RESULTS
High-throughput micro-neutralization assay development
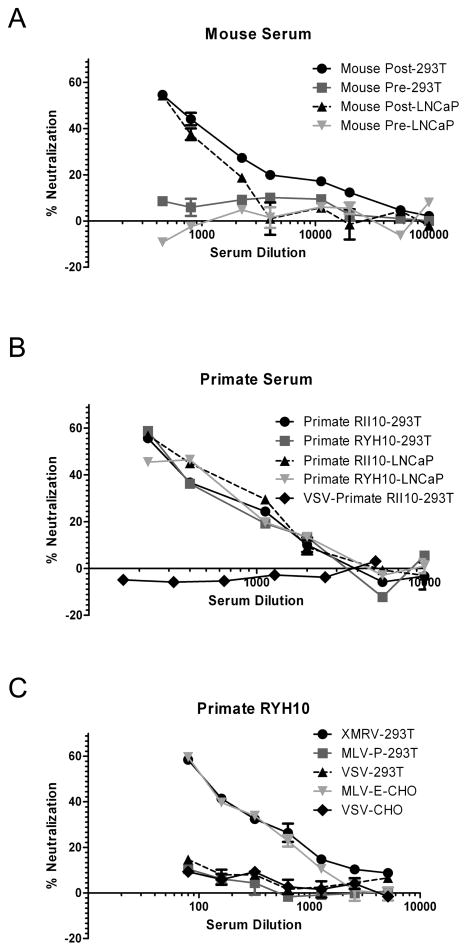
XMRV pseudoviruses [MLV-luc(XMRV Env)] were generated using a MLV-based retroviral backbone. These pseudoviruses infected both 293T and LNCaP cells. As expected from previous studies42, levels of infection mediated by the XMRV envelope were somewhat lower compared to control envelopes. For example, on 293T/17 cells, infection of unconcentrated MLV-luc(XMRV Env) was about equal to VSV-G pseudotyped virus stocks diluted ten-fold (67714 and 63742 relative light units, respectively). On both cell types MLV-luc(XMRV Env) was neutralized by sera from mice (Fig. 1A) and rhesus macaques (Fig. 1B) challenged with XMRV, whereas no clear neutralization was observed with pre-immune sera. Similar results were obtained with HIV-luc(XMRV Env) (data not shown). Moreover, ecotropic MLV pseudoviruses [MLV-luc(MLV-E Env)] were also neutralized by sera from rhesus macaques challenged with XMRV (Fig. 1C). However, LacZ encoding polytropic MLV (MLV-P), or HIV-luc(VSV G) pseudoviruses (Fig. 1B, 1C) were not neutralized.
Fig. 1. Detection of XMRV Env neutralizing antibodies in positive controls.
MLV-luc(XMRV Env) pseudovirus infection of 293T/17 and prostate LNCaP cells was neutralized by sera from both mice (A) and rhesus macaques (B) challenged with XMRV, whereas no clear neutralization was observed with pre-immune sera, or HIV-luc(VSV G) pseudoviruses. (C) MLV-luc(MLV-E Env) pseudoviruses were neutralized by sera from rhesus macaques challenged with XMRV in mCAT-1 expressing CHO cells (CERD9 cells), but no clear neutralization of LacZ encoding MLV-P or HIV-luc(VSV G) pseudoviruses was observed in 293T/17 cells. Infection of pseudoviruses with firefly luciferase reporter was detected with Bright-Glo™ Luciferase Assay System (Promega), whereas infection of LacZ encoding MLV-P was measured using Galacto-Light Plus System for detection of β-Galactosidase (Applied Biosystems). Absolute values for the no sera controls were: MLV-luc(XMRV Env) gave 55810 RLU on 293T cells and 20213 RLU on LNCaP cells; HIV-luc(VSV-G) gave 65961 RLU on 293T cells and 51677 RLU on CHO cells; MLV-P gave 32356 RLU on 293T cells and MLV-luc(MLV-E Env) gave 41771 RLU on CHO cells. Results are presented as percentage of neutralization and shown as mean ± S.D. of triplicate measurements. A representative experiment of at least two experiments is shown.
To develop a reliable high-throughput assay system for the screening of large numbers of samples for XMRV infection, we generated a cell-based XMRV micro-neutralization assay system based on the internally-controlled dual envelope pseudovirus (DEP) assay we recently developed to screen for small molecule inhibitors22, which has been proven to be a rapid, safe, sensitive, and specific high-throughput system for antiviral drug discovery targeting viral entry. We adopted a similar approach here for XMRV micro-neutralization assay. The assays were performed in 96-well plate format with the aid of liquid dispensing equipment for high-throughput applications. After preliminary experiments, a combination of MLV-luc(XMRV Env) and HIV-ren(Lassa GP), which showed no clear interference between the two envelopes, was chosen for the sera screening. This combination proved to give very robust and reproducible results. A combination of MLV-luc(XMRV Env) and HIV-ren(Lassa GP) from three 96-well plates indicated that the inter-plate coefficient of variation 43 was 8.2% and 5.2% for MLV-luc(XMRV Env) and HIV-ren(Lassa GP), respectively. A set of 20 sera samples indicated that for the intra-assays, the CV of every sample in triplicate was within 5% and for the inter-assays, the CV of every sample from three plates was within 12%, for both MLV-luc(XMRV Env) and HIV-ren(Lassa GP) (data not shown).
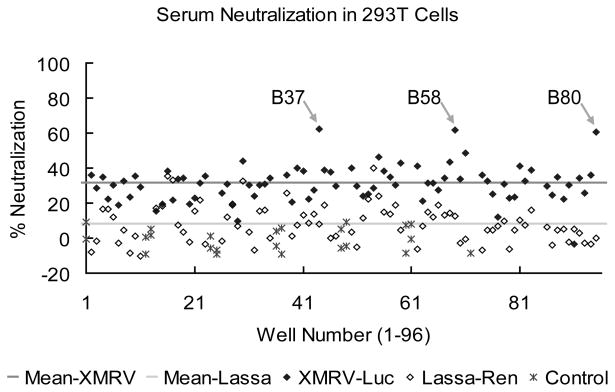
Generally, sera showed relatively higher levels of neutralization of XMRV Env pseudoviruses (~30%) than the Lassa GP control (~ 8%) (Fig. 2). Similar results were obtained with sera at 240-fold dilutions, individual virus alone, and in LNCaP cells (data not shown). Despite this higher level of background neutralization, neutralization with a number of sera was noticeably more pronounced. For example, in Fig. 2, three (B37, B58, and B80) of over 80 donor sera showed approximately 50% reduction in XMRV Env, but not Lassa GP, mediated viral infection in 293T/17 cells.
Fig. 2. XMRV Env neutralizing antibody in blood donor sera using a cell-based XMRV micro-neutralization assay system.
Shown is an example screen of 80 donor serum samples (80-fold dilutions) for XMRV neutralization with virus combinations of MLV-luc(XMRV Env) and HIV-ren(Lassa GP) in a 96-well plate format. Three (B37, B58, and B80) out of a total of over 80 donor sera showed approximately 50% reduction in XMRV Env-, but not Lassa GP-, mediated viral infection in 293T/17 cells.
Screening of blood donors
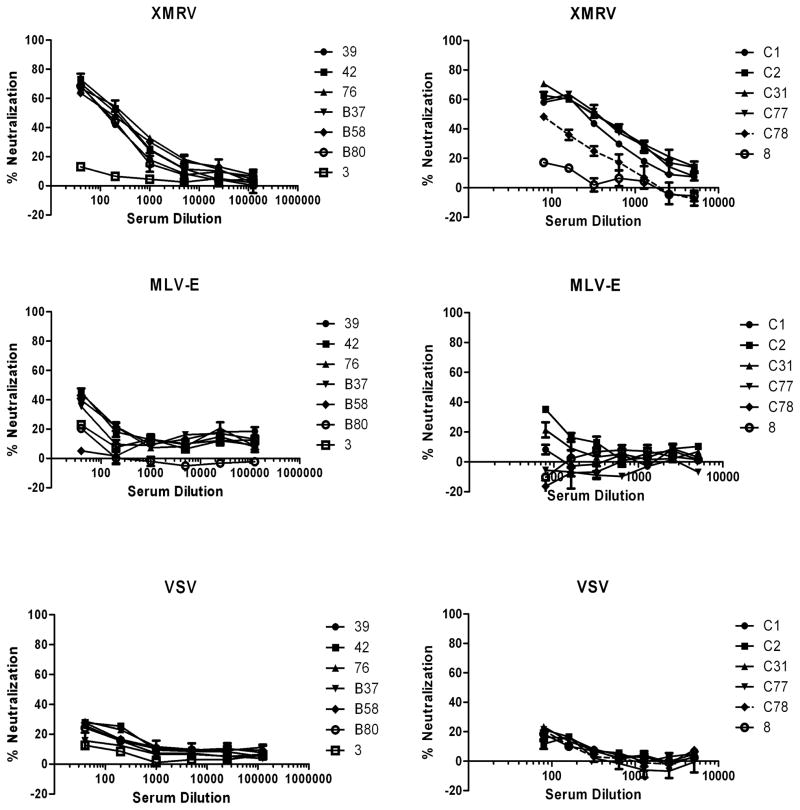
We used this assay to screen a total of 354 blood donor sera collected within the United Blood Service region of Reno/Lake Tahoe. The Reno facility was chosen due to the collection territory including regions of Northern Nevada and California known to have clusters of CFS 44,45,46. Patients from CFS clusters, including the Reno/Lake Tahoe area, formed the majority of subjects in the original demonstration of the presence XMRV in blood8. 23 sera gave over 50% reduction in XMRV Env, but not Lassa GP, mediated viral infection at either 80- or 240-fold dilutions. All 23 serum samples showed a dose-dependent neutralization of XMRV pseudoviruses (~ 60% neutralization at 80-fold dilution), but unlike the mouse and macaque antisera, the blood donor sera demonstrated very limited neutralization for MLV-E pseudoviruses (<50% at 80-fold dilution) (Fig. 3). No clear neutralization was detected for VSV pseudoviruses (Fig. 3) and LacZ encoding polytropic MLV (data not shown).
Fig. 3. Dose response curves with selected blood donor sera.
Neutralization of infection of HIV-luc(XMRV Env) and HIV-luc(VSV G) pseudoviruses with serially diluted donor sera samples were detected in 293T/17 cells and HIV-luc(MLV-E Env) in mCAT-1 expressed CHO cells (CERD9 cell). Results are presented as percentage of neutralization and shown as mean ± S.D. of triplicate measurements. A representative of at least two experiments is shown.
Confirmatory testing
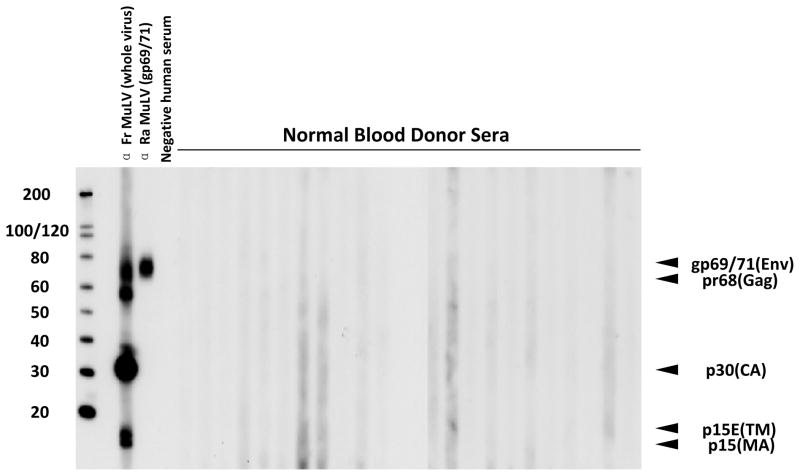
The 23 moderately neutralizing sera (> 50%) as well as 14 additional poor neutralizers (~ 30%–50%) and 12 donors with no clear neutralizing ability (< 30%) were further assessed with a recently developed WB assay36,37 using purified, denatured XMRV antigen from XMRV-infected DU145 prostate cells (C7). All 50 of the tested blood donor sera were WB-negative (Fig. 4).
Fig. 4. Absence of XMRV/MLV antibodies in blood donor sera by Western blot (WB) analysis.
Purified, denatured XMRV antigen from XMRV-infected DU145 prostate cells (C7) was used for WB detection of anti-XMRV/MLV antibodies in selected donor sera samples. Results of positive control anti-sera to purified XMRV antigen and 24 normal donor sera samples (B58, B80, E6, E8, E10, D17, D40, C5, C20, C30, C33, C35, C45, C47, C49, C50, C51, C67, 3, 4, 5, 6, 7, 8, from left to right) are shown; locations of reactivity to specific viral proteins are indicated. Env (gp69/71), envelope; TM (p15E), transmembrane; Gag (pr68); MA (p15), matrix; CA (p30), capsid. Molecular weight markers (kD) are provided on the left of the WB.
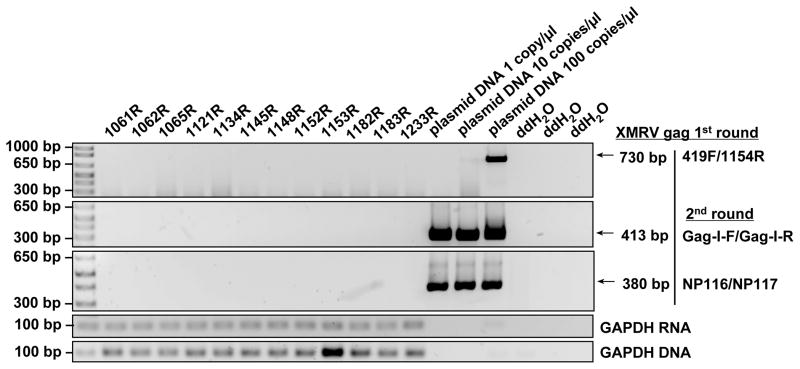
In order to further confirm whether there was any evidence of XMRV or other MLV infection in these individuals that would lead to a positive serological response, we performed PCR assays and virus cultures that would detect both specifically XMRV and more broadly other MLVs. Whole blood samples of the selected donors, were tested by qRT-PCR using primer sets located in either XMRV integrase 41 or gag. No positive signal was seen in any sample with either primer set (data not shown). Plasma samples of the 23 selected donors, were also tested and found negative by nested RT-PCR using generic MLV primers previously shown to detect both XMRV and the broader family of xenotropic and polytropic MLVs40 (Fig. 5).
Fig. 5. Absence of XMRV gag sequences in blood donor plasma by nested RT-PCR.
A representative result of 12 donor samples is shown with positive controls containing 1 to 100 copies/μl of a plasmid harboring a cloned fragment of XMRV gag37 and negative water controls. First round PCR amplification used primer pair 419F and 1154R and second round PCR amplification used primer pairs Gag-I-F and Gag-I-R, or NP116 and NP117. GAPDH RNA and DNA PCR results for the same samples are shown in the bottom two panels.
To test for the presence of infectious MLVs in donor plasma, the indicator cell line DERSE was used. As a positive control, culture supernatant of the XMRV-producing prostate cancer cell line 22Rv1 (containing roughly RNA 109 copies/ml) was utilized as an inoculum. Whereas cells inoculated with 22Rv1 supernatants showed a concentration-dependent GFP expression on day 7 and spread of the virus on day 21 as previously described39. no GFP expression could be observed in any of the cells inoculated with donor plasma from the 23 seroreactive persons, even when spin-infection was used to enhance the potential infection efficiency (data not shown).
DISCUSSION
Determining whether serological evidence of immune responses to gammaretroviruses in humans8,21. is an indication of authentic infection or just non-specific cross-reactivity is an important final step in the XMRV saga. In this study, we generated a robust, high-throughput micro-neutralization assay for the screening of large numbers of subjects for serological evidence of XMRV and MLV infection based on the DEP assay system we recently developed22. This assay includes an internal control pseudovirus which is very useful for avoiding nonspecific inhibition and also controls for cytotoxicity. This method provides a reproducible high-throughput micro-neutralization research assay for large scale testing for evidence of XMRV and MLV infection.
Currently, enzyme immuno-assays (EIAs) and Western blot (WB) are the two most common serological methods utilized for viral diagnosis47,48. WB is limited to the recognition of linear epitopes and is prone to high-background rates, while EIA can be restricted by the quality of the antigens, antibodies and detection methods. Instead of directly detecting the existence of antiviral antibodies in the sera, the DEP-based micro-neutralization assay is based on the ability of a serum to neutralize pseudovirus infection. Compared with standard assays such as EIAs, the micro-neutralization assay has fewer steps and can be performed by automated liquid handling equipment, which may generate less standard deviation. The disadvantage is a two-day incubation period during the assay, which impacts the clinical usefulness of the assay.
A recent study identified neutralizing activity against XMRV in about 14% of blood donor samples10. although in this instance many of these sera neutralized control viruses in addition to XMRV. In contrast, while we identified 23/354 blood donors (6.5%) able to moderately neutralize XMRV Env-mediated infection, control and other MLV envelopes were poorly or not at all neutralized. None of the samples tested showed any evidence of a serologic response to XMRV by WB testing. Furthermore, all 23 seroreactive samples were negative for XMRV and MLV sequences using PCR or virus culture. These PCR and culture assays were designed to detect a broad range of gammaretroviruses, as well as XMRV specifically, thus, excluding XMRV/MLV and other gammaretroviruses as a source of the non-specific reactivity. The finding that neutralization by the 23 blood donors was specific to XMRV envelopes, but not other MLV envelopes was surprising. Pairwise comparison of the amino acid (aa) sequence of the envelope region between XMRV, and MLV-P or MLV-E shows the aa similarity is about 89% and 68% respectively.
Given that the true XMRV neutralizing responses raised in animals were more broadly neutralizing (Fig. 1), this result strongly argues against specific neutralization, but rather suggests the moderate neutralization observed was mediated by other non-specific means. This could be cross-reactive antibodies raised against endogenous retroviral elements, completely unrelated proteins, or other non-antibody serum factors. Human serum potently inhibits XMRV14, however, this is largely complement-driven, and in our assay serum complement was inactivated by heating and did not influence our test results. The relatively high level of non-specificity is greater than that seen with other microneutralization assays 20,49, and is partly due to the lack known of human positive cases that can be used in order to accurately set cut-offs for defining specific neutralization. Our results likely also explain other reported XMRV neutralization results in human samples 21.
In addition to the initial association of XMRV to CFS made by Lombardi et al.8, a second publication by Lo and colleagues40, based only on PCR analysis, also yielded a strong association between CFS and MLV-like viruses40. These subsequent viruses demonstrated a far greater degree of sequence variation than XMRV, with the majority of sequences resembling polytropic MLV (P-MLV). Although Lo et al. reported very stringent measures to minimize contamination40, the most parsimonious explanation, given the extent of reported contamination of laboratory reagents, is that their PCR results are false positives resulting from reagent contamination. Indeed, Lo et al. used Platinum Taq (Invitrogen) for PCR amplification, which several groups have convincingly demonstrated is contaminated with mouse DNA14,15,50 due to the use of a mouse monoclonal antibody in the enzyme mix. Furthermore, recent detailed phylogenetic analysis of the longitudinal polytropic MLV sequences reported by Lo et al. showed that these sequences are inconsistent with retroviral evolution51. Nonetheless, the findings of Lo et al. raised the hypothesis that while XMRV itself is clearly a laboratory contaminant, the serological responses detected in Lombardi et al. may be due to infection by other MLVs or gammaretroviruses. The serological assay used by Lombardi et al. relies on antibody binding to the MLV spleen focus-forming virus (SFFV) Env expressed on the surface of cells. The logic of this assay is that conformationally-dependent cross-reactive epitopes shared between this mouse gammaretrovirus and XMRV would bind XMRV antibodies which would then be detected in a flow cytometry-based assay. However, it is likely that, as with our micro-neutralization assay, mammalian cell culture-based expression of an unrelated retrovirus Env would be highly prone to non-specific cross-reactivity that can confound the testing and which requires clarification by WB analysis using purified antigen. Indeed, when the Lombardi et al. flow-based assay was used by two laboratories on plasma specimens in a blinded study, high levels of non-specific reactivity was observed15.
In conclusion, we developed a robust, high-throughput micro-neutralization assay in order to conduct studies looking for evidence of infection with XMRV and MLV. Although a small proportion of blood donors demonstrated the ability to block XMRV-mediated infection, we found no evidence that this inhibition was mediated by specific antibodies elicited by exposure to XMRV or related MLVs. It is likely that this moderate neutralization is mediated through another, non-specific mechanism. Our findings also explain further the highly non-reproducible and non-specific serological responses detected with other assays8,17. In addition, this micro-neutralization assay system can be easily adapted to screen donor samples against other viruses with careful selection of matching partner virus envelopes, which will provide important information for neutralizing antibody responses and infectious disease profiles.
Acknowledgments
SOURCE(S) OF SUPPORT
This work was partially funded by a R21 grant from the National Heart, Lung, and Blood Institute (NHLBI) to GS (1R21HL109761).
We would like to thank Drs. KyeongEun Lee and Vineet KewalRamani (NCI) for kindly providing the DERSE.Li-G cells and Dr. Indira Hewlett (FDA) for advice on virus culture protocols. We would like to thank Dr John Hackett (Abbott Diagnostics) for macaque serum. GS was supported by Grant Number R21HL109761 from the National Heart, Lung, And Blood Institute. The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of their sponsoring institutions.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
References
- 1.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106:16351–6. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Fischer N, Hellwinkel O, Schulz C, Chun FK, Huland H, Aepfelbacher M, Schlomm T. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J Clin Virol. 2008;43:277–83. doi: 10.1016/j.jcv.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Hohn O, Krause H, Barbarotto P, Niederstadt L, Beimforde N, Denner J, Miller K, Kurth R, Bannert N. Lack of evidence for xenotropic murine leukemia virus-related virus(XMRV) in German prostate cancer patients. Retrovirology. 2009;6:92. doi: 10.1186/1742-4690-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hue S, Gray ER, Gall A, Katzourakis A, Tan CP, Houldcroft CJ, McLaren S, Pillay D, Futreal A, Garson JA, Pybus OG, Kellam P, Towers GJ. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology. 2010;7:111. doi: 10.1186/1742-4690-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garson JA, Kellam P, Towers GJ. Analysis of XMRV integration sites from human prostate cancer tissues suggests PCR contamination rather than genuine human infection. Retrovirology. 2011;8:13. doi: 10.1186/1742-4690-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ, Jr, Coffin JM, Pathak VK. Recombinant origin of the retrovirus XMRV. Science. 2011;333:97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–9. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 9.Erlwein O, Kaye S, McClure MO, Weber J, Wills G, Collier D, Wessely S, Cleare A. Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS One. 2010;5:e8519. doi: 10.1371/journal.pone.0008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groom HC, Boucherit VC, Makinson K, Randal E, Baptista S, Hagan S, Gow JW, Mattes FM, Breuer J, Kerr JR, Stoye JP, Bishop KN. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology. 2010;7:10. doi: 10.1186/1742-4690-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunstman KJ, Bhattacharya T, Flaherty J, Phair JP, Wolinsky SM. Absence of xenotropic murine leukemia virus-related virus in blood cells of men at risk for and infected with HIV. Aids. 2010;24:1784–5. doi: 10.1097/qad.0b013e32833b76fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Switzer WM, Jia H, Hohn O, Zheng H, Tang S, Shankar A, Bannert N, Simmons G, Hendry RM, Falkenberg VR, Reeves WC, Heneine W. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology. 2010;7:57. doi: 10.1186/1742-4690-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Kuppeveld FJ, de Jong AS, Lanke KH, Verhaegh GW, Melchers WJ, Swanink CM, Bleijenberg G, Netea MG, Galama JM, van der Meer JW. Prevalence of xenotropic murine leukaemia virus-related virus in patients with chronic fatigue syndrome in the Netherlands: retrospective analysis of samples from an established cohort. Bmj. 2010;340:c1018. doi: 10.1136/bmj.c1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knox K, Carrigan D, Simmons G, Teque F, Zhou Y, Hackett J, Jr, Qiu X, Luk KC, Schochetman G, Knox A, Kogelnik AM, Levy JA. No evidence of murine-like gammaretroviruses in CFS patients previously identified as XMRV-infected. Science. 2011;333:94–7. doi: 10.1126/science.1204963. [DOI] [PubMed] [Google Scholar]
- 15.Shin CH, Bateman L, Schlaberg R, Bunker AM, Leonard CJ, Hughen RW, Light AR, Light KC, Singh IR. Absence of XMRV retrovirus and other murine leukemia virus-related viruses in patients with chronic fatigue syndrome. J Virol. 2011;85:7195–202. doi: 10.1128/JVI.00693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman RH, Das Gupta J, Lombardi VC, Ruscetti FW, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Mikovits JA. Partial Retraction. Science. 2011 doi: 10.1126/science.1212182. [DOI] [PubMed] [Google Scholar]
- 17.Simmons G, Glynn SA, Komaroff AL, Mikovits JA, Tobler LH, Hackett J, Jr, Tang N, Switzer WM, Heneine W, Hewlett IK, Zhao J, Lo SC, Alter HJ, Linnen JM, Gao K, Coffin JM, Kearney MF, Ruscetti FW, Pfost MA, Bethel J, Kleinman S, Holmberg JA, Busch MP. Failure to Confirm XMRV/MLVs in the Blood of Patients with Chronic Fatigue Syndrome: A Multi-Laboratory Study. Science. 2011 doi: 10.1126/science.1213841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanas AC, Johnson BK, Simpson DI. Antigenic relationships of alphaviruses by a simple micro-culture cross-neutralization method. J Gen Virol. 1976;32:295–300. doi: 10.1099/0022-1317-32-2-295. [DOI] [PubMed] [Google Scholar]
- 19.Assays for neutralizing antibody to influenza viruses. Report of an informal scientific workshop, Dresden, 18-19 March 2003. Wkly Epidemiol Rec. 2003;78:290–3. [PubMed] [Google Scholar]
- 20.Temperton NJ, Hoschler K, Major D, Nicolson C, Manvell R, Hien VM, Ha do Q, de Jong M, Zambon M, Takeuchi Y, Weiss RA. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Other Respi Viruses. 2007;1:105–12. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold RS, Makarova NV, Osunkoya AO, Suppiah S, Scott TA, Johnson NA, Bhosle SM, Liotta D, Hunter E, Marshall FF, Ly H, Molinaro RJ, Blackwell JL, Petros JA. XMRV infection in patients with prostate cancer: novel serologic assay and correlation with PCR and FISH. Urology. 2010;75:755–61. doi: 10.1016/j.urology.2010.01.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Agudelo J, Lu K, Goetz DH, Hansell E, Chen YT, Roush WR, McKerrow J, Craik CS, Amberg SM, Simmons G. Inhibitors of SARS-CoV entry - Identification using an internally-controlled dual envelope pseudovirion assay. Antiviral Res. 2011 doi: 10.1016/j.antiviral.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci U S A. 1999;96:927–32. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tailor CS, Nouri A, Kabat D. Cellular and species resistance to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J Virol. 2000;74:9797–801. doi: 10.1128/jvi.74.20.9797-9801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Paul R, Burgeson RE, Keene DR, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991;65:6468–77. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakuma T, Tonne JM, Squillace KA, Ohmine S, Thatava T, Peng KW, Barry MA, Ikeda Y. Early events in retrovirus XMRV infection of the wild-derived mouse Mus pahari. J Virol. 2011;85:1205–13. doi: 10.1128/JVI.00886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X, Swanson P, Luk KC, Tu B, Villinger F, Das Gupta J, Silverman RH, Klein EA, Devare S, Schochetman G, Hackett J., Jr Characterization of antibodies elicited by XMRV infection and development of immunoassays useful for epidemiologic studies. Retrovirology. 2010;7:68. doi: 10.1186/1742-4690-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert JM, Bates P, Varmus HE, White JM. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;68:5623–8. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102:11876–81. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A. 2004;101:4240–5. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons G, Wool-Lewis RJ, Baribaud F, Netter RC, Bates P. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol. 2002;76:2518–28. doi: 10.1128/jvi.76.5.2518-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvador B, Zhou Y, Michault A, Muench MO, Simmons G. Characterization of Chikungunya pseudotyped viruses: Identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology. 2009;393:33–41. doi: 10.1016/j.virol.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–44. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 34.Amberg SM, Netter RC, Simmons G, Bates P. Expanded tropism and altered activation of a retroviral glycoprotein resistant to an entry inhibitor peptide. J Virol. 2006;80:353–9. doi: 10.1128/JVI.80.1.353-359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–33. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satterfield BC, Garcia RA, Jia H, Tang S, Zheng H, Switzer WM. Serologic and PCR testing of persons with chronic fatigue syndrome in the United States shows no association with xenotropic or polytropic murine leukemia virus-related viruses. Retrovirology. 2011;8:12. doi: 10.1186/1742-4690-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Switzer WM, Jia H, Hohn O, Zheng H, Tang S, Shankar A, Bannert N, Simmons G, Hendry RM, Falkenberg VR, Reeves WC, Heneine W. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology. 2010;7:57. doi: 10.1186/1742-4690-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, Derisi JL, Chow SA, Silverman RH. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A. 2007;104:1655–60. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steffen I, Tyrrell DL, Stein E, Montalvo L, Lee T-H, Zhou Y, Lu K, Switzer WM, Tang S, Jia H, Hockman D, Santer DM, Logan M, Landi A, Law J, Houghton M, Simmons G. No Evidence for XMRV Nucleic Acids, Infectious Virus or Anti-XMRV Antibodies in Canadian Patients with Chronic Fatigue Syndrome. PLoS ONE. 2011;6(11):e27870. doi: 10.1371/journal.pone.0027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo SC, Pripuzova N, Li B, Komaroff AL, Hung GC, Wang R, Alter HJ. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc Natl Acad Sci U S A. 107:15874–9. doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Kearney MF, Lee K, Bagni RK, Wiegand A, Spindler J, Maldarelli F, Pinto PA, Linehan WM, Vocke CD, Delviks-Frankenberry KA, deVereWhite RW, Del Prete GQ, Mellors JW, Lifson JD, KewalRamani VN, Pathak VK, Coffin JM, Le Grice SFJ. Nucleic Acid, Antibody, and Virus CultureMethods to Detect Xenotropic MLV-Related Virus in Human Blood Samples. Adv Virol. 2011 doi: 10.1155/2011/272193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakuma T, De Ravin SS, Tonne JM, Thatava T, Ohmine S, Takeuchi Y, Malech HL, Ikeda Y. Characterization of retroviral and lentiviral vectors pseudotyped with xenotropic murine leukemia virus-related virus envelope glycoprotein. Hum Gene Ther. 2010;21:1665–73. doi: 10.1089/hum.2010.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cvitanich C, Pallisgaard N, Nielsen KA, Hansen AC, Larsen K, Pihakaski-Maunsbach K, Marcker KA, Jensen EO. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc Natl Acad Sci U S A. 2000;97:8163–8. doi: 10.1073/pnas.090468497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daugherty SA, Henry BE, Peterson DL, Swarts RL, Bastien S, Thomas RS. Chronic fatigue syndrome in northern Nevada. Rev Infect Dis. 1991;13 (Suppl 1):S39–44. doi: 10.1093/clinids/13.supplement_1.s39. [DOI] [PubMed] [Google Scholar]
- 45.Holmes GP, Kaplan JE, Stewart JA, Hunt B, Pinsky PF, Schonberger LB. A cluster of patients with a chronic mononucleosis-like syndrome. Is Epstein-Barr virus the cause? JAMA. 1987;257:2297–302. [PubMed] [Google Scholar]
- 46.DeFreitas E, Hilliard B, Cheney PR, Bell DS, Kiggundu E, Sankey D, Wroblewska Z, Palladino M, Woodward JP, Koprowski H. Retroviral sequences related to human T-lymphotropic virus type II in patients with chronic fatigue immune dysfunction syndrome. Proc Natl Acad Sci U S A. 1991;88:2922–6. doi: 10.1073/pnas.88.7.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen K, Bryson YJ. Diagnosis of HIV infection in children. Pediatr Clin North Am. 2000;47:39–63. doi: 10.1016/s0031-3955(05)70194-2. [DOI] [PubMed] [Google Scholar]
- 48.Suligoi B, Rodella A, Raimondo M, Regine V, Terlenghi L, Manca N, Casari S, Camoni L, Salfa MC, Galli C. Avidity Index for anti-HIV antibodies: comparison between third- and fourth-generation automated immunoassays. J Clin Microbiol. 2011;49:2610–3. doi: 10.1128/JCM.02115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Temperton NJ, Chan PK, Simmons G, Zambon MC, Tedder RS, Takeuchi Y, Weiss RA. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. 2005;11:411–6. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuke PW, Tettmar KI, Tamuri A, Stoye JP, Tedder RS. PCR master mixes harbour murine DNA sequences. Caveat emptor! PLoS One. 2011;6:e19953. doi: 10.1371/journal.pone.0019953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katzourakis A, Hue S, Kellam P, Towers GJ. Phylogenetic analysis of murine leukemia virus sequences from longitudinally sampled chronic fatigue syndrome patients suggests PCR contamination rather than viral evolution. J Virol. 2011;85:10909–13. doi: 10.1128/JVI.00827-11. [DOI] [PMC free article] [PubMed] [Google Scholar]